Abstract
The evolution of protein interactions cannot be deciphered without a detailed analysis of interaction interfaces and binding modes. We performed a large-scale study of protein homooligomers in terms of their symmetry, interface sizes, and conservation of binding modes. We also focused specifically on the evolution of protein binding modes from nine families of homooligomers and mapped 60 different binding modes and oligomerization states onto the phylogenetic trees of these families. We observed a significant tendency for the same binding modes to be clustered together and conserved within clades on phylogenetic trees; this trend is especially pronounced for close homologs with 70% sequence identity or higher. Some binding modes are conserved among very distant homologs, pointing to their ancient evolutionary origin, while others are very specific for a certain phylogenetic group. Moreover, we found that the most ancient binding modes have a tendency to involve symmetrical (isologous) homodimer binding arrangements with larger interfaces, while recently evolved binding modes more often exhibit asymmetrical arrangements and smaller interfaces.
Keywords: homooligomer, protein interaction, protein binding mode, evolutionary conservation, homooligomer symmetry
Introduction
Many soluble and membrane-bound proteins form homooligomeric complexes in a cell, although their oligomerization states are often difficult to characterize.1–8 For example, more than three-fourths of all entries in the Protein Quaternary Structure database are homooligomers,9 while the BRENDA Enzyme Database† contains 70% multimeric enzymes, most of them representing homooligomers. It is difficult to overestimate the functional importance of protein oligomerization, which can be used to regulate the activity of many proteins such as enzymes, ion channel proteins, receptors, and transcription factors. Indeed, it has been suggested that large assemblies consisting of many identical subunits have advantageous regulatory properties as they can undergo sensitive phase transitions.10 Oligomerization can also provide sites for allosteric regulation, generate new binding sites at dimer interfaces to increase specificity, and increase diversity in the formation of regulatory complexes.11–16 In addition, oligomerization allows proteins to form large structures without increasing genome size and provides stability, while the reduced surface area of the monomer in a complex can offer protection against denaturation.10,17,18
Recently, analysis of high-throughput protein–protein interaction networks found that there are significantly more self-interacting proteins than expected by chance,19 and that the efficiency of co-aggregation between different protein domains decreases with decreasing sequence identity.20 Several explanations were proposed to account for these observations of self-attraction, including stability and foldability arguments.21,22 It was found, for example, that predictions of energy distributions of homodimers are shifted toward lower energies compared to those of heterodimers.23 The physical effect of a statistically enhanced self-attraction was further modeled to show that interactions between identical random surfaces are stronger than attractive interactions between different random surfaces of the same size.24,25
Stability requirements are important, but are not the only requirements governing protein evolution. Protein evolution optimizes the biological function of a protein and might not necessarily lead to optimal stability or foldability, especially if these properties are antagonistic with functional constraints. Different evolutionary scenarios of protein oligomerization have been discussed in the literature. Some of them propose evolutionary pathways that follow kinetic scenarios of two-state or three-state folding or domain swapping.26–29 At the same time, duplication of homodimers may lead to oligomers of paralogs and may create new protein complexes in evolution.30 Although oligomerization plays an important functional role, the formation of multiple oligomerization interfaces and symmetry requirements puts additional constraints on the evolution of constituent monomers and on the complex itself.
Homooligomers provide convenient systems for studying the evolution of protein interactions using only one phylogenetic tree, thus avoiding the ambiguity of finding corresponding branches between different phylogenetic trees for heterooligomeric complexes. At the same time, the evolution of protein interactions cannot be decoded without a detailed analysis of interaction interfaces and binding modes. This in turn requires information on the atomic details of interacting residues for different and diverse members of a given protein family. In this article, we analyze the general principles of the evolution of homooligomers in terms of their symmetry, interface sizes, and conservation of binding modes, and focus specifically on the evolution of the binding modes of nine homooligomer families. We successfully map different binding modes and oligomerization states on phylogenetic trees and trace their evolution. First, we find that binding modes have a tendency to be conserved between proteins from the same homooligomeric family sharing more than 50% sequence identity, with the trend being more pronounced for close homologs of above 70% identity. This result is important for inferring protein binding modes from known complexes to homologs/interlogs with unannotated interaction modes or binding sites. Second, we show that the most ancient binding modes have a tendency to involve symmetrical larger interfaces, while the more recent binding modes exhibit more asymmetrical smaller interfaces.
Results
Large-scale analysis of homooligomer properties
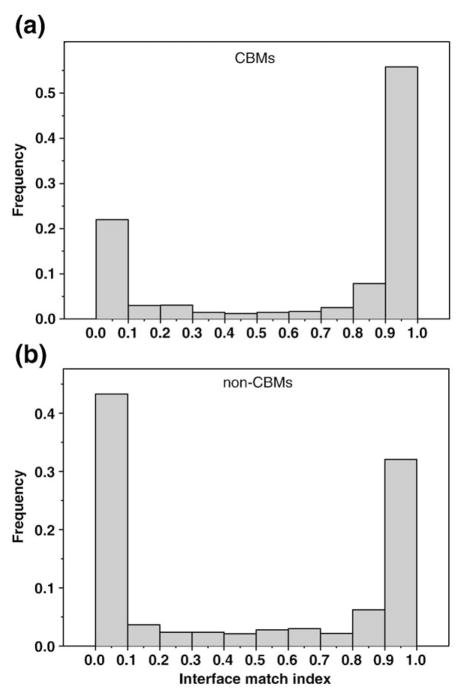
First, we performed a large-scale analysis of conserved binding modes in all homooligomeric structures from the Conserved Binding Mode (CBM) database (1141 homooligomeric families). We found that 64% of families have just one binding mode per family, which might reflect the fact that the majority of all homooligomers are homodimers with one predominant binding arrangement (Fig. S1). There were only 36 homooligomeric families with more than five different binding modes per family. Analysis of the degree of interface similarity in conserved binding modes measured by the interface match index (IMI) shows a bimodal distribution of IMI in the data set of 1141 homooligomeric families with predominant occurrences of symmetrical or isologous interfaces (IMI close to 1) compared to asymmetrical ones (Fig. 1a). This is consistent with a previous observation31 and is the result of a predominant number of complexes with C2 and D2 symmetry types. Interestingly, the distribution of the IMI for binding modes that are not conserved (nonconserved binding modes) shows quite a different situation. As can be seen from Fig. 1b, for nonconserved binding modes, the peak at low IMI is predominant (Fig. 1b). Possible reasons for the strong tendency towards symmetry in conserved binding modes will be discussed later in the article.
Fig. 1.
The histogram of IMI for homooligomers from the overall database for conserved binding modes (a) and nonconserved binding modes (b).
Conservation of binding modes in relation to sequence similarity
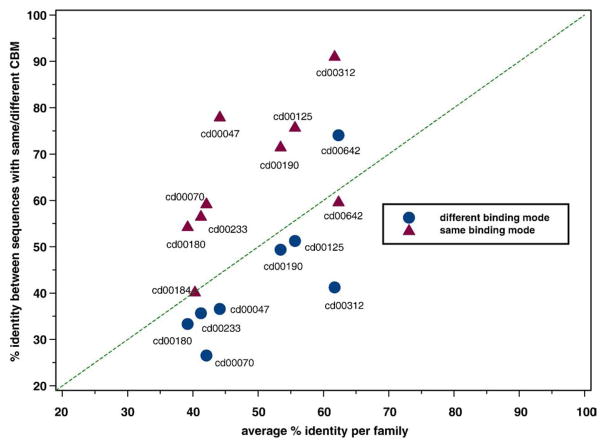
Evolutionary analysis of conserved binding modes was performed on nine example families of homooligomers. First, we analyzed how the conservation of the geometry of binding modes relates to evolutionary distance. Figure S3 shows sequence similarity among protein chains mapped on a phylogenetic tree and sharing the same conserved binding mode. The conserved binding modes from our data set span a wide range of sequence identity; one peak corresponds to evolutionarily older binding modes with about 30% sequence identity, while the other peak corresponds to binding modes with more than 70% sequence identity. If we compare two distributions of sequence identity between sequences sharing the same conserved binding mode and sequences having different conserved binding modes, the difference is found to be statistically significant (P <10−10), with the distribution for sequences sharing the same conserved binding modes shifted toward higher sequence similarity levels. This is also evident in Fig. 2, which shows an average similarity between sequences with the same conserved binding modes and sequences with different conserved binding modes plotted versus the average percent identity per family. As can be seen from this figure, pairs of sequences sharing the same conserved binding mode (triangles) are positioned on the graph higher than the slanted line, quantifying the average level of sequence similarity in the family (sequence similarity around the diagonal would be achieved if conserved binding modes were scattered randomly on the phylogenetic tree). At the same time, those data points corresponding to different conserved binding modes are all located below the diagonal line, except for one. A similar trend is seen if we purge redundant sequences (Fig. S4). This implies that binding modes are clustered together and pretty well conserved within the clades on phylogenetic trees.
Fig. 2.
Average sequence identity between sequences with the same (red triangles) or different (blue circles) conserved binding modes within a given family plotted against the average sequence identity of a family (for cd00184, all chains have at least one common conserved binding mode; no blue circle).
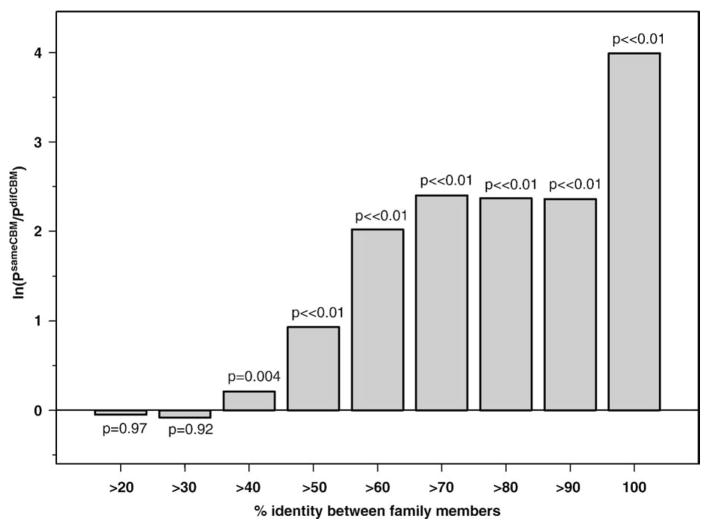
To find whether this conservation is maintained at lower levels of sequence similarity, we divided all sequence pairs into bins based on their sequence identity (those that have higher than 20% identity, those that have higher than 30% identity, and so on, up to the bin containing identical sequences). Figure 3 shows a logarithm of probability ratio for finding the same/different conserved binding modes on a pair of family members sharing similarity above the specified sequence identity level. As shown in Fig. 3, below 50% identity, the probabilities of finding sequences with the same or different binding modes are almost equal (in fact, the probability of finding different conserved binding modes may even be higher such that the logarithm is negative), whereas above this threshold, there is statistically significant enrichment for sequence pairs with the same binding modes. Interestingly, the probabilities of finding sequences with the same binding modes above 70% and 100% identities are 1 order of magnitude higher and almost 2 orders of magnitude higher, respectively, than the probability of finding sequences with different binding modes. The statistically significant association between sequence identity bins above 50% identity and the same/different conserved binding mode categories was also confirmed with the Fisher Exact Test, with larger counts (P ≪ 0.01).32
Fig. 3.
Logarithm of probability ratio for finding the same or different conserved binding modes on a pair of family members from a given bin of sequence identity between them. The first bin includes all members from nine families with more than 20% identity between them; the last bin includes sequence-identical family members. The probabilities of observing a given number (or higher) of sequence pairs with the same binding modes purely by chance were calculated from the binomial distribution and shown above each bar.
Conservation of binding mode symmetries and interface properties
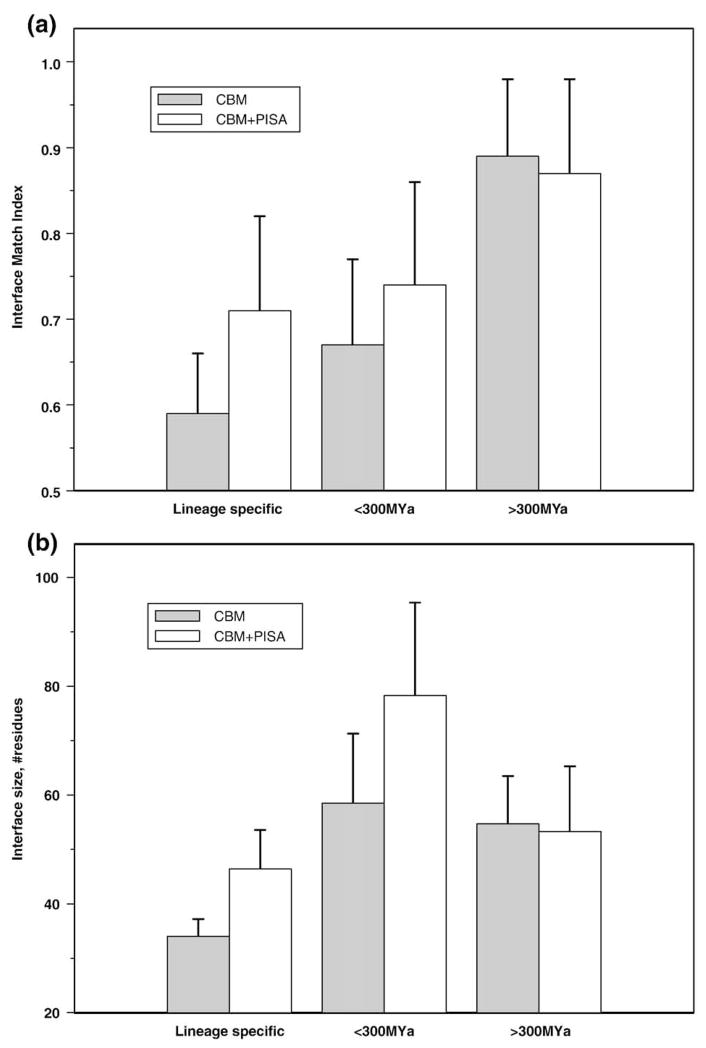
We analyzed different properties of interfaces with respect to the taxonomic diversity of family members with the constituent binding modes. As can be seen from Fig. 4a (gray bar), the IMI is the highest (tendency for isologous interfaces) for evolutionarily older binding modes and decreases for binding modes that were developed in evolution more recently. The difference between interface match indices in “ancient” and lineage-specific categories is statistically significant (the null hypothesis on the equality of mean values was rejected with P<0.01). This trend is less obvious with PISA, which was used as another reference point for identifying interactions predicted to be biological (Fig. 4a, empty box); the sample size could be an issue in this case, since PISA did not provide assignments for a number of structures from our data set.
Fig. 4.
The average IMI (a) and interface size (b) are plotted versus evolutionary age for each conserved binding mode. Evolutionary age is defined as divergence time between the most remotely related species with a given binding mode. Three categories are shown: “>300MYa”—corresponding to those binding modes found in species that diverged from each other more than 300MYa “<300MYa”—binding modes found in more than one species diverged less than 300MYa; and “lineage specific”—binding modes found in one species only. Interactions verified by conserved binding mode analysis are shown in gray (38, 10, and 10 data points in each bin, respectively). Empty boxes show results for those structures where PISA provided an oligomeric state assignment and was in agreement with conserved binding modes (13, 6, and 9, observations in each bin, respectively).
Figure 4b shows interface sizes (measured as the number of residues on both sides of the interface) for different evolutionary age categories. Interface size was found to be significantly smaller for lineage-specific binding modes compared to the more ancient binding modes (P<0.002). This observation was also confirmed using PISA. In accordance with this observation, it was elegantly demonstrated recently on examples of 52 complexes that the largest interface is maintained consistently during complex (dis) assembly, which mimics evolutionary pathways.33 It should be noted that there is a possibility that conserved binding modes from a lineage-specific category actually occur in more species, but that those structures are not yet present in the Protein Data Bank (PDB). Nevertheless, we would not expect a bias toward nonsymmetrical or smaller interfaces to be systematic.
Evolution of oligomeric symmetries
Most homooligomeric proteins form compact complexes with subunits related to each other by point group symmetry. We extracted information about crystallographic point group symmetries from the 3DComplex database,34 which provided us with quaternary structure symmetry assignments for 52 out of 144 structures used in our analysis. Half of them represent dimers with isologous interfaces from cyclic and dihedral C2 and D2 symmetry complexes, and most others are from C3, D3, and D5 symmetry complexes. We mapped group symmetry assignments of structures on the phylogenetic trees. Despite missing symmetry assignments, there is a trend to conserve group symmetry within the phylogenetic clade on the tree, with some cases of conservation going back in time (conservation of C2 symmetry for diverse homologs with ~20–40% identity for cd00070 and cd00312, cd00184, and cd00642 families). This is consistent with previous studies that showed that, in this range of sequence similarity, symmetry type is conserved in approximately 70% of cases.33
Crystallographic point group symmetries, evolutionary age, and IMI are given in Supplementary Materials, Tables S1–S9. Below, we present an analysis of the evolution of binding modes and symmetry types on three examples: the galectin, serine/threonine protein kinase, and esterase families.
Examples of binding mode evolution
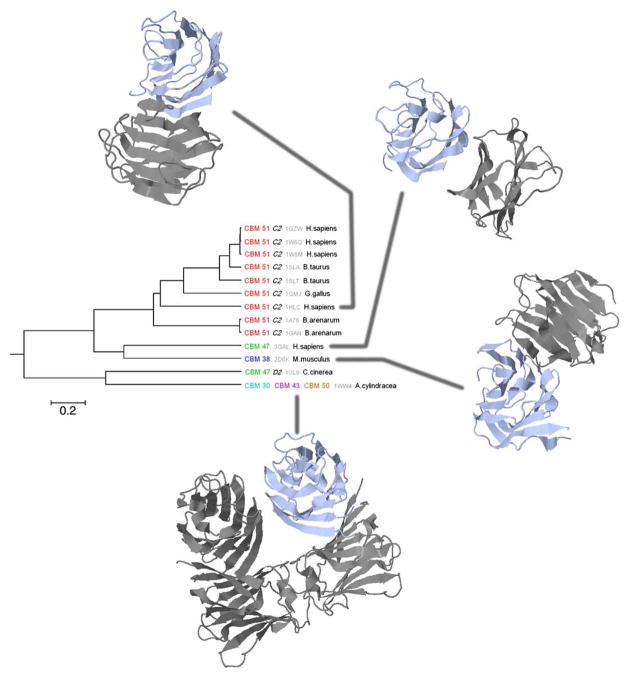
Galectin family
Oligomerization is very important for the functioning of the galectin family (cd00070), which binds β-galactosides, and is involved in all processes related to cell adhesion. For example, the tetrameric structure allows for a precise positioning of glyco-ligands on galectins and provides selective binding of certain ligands to galectin molecules. Oligomerization has also been shown to increase binding affinity and to allow the separation of binding sites into horizontal and vertical orientations with respect to the cell surface.35
Figure 5 shows the phylogenetic tree for the galectin family (cd00070); each branch of the tree is labeled by the PDB code of the corresponding structure, conserved binding mode identifiers, symmetry type (if available), and organism name. Six different binding modes can be seen on the tree, with three appearing individually and the other three appearing together in the same structure. A summary of the binding mode features is given in Table S1.
Fig. 5.
Phylogenetic tree for the galectin family (cd00070) with conserved binding mode identifiers and taxonomy annotations.
The most prevalent, CBM 51, covers a wide range of species (human, cattle, chicken, and toad). Its interface consists of four anti-parallel strands—two from each protomer, forming a dense network of interactions. Two other binding modes (CBMs 47 and 38) represent different spatial arrangements of two protomers, where the tips of several strands come into close proximity with each other. After mapping the symmetry assignments on the tree, one can see that majority of proteins, represent dimers with C2 symmetry, whereas one structure (from fungus) is annotated as a tetramer with D2-type symmetry.
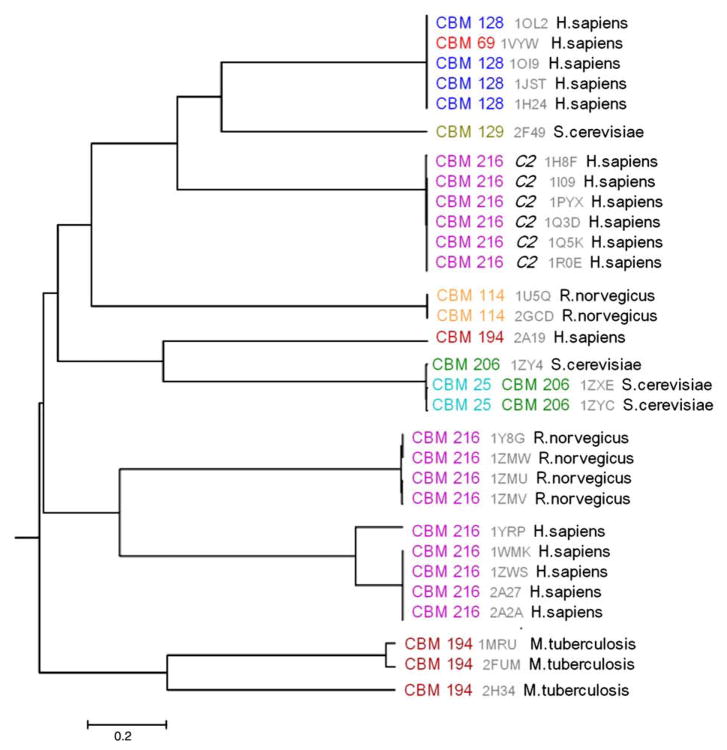
Serine/threonine protein kinases
Figure 6 shows the phylogenetic tree for the serine/threonine protein kinase family (cd00180). The enzymatic activity of these protein kinases is controlled by phosphorylation of specific residues in the activation segment of the catalytic domain. It has been proposed that protein kinases can be regulated through homooligomerization. In particular, ligand binding can promote dimerization of a catalytic domain, which in turn induces autophosphorylation. The monomeric state is inactive, probably due to the displacement of helix αC, which is thought to connect dimerization with the catalytic function of protein kinases.36,37 Table S2 shows the features of the conserved binding modes for this family. Eight binding modes appear on the phylogenetic tree for serine/threonine kinases. Some of them, such as CBMs 216 and 194, comprise diverse sequences, while yeast and rats seem to develop specific binding modes that are not seen on the tree anywhere else.
Fig. 6.
Phylogenetic tree for the serine/threonine protein kinases (cd00180) with conserved binding mode identifiers and taxonomy annotations.
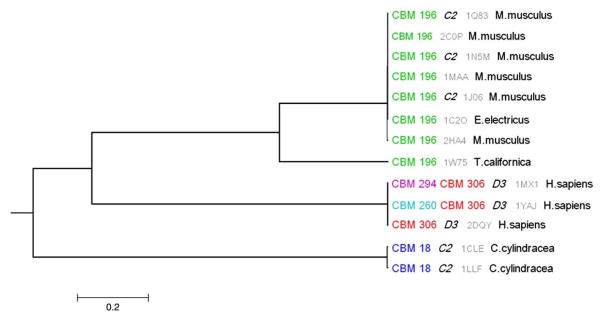
Esterases and lipases
Figure 7 shows the phylogenetic tree for the esterase/lipase family (cd00312), which includes esterases and lipases that act on carboxylic esters. The phylogenetic tree shows that each taxonomic group has characteristic binding modes. For example, CBM 18 is specific for yeast, while CBMs 260, 294, and 306 only occur in human proteins. Inspection of symmetry types assigned to different branches on the tree shows C2 symmetries in yeast and mice and D3 symmetries in humans. The most parsimonious scenario suggests the evolution of these oligomers from C2 dimers, with subsequent acquisition of D3 arrangements by humans. Indeed, it has been shown that many enzymes from this family are regulated through dimerization or higher-order trimer–hexamer transitions,38 while cholesterol esterases from yeast apparently function as dimers, where the active site is positioned on the dimer interface.39 In addition, recent studies propose an evolutionary scenario where homooligomers with dihedral symmetry evolve through their cyclic intermediates.33,40
Fig. 7.
Phylogenetic tree for the esterase/lipase family (cd00312) with conserved binding mode identifiers and taxonomy annotations.
Discussion
We have explored the evolutionary patterns of conserved binding modes and oligomeric symmetries for a spectrum of homooligomeric families and have identified aspects of the interplay between evolution and protein binding. The vast majority of homologous families of homooligomers exhibit just one binding mode conserved within the family, while a larger variety of binding modes exist for other families. These families are the subject of our study. Our analysis of nine families and 60 different binding modes from 144 structures shows that binding modes are usually well conserved within phylogenetic clades, and that protein chains from the same family of homooli-gomers with more than 50% sequence identity have a significantly higher tendency to have the same binding mode than random assignments. Moreover, for proteins above 70% sequence identity, the probability of sharing the same binding mode increases significantly. Many protein interaction prediction methods rely on evolutionary relationships and look for sequence similarity between unannotated proteins and proteins with known interactions, the so-called interolog mapping.41,42 It has been suggested that interaction partners can be reliably inferred only for close homologs,43,44 while inference of protein binding modes is still a topic of ongoing research.45,46
While it is tempting to draw general conclusions about the conservation of protein binding modes, one should keep in mind that these trends are family specific; although some binding modes are conserved among very distant homologs, pointing to an old evolutionary origin, others are specific for a certain phylogenetic clade. Moreover, there might be differences in the way protein complexes have evolved in major taxonomic groups, and novel interactions might be acquired rapidly in evolution through domain recombinations.47 Even though the sequence–structure gap continuously closes with the progress of the structural genomics initiative, the task of mapping protein structural complexes on phylogenetic trees remains extremely complex given that many family representatives do not have structures and those that do often lack interacting partners. This highlights the fact that structure-based phylogenetic studies cannot currently match the statistical power of sequence-based approaches—this is the tradeoff for gaining insights into the atomic details of interactions from structures.
Our analysis also shows that there is a prevalence of symmetrical homodimer binding modes in PDB, which mostly come from cyclic C2 symmetry dimers and dihedral D2 tetramers. The IMI of two randomly docked protomers of the same type depends on the ratio of the binding interface size to the overall surface area. For most nonbiological interfaces covering a moderate amount of surface area, this ratio is expected to be low. Despite this, we observe a bias towards high values of IMI for biologically relevant conserved binding modes. From the analysis of homooligomeric families, we also find that there is a tendency for evolutionarily older binding modes to contain symmetrical binding arrangements, a finding consistent with a recent study.48 The binding modes that correspond to proteins that diverged less than 300 MYrs ago exhibit a mixture of both symmetrical and asymmetrical arrangements, with a tendency to increase asymmetry for more recent binding modes. In addition, the interfaces for lineage-specific binding modes, which correspond to relatively recently acquired binding arrangements, tend to be smaller and probably less stable compared to more evolutionarily established modes. Interestingly, the most recent study that modeled the thermodynamics and kinetics of the self-assembly of D2 tetramers similarly demonstrated that newer interactions might be weaker.40
It has been discussed earlier that symmetrical arrangements can be evolutionarily advantageous for reasons of stability, folding, and function.21–23,25 The role of symmetry has also been considered with regard to domain swapping49 and protein folding.50 The energy landscape of a symmetrical oligomer was argued to be smoother, resulting in faster folding.50 We also argue that symmetrical arrangements would be favorable for preventing protein aggregation, since asymmetrical arrangements might expose interaction-prone interfaces and result in infinitely long polymers. Homooligomers, indeed, were shown to have a lower propensity for aggregation compared to heterooligomers.51 At the same time, we recently showed that intrinsic disorder in symmetrical homooligomers might also have a pronounced functional importance. Symmetrical arrangements might keep disordered regions close together in space to form joint binding interfaces or to regulate the accessibility of binding partners.14
It is intriguing to see how protein families can develop a variety of binding orientations within a relatively short period of time while keeping the essential evolutionarily conserved binding arrangements. Such variations enable the families to accommodate different functional specificities and regulatory mechanisms within the framework of their general function. With recent advancements in the structural identification of protein complexes, one would anticipate that protein binding and binding mode evolution will be extensively studied and their mechanisms will be revealed.
Materials and Methods
Defining conserved binding modes
Oligomeric interfaces were taken from structural complexes from the PDB database,52 and their biological relevance was confirmed by using the CBM database‡53 and the PISA algorithm§.54 In the last 3 years, the CBM database has grown from its original release, which contained 1416 different conserved binding modes, to 3525 conserved binding modes in the most recent version.
The CBM database defines binding modes between two protein domains using domain families from the Conserved Domain Database (CDD) (in the current study, we used version 2.10).55 The definition and construction of a binding mode begin with a distance threshold to determine which residues from two different domain occurrences are close enough to be considered interacting. Any residue from one domain with an atom (excluding hydrogen) within 6 Å of an atom on the other domain is identified as part of the binding surface; the positions of all such residues at the surface constitute the binding mode for modes with at least five such residue–residue pairs. In determining whether a binding mode is conserved, at least two occurrences of the interacting domain pair from different structures (with different cell constants) must contain 50% of the interface residues in the same positions as determined by Vector Alignment Search Tool.56 This criterion filters out spurious crystal packing interactions and retains biologically relevant interactions and complexes. The CBM database organizes pairwise protein domain–domain interactions by binding orientation and lists their properties, such as the domain families for each interface, the chains involved, the residues on each chain, the PDB structures, and their taxonomies. Homodimers are defined as domain interactions where both domains in a pair belong to the same CDD family. The PISA algorithm is a new method used for the automatic detection of macromolecular assemblies within PDB entries that are the results of X-ray diffraction experiments.54 It is used to validate oligomeric states and interfaces between different protein chains based on stability calculations of multimeric states inferred from crystalline states. PISA provided oligomeric assignments for complexes containing 31 of the 58 conserved binding modes used in this study. Information about PISA assignments is available in Supplementary Materials.
Quaternary structure symmetries were derived from the 3DComplex database.34 This database analyzes the topological arrangements of subunits in structural complexes and compares the biological unit assignments from PDB with the Protein Quaternary Structure database.9
Mapping of binding modes on phylogenetic trees of domain families
To study the evolution of binding modes, we chose domain families with different homooligomerization states, several conserved binding modes, and a variety of taxonomic groups. First, we started with all homooligomers from manually curated CDD families and selected the ones that have at least five distinct conserved binding modes (36 CDD families). After purging the redundancy between different families, removing cases with overwhelming numbers of family members (such as immunoglobulins), and manually inspecting the phylogenetic trees, we obtained nine manually curated CDD families, which covered 144 different structures and 60 different conserved binding modes (58 interchain and 2 intrachain conserved binding modes; the 2 intrachain conserved binding modes were mapped on the trees but were excluded from the quantitative analysis). It should be mentioned that the task of obtaining a sufficiently diverse set of binding modes mapped onto phylogenetic trees is a difficult one, since many family representatives do not have structures, and those that do often lack interacting partners. The selected families, as well as information on conserved binding modes, taxonomy, sequence spans, and other parameters, are given in Supplementary Materials.
To ensure adequate mapping between the CBM database and the CDD database, we merged additional sequences with known structures from the CBM database to the CDD domains using RPS-BLAST57 and purged identical sequences. Phylogenetic trees of homooligomers were constructed from first chains “A” listed in PDB files using the UPGMA algorithm58 and the amino acid substitution model of Jones et al.59 Some trees were manually rerooted, when necessary, to adhere to correct ancient evolutionary branching.
All conserved binding modes were categorized in terms of the symmetry of homodimers, their sequence, and their taxonomic diversity. The symmetry of a homodimer interface was estimated by the IMI, which was defined as the number of equivalent interfacial positions in structure–structure alignment on both subunits divided by the overall number of residues on both sides of the interface. Let X be the set containing the positions of residues on the first interface, and let Y be the set containing the positions of residues on the second interface. Note that the intersection of X and Y is the set of residue positions on the first interface that appear also on the second interface, and vice versa. The IMI can then be defined as twice the cardinality (number of members) of X∩Y (counting matching residue positions on both sides) divided by the sum of the cardinality of X plus the cardinality of Y. Hence:
where |X| denotes the cardinality of set X and |Y| denotes the cardinality of set Y.
We have also subdivided all conserved binding modes into categories based on their taxonomic diversity and tried to find the relationship between the evolutionary age of a given binding mode and its different properties, including symmetry, interface size, and oligomeric state. The first category of conserved binding modes represents the most ancient binding modes in our data set, corresponding to those conserved binding modes found in species that diverged from each other more than 300 MYrs ago (approximate time of divergence between Reptilians and Mammalians). The second category includes conserved binding modes that are found in more than one species that diverged less than 300 MYrs ago. Finally, the third category constitutes conserved binding modes that are found in one-species-only lineage-specific binding modes.
Supplementary Material
Acknowledgments
The authors thank Chris Lanczycki for help with the CDtree software. The work was supported by the Intramural Research Program of the National Library of Medicine at the National Institutes of Health/Department of Health and Human Services. J.E.D. also thanks the Oak Ridge Institute for Science and Education for the visiting fellowship.
Abbreviations used
- CBM
Conserved Binding Mode
- IMI
interface match index
- PDB
Protein Data Bank
- CDD
Conserved Domain Database
Footnotes
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at doi:10.1016/j.jmb.2009.10.052
References
- 1.Mon od J. In: Symmetry and Function of Biological Systems at the Macromolecular Level. Engstrom ASB, editor. Wiley; New York, NY: 1969. [Google Scholar]
- 2.Bahadur RP, Chakrabarti P, Rodier F, Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- 3.Ali MH, Imperiali B. Protein oligomerization: how and why. Bioorg Med Chem. 2005;13:5013–5020. doi: 10.1016/j.bmc.2005.05.037. [DOI] [PubMed] [Google Scholar]
- 4.Ponstingl H, Kabir T, Gorse D, Thornton JM. Morphological aspects of oligomeric protein structures. Prog Biophys Mol Biol. 2005;89:9–35. doi: 10.1016/j.pbiomolbio.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 5.Janin J, Rodier F. Protein–protein interaction at crystal contacts. Proteins. 1995;23:580–587. doi: 10.1002/prot.340230413. [DOI] [PubMed] [Google Scholar]
- 6.Carugo O, Argos P. Protein–protein crystal-packing contacts. Protein Sci. 1997;6:2261–2263. doi: 10.1002/pro.5560061021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasgupta S, Iyer GH, Bryant SH, Lawrence CE, Bell JA. Extent and nature of contacts between protein molecules in crystal lattices and between subunits of protein oligomers. Proteins. 1997;28:494–514. doi: 10.1002/(sici)1097-0134(199708)28:4<494::aid-prot4>3.0.co;2-a. [DOI] [PubMed] [Google Scholar]
- 8.Saha RP, Bahadur RP, Chakrabarti P. Interresidue contacts in proteins and protein–protein interfaces and their use in characterizing the homodimeric interface. J Proteome Res. 2005;4:1600–1609. doi: 10.1021/pr050118k. [DOI] [PubMed] [Google Scholar]
- 9.Henrick K, Thornton JM. PQS: a Protein Quaternary Structure file server. Trends Biochem Sci. 1998;23:358–361. doi: 10.1016/s0968-0004(98)01253-5. [DOI] [PubMed] [Google Scholar]
- 10.Goodsell DS, Olson AJ. Structural symmetry and protein function. Annu Rev Biophys Biomol Struct. 2000;29:105–153. doi: 10.1146/annurev.biophys.29.1.105. [DOI] [PubMed] [Google Scholar]
- 11.Hattori T, Ohoka N, Inoue Y, Hayashi H, Onozaki K. C/EBP family transcription factors are degraded by the proteasome but stabilized by forming dimer. Oncogene. 2003;22:1273–1280. doi: 10.1038/sj.onc.1206204. [DOI] [PubMed] [Google Scholar]
- 12.Marianayagam NJ, Sunde M, Matthews JM. The power of two: protein dimerization in biology. Trends Biochem Sci. 2004;29:618–625. doi: 10.1016/j.tibs.2004.09.006. [DOI] [PubMed] [Google Scholar]
- 13.Jaenicke R, Lilie H. Folding and association of oligomeric and multimeric proteins. Adv Protein Chem. 2000;53:329–401. doi: 10.1016/s0065-3233(00)53007-1. [DOI] [PubMed] [Google Scholar]
- 14.Fong J, Shoemaker BA, Garbuzynskiy SO, Lobanov MY, Galzitskaya OV, Panchenko AR. Intrinsic disorder in protein interactions: insights from a comprehensive structural analysis. PLoS Comput Biol. 2009;5:e1000316. doi: 10.1371/journal.pcbi.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ofran Y, Rost B. Analysing six types of protein–protein interfaces. J Mol Biol. 2003;325:377–387. doi: 10.1016/s0022-2836(02)01223-8. [DOI] [PubMed] [Google Scholar]
- 16.Zhao H, Naganathan S, Beckett D. Thermodynamic and structural investigation of bispecificity in protein–protein interactions. J Mol Biol. 2009;389:336–348. doi: 10.1016/j.jmb.2009.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Miller S, Lesk AM, Janin J, Chothia C. The accessible surface area and stability of oligomeric proteins. Nature. 1987;328:834–836. doi: 10.1038/328834a0. [DOI] [PubMed] [Google Scholar]
- 18.Jones S, Thornton JM. Protein–protein interactions: a review of protein dimer structures. Prog Biophys Mol Biol. 1995;63:31–65. doi: 10.1016/0079-6107(94)00008-w. [DOI] [PubMed] [Google Scholar]
- 19.Ispolatov I, Yuryev A, Mazo I, Maslov S. Binding properties and evolution of homodimers in protein–protein interaction networks. Nucleic Acids Res. 2005;33:3629–3635. doi: 10.1093/nar/gki678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wright CF, Teichmann SA, Clarke J, Dobson CM. The importance of sequence diversity in the aggregation and evolution of proteins. Nature. 2005;438:878–881. doi: 10.1038/nature04195. [DOI] [PubMed] [Google Scholar]
- 21.Cornish-Bowden AJ, Koshland DE., Jr The quaternary structure of proteins composed of identical subunits. J Biol Chem. 1971;246:3092–3102. [PubMed] [Google Scholar]
- 22.Blundell TL, Srinivasan N. Symmetry, stability, and dynamics of multidomain and multi-component protein systems. Proc Natl Acad Sci USA. 1996;93:14243–14248. doi: 10.1073/pnas.93.25.14243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lukatsky DB, Zeldovich KB, Shakhnovich EI. Statistically enhanced self-attraction of random patterns. Phys Rev Lett. 2006;97:178101. doi: 10.1103/PhysRevLett.97.178101. [DOI] [PubMed] [Google Scholar]
- 24.Lukatsky DB, Shakhnovich BE, Mintseris J, Shakhnovich EI. Structural similarity enhances interaction propensity of proteins. J Mol Biol. 2007;365:1596–1606. doi: 10.1016/j.jmb.2006.11.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Andre I, Strauss CE, Kaplan DB, Bradley P, Baker D. Emergence of symmetry in homooligomeric biological assemblies. Proc Natl Acad Sci USA. 2008;105:16148–16152. doi: 10.1073/pnas.0807576105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bennett MJ, Schlunegger MP, Eisenberg D. 3D domain swapping: a mechanism for oligomer assembly. Protein Sci. 1995;4:2455–2468. doi: 10.1002/pro.5560041202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.D’Alessio G. Oligomer evolution in action? Nat Struct Biol. 1995;2:11–13. doi: 10.1038/nsb0195-11. [DOI] [PubMed] [Google Scholar]
- 28.Xu D, Tsai CJ, Nussinov R. Mechanism and evolution of protein dimerization. Protein Sci. 1998;7:533–544. doi: 10.1002/pro.5560070301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tiana G, Broglia RA. Design and folding of dimeric proteins. Proteins. 2002;49:82–94. doi: 10.1002/prot.10196. [DOI] [PubMed] [Google Scholar]
- 30.Pereira-Leal JB, Levy ED, Kamp C, Teichmann SA. Evolution of protein complexes by duplication of homomeric interactions. Genome Biol. 2007;8:R51. doi: 10.1186/gb-2007-8-4-r51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuchiya Y, Kinoshita K, Nakamura H. Analyses of homooligomer interfaces of proteins from the complementarity of molecular surface, electrostatic potential and hydrophobicity. Protein Eng Des Sel. 2006;19:421–429. doi: 10.1093/protein/gzl026. [DOI] [PubMed] [Google Scholar]
- 32.Langsrud O, Jørgensen K, Ragni Ofstad R, Næs T. Analyzing designed experiments with multiple responses. J Appl Stat. 2007;34:1275–1296. [Google Scholar]
- 33.Levy ED, Boeri Erba E, Robinson CV, Teichmann SA. Assembly reflects evolution of protein complexes. Nature. 2008;453:1262–1265. doi: 10.1038/nature06942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Levy ED, Pereira-Leal JB, Chothia C, Teichmann SA. 3D complex: a structural classification of protein complexes. PLoS Comput Biol. 2006;2:e155. doi: 10.1371/journal.pcbi.0020155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Walser PJ, Haebel PW, Kunzler M, Sargent D, Kues U, Aebi M, Ban N. Structure and functional analysis of the fungal galectin CGL2. Structure. 2004;12:689–702. doi: 10.1016/j.str.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 36.Wehenkel A, Fernandez P, Bellinzoni M, Catherinot V, Barilone N, Labesse G, et al. The structure of PknB in complex with mitoxantrone, an ATP-competitive inhibitor, suggests a mode of protein kinase regulation in mycobacteria. FEBS Lett. 2006;580:3018–3022. doi: 10.1016/j.febslet.2006.04.046. [DOI] [PubMed] [Google Scholar]
- 37.Dar AC, Dever TE, Sicheri F. Higher-order substrate recognition of eIF2alpha by the RNA-dependent protein kinase PKR. Cell. 2005;122:887–900. doi: 10.1016/j.cell.2005.06.044. [DOI] [PubMed] [Google Scholar]
- 38.Bencharit S, Morton CL, Hyatt JL, Kuhn P, Danks MK, Potter PM, Redinbo MR. Crystal structure of human carboxylesterase 1 complexed with the Alzheimer’s drug tacrine: from binding promiscuity to selective inhibition. Chem Biol. 2003;10:341–349. doi: 10.1016/s1074-5521(03)00071-1. [DOI] [PubMed] [Google Scholar]
- 39.Ghosh D, Wawrzak Z, Pletnev VZ, Li N, Kaiser R, Pangborn W, et al. Structure of uncomplexed and linoleate-bound Candida cylindracea cholesterol esterase. Structure. 1995;3:279–288. doi: 10.1016/s0969-2126(01)00158-7. [DOI] [PubMed] [Google Scholar]
- 40.Villar G, Wilber AW, Williamson AJ, Thiara P, Doye JP, Louis AA, et al. Self-assembly and evolution of homomeric protein complexes. Phys Rev Lett. 2009;102:118106. doi: 10.1103/PhysRevLett.102.118106. [DOI] [PubMed] [Google Scholar]
- 41.Matthews LR, Vaglio P, Reboul J, Ge H, Davis BP, Garrels J, et al. Identification of potential interaction networks using sequence-based searches for conserved protein–protein interactions or “interologs”. Genome Res. 2001;11:2120–2126. doi: 10.1101/gr.205301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shoemaker BA, Zhang D, Thangudu RR, Tyagi M, Fong JH, Marchler-Bauer A, et al. Inferred Biomolecular Interaction Server—a Web server to analyze and predict protein interacting partners and binding sites. Nucleic Acids Res. 2009;37:1–7. doi: 10.1093/nar/gkp842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu H, Luscombe NM, Lu HX, Zhu X, Xia Y, Han JD, et al. Annotation transfer between genomes: protein–protein interologs and protein–DNA regulogs. Genome Res. 2004;14:1107–1118. doi: 10.1101/gr.1774904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mika S, Rost B. Protein–protein interactions more conserved within species than across species. PLoS Comput Biol. 2006;2:e79. doi: 10.1371/journal.pcbi.0020079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J Mol Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 46.Xu Q, Canutescu AA, Wang G, Shapovalov M, Obradovic Z, Dunbrack RL., Jr Statistical analysis of interface similarity in crystals of homologous proteins. J Mol Biol. 2008;381:487–507. doi: 10.1016/j.jmb.2008.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dessailly BH, Reid AJ, Yeats C, Lees JG, Cuff A, Orengo CA. The evolution of protein functions and networks: a family-centric approach. Biochem Soc Trans. 2009;37:745–750. doi: 10.1042/BST0370745. [DOI] [PubMed] [Google Scholar]
- 48.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein–protein interactions: a compendium of interface geometry. PLoS Comput Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bennett MJ, Choe S, Eisenberg D. Domain swapping: entangling alliances between proteins. Proc Natl Acad Sci USA. 1994;91:3127–3131. doi: 10.1073/pnas.91.8.3127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wolynes PG. Symmetry and the energy landscapes of biomolecules. Proc Natl Acad Sci USA. 1996;93:14249–14255. doi: 10.1073/pnas.93.25.14249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chen Y, Dokholyan NV. Natural selection against protein aggregation on self-interacting and essential proteins in yeast, fly, and worm. Mol Biol Evol. 2008;25:1530–1533. doi: 10.1093/molbev/msn122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Henrick K, Feng Z, Bluhm WF, Dimitropoulos D, Doreleijers JF, Dutta S, et al. Remediation of the Protein Data Bank archive. Nucleic Acids Res. 2007;36:D426–D433. doi: 10.1093/nar/gkm937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Shoemaker BA, Panchenko AR, Bryant SH. Finding biologically relevant protein domain interactions: conserved binding mode analysis. Protein Sci. 2006;15:352–361. doi: 10.1110/ps.051760806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J Mol Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 55.Marchler-Bauer A, Anderson JB, Derbyshire MK, DeWeese-Scott C, Gonzales NR, Gwadz M, et al. CDD: a Conserved Domain Database for interactive domain family analysis. Nucleic Acids Res. 2007;35:D237–D240. doi: 10.1093/nar/gkl951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Madej T, Gibrat JF, Bryant SH. Threading a database of protein cores. Proteins. 1995;23:356–369. doi: 10.1002/prot.340230309. [DOI] [PubMed] [Google Scholar]
- 57.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sneath PH, Sokal RR. Numerical Taxonomy. Freeman; San Francisco, CA: 1973. [Google Scholar]
- 59.Jones DT, Taylor WR, Thornton JM. The rapid generation of mutation data matrices from protein sequences. Comput Appl Biosci. 1992;8:275–282. doi: 10.1093/bioinformatics/8.3.275. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.