Abstract
Many specific sequence DNA binding proteins locate their target sequence by first binding to DNA nonspecifically, then linearly diffusing or hopping along DNA until either the protein dissociates from the DNA or it finds the recognition sequence. We have devised a method for measuring 1-dimensional diffusion along DNA based on the ratio of the dissociation rates of EcoRI from DNA fragments containing one and two specific binding sites. Our extensive measurements of dissociation rates and specific-nonspecific relative binding constants of the restriction nuclease EcoRI enable us to determine the diffusion rate of nonspecifically bound protein along the DNA. By varying the distance between the two binding sites we confirm a linear diffusion mechanism. The sliding rate is relatively insensitive to salt concentration and osmotic pressure indicating the protein moves smoothly along the DNA probably following the helical phosphate-sugar backbone of DNA. We calculate a diffusion coefficient for EcoRI of 3 × 104 bp2sec−1. EcoRI is able to diffuse ~150 base pairs on average along DNA in 1 second. This diffusion rate is about 2000-fold slower than the diffusion of the free protein in solution. A factor of 40–50 can be accounted for by a rotational friction resulting from following the helical path of the DNA backbone. Two possibilities could account for remaining activation energy: the salt bridges between the DNA and protein are transiently broken or the water structure at the protein-DNA interface is disrupted as the two surfaces move past one another.
Introduction
The initial observation that the association rate of lac repressor binding to its operator DNA was significantly faster that diffusion rate limited led to the realization that association occurs in 2 steps 1; 2; 3; 4; 5; 6. Many sequence specific DNA binding proteins will initially bind nonspecifically to DNA and subsequently move along the DNA helix until either the protein finds a recognition sequence or dissociates. This combination of one and three dimensional searching can lead to association rates that are much faster than expected for only a three dimensional search 1; 7; 8; 9, although whether experimentally observed rates are faster due to one dimensional searching has been recently questioned 10. After nonspecific association, the extent of DNA the protein can explore depends on the rates of both diffusion along the DNA and dissociation of the nonspecifically bound complex. Initially, estimates of the length of DNA explored have come from the sensitivity of the association rates to DNA length 6. The ability of restriction endonucleases to cleave DNA after finding their recognition sequences has proven particularly useful for measuring the DNA length dependent rates of association 11; 12; 13; 14. These rates depend on association and dissociation rates of protein to nonspecific DNA and on one dimensional diffusion coefficients. Determining the encounter rates between free protein and DNA of different lengths and the dissociation rate of nonspecifically bound complex can, however, be problematic.
Recently there have been several direct single molecule measurements of one dimensional diffusion rates using fluorescently labeled protein 2; 15; 16; 17; 18; 19; 20; 21. This approach avoids the problems inherent in knowing association and nonspecific complex dissociation rates. The protein must stay on the DNA long enough and move far enough for a precise measurement.
Three mechanisms for the movement of proteins along DNA have been postulated 3. Sliding is the base pair by base pair diffusion along DNA; the protein remains in contact with the helix. Several crystal and NMR structures of nonspecific DNA-protein complexes (e.g., 22; 23; 24) and our own measurements of the difference in sensitivity to water activity between specific and nonspecific binding modes for several DNA-protein complexes 25; 26; 27; 28 show that substantial water is present at the DNA-protein interface of nonspecific complexes that can `lubricate' movement along the helix 22. Hopping is the transient dissociation and rebinding of the protein to the same helix 29; 30. Hopping would be expected to have a significant salt dependence since transient dissociation breaks DNA-protein charge interactions. Hopping can span a few to many base pairs in one movement. Direct transfer is the reaction of free DNA with a complex and consequent exchange of the protein from one helix to another.
Our approach here is to measure the ratio of specific site dissociation rates between a DNA fragment with a single EcoRI binding site and another fragment with two binding sites separated by variable lengths. The probability of reaching the second site after dissociation from the first affects the dissociation rate of the two-site DNA fragment relative to the one-site DNA fragment. By measuring the ratios of dissociation rates we eliminate the transition kinetics between specific and nonspecific binding modes at the recognition site. In order to better fit the one dimensional diffusion rate, we vary the nonspecific complex dissociation rate over an 8000-fold range by varying the salt concentration. The overall specific site dissociation rate is held approximately constant by appropriately adjusting the osmotic pressure that varies the transition kinetics between the specific and nonspecific binding modes at the specific site. We use our previous measurements 31 of the nonspecific complex dissociation rate to calculate the sliding rate or one dimensional diffusion coefficient from fits to the data using the theoretical framework of Belotserkovskii and Zarling 32. Our results also indicate that the sliding rate is insensitive to salt concentration and osmotic pressure suggesting that EcoRI follows the helical backbone as it diffuses. We find that the sliding rate of EcoRI along DNA is about 2000-fold slower than expected if only translational flow friction is considered. This is consistent with measurements on several other DNA-protein complexes. At least part of the extra friction can likely be attributed to a rotational flow friction as the protein follows the helical path of DNA. The remainder is perhaps due to the transient breaking and remaking of DNA-protein charge pairs or to disruption of the water structuring at the protein-DNA interface.
Theory
We use the theoretical framework described by Belotserkovskii and Zarling 32 for random walks with a probability for irreversible dissociation. Probabilities of dissociation are calculated, not rates. We work, however, under conditions such that the dissociation rate of nonspecifically bound complex is much faster than the transition between specific and nonspecific binding modes (at any time most all complexes are specific). We previously measured the nonspecific dissociation rate for an oligonucleotide as 50/min at 90 mM NaCl, pH 7.0, and 20 °C; whereas the dissociation rate of a specific site oligonucleotide complex under the same experimental conditions is ~ 0.005/min 31; 33. This means that ratios of dissociation probabilities once the complex is nonspecifically bound can substitute for ratios of rates.
Within the Belotserkovskii and Zarling framework, the parameters ksl and knsp,diss are the rate constants for moving one base pair along the DNA (equally probable in either direction) and for dissociation of the nonspecifically bound complex, respectively. The dissociation rate constant from the ends, k'nsp,diss, may be different from interior sequences. We assume that the specific recognition sites are absorbing barriers and that the random walk begins one base pair from a site binding.
The probability that a protein starting next to a binding site and with an end in the opposite direction l bp away will rebind before dissociating can be calculated as:
| (1) |
| (2) |
The probability that a protein starting next to a binding site with another binding site in the opposite direction from the first and ΔL bp away will find one or the other binding sites before dissociating is:
| (3) |
The dissociation probability for the single site fragment with distances l1 and l2 bp from the binding site to the ends is:
| (4) |
Similarly, the dissociation probability for the two site fragment with l3 bp from the end to the first binding site, ΔL bp between sites, and l4 bp between the second binding site and the far end is:
| (5) |
The ratio of specific complex off-rates is then given by,
| (6) |
For the DNA fragments used and knsp,diss, values examined, this ratio depends primarily on the distance ΔL between sites and the ratio of nonspecific complex dissociation and sliding rates through the parameter a (equation (2)); the parameter c (equation (2)) affects fits to the data minimally for the DNA fragments used; a value of c = 1 will be assumed for the initial fits to data with 360 bp DNA fragments. A better value will be estimated from comparing specific site dissociation rates for 360 bp fragments and 30 bp oligonucleotides.
Results
Figure 1 illustrates the basic principle in comparing dissociation rates of DNA fragments that contain one and two specific binding sites. The initial step is a transition between specific and nonspecific binding modes. The nonspecifically bound protein then begins a random walk and can either dissociate, return to the original binding site, or diffuse to the second binding site for the two-site fragment. If the protein begins the random walk to the left, then if binding sites are absorbing barriers the one and two binding site complexes have equal probabilities of either dissociating or returning to the original binding site. If, however, the protein begins the random walk to the right such that it is bracketed by the recognition sequences of the 2-site fragment, then the dissociation rates of the two fragments are not necessarily the same. If the nonspecific dissociation rate, kdiss,nsp, is much faster than the one-dimensional diffusion rate, ksl, then the probability that the protein reaches the other site before dissociating is quite small. The dissociation rates of the one and two site DNA fragments are once again the same. If, however, the one-dimensional diffusion rate is much faster than nonspecific dissociation, then the probability is very high that the protein will reach the other site before dissociating. In this case the two-site fragment will dissociate half as fast as the one-site fragment; only those complexes that begin the random walk to the left will have a chance of dissociating. The ratio of the one-site to two-site dissociation rates will vary between 1 and 2 depending on the relative rates of one-dimensional diffusion and nonspecific complex dissociation. The equations describing the dependence of the ratio of dissociation rate constants on ksl and knsp,diss are given in the Theory section.
Figure 1.
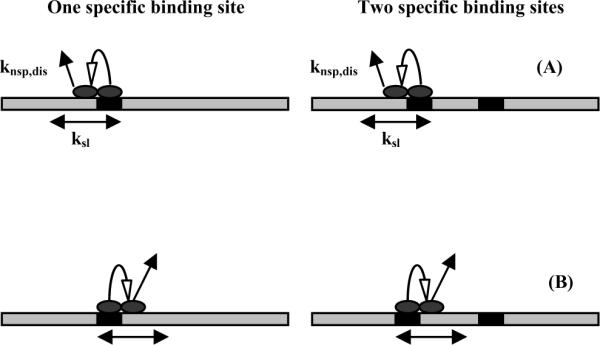
A schematic representation illustrating the basis for the method for measuring sliding rates. The dissociation reaction starts with a transition from specific to nonspecific binding modes at the recognition sites (the black boxes), followed by a sliding translation to either the left (1a) or right (1b). If recognition sites are considered absorbing barriers and if the walks start from the left, then the probabilities that EcoRI will dissociate from one- and two-site DNA fragments are the same. Starting from the right, however, probabilities will be the same only if the dissociation rate is much faster than the linear diffusion rate. If the sliding rate is much faster than the dissociation rate, the ratio of the dissociation probabilities for the two-site and one-site DNA fragments is 0. The protein will find the second site before dissociating. The ratio of off-rates for complexes with the one-site and two-site DNA fragments will, therefore, vary between 2 and 1 depending on the ratio of the nonspecific complex dissociation rate and the sliding rate.
The nonspecific dissociation rate can be controlled so that the probability of reaching the second site can be varied allowing a better fit to the equations. We have extensive measurements of the osmotic pressure and salt concentration dependence of the relative nonspecific-specific binding constant and the specific site dissociation rate from previous work. The fundamental Gibbs-Duhem equation relates the dependence on osmotic pressure, d ln(K)/d∏or d ln(k)/d∏, to the difference in the number of sequestered or bound water molecules between initial and final or transition states; while the salt sensitivity, d ln(K)/d ln[salt] or d ln(k)/ d ln[salt], primarily gives a difference in the number of thermodynamically bound ions 34. We have found that these dependencies are linear over a wide range of salt concentrations and betaine osmotic pressures for the relative specific-nonspecific binding constant and the specific site dissociation rate 31; 33. The relative specific-nonspecific binding constant and the specific site dissociation rate have about the same large osmotic pressure dependence, but the specific site dissociation rate has much larger salt concentration sensitivity than the relative binding constant. As in Figure 1, the dissociation rate of EcoRI from a specific site DNA fragment can be divided into two processes: a transition between specific and nonspecific binding modes that will have the same osmotic pressure and salt concentration dependences as the relative specific-nonspecific binding constant and a dissociation of nonspecifically bound protein. We assume that the difference in osmotic and salt dependencies between the relative specific-nonspecific binding constant and the specific sequence dissociation rate then is the difference in sequestered water and salt binding between the nonspecific complex and the dissociation transition state of the nonspecific complex, i.e., the osmotic and salt sensitivity of knsp,diss. We can, therefore, vary knsp,diss widely through the salt concentration but maintain a conveniently measured overall specific site off rate by adjusting the osmotic pressure to control the equilibrium between specific and nonspecific binding. The knsp,diss at any salt concentration (in mM) and glycine betaine osmotic pressure (measured as an osmolal concentration) can be related to k0nsp,diss at 90 mM NaCl and 0 osmolal glycine betaine (20 °C and pH 7.0) by:
| (7) |
The parameter (a) of equations (2) and ultimately (6) can be rewritten in terms of the ratio of nonspecific complex dissociation rates, knsp,diss/k0nsp,diss, as:
| (8) |
The only fitting parameter in equation (6) is then (k0nsp,diss/ksl).
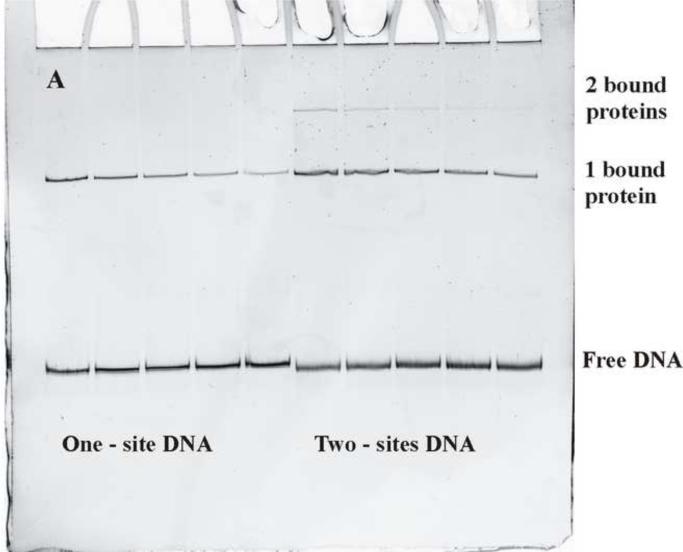
Figure 2a shows a gel mobility shift assay for measuring dissociation rates. At time t = 0, oligonucleotide containing the EcoRI recognition sequence is added to the reaction mixture of protein and DNA fragment in large excess. The molar ratio of oligonucleotide to DNA fragment was typically ~500. Protein that dissociates is efficiently removed from further reaction with DNA fragment. The experimental conditions (90 mM NaCl, pH 7.0, and 20°C with no added Mg2+) are such that EcoRI binds essentially stoichiometrically to DNA. The dissociation reaction is stopped by adding triethylene glycol that both lowers the salt concentration and greatly increases the osmotic pressure, both of which substantially decrease (by more than 100-fold) the specific site dissociation rate. The five lanes on the left illustrate the increasing loss of complex with increasing time of incubation with oligonucleotide for the one-site fragment. The five lanes on the right show the corresponding dissociation of protein from the two-site fragment with 21 bp between sites (Δ21). It is apparent that the two-site fragment complex dissociates more slowly. The two-site fragment shows a small fraction of fragment with two bound proteins.
Figure 2.
Dissociation of EcoRI from DNA with one or two recognition site. (a) A typical polyacrylamide gel shows the loss of EcoRI binding to the one-site DNA fragment (the first five lanes) with time and to the two-site DNA fragment (the last five lanes). The one protein bound complex and free DNA bands are indicated. The two-site DNA fragment has an additional weak band of complex with two bound proteins. A large excess of specific site oligonucleotide is added to the EcoRI/DNA fragment mixture to initiate dissociation. Incubation times are 0, 15, 30, 45, and 60 min from left to right. The dissociation reaction is stopped by adding triethylene glycol.
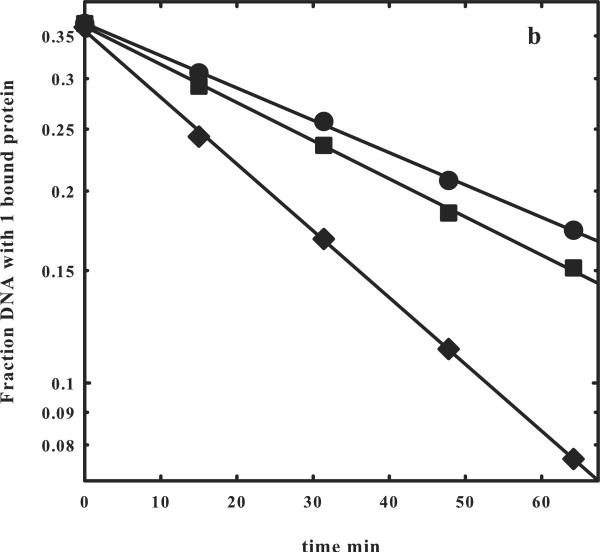
(b) The logarithmic dependences of the fraction of one bound protein complex on time are shown for the one- (◆) and two- (●) site DNA fragments shown in (a). The slopes give the off rates for EcoRI dissociating from the fragments into solution, koff,1-site or koff,2-site. A correction to the fraction of one bound protein complex to account for the contribution from dissociation of the two-site fragment with two bound proteins can be applied using equation (9). The corrected data is shown (■). The observed ratio koff,1-site/koff,2-site = 1.93 indicates that the EcoRI that dissociates from one specific site and begins a random walk in the direction of the other site will almost certainly find the other before dissociating from the fragment into solution (Figure 1b).
Figure 2b shows the semilog plots for the dissociation kinetics of complexes with one bound protein for the one and two- site fragments shown in Figure 2a. The kinetics for the two-site fragment can be corrected for the dissociation of complex with two bound proteins as described by equation (9). The ratio of the one- and two- site dissociation rates is 1.9, indicating that the protein is able to diffuse the 21 bp separation between sites with high probability before dissociating.
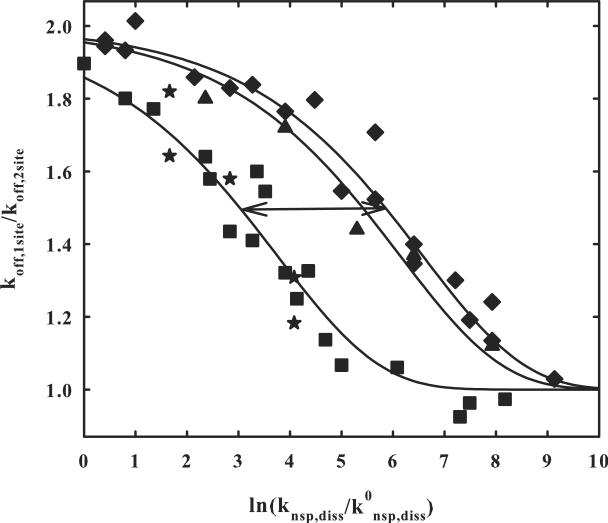
Figure 3 shows the ratio of one- and two-site dissociation rates for two site separations of 21, 26, and 84 bp as a function of the normalized nonspecific dissociation rate, ln(k0nsp,diss/knsp,diss), as the salt and osmotic pressures are varied (equation (7)). The ratios do vary between 1 and 2 as predicted. The solid lines are fits of the data to equation (6) with ln(k0nsp,diss/ksl) = −12.6. At kdiss,1-site/kdiss,2-site = 1.5, the nonspecific dissociation rate is about 16-fold faster for the Δ21 bp 2-site separation than for the Δ84 bp as would be expected for simple diffusion and the 4-fold difference in distance between sites. The best fit of equations (6) to the 21 bp spacing data alone is ln(k0nsp,diss/ksl) = −12.7 ± 0.2 and −12.6 ± 0.2 for the 84 bp distance data alone. There is not enough data to fit the 26 bp spacing data independently and accurately. The good fit of the solid line to the 26 bp spacing data with ln(k0nsp,diss/ksl) = −12.6, however, indicates that it is not important if the recognition sites of the two-site DNA are on the same or opposite sides of the helix. The second recognition site is found with the same probability after dissociation from the first. The `star' points in the Δ84 two-site fragment data show the result of only varying the salt concentration while holding the osmotic pressure constant. The points lie on the same curve within experimental error.
Figure 3.
The dependence of the ratio of the dissociation rates of EcoRI from DNA with one and two binding sites on changes in the nonspecific dissociation rate. The ratio of one- and two-site fragment dissociation rate constants determined in figure 2 is shown as a function of ln(knsp,diss/k0nsp,diss); k0nsp,diss is the nonspecific complex dissociation rate at 90 mM NaCl, 0 osmolal, pH 7.0, and 20 °C and knsp,diss is the nonspecific complex dissociation rate at the salt and osmolyte concentrations used. From our previous data, ln(knsp,diss/k0nsp,diss) is given by equation (7). The salt concentration is varied from 90 to 360 mM NaCl and the betaine glycine osmolyte concentration from 0 to 4 osmolal to give the range examined.
The ratios of dissociation rates for the fragment with two sites a separated by 84 bp is given by (■). The `star' (★) data points for the Δ84 bp two site fragment were obtained keeping the osmolal concentration of betaine glycine constant and only varying the salt concentration. Dissociation rate ratios for the Δ21 bp two site fragment are shown by (◆) and for the Δ26 two site fragment by (▲). The two sites are on the same side of the DNA helix for Δ84 and Δ21, but on opposite sides for Δ26. The solid lines are calculations of equation (6) with ln(k0nsp,diss/ksl) = −12.6. The arrow represents a 16-fold change in knsp,diss for the 4-fold change in spacing between sites as expected for a diffusion process. A complete data set that includes osmolal and salt concentrations, koff,1-site, and koff,2-site is given in supplementary materials for the Δ21 and Δ84 two site DNA fragments, as well as a graph of fitting errors.
Discussion
The one-dimensional movement of nonspecifically bound protein along DNA can greatly accelerate specific site association kinetics. There have been now several theoretical estimates of the optimal combination of one and three dimensional diffusion 7; 8; 9. This, of course, depends sensitively on the one dimensional diffusion rate. In the cell, so many proteins are bound to DNA blocking linear movement that only small lengths of DNA are essentially accessible to be scanned. There is no advantage to sliding ranges of 1000s of base pairs.
A wide range of one dimensional diffusion coefficients of proteins moving along DNA have been measured using single molecule techniques. Some, as the DNA repair enzyme oxoguanine DNA glycosylase 1 (Ogg1)19 and p5320, have one-dimensional diffusion coefficients almost as large as the three-dimensional diffusion coefficients in dilute solution. There are only small additional energy barriers to movement along DNA. Others, as EcoRV 15, the LacI repressor 17, and the βclamp 16, move significantly more slowly (at least 1000-fold) along DNA than free in solution indicating significant energy barriers.
In addition to one-dimensional diffusion or sliding other mechanisms commonly considered include `hopping' and direct DNA-to-DNA transfer. Hopping is the dissociation and rebinding of protein to the same DNA, i.e., dissociation is only transient 3. Direct transfer is due to the collision of a protein-DNA complex with DNA either on the same fragment or another 3. The protein is transferred from the first DNA site to the second. By using high concentrations of specific site oligonucleotides to act as a sink for dissociated protein we accentuate the contribution from one-dimensional sliding. The specific site dissociation rate does not depend on the oligonucleotide concentration added over a 100 to 1000 range of ratios between specific site concentrations of oligonucleotide and DNA fragment. This indicates that direct transfer does not contribute significantly to our measurements of movement along DNA compared with one-dimensional diffusion. It would also strongly suggest that the contribution from hopping is not significant.
Many DNA binding proteins are not highly stringent for the specific recognition sequence, but can bind to related noncognate sequences more strongly than to completely nonspecific sequences. The sliding rate at a particular site is likely related to the binding constant at that site. Sliding rates could be strongly sequence dependent. In this respect, the binding specificity of EcoRI is extraordinary. In 90 mM NaCl, pH 7.0, and at 20°C, the ratio of equilibrium constants for binding to a specific recognition sequence and to a nonspecific 30bp oligonucleotide 25; 31 is ~2 × 104. A 30 bp oligonucleotide containing the `star' sequence TAATTC that differs by a single base pair from the recognition sequence binds EcoRI only two-fold more strongly than the nonspecific oligonucleotide 31. Other `star' sequences with single base pair changes from the recognition sequence bind even more weakly 35. Sequences with two or more base pair changes from the recognition sequence are virtually indistinguishable from nonspecific DNA 35. The DNA fragments we use here do not contain any `star' sequences between the two sites. We expect that EcoRI sliding rates under our experimental conditions are not particularly sequence dependent.
The ratio of specific site dissociation rates for the one-site fragment and the two-site fragments with ΔL = 84, 26, and 21 bp shown in Figure 3 can all be well fit by equation (6) with ln(k0nsp,diss/ksl) = −12.6 ± 0.2. This would indicate that the assumptions underlying the equations are correct. It was assumed that a recognition site is an absorbing barrier, i.e., that the protein does not pass over or miss a site. It does not matter if the site is on the same side of the helix (Δ21) or the opposite (Δ26); EcoRI has the same high probability of finding the site if the protein diffuses to it. This also indicates that the protein is not transiently dissociating and taking hops of several base pairs on the DNA. Equation (6) also assumes that ksl does not have a significant salt concentration or osmotic pressure dependence. The salt concentrations and osmotic pressures are changed significantly over the range of ln(knsp,diss/k0nsp,diss) from 1 to 9 (an 8000-fold change in rate) in Figure 3. The salt concentration was varied from 90 to 360 mM and the osmotic pressure (to maintain an approximately constant koff) from 0 to 4 osm. The data fits then indicate that ksl does not depend significantly on salt or osmotic pressure, suggesting that the motion along the helix maintains approximately the same cushion of water at the protein-DNA interface and remains in close contact with the DNA-phosphate backbone. This would be consistent with previous suggestions that the nonspecifically bound protein follows the helical path of DNA. In agreement with the EcoRI results here, the one-dimensional diffusion coefficients of Ogg1 19 and p53 20 have little salt concentration sensitivity. A non-negligible salt concentration dependence has been reported, however, for EcoRV sliding 15 with added Mg2+.
We have previously estimated the nonspecific complex dissociation rate, k0nsp,diss, from the effect of added nonspecific 30 bp oligonucleotides on the specific sequence association rate under conditions that there was little free protein 9. The protein was either bound to the specific sequence fragment or to nonspecific oligonucleotide. Under standard conditions (0 osm, 90 mM salt, 20°C, and pH 7.0), k0nsp,diss = 0.8 sec−1. The ratio of this value and k0sp,diss for a 30 bp specific site oligonucleotide was consistent with our previously measured value for the ratio of specific and nonspecific site binding constants. The association and dissociation rates from the 30 bp specific site oligonucleotide are both ~ four-fold slower than for the 360 bp fragments, reflecting the contribution of ends to dissociation kinetics. Dissociation rates from DNA ends can be much different than from internal sequences. We can calculate a ratio of dissociation rates for the specific site 30 and 360 bp DNA fragments, varying the relative dissociation rate from the ends using the formalism of Belotserkovskii and Zarling and equation (4). If there was no difference in dissociation rates from internal and end sequences (c = 1 in equation (2)), the 30 bp oligonucleotide off rate is calculated to be ~11-fold slower than the 360 bp fragment using the k0nsp,diss/ksl ratio determined from the fit to Figure 3. In order to account for the observed ratio of 4, the dissociation rate would have to be ~ 40-fold faster from ends than from internal sequences. The dissociation rate of the 30 bp nonspecific oligonucleotide can be corrected for end dissociation to give k0nsp,diss = 0.2 sec−1. Including 40-fold difference between ends and internal sequences in the calculation of ratio of one and two site dissociation rates for the 360 bp DNA fragments does not significantly change fits shown in Figure 3 or the best fitting value of ln(k0nsp,diss/ksl).
With k0nsp,diss = 0.2 sec−1, we calculate that ksl = 6.0 × 104 bp2/sec, corresponding to a diffusion rate along the helix of ~ 3.5 × 10−11 cm2/sec. Under standard conditions of 90 mM salt, 20°C, and pH 7.0, in the absence of Mg2+, EcoRI can scan ~ 400 bp, on average, before dissociating. The diffusion coefficient is ~ 2000-fold smaller than expected if the translational frictional drag of the solution was limiting, similar to the results for EcoRV. If proteins follow the helical path of DNA, as seems likely for EcoRI, then at least part of the extra friction comes from the rotational motion as first pointed out by Schurr 36 and further developed by Bagchi et al 37. Using these latter, the rotational friction slows the movement of EcoRI along DNA by a factor of 40–50, leaving only an additional factor of 40–50 to account for. This is somewhat surprising given the ~10 charge-charge interactions between DNA and protein in the nonspecific complex. If the remaining factor of 40–50 is due to breaking and remaking these DNA-protein `salt bridges', the activation energy barrier would only be ~ 0.4 kT (~250 cal) for each. Additionally, the 110 waters at the DNA-protein interface could be structured with an energy penalty for perturbing this structuring as the protein moves.
As a final check on the measured sliding rate, we can also estimate the absolute dissociation rate constant of the 360 bp, single specific site DNA fragment. Figure 4 illustrates the steps involved in arriving at the initial state considered in Figure 1 with the protein bound nonspecifically next to the specific site. There is first a transition between specific and nonspecific binding modes at the specific site, followed by a sliding step in either direction. Since the nonspecific binding mode has ~110 water molecules sequestered at the DNA-protein interface, we do not expect that the nonspecific mode binding constant will have a significant dependence on sequence. If the rate of returning to the specific binding mode is much faster than the sliding rate, as seems likely, then the rate of formation of the initial state is 2ksl Ksp-nsp, where Ksp-nsp is the ratio of nonspecific and specific site association constants. The overall dissociation rate is then 2ksl Ksp-nsp Pdiss,1-site, where Pdiss,1-site is the probability that the protein will dissociate before rebinding to the specific site and is given by equation (4). Under our standard experimental conditions, pH 7.0 and 90 mM NaCl, the calculated Pdiss,1-site is 7 × 10−4 for ln(k0 nsp,diss/ksl) = −12.6. As mentioned above, the relative binding constant for the competition between a 360 bp, specific site fragment and a 30 bp nonspecific oligonucleotide is ~ 1 /(2 × 104). If we consider that the 30 bp nonspecific oligonucleotide contains ~25 possible sites, then Ksp-nsp for each nonspecific site ~ 0.5×10−4/25. The specific site dissociation rate constant, koff, for the 360 bp specific site fragment can then be estimated as ~ 1.7×10−4 sec−1, compared with our measured value of ~ 2×10−4 sec−1. The agreement is quite reasonable and supports our analysis.
Figure 4.
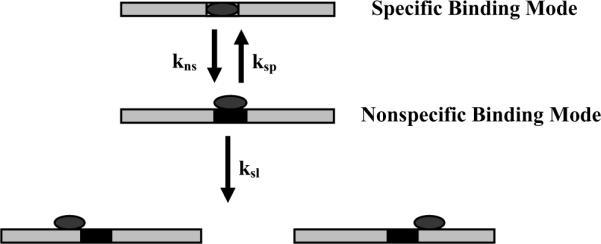
A schematic representation of the kinetic steps involved in the transition from a specifically bound protein to a nonspecific complex abutting the specific site.
Conclusions
By measuring the ratio of dissociation rates of complexes of protein with DNA fragments with one and two specific recognition binding sites, we are able to able to determine the ratio of nonspecific complex dissociation and linear diffusion rates. At 90 mM NaCl, pH 7.0, and 20 °C, EcoRI is able to cover ~ 400 bp, on average, before dissociating. The sliding rate is insensitive to salt and osmolyte concentrations, suggesting that the protein follows the sugar-phosphate backbone of DNA. The diffusion rate is about 2000-fold slower than the diffusion of free protein in solution. A factor of 40–50 can be accounted for by a rotational flow friction if the protein follows the helical path of DNA. The rest is likely due to the transient disruption either of DNA-protein charges interactions or of the water structuring at the protein-DNA interface.
Methods and Materials
Materials
Betaine glycine and triethylene glycol were purchased from Fluka Chemical Co. and used without further purification.
The DNA fragment containing one EcoRI site was prepared from pNEB193 (New England Biolabs) as described below. DNA fragments with two EcoRI sites were constructed by inserting oligonucleotides into pNEB193 cleaved in the polylinker region at two restriction endonuclease sites. The sequence of the oligonucleotide used to construct the two-site DNA with a 21 bp separation between sites (Δ21) was CAGTGAATTCGAGCGCGGGG. It was inserted into the KpnI/BamHI gap of cleaved pNEB193. The sequence of Δ26 two-site oligonucleotide was CGGGCCAGTGAATTCGAGCG and was also inserted into the KpnI/BamHI gap. The Δ84 two-site DNA was constructed with the oligonucleotide GGCGAGGGCCAGTGAATTCG that was inserted into the PstI/HindIII gap. The EcoRI recognition site is underlined. The second EcoRI sites have the same three bp flanking sequences on either side of the site as the native EcoRI site in pNEB193 to ensure both sites bind EcoRI equally well. E. coli (New England Biolabs turbo competent cells) were transformed with the cloned plasmids and individual colonies containing the correct plasmid isolated using standard techniques. Plasmids were isolated from 500 ml cultures using the Sigma GenElute HP Plasmid Megaprep kit. The one- and two-site plasmids were then digested with PvuII and the 360 bp fragments isolated from an agarose gel during electrophoresis. The fragments were then amplified using standard PCR protocols.
The specific sequence oligonucleotide was described in Sidorova and Rau 31. Purified EcoRI was a kind gift of Dr. L. Jen-Jacobson.
Dissociation reaction
The dissociation reaction mixture contained 20 mM Imidazole (7.0), 1 mM EDTA, 2 mM DTT, and 100 μg/ml acetylated BSA (Ambion). The NaCl concentration varied between 90–360 mM and the betaine glycine concentration between 0–4 osmolal (0–24 % w/w). The DNA fragment concentration was 2 nM and the EcoRI concentration was ~0.9 nM. The enzyme was allowed to equilibrate with the DNA fragment for ~ 30 min at 20 °C. The dissociation reaction was initiated by adding oligonucleotide containing the recognition site for EcoRI to a final concentration of 1 μM. The total reaction volume was 30 μl. The dissociation rate is insensitive to oligonucleotide/fragment concentration ratios between ~100–1000. The reaction was stopped at various times by adding 10 μl triethylene glycol as described in Rau 27.
Gel electrophoresis and quantitation
Samples were loaded onto a 12% polyacrylamide gel in 1×TAE and electrophoresed at 180 v for ~14 hrs. The gel was stained with Sybr Green I (Invitrogen) and imaged with an FLA3000 (Fujifilm Life Sciences) laser scanner. Intensities of free DNA and protein-DNA complex bands were analyzed using Multigauge (Fujifilm Life Sciences) and SigmaPlot 10 (Systat Software).
Three bands are seen in the electrophoresis of the two-site fragment with EcoRI (Figure 2a): free DNA, the complex with one bound protein, and, in the uppermost band, the complex with two bound proteins. The initial fractions of free DNA and complexes with one and two bound proteins were consistent with a simple binomial distribution of independent binding sites. The dissociation of the complex with two bound proteins to form a complex with one bound protein complicates analysis of the real dissociation rate of the complex with one bound protein. The dissociation rate of the complex with two bound proteins can be measured from the loss of intensity of the upper band. If the fraction of DNA present as complex with one protein is f1b, the initial fraction of complex with two bound proteins is f02b, and the rate constants for the loss of complexes with one and two bound proteins are k1 and k2, respectively, then the fraction of DNA with one bound protein correcting for the contribution of the complex with two bound proteins, f1b,corr is:
| (9) |
Iteration with the new k1 changes the correction negligibly. The ratio of the dissociation rate of the one-site complex and the rate of loss of the two-site fragment with two bound proteins varied between 1–1.8 suggesting that a bound protein is a reflecting barrier as concluded by Jeltsch et al. 11
Supplementary Material
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Berg OG, Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids: Models and theory. Biochem. 1981;20:6929–6948. doi: 10.1021/bi00527a028. [DOI] [PubMed] [Google Scholar]
- 2.Gorman J, Greene EC. Visualizing one-dimensional diffusion of proteins along DNA. Nature Struct. Molec. Biol. 2008;15:768–774. doi: 10.1038/nsmb.1441. [DOI] [PubMed] [Google Scholar]
- 3.Halford SE, Marko JF. How do site-specific DNA-binding proteins find their targets? Nucl. Acids Res. 2004;32:3040–3052. doi: 10.1093/nar/gkh624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Winter RB, Berg OG, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 3. The Escherichia coli lac repressor-operator interaction: Kinetic measurements and conclusions. Biochem. 1981;20:6961–6977. doi: 10.1021/bi00527a030. [DOI] [PubMed] [Google Scholar]
- 5.Winter RB, von Hippel PH. Diffusion-driven mechanisms of protein translocation on nucleic acids. 2. The Escherichia coli repressor-operator interaction: Equilibrium measurements. Biochem. 1981;20:6948–6960. doi: 10.1021/bi00527a029. [DOI] [PubMed] [Google Scholar]
- 6.Berg OG, von Hippel PH. Diffusion-controlled macromolecular interactions. Annu. Rev. Biophys. Biophys. Chem. 1985;14:131–160. doi: 10.1146/annurev.bb.14.060185.001023. [DOI] [PubMed] [Google Scholar]
- 7.Coppey M, Benichou O, Voituriez R, Moreau M. Kinetics of target site localization of a protein on DNA: A stochastic approach. Biophys. J. 2004;87:1640–1649. doi: 10.1529/biophysj.104.045773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cherstvy AG, Kolomeisky AB, Kornyshev AA. Protein-DNA interactions: Reaching and recognizing the targets. J. Phys. Chem. B. 2008;112:4741–4750. doi: 10.1021/jp076432e. [DOI] [PubMed] [Google Scholar]
- 9.Slutsky M, Mirny LA. Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. Biophys. J. 2004;87:4021–4035. doi: 10.1529/biophysj.104.050765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Halford SE. An end to 40 years of mistakes in DNA-protein association kinetics. Biochem. Soc. Trans. 2009;37:343–348. doi: 10.1042/BST0370343. [DOI] [PubMed] [Google Scholar]
- 11.Jeltsch A, Alves J, Wolfes H, Maass G, Pingoud A. Pausing of the restriction endonuclease EcoRI during linear diffusion on DNA. Biochem. 1994;33:10215–10219. doi: 10.1021/bi00200a001. [DOI] [PubMed] [Google Scholar]
- 12.Jack WE, Terry BJ, Modrich P. Involvement of outside DNA sequences in the major kinetic path by which EcoRI endonuclease locates and leaves its recognition sequence. Proc. Natl. Acad. Sci. USA. 1982;79:4010–4014. doi: 10.1073/pnas.79.13.4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeltsch A, Pingoud A. Kinetic characterization of linear diffusion of the restriction endonuclease EcoRV on DNA. Biochem. 1998;37:2160–2169. doi: 10.1021/bi9719206. [DOI] [PubMed] [Google Scholar]
- 14.Ehbrecht HJ, Pingoud A, Urbanke C, Maass G, Gualerzi C. Linear diffusion of restriction endonucleases on DNA. J. Biol. Chem. 1985;260:6160–6166. [PubMed] [Google Scholar]
- 15.Bonnet I, Biebricher A, Porte P-L, Loverdo C, Benichou O, Voituriez R, Escude C, Wende W, Pingoud A, Desbiolles P. Sliding and jumping of single EcoRV restriction enzymes on non-cognate DNA. Nucl. Acids Res. 2008;36:4118–4127. doi: 10.1093/nar/gkn376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laurence TA, Kwon Y, Johnson A, Hollars CW, O'Donnell M, Camarero JA, Barsky D. Motion of a DNA sliding clamp observed by single molecule fluorescence spectroscopy. J. Biol. Chem. 2008;283:22895–22906. doi: 10.1074/jbc.M800174200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang YM, Cox EC, Austin R. Single molecule measurements of repressor protein 1D diffusion on DNA. Phys. Rev. Lett. 2006;97:048302. doi: 10.1103/PhysRevLett.97.048302. [DOI] [PubMed] [Google Scholar]
- 18.Gorman J, Chowdhury A, Surtees JA, Shimada J, Reichman DR, Alani E, Greene EC. Dynamic basis for one-dimensional DNA scanning by the mismatch repair complex Msh2–Msh6. Molec. Cell. 2007;28:359–370. doi: 10.1016/j.molcel.2007.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Blainey PC, van Oijen AM, Banerjee A, Verdine GL, Xie XS. A base-excision DNA-repair protein finds intrahelical lesion bases by fast sliding in contact with DNA. Proc. Natl. Acad. Sci. USA. 2006;103:5752–5757. doi: 10.1073/pnas.0509723103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tafvizi A, Huang F, Leith JS, Fersht AR, Mirny LA, van Oijen AM. Tumor suppressor p53 slides on DNA with low friction and high stability. Biophys. J. 2008;95:L01–L03. doi: 10.1529/biophysj.108.134122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Komazin-Meredith G, Mirchev R, Golan DE, van Oijen AM, Coen DM. Hopping of a processivity factor on DNA revealed by single molecule assays of diffusion. Proc. Natl. Acad. Sci. USA. 2008;105:10721–10726. doi: 10.1073/pnas.0802676105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Viadiu H, Aggarwal AK. Structure of BamHI bound to nonspecific DNA: A model for sliding. Molec. Cell. 2000;5:889–895. doi: 10.1016/s1097-2765(00)80329-9. [DOI] [PubMed] [Google Scholar]
- 23.Kalodimos CG, Biris N, Bonvin AMJJ, Levandoski MM, Guennuegues M, Boelens R, Kaptein R. Structure and flexibility adaptation in nonspecific and specific protein-DNA complexes. Science. 2004;305:386–389. doi: 10.1126/science.1097064. [DOI] [PubMed] [Google Scholar]
- 24.Winkler FK, Banner DW, Oefner C, Tsernoglou D, Brown RS, Heathman SP, Bryan RK, Martin PD, Petratos K, Wilson KS. The crystal structure of EcoRV endonuclease and of its complexes with cognate and non-cognate DNA fragments. EMBO J. 1993;12:1781–1795. doi: 10.2210/pdb4rve/pdb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidorova NY, Rau DC. Differences in water release for the binding of EcoRI to specific and nonspecific DNA sequences. Proc. Natl. Acad. Sci. USA. 1996;93:12272–12277. doi: 10.1073/pnas.93.22.12272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sidorova NY, Muradymov S, Rau DC. Differences in hydration coupled to specific and nonspecific competitive binding and to specific DNA binding of the restriction endonuclease BamHI. J. Biol. Chem. 2006;281:35656–35666. doi: 10.1074/jbc.M608018200. [DOI] [PubMed] [Google Scholar]
- 27.Rau DC. Sequestered water and binding energy are coupled in complexes of lambda Cro repressor with non-consensus binding sequences. J. Mol. Biol. 2006;361:352–361. doi: 10.1016/j.jmb.2006.06.036. [DOI] [PubMed] [Google Scholar]
- 28.Garner MM, Rau DC. Water release associated with specific binding of gal repressor. EMBO J. 1995;14:1257–1263. doi: 10.1002/j.1460-2075.1995.tb07109.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Halford SE. Hopping, jumping and looping by restriction enzymes. Biochem. Soc. Trans. 2001;29:824–834. doi: 10.1042/bst0290363. [DOI] [PubMed] [Google Scholar]
- 30.Iwahara J, Zweckstetter M, Clore GM. NMR structural and kinetic characterization of a homeodomain diffusing and hopping on nonspecific DNA. Proc. Natl. Acad. Sci. USA. 2006;103:15062–15067. doi: 10.1073/pnas.0605868103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sidorova NY, Rau DC. Differences between EcoRI nonspecific and `star' sequence complexes revealed by osmotic stress. Biophys. J. 2004;87:2564–2576. doi: 10.1529/biophysj.104.042390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Belotserkovskii BP, Zarling DA. Analysis of a one-dimensional random walk with irreversible losses at each step: Applications for protein movement on DNA. J. Theor. Biol. 2004;226:195–203. doi: 10.1016/j.jtbi.2003.08.013. [DOI] [PubMed] [Google Scholar]
- 33.Sidorova NY, Rau DC. Linkage of EcoRI dissociation from its specific DNA recognition site to water activity, salt concentration, and pH: Separating their roles in specific and non-specific binding. J. Mol. Biol. 2001;310:801–816. doi: 10.1006/jmbi.2001.4781. [DOI] [PubMed] [Google Scholar]
- 34.Parsegian VA, Rand RP, Rau DC. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc. Natl. Acad. Sci. USA. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lesser DR, Kurpiewski MR, Jen-Jacobson L. The energetic basis of specificity in the EcoRI endonuclease-DNA interaction. Science. 1990;250:776–786. doi: 10.1126/science.2237428. [DOI] [PubMed] [Google Scholar]
- 36.Schurr JM. The one-dimensional diffusion coefficient of proteins absorbed on DNA. Hydrodynamic considerations. Biophys. Chem. 1978;9:413–414. [PubMed] [Google Scholar]
- 37.Bagchi B, Blaney PC, Xie XS. Diffusion constant of a nonspecifically bound protein undergoing curvilinear motion along DNA. J. Phys. Chem. B. 2008;112:6282–6284. doi: 10.1021/jp077568f. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.