Abstract
Comprehensive mutliple sequence alignments of the multi-subunit DNA-dependent RNA polymerase large subunits, including the bacterial β and β′ subunits and their homologs from archaebacterial RNA polymerases, eukaryotic RNA polymerases I, II, and III, nuclear-cytoplasmic large double-stranded DNA virus RNA polymerases, and plant plastid RNA polymerases, were created (Lane & Darst). The alignments were used to delineate sequence regions shared among all classes of mutli-subunit RNA polymerases, defining common, fundamental RNA polymerase features as well as identifying highly conserved positions. Here, we present a systematic, detailed structural analysis of these shared regions and highly conserved positions in terms of the RNA polymerase structure, as well as the RNA polymerase structure/function relationship, when known.
Keywords: Evolution, RNA polymerase, Sequence analysis
Introduction
All transcription in cellular organisms is driven by a large, mutli-subunit molecular machine, the DNA-dependent RNA polymerase (RNAP) 1. In its simplest bacterial form, the enzyme comprises five subunits with a total molecular mass of around 400 kDa. The core component (subunit composition α2ββ′ω) is evolutionarily conserved, particularly evident among the large subunit (bacterial β and β′ subunits) homologs 2–6. Comprehensive mutliple sequence alignments (MSAs) of the multi-subunit DNA-dependent RNA polymerase (RNAP) large subunits from bacteria (bRNAP), plant plastids (pRNAP), archaea (aRNAP), eukaryotes (eRNAP I, II, and III), as well as nuclear-cytoplasmic large double-stranded DNA viruses (vRNAP), were created 4. The alignments were used to delineate sequence regions common to all classes of mutli-subunit RNAPs and to all bRNAPs, to delineate bacterial lineage-specific domain insertions, and to analyze other aspects of the large subunit gene organization 4.
The complete delineation of shared sequence regions defines common, fundamental RNAP features. In addition to delineating shared sequence regions, the MSAs allowed the identification of highly conserved positions. These common features and specific residues required for RNAP function are best appreciated in the context of RNAP structures 6–8, and the RNAP structure/function relationship, when known. Here, we present a systematic, detailed structural analysis of these shared regions and highly conserved positions.
Results
This structure/function analysis of the RNAP shared regions and conserved residues is presented in the context of the Thermus thermophilus bRNAP ternary elongation complex (TEC) structure 9; PDB ID 2O5J). This structure was assembled on a synthetic scaffold containing 14 bp of downstream dsDNA, 9 bp of the RNA/DNA hybrid, and 7 single-stranded nucleotides of the upstream RNA transcript (see Figs. 1–10). The nucleic acids are in the post-translocated state, so the binding site for the nucleotide substrate (the +1 position) is available and is occupied by the non-hydrolyzable nucleotide analog AMPPCP. The TEC was analyzed since this is the stage of the transcription cycle that is most common in terms of the structure/function relationship between bRNAP 9; 10 and eRNAP II 11–16, the two cases where TEC structural information is available. This is illustrated by the fact that the protein/nucleic acid interactions and functional features of the shared RNAP regions are generally conserved in all of the bRNAP and eRNAP TEC structures.
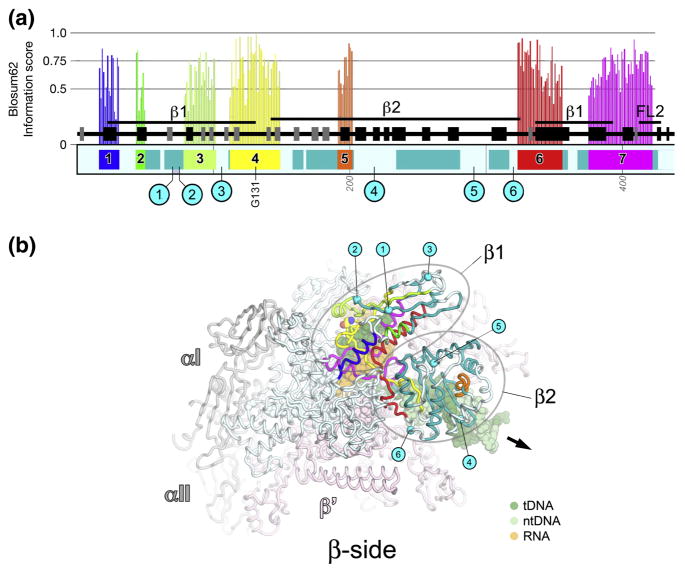
Fig. 1. βa1-a7; β1 and β2 domains.
A. Schematic representation of the sequence context of βa1-a7 in Thermus RNAP. Regions shared among all RNAPs are color-coded and labeled: βa1, blue; βa2, green; βa3, yellow-green;β a4, yellow; βa5, orange; βa6, red; βa7, magenta. Additional regions shared among bRNAP are colored teal. Above each region shared among all RNAPs is a histogram showing the Blosum62 information score (scale on the left) for each residue, as determined by the program PFAAT 17. The secondary structure is shown directly above the sequence bar (helices, black rectangles; β-strands, grey rectangles). Important structural features discussed in the text are denoted above that. Below the sequence bar, small grey numbers (vertically oriented 100, 200, etc.) denote the residue numbering of the Thermus β-subunit. The approximate insertion points of the lineage-specific insertions are denoted by cyan circles and labeled according to Lane & Darst 4. The position of highly conserved βGly131 (within β a4) is denoted.
B. β-side view of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β′, light pink; β as in (A). The nucleic acids of the TEC are shown in CPK format and color-coded as shown in the key. Highly conserved βGly131 is shown in CPK format. The approximate insertion points for the lineage-specific insertions are denoted by cyan spheres and labeled as in (A). The thick black arrow points in the downstream direction (the direction of RNAP transcription).
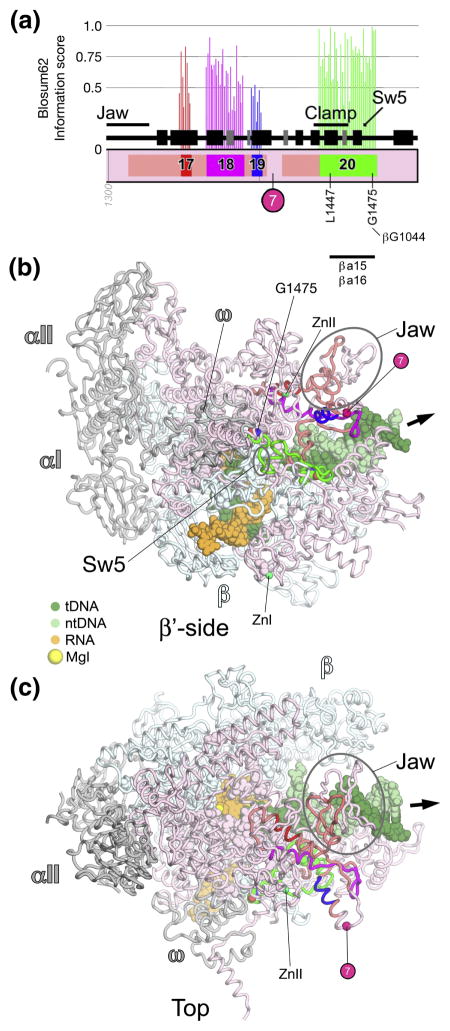
Fig. 10. β′a17-a20; clamp, Sw5.
A. Schematic representation of the sequence context of β′a17-a20 in Thermus RNAP. As in Fig. 5A, except: β′a17, red; β′a18, magenta; β′a19, blue; β′a20, green. The approximate insertion point of β′In7 is denoted by a magenta circle. At the bottom, the horizontal line denotes a segment of β′a20 that participates in conserved interactions with other shared regions.
B, C. Views of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β, cyan; β′ as in (A). The nucleic acids of the TEC are shown in CPK format and color-coded as shown in the key. Highly conserved residues are shown in CPK format. Zn2+ ions are shown as green spheres. The approximate insertion point for β′In7 is denoted by a magenta sphere. The thick black arrow points in the downstream direction (the direction of RNAP transcription).
B. β′-side view.
C. Top view.
In the following analysis, we systematically focus on the shared regions and conserved residues beginning at the N-terminus of the β subunit, through to the C-terminus of β′. All of the Figures are organized similarly. Part A of each Figure shows a schematic representation of the relevant portion of the β (light blue) or β′ (light pink) primary sequence of Thermus RNAP (the complete schematics for each subunit are contained in Fig. S1). Regions shared among all RNAPs are color-coded and labeled 4. Additional regions shared among bRNAP are colored teal (β) or darker pink (β′). Above each region shared among all RNAPs is a histogram showing the Blosum62 information score (scale on the left) for each residue, as determined by the program PFAAT 17. The secondary structure is shown directly above the sequence bar (helices, black rectangles; β-strands, grey rectangles). Important structural features discussed in the text are denoted above that. Below the sequence bar, small grey numbers (vertically oriented 100, 200, etc.) denote the numbering of the Thermus subunits. The approximate insertion points of the lineage-specific insertions are denoted and labeled according to Lane & Darst 4 (cyan circles for β insertions, magenta circles for β′ insertions). At the bottom of the schematic, horizontal lines denote segments of the shared regions that participate in conserved interactions with other shared regions. Finally, the identity and positions of very highly conserved residues, with a Blosum62 information score ≥ 0.98 among all RNAPs, are denoted (vertically oriented single-letter amino acid code and amino acid position in Thermus RNAP). Using this cutoff, 34 β residues (out of 895 aligned residues) and 38 β′ residues (out of 895 aligned residues) are included, roughly 4% of the aligned residues for each subunit (Table S1). The average sequence identity among these 72 total positions is 99.6%, corresponding to 4 substitutions in an alignment of 1000 sequences.
Additional parts of each Figure illustrate one or more views of the TEC. In all the structural views, the RNAP is shown as a backbone worm, unless otherwise noted. The various views correspond roughly to standard views as defined in 4, Figs. 5 and 6. The highly conserved residues in β and β′ are shown in CPK format. The αI, αII, and ω subunits are colored grey. The β and β′ subunits are color-coded according to the schematic. In the relevant regions of β or β′ encompassed by the schematic, the points of insertion for the lineage-specific insertions are shown as cyan (β) or magenta (β′) spheres. Zn2+ ions are shown as green spheres. The nucleic acids of the TEC are shown in CPK or ribbon format, depending on the view. The template and nontemplate DNA strands are colored dark green and light green, respectively. The RNA transcript is colored orange. A thick black arrow points in the downstream direction (the direction of RNAP transcription). The Mg2+ ions in the RNAP active site (MgI and MgII, when shown) are shown as yellow spheres. The incoming nucleotide substrate, when shown, is shown in stick format and is colored blue. Other features specific to each Figure are described in the corresponding figure legends.
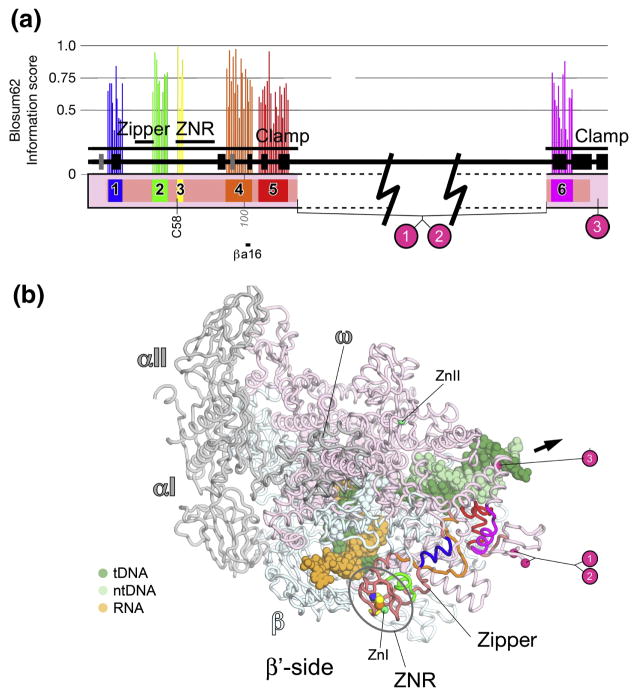
Fig. 5. β′a1-a6; clamp.
A. Schematic representation of the sequence context of β′a5-a6 in Thermus RNAP. Regions shared among all RNAPs are color-coded and labeled: β′a1, blue; β′a2, green; β′a3, yellow; β′a4, orange; β′a5, red; β′a6, magenta. Additional regions shared among bRNAPs are colored dark pink. Above each region shared among all RNAPs is a histogram showing the Blosum62 information score (scale on the left) for each residue, as determined by the program PFAAT 17. The secondary structure is shown directly above the sequence bar (helices, black rectangles; β-strands, grey rectangles). Important structural features discussed in the text are denoted above that. Below the sequence bar, small grey numbers (vertically oriented 100, 200, etc.) denote the residue numbering of the Thermus β′-subunit. The approximate insertion points of the lineage-specific insertions are denoted by cyan circles and labeled according to Lane & Darst 4. At the bottom, the horizontal line denotes a segment of β′a4 that participates in conserved interactions with another shared region, βa16. The approximate insertion points of the lineage-specific insertions are denoted by magenta circles and labeled according to Lane & Darst 4. The position of highly conserved β′C58 is denoted.
B. β′-side view of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β, light cyan; β′, as in (A). The nucleic acids of the TEC are shown in CPK format and color-coded as shown in the key. Zn2+ ions are shown as green spheres. Highly conserved β′Cys58 is shown in CPK format. The approximate insertion points for the lineage-specific insertions are denoted by magenta spheres and labeled as in (A). The thick black arrow points in the downstream direction (the direction of RNAP transcription).
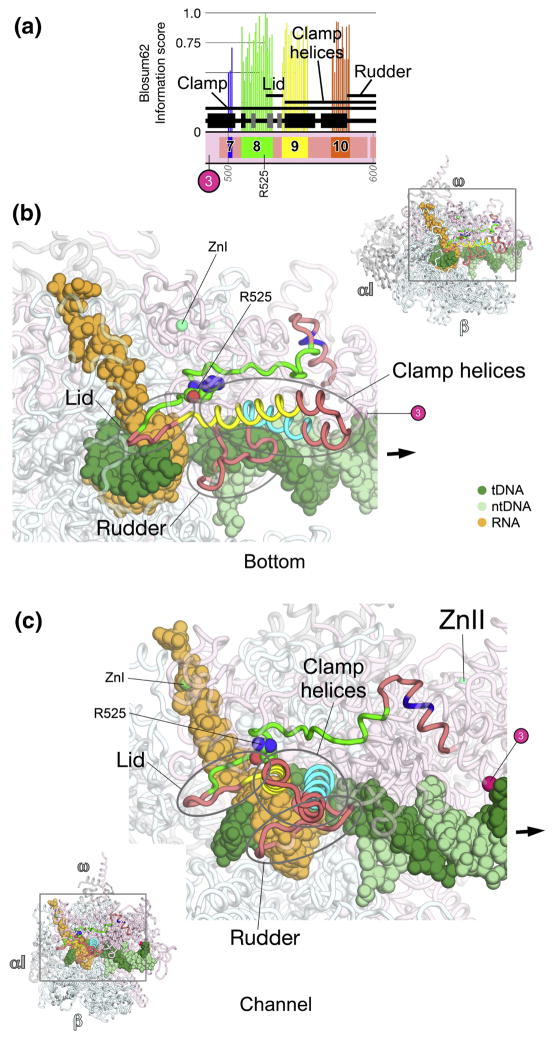
Fig. 6. β′a7-a10; clamp, lid, clamp helices.
A. Schematic representation of the sequence context of β′a7-a10 in Thermus RNAP. As in Fig. 5A, except: β′a7, blue; β′a8, green; β′a9, yellow; β′a10, orange. The approximate insertion point of β′In3 is denoted by a magenta circle. The position of highly conserved β′R525 (within β′a8) is denoted.
B, C. Views of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β, cyan; β′ as in (A). The nucleic acids of the TEC are color-coded as shown in the key. Zn2+ ions are shown as green spheres. The approximate insertion point for β′In3 is denoted by a magenta sphere. Highly conserved β′R525 is shown in CPK format. The thick black arrow points in the downstream direction (the direction of RNAP transcription).
B. (Inset, upper-right) Bottom view. The boxed region is magnified below. (Magnified view) In this view, most of β and β′ are transparent for clarity.
C. (Inset, lower-left) Channel view. The boxed region is magnified above. (Magnified view) In this view, most of β and β′ are transparent for clarity.
bRNAP β subunit
βa1-a7; β1 and β2 domains (Fig. 1)
βa1-a7 form the β1 and β2 domains (called the protrusion and lobe, respectively, in Saccharomyces cerevisiae eRNAP II; 8; Fig. 1). The β1 domain covers the RNA/DNA hybrid within the RNAP active site channel. The β2 domain covers the downstream double-stranded DNA (Fig. 1B). A channel between the β1 and β2 domains guides the single-strand of the non-template DNA within the transcription bubble 18. The β1 and β2 domains are relatively rich in lineage-specific insertions; the β1 and β2 domains make up roughly 1/3 the sequence of β, but contain 1/2 (6 of 12) of the β insertions 4.
The β1 domain includes βa1-a3.β a4 (yellow; Fig. 1) starts in the β1 domain, traverses across the RNA/DNA hybrid, and initiates the β2 domain. Much of the β2 domain is structurally conserved among bacterial sequences (βb4-b9) 4. Most aRNAP and eRNAPs have sequence here, but extremely low sequence conservation, and the presence of many insertions and deletions throughout this region, make confident alignment impossible. βa6 (red; Fig. 1) starts in the β2 domain and traverses back to the β1 domain, entering into a long α-helix within the β1 domain. The N-terminal α-helix of βa7 (magenta; Fig. 1) completes the β1 domain.
The β1 domain interacts with the transcription-repair coupling factor (TRCF, also called Mfd), positioning TRCF to interact with the upstream double-stranded DNA 19; 20. Substitutions at Escherichia coli (Eco) bRNAP β117, 118, or 119 (corresponding to Thermus β108/109/110), between βa3 and β a4, disrupt the TRCF/RNAP protein/protein interaction 19; 20. In Saccharomyces cerevisiae (Sce) eRNAP II, the protrusion interacts with the Rpb12 subunit 8.
A large deletion encompassing most of the β2 domain of Eco RNAP as well as βIn4 (Eco βΔ[186–433], corresponding to Thermus β [174–311]) resulted in dramatic alterations in promoter melting behavior 21. In Sce RNAPII, the lobe interacts with the Rpb9 subunit 8.
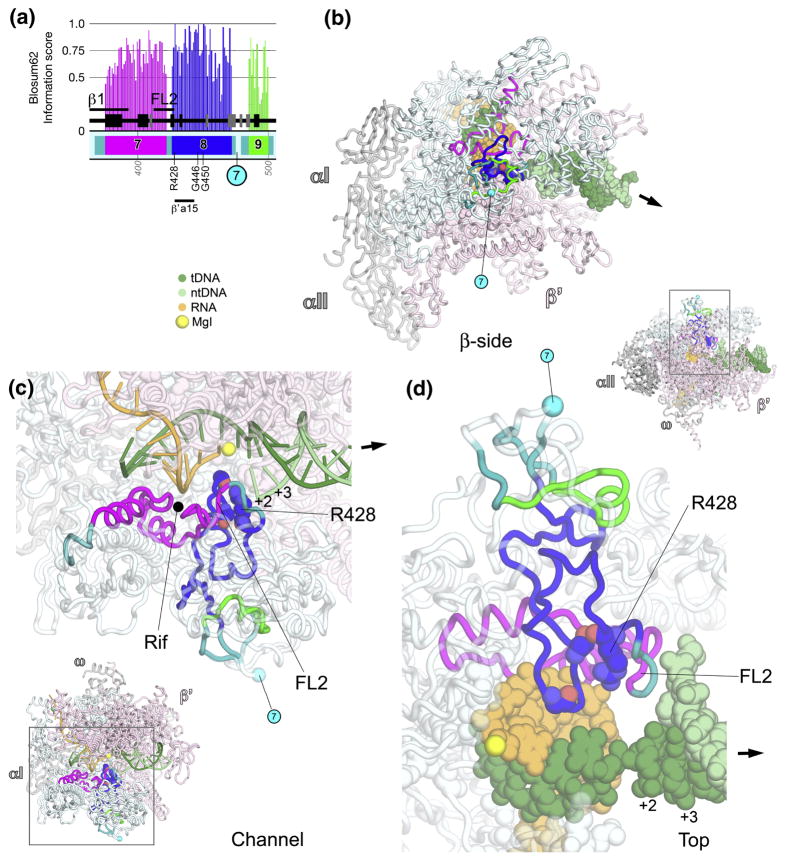
βa7-a9; fork-loop 2 (Fig. 2)
Fig. 2. βa7-a9; fork-loop 2.
A. Schematic representation of the sequence context of βa7-a9 in Thermus RNAP. As in Fig. 1A, except: βa7, magenta; βa8, blue; βa9, green. The approximate insertion point for βIn7 is shown as a cyan circle. At the bottom, the horizontal line denotes a segment of βa8 that participates in conserved interactions with another shared region, β′a15. The positions of highly conserved residues are denoted.
B–D. Views of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β′, light pink; β as in (A). The active-site MgI is shown as a yellow sphere (C, D). The nucleic acids of the TEC are color-coded as shown in the key. Highly conserved residues are shown in CPK format. The approximate insertion point for βIn7 is denoted by a cyan sphere and labeled as in (A). The thick black arrow points in the downstream direction (the direction of RNAP transcription).
B. β-side view. The nucleic acids of the TEC are shown in CPK format.
C. (Inset, lower-left) Channel view. The boxed region is magnified above. (Magnified view) In this view, most of β and β′ are transparent for clarity. The nucleic acids of the TEC are shown as backbone worms.
D. (Inset, upper-right) Top view. The boxed region is magnified below. (Magnified view) In this view, most of β and β′ are transparent for clarity. The nucleic acids of the TEC are shown in CPK format.
βa7 forms a shallow channel on the floor of the RNAP active-site channel that accommodates the RNA transcript from about −3 to −6 (Fig. 2C). βa7 harbors almost all of the residues that interact with rifamycins (Rif), as well as almost all known Rif-resistant mutations (Fig. 2C) 22. Rifs are among the most potent and broad-spectrum antibiotics against bacterial pathogens, and are a key component of anti-tuberculosis therapy. Rifs bind in the shallow pocket and inhibit bacterial RNAP by sterically preventing synthesis of RNA transcripts > 2–3 nt in length 22–24.
Fork-loop 2 (FL2) 8 comprises a loop containing the last 11 residues of βa7 and the first 3 residues of βa8, including absolutely conserved βArg428 (Figs. 2C, 2D). The bacterial FL2 harbors a conserved 4-residue insert between βa7 and βa8 that is missing in aRNAP and eRNAP. This 4-residue bacterial-specific insert harbors 3 residues (β423, 424, and 425) that interact with the bacterial-specific inhibitor streptolydigin, and substitutions at these positions give rise to streptolydigin resistance 9; 25; 26.
FL2 appears to maintain the downstream edge of the transcription bubble in the TEC by sterically blocking the DNA duplex, interfering with the nontemplate DNA strand upstream of position +3, and preventing reassociation of the separated DNA strands (Figs. 2C, 2D) 10; 11; 16; 18. Substitutions and deletions in the aRNAP FL2 indicate that FL2 is strictly required for initiation and elongation 27; 28, demonstrating an essential role for FL2 in downstream DNA unwinding during elongation. A segment of βa8 immediately following FL2, including absolutely conserved βArg428, interacts with highly conserved residues within the bridge helix (BH) of β′a15 (see Fig. 9). Following this region that interacts with β′, two residues of βa8, absolutely conserved βGly450, along with Leu451, interact with streptolydigin 9; 25; 26.
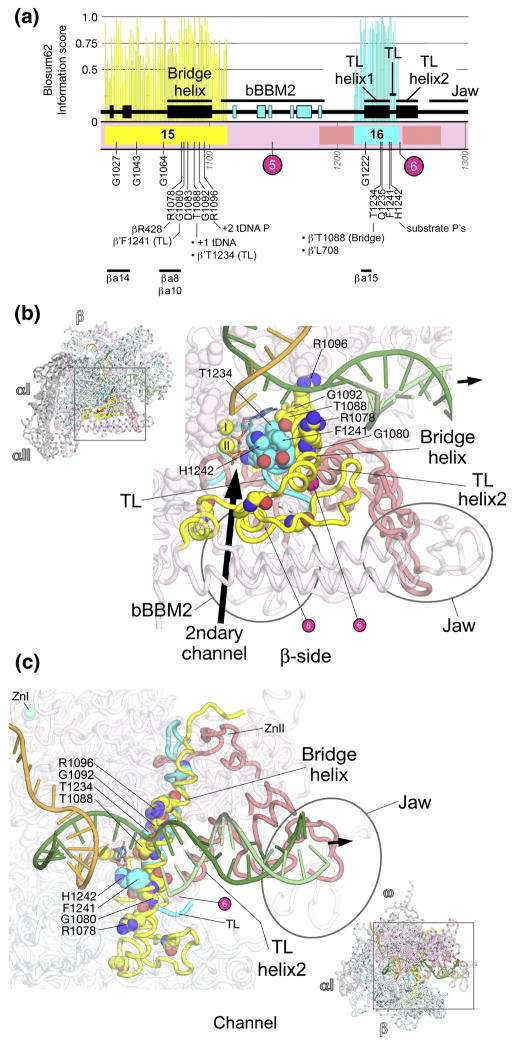
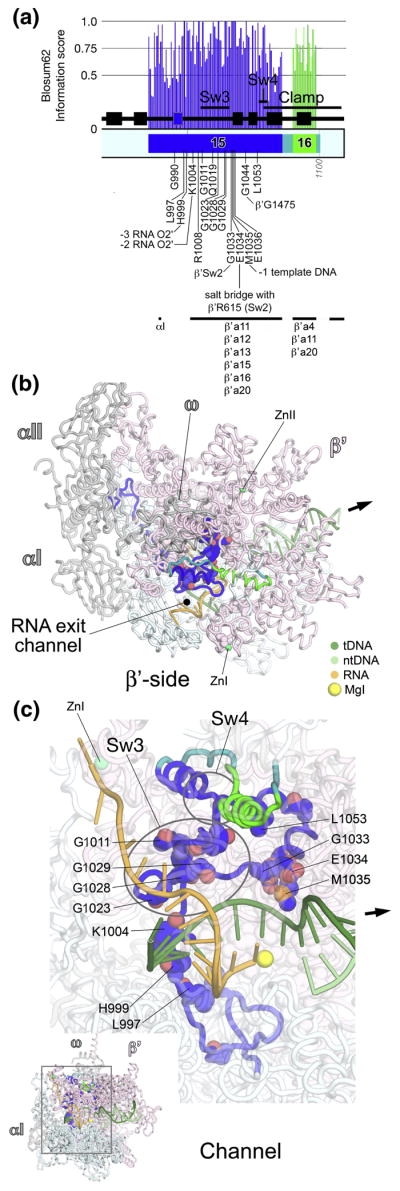
Fig. 9. β′a15-16; bridge helix, trigger-loop.
A. Schematic representation of the sequence context of β′a15-a16 in Thermus RNAP. As in Fig. 5A, except: β′a15, yellow; β′a16, cyan. The positions and roles of highly conserved residues are denoted. The approximate insertion points of lineage-specific insertions are denoted by magenta circles and labeled according to Lane & Darst 4. At the bottom, the horizontal lines denote segments that participate in conserved interactions with other shared regions. The secondary structural elements (4 β-strands, 2 α-helices) of a BBM2 domain shared among many bRNAPs (bBBM2) are colored cyan. The ‘jaw’ domain comprises another SBHM fold that is shared among many bRNAPs.
B, C. Views of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β, cyan; β′ as in (A). The nucleic acids of the TEC are shown as backbone worms and color-coded as shown in the key. Highly conserved residues are shown in CPK format. The active site MgI and MgII are shown as yellow spheres. The incoming nucleotide substrate is shown in stick format with carbon atoms colored blue. The approximate insertion points for lineage-specific insertions are denoted by magenta spheres and labeled as in (A). The thick black arrow points in the downstream direction (the direction of RNAP transcription).
B. (Inset, upper-left) β-side view. The boxed region is magnified below. (Magnified view) In this view, β is not shown, and most of β′ is transparent for clarity.
C. (Inset, lower-right) Channel view. The boxed region is magnified above. (Magnified view) In this view, most of β and β′ are transparent for clarity.
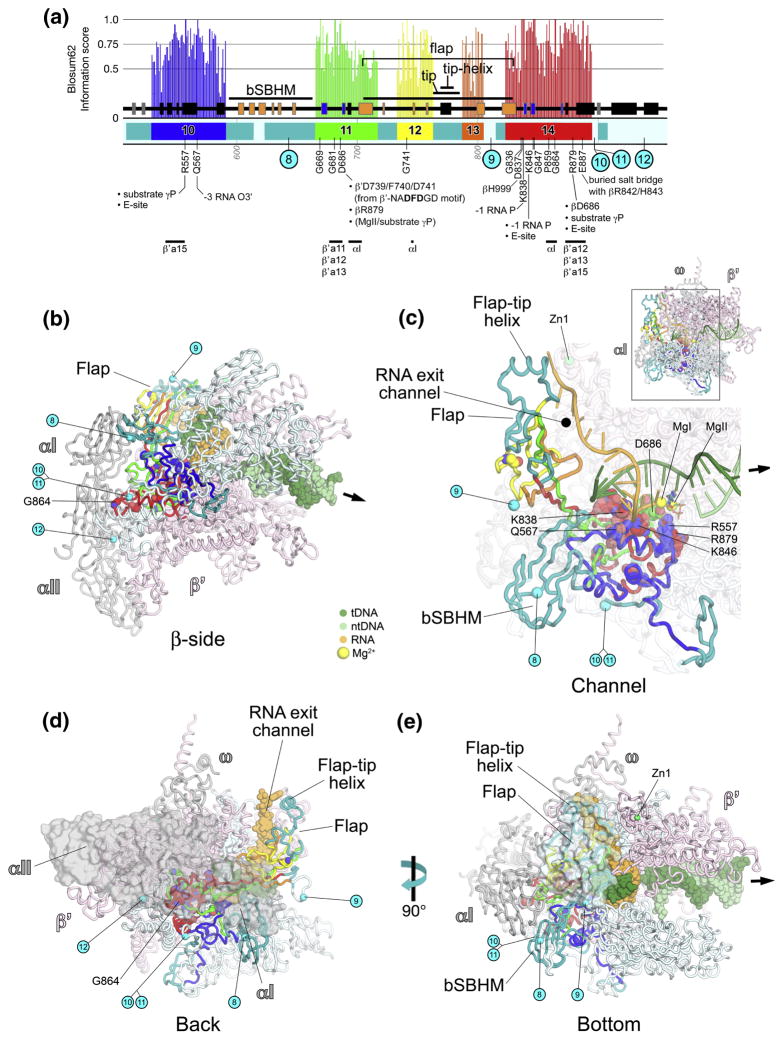
βa10-a14; catalytic center, flap (Fig. 3)
Fig. 3. βa10-a14; catalytic center, flap domain.
A. Schematic representation of the sequence context of βa10-a14 in Thermus RNAP. As in Fig. 1A, except: βa10, blue; βa11, green; βa12, yellow; βa13, orange; βa14, red. The positions and roles of highly conserved residues are denoted. The approximate insertion points of lineage-specific insertions are denoted by cyan circles and labeled according to Lane & Darst 4. At the bottom, the horizontal lines denote segments that participate in conserved interactions with other shared regions. The core secondary structural elements of evolutionarily conserved domains 31 are denoted by color-coding. The 6 β-strands of an SBHM domain shared among bRNAPs (bSBHM) are colored orange, as are the 6 β-strands of the SBHM domain comprising the base of the flap domain, which is shared among all RNAPs. The first 5 β-strands (out of 6) of the βDPBB (Double-psi β-Barrel; see Discussion and Figs. S4 and S5) are colored blue.
B–E. Views of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β′, light pink; β as in (A). The nucleic acids of the TEC are color-coded as shown in the key. Zn2+ ions are shown as green spheres (C, E). Highly conserved residues are shown in CPK format. The approximate insertion points for lineage-specific insertions are denoted by cyan spheres and labeled as in (A). The thick black arrow points in the downstream direction (the direction of RNAP transcription).
B. β-side view. The nucleic acids of the TEC are shown in CPK format.
C. (Inset, upper-right) Channel view. The boxed region is magnified below. (Magnified view) In this view, most of β and β′ are transparent for clarity. The nucleic acids of the TEC are shown as backbone worms. The active-site MgI and MgII are shown as yellow spheres (C). The incoming nucleotide substrate is shown in stick format, with carbon atoms colored blue.
D. Back view. The α subunits are shown as a molecular surface instead of a backbone worm. The nucleic acids of the TEC are shown in CPK format.
E. Bottom view. The flap includes a transparent molecular surface. The nucleic acids of the TEC are shown in CPK format.
βa10-a14 form the heart of the β subunit, comprising critical elements of the RNAP active site (βa10, a11, and a14), the flap domain (βa11-a14), and providing critical interactions with the αI (aRNAP subunit D 7; Rbp3 in eRNAP IIa) 8 subunit (βa14) 29.
βa10 makes interactions with highly conserved residues in the BH of β′a15, then forms structural elements supporting the RNAP active site, including two absolutely conserved residues: βArg557 interacts with the γ-phosphate of the incoming nucleotide substrate; βGln567 interacts with the RNA transcript backbone at the −3 position (Fig. 3C) 9; 10; 14; 15. βArg557 also participates in the entry (E) site, where nucleotide substrates bind prior to binding in the active site for catalysis 15. Inserted between βa10 and βa11 is a surface-exposed sandwich-barrel hybrid motif (SBHM) 30 domain shared among bRNAPs but missing in aRNAP and eRNAP (bSBHM; Figs. 3C, 3E) 4; 31.
βa11 forms structural elements that support the RNAP active site, making interactions with β′a11 and β′a12 (which contain the core elements of the RNAP active site; see Fig. 7). βa11 includes absolutely conserved βAsp686, which is involved in a network of critical interactions. βAsp686 interacts with:
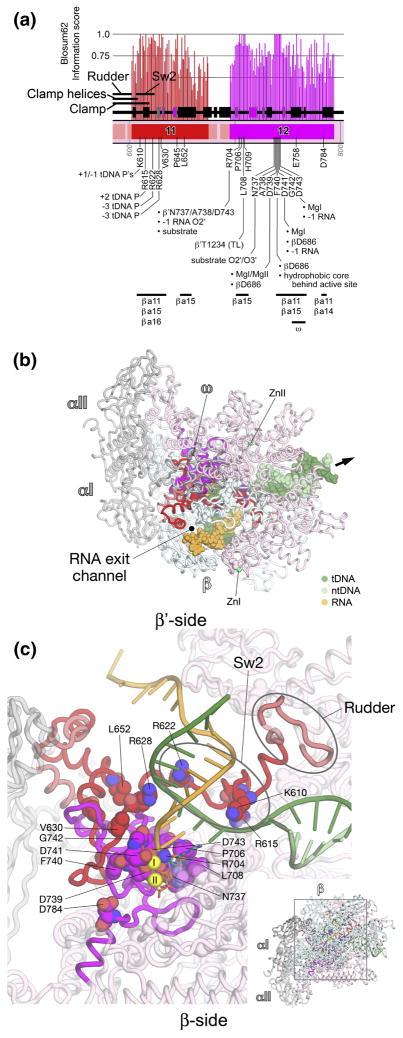
Fig. 7. β′a11-a12; Sw2, catalytic center.
A. Schematic representation of the sequence context of β′a11-a12 in Thermus RNAP. As in Fig. 5A, except: β′a11, red; β′a12, magenta. The positions and roles of highly conserved residues are denoted. At the bottom, the horizontal lines denote segments that participate in conserved interactions with other shared regions. The 6 β-strands of the β′DPBB (see Discussion and Figs. S4 and S6) are colored magenta.
B, C. Views of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β, cyan; β′ as in (A). The nucleic acids of the TEC are color-coded as shown in the key. Highly conserved residues are shown in CPK format. The thick black arrow points in the downstream direction (the direction of RNAP transcription).
B. β′-side view. Zn2+ ions are shown as green spheres. The nucleic acids of the TEC are shown in CPK format.
C. (Inset, lower-right) β-side view. The boxed region is magnified above. (Magnified view) In this view, β is not shown, and most of β′ is transparent for clarity. MgI and MgII in the active site are shown as yellow spheres. The incoming nucleotide substrate is shown in stick format, with carbon atoms colored blue. The nucleic acids of the TEC are shown as backbone worms.
β′Asp739/Phe740/Asp741 of the absolutely conserved β′-NADFDGD motif (β′a12; see Fig. 7),
The γ-phosphate of the incoming nucleotide substrate as well as MgII in the active site (Fig. 3C).
Absolutely conserved βArg879 (from βa14), which also interacts with the substrate γ-phosphate (Fig. 3C).
After βAsp686, βa11 participates in more of the structural core behind the RNAP active site, interacts with the αI subunit (Figs. 3A, 3B, 3D) 29, then enters into a long β-strand that marks the beginning of the flap domain (called the ‘wall’ in eRNAP II; Fig. 3C).
The last β-strand of βa11, then βa12, βa13, and the first β-strand of βa14 comprise the structural core of the flap domain, which is itself another SBHM domain with large inserts in the loops connecting the core SBHM β-strands 4; 31. The flap domain plays multiple roles in each phase of the transcription cycle; initiation, elongation, and termination.
The flap forms an independent, flap-like structural domain that gives rise to a narrow channel between itself and the RNAP 6. During transcription elongation, this channel accommodates the upstream, single-stranded RNA transcript after it leaves the RNA/DNA hybrid (from −10 to −16), and has thus been called the RNA exit channel (Figs. 3C, 3D, 3E) 6; 10; 18.
The flap-tip helix (connecting βa12 and βa13) is a critical structural element of bRNAP that is not conserved with aRNAP and eRNAP (Figs. 3A, 3C, 3D, 3E), and it plays essential, bacterial-specific roles in initiation, termination, and other regulatory functions. In bacterial transcription initiation, most promoters are defined by two conserved DNA sequence hexamers, the −10 and −35 elements, positioned roughly 10 and 35 bp upstream of the transcription start site, respectively 32. An interaction between the flap-tip helix and domain 4 of the promoter-specificity subunit σ(σ4) is essential for initiation from such promoters (called –10/-35 promoters) because it positions σ4 for proper interaction with the −35 element 33; 34.
Structural elements that form in the nascent RNA transcript play key regulatory roles in bacterial transcription 35. Specifically, stem-loop hairpins in the RNA can induce pausing of the elongating RNAP, or can cause termination (release of the transcript and DNA template from the RNAP enzyme), depending, among other things, on the spacing of the hairpin from the RNA 3′-end. The RNA hairpin forms in the RNA exit channel underneath the flap domain 18. A pause hairpin contacts residues within the flap-tip helix 36, and the flap-tip is required for hairpin-induced pausing 37. The pause hairpin/flap-tip interaction may inhibit the rate of nucleotide addition at the RNAP active site (more than 50 Å away) through an allosteric mechanism that depends on the direct connection between the flap and the RNAP catalytic center through βa11 and βa14 (Fig. 3C). A number of bacterial-specific transcriptional regulators interact with the flap through the flap-tip helix, including bacteriophage T4 AsiA 38 and gp33 39, and the bacteriophage λ Q protein 39.
βa14 begins with the last β-strand of the flap domain, and then continues directly to participate in the RNAP active site, where absolutely conserved residues βLys838 and βLys846 interact with the backbone of the RNA transcript at the –1/-2 positions (Fig. 3C) 10. βLys846 also participates in the E-site 15. βa14 then travels towards the back of the RNAP, where two absolutely conserved residues appear to be important for interactions with the αI subunit (βPro859 and βGly684; Figs. 3B, 3D) 29. βa14 then makes its way back to the RNAP active site and participates in interactions with β′a12, β′a13, and β′a15. Here, absolutely conserved βArg879 interacts with the substrate γ-phosphate (Fig. 3C), and also participates in the E-site 15. Finally, absolutely conserved βGlu887 makes a buried salt bridge with highly conserved residues βArg842 and βHis843 (also within βR14) that may play a structural role.
βa15-a16; RNA/DNA hybrid interactions, RNA exit channel, clamp (Fig. 4)
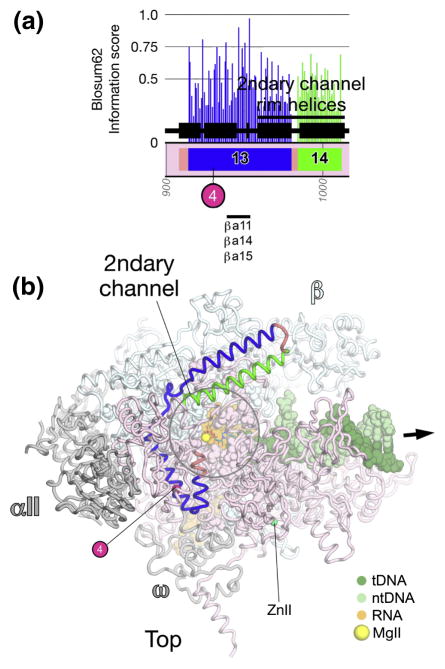
Fig. 4. βa15-a16; RNA/DNA hybrid interactions, RNA exit channel, clamp.
A. Schematic representation of the sequence context of βa15-a16 in Thermus RNAP. As in Fig. 1A, except: βa15, blue; βa16, green. The positions and roles of highly conserved residues are denoted. At the bottom, the horizontal lines denote segments that participate in conserved interactions with other shared regions. The 6th β-strand of the βDPBB (see Discussion and Figs. S4 and S5) is colored blue.
B, C. Views of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β′, light pink; β as in (A). The nucleic acids of the TEC are color-coded as shown in the key. Zn2+ ions are shown as green spheres. Highly conserved residues are shown in CPK format. The thick black arrow points in the downstream direction (the direction of RNAP transcription).
B. β′-side view. The nucleic acids of the TEC are shown as backbone worms.
C. (Inset, lower-left) Channel view. The boxed region is magnified above. (Magnified view) In this view, most of β and β′ are transparent for clarity. The nucleic acids of the TEC are shown as backbone worms. The active-site MgI is shown as a yellow sphere.
βa15 begins near the back of the RNAP, where, like βa14, it makes critical interactions with the αI subunit (Figs. 4A, 4B) 29. βa15 then participates in the structural core behind the RNAP active site, where absolutely conserved βLeu997 (Fig. 4C) interacts with and helps position three other absolutely conserved residues that interact with the RNA transcript; βGln567 from βa10, and βLys838 and βLys846 from βa14 (Figs. 3C, S2).
Further along, absolutely conserved βHis999 and βLys1004 (Fig. 4C) point towards the RNA/DNA hybrid and interact with the RNA transcript in some TEC structures 11; 14, while they point away from the RNA/DNA hybrid into the RNAP structural core in others 10. This region of the RNAP has been known to be conformationally flexible since an initiating nucleotide analog crosslinked to His999 can be extended by the RNAP into an RNA chain up to 9 nucleotides in length 40.
The N-terminal portion of βa15, up to about βLys1004, resides within the β side of the RNAP active site channel. βa15 then traverses to the β′ side of the active site channel (Fig. 4C). Absolutely conserved residues constitute Switch 3 (Sw3; βGly1011, Gln1019, Gly1023, Gly1028, Gly1029). These residues line the RNA exit channel (βGly1011), and line the path of the template DNA around –3/-4 (βGly1023, Gly1028, Gly1029). Following Sw3, absolutely conserved βGly1033 lines the path of the template DNA around the −2 position and interacts with β′ residues within Sw2. Absolutely conserved βGlu1034 interacts with absolutely conserved β′Arg615 within Sw2, and βMet1035 interacts with the template DNA at the −1 position (Fig. 4C). The C-terminal portion of βa15 as well as βa16 then interact extensively with the β′ subunit, and make up a core structural element of the clamp (Fig. 4). The antibiotic myxopyronin (Myx) interacts with residues within βa15 and βa16, including absolutely conserved βGly1033, Glu1034, and Leu1053 41; 42.
bRNAP β′ subunit
β′a1-a6; clamp (Fig. 5)
β′a1-a6, which all lie within the clamp, comprise pieces of the β′ N-terminus linked by segments that are conserved only among bacteria (Fig. 5). A structural element termed the ‘zipper’ 8, which lies between β′a1 and β′a2, is shared among all bacteria, but has very low sequence conservation among aRNAP and eRNAPs. In addition, there are many small insertions and deletions in this region, making a confident alignment impossible.
One absolutely conserved residue in β′a3 (β′Cys58) participates in chelating a Zn2+. The presence of a Zn-ribbon (ZNR) near the N-terminus of β′ is a shared feature of all multi-subunit RNAPs 4; 31; 43.
β′a7-a10; clamp, lid, clamp helices (Fig. 6)
β′a7-a10 all lie within the clamp, and make up two important RNAP structural elements, the lid 8 and the clamp helices (also called the coiled-coil) 6. The shared regions are linked by regions conserved only among bacteria.
The lid is an extended β-hairpin and connecting loop (Figs. 6B, 6C). The tip of the lid interacts with the flap to topologically enclose the RNA transcript at the base of the RNA exit channel in the TEC (Fig. 6B). Likewise, in the bRNAP holoenzyme (the catalytic core RNAP with the promoter-specificity σ subunit), the lid topologically encloses the extended linker between the σ3 and σ4 domains, which occupies the RNA exit channel in the initiating form of the enzyme 44; 45.
The stem of the lid comprises the C-terminal part of β′a8 and the N-terminal part of β′a9. The connecting tip is conserved among bacteria but varies in length among aRNAP and eRNAPs, and has very low sequence conservation, making confident alignment impossible. Absolutely conserved β′Arg525 within β′a8 sits at the base of the lid-stem, making multiple interactions that likely stabilize the structure.
Structurally, the lid seems to serve as a wedge to part the RNA and DNA strands, interacting with the RNA transcript around the −8, −9, and −10 positions at the upstream edge of the RNA/DNA hybrid 10; 14. It forms a barrier to maintain the separation of the strands and guide the RNA into the exit channel.
RNAPs harboring structure-based deletions of the lid have been investigated for both bRNAP and aRNAP. The lid plays an important role in initiation. The Δlid-bRNAP forms unstable promoter open complexes and has dramatically reduced activity during σ70-dependent initiation from both −35-dependent and extended −10 promoters 46; 47. The lid is also required for aRNAP initiation 27; 28.
During transcript elongation, RNA displacement, and termination all occurred normally with Δlid-RNAP. When transcribing single-stranded DNA templates, wild-type RNAP normally stalls after the synthesis of a short RNA transcript, but the Δlid-RNAP formed persistent RNA/DNA hybrids 27; 28; 46; 47.
β′a9 and β′a10 comprise two α-helices, the clamp helices, that from an anti-parallel, coiled-coil like structure that serves as a major binding platform for the initiaiton-specific σ subunit in bRNAP 44; 45; 48; 49. The clamp helices may serve as a binding platform for other bRNAP regulators as well, such as RfaH 50. The helices and loop connecting the two anti-parallel helices is conserved in length among bRNAPs, but varies in aRNAP and eRNAPs. The coiled-coil tip has very low sequence conservation, making confident alignment impossible.
Inserted into the second clamp α-helix is a structural feature called the rudder 6, comprising two AT-hook-like modules 31. The rudder is shared among all bRNAPs, but diverges among aRNAP and eRNAPs. There are many small insertions and deletions in this region, making confident alignment impossible. In functional studies of rudder deletions in bRNAP, the major defect of the Δrudder-RNAP was an inability to form stable TECs, leading to the conclusion that the rudder plays a role in stabilizing unwound DNA beyond the RNA/DNA hybrid 51. This is consistent with TEC structures, where the rudder interacts with DNA at the –9/-10 positions, at the upstream edge of the RNA/DNA hybrid (Figs. 6B, 6C), preventing reassociation with the RNA transcript 10; 14.
β′a11-a12; Sw2, catalytic center (Fig. 7)
β′a11-a12 form the heart of the β′ subunit, making up critical elements of the RNAP active site, including the universally conserved β′-NADFDGD motif that chelates MgI (β′a12; Fig. 7). β′a11-a12 harbor a large number of absolutely conserved residues that make critical interactions with: i) the template DNA, ii) the RNA transcript, iii) the nucleotide substrate, iv) MgI, v) MgII, vi) other absolutely conserved elements of β′ in close proximity to the active site, vii) absolutely conserved elements of the β subunit in close proximity to the active site.
β′a11 starts immediately where the rudder ends, where β′a11 makes up Sw2 (Fig. 7C). Sw2 harbors 3 absolutely conserved residues that make critical contacts with the template DNA. β′Lys610 interacts with the template DNA phosphate backbone at −1 and +1, β′Arg615 interacts with the template DNA at +2, and β′Arg622 contacts the template DNA phosphate backbone at −3 (Fig. 7C) 10; 14. Substitution of β′Arg615 to Ala renders aRNAP totally inactive in elongation 27. Highly conserved residues in Sw2 make critical contacts with the antibiotic Myx (β′Phe614, Leu619, Gly620, Lys621) 41; 42.
Following Sw2, absolutely conserved β′Arg628 interacts with the template DNA at the −3 position (Fig. 7C). Absolutely conserved β′Val630 and Leu652 participate in the hydrophobic core immediately behind the active center cleft. Finally, β′a11 ends with an α-helix that lines one side of the RNA exit channel (Figs. 7B, 7C).
β′a12 contains the core components of the RNAP catalytic center, and is rich in absolutely conserved residues. β′Arg704 interacts simultaneously with the nucleotide substrate (O4′), the 2′-OH of the RNA transcript at −1, and β′Asn737, Ala738, and Asp743 of the β′-NADFDGD motif (Fig. 7C). β′Pro706 lines the path for the template DNA around +1. β′Leu708 interacts with absolutely conserved β′Thr1234 in trigger-loop (TL) helix1 (see Fig. 9).
After forming part of the structural scaffold immediately behind the active center, β′a12 enters into a loop that harbors the β′-NADFDGD motif, a string of 7 absolutely conserved residues that constitutes the RNAP active center (Fig. 7C). Within the β′-NADFDGD motif: β′Asn737 interacts with O2′ and O3′ of the nucleotide substrate; β′Ala738 makes van der Waals contacts with absolutely conserved β′Arg704; β′Asp739 interacts with MgI and MgII, and interacts with absolutely conserved βAsp686 (see Fig. 3); β′Phe740 interacts with βAsp686 (Fig. 3) and participates in the hydrophobic core immediately behind the active site; β′Asp741 interacts with MgI, the RNA transcript at the −1 position, and βAsp686; β′Asp743 interacts with MgI and the RNA transcript at −1. Substitution of any one of the three conserved Asp residues of the NADFDGD motif abrogates all catalytic activities of the RNAP 52; 53. Finally, following the β′-NADFDGD motif, absolutely conserved β′Glu758 (hidden in Fig. 7C) and β′Asp784 make buried polar contacts that likely play a structural role (Fig. 7C). The C-terminal part of β′a12 (about residues β′777–790) forms a part of the secondary channel, where nucleotide substrates likely access the RNAP active site 6; 18.
β′a11-12 make extensive interactions with βa11 and βa14-a16, which together form the central core of the RNAP active site. β′a12 also makes the only significant interactions of a shared region with the bRNAP ω subunit (Fig. 7A; corresponding to aRNAP subunit K or eRNAP ABC23/Rpb6) 54.
β′a13-a14; secondary-channel rim helices (Fig. 8)
Fig. 8. β′a13-a15; secondary-channel rim helices.
A. Schematic representation of the sequence context of β′a13-a14 in Thermus RNAP. As in Fig. 5A, except: β′a13, blue; β′a14, green. The approximate insertion point of β′In4 is denoted by a magenta circle. At the bottom, the horizontal line denotes a segment of β′a13 that participates in conserved interactions with other shared regions.
B. Top view of the Tth bRNAP TEC structure (PDB 2O5J) 9. The RNAP is shown as a backbone worm and color-coded as follows: αI, αII, ω, grey; β, light cyan; β′, as in (A). The nucleic acids of the TEC are shown in CPK format and color-coded as shown in the key. MgII in the active site are shown as yellow spheres. The incoming nucleotide substrate is shown in stick format, with carbon atoms colored blue. Zn2+ ions are shown as green spheres. The approximate insertion point for β′In4 is denoted by a magenta sphere and labeled as in (A). The thick black arrow points in the downstream direction (the direction of RNAP transcription).
β′a13 begins with a short helical hairpin with an extended second α-helix (Fig. 8). The extended α-helix and a loop traverse from the β′-side to the β-side of the active site channel. The last α-helix of β′a13 and the first α-helix of β′a14 pack in an antiparallel manner to form a structural element that has been called the secondary-channel rim helices (Fig. 8). The secondary channel (called the funnel in eRNAP II) 8 provides the only direct pathway for the nucleotide substrate to reach the RNAP active site from the bulk solution 6; 15. The single-stranded 3′ segment of the RNA transcript formed during backtracking 55–57 is also extruded out through the secondary channel 18. The secondary channel also provides direct access to the RNAP active center for extrinsic factors that modulate various aspects of RNAP function 58, such as eukaryotic TFIIS 59, or prokaryotic Gre-factors 60–62 and DksA 63. The secondary channel rim helices serve as a binding platform for the Gre-factors 61. Sce RNAP II Arg726 (corresponding to Thermus β′Thr1000) of β′a14 is critical for the binding of the eRNAP II-specific inhibitor α-amanitin 13; 64; 65.
β′a15-a16; bridge helix, trigger-loop (Fig. 9)
β′a15-a16 form two additional structural elements that are central to the RNAP catalytic activity, the BH (β′a15) and the TL (β′R16), both containing many absolutely conserved residues (Fig. 9A). The N-terminal region of β′a15 forms one wall of the secondary channel, continues into a structural element directly behind the secondary-channel rim helices, then enters into a long α-helix that traverses from the β side of the RNAP active site channel, across the middle of the channel, back to the β′ side of the channel. This α-helix has been called the BH 8, since it bridges across the β and β′ sides of the active site channel.
Because the BH has been observed in either straight or kinked conformations in different crystal structures of RNAP 6; 8, it was proposed that the BH cycles between kinked and straight conformations, and that this was an integral part of the enzyme’s catalytic cycle 8. From a study where the functional properties of 367 site-directed mutants in the BH of aRNAP were characterized, Tan et al. 66 concluded that localized BH kinking forms a normal part of the RNAP nucleotide addition cycle. This conclusion is supported by additional structural studies 12; 25; 26.
Tan et al. 66 found that three absolutely conserved residues in the BH cannot be substituted without dramatic consequences for RNAP catalytic activity, β′Thr1088, Gly1092, and Arg1096. β′Thr1088 interacts with the template DNA at +1, as well as with absolutely conserved β′Thr1234 in TL helix1 (see below). A Gly residue appears to be required at position 1092; any other side chain at this position would interfere with the path of the template DNA at the +1 position (Fig. 9B). β′Arg1096 interacts with the template DNA at +2 (Figs. 9B, 9C).
The BH interacts extensively with two other α-helices, trigger-loop (TL) helices 1 and 2, to form a sort of three-helix bundle (Fig. 9). β′a16 comprises essentially TL helix1 and the first part of the TL, the loop connecting TL helices 1 and 2 (Fig. 9). TL helix 2 is shared among all RNAPs except possibly pRNAPs. This region shows weak sequence homology, and the pRNAPs have a roughly 500 amino acid insertion at this point (β′In6; Fig. 9) 4, making confident alignment impossible. Thus, we cannot rule out that TL helix 2 is a structural feature shared among all RNAPs, but we also cannot rule out confidently that the pRNAPs have a different structure in this region.
The TL tends to be unstructured in unliganded RNAP 6; 8, but was revealed in a structured conformation where it interacts with the correct (matched) incoming nucleotide substrate in the TEC 12. Thus, the TL is a mobile structural element that makes many direct contacts with the NTP substrate in the RNAP active center, detecting the topology of a correct RNA/DNA hybrid base pair 9; 12. A network of contacts between the tip and various parts of the rNTP promote substrate recognition, enzyme fidelity, and catalysis, including an interaction between absolutely conserved β′His1242 and the substrate 9; 12; 65; 67; 68.
The BH and β′a16 (TL helix1 and TL) appear to work in concert, conformational changes in one structural element likely influence the conformation of the others. Supporting this notion, many of the absolutely conserved residues in β′a15 and β′a16 interact with each other: Absolutely conserved β′Gly1080 and Thr1088 (both of the BH) interact with absolutely conserved β′Phe1241 (TL) and Thr1234 (TL helix1; Figs. 9B, 9C). These residues interact with other absolutely conserved residues in the vicinity of the active site as well. Absolutely conserved β′Arg1078 (BH) interacts with absolutely conserved βArg428 (FL2, βR8, see Figs. 2C, 2D). An interesting, absolutely conserved three-way interaction occurs between β′Leu708 (β′a12, see Fig. 7C), β′Thr1088 (BH), and β′Thr1234 (TL helix1; Fig. S3).
Many residues within β′a15-a16 are important for streptolydigin binding. These include BH residues β′Leu1086, Ala1089, Ser1091, and absolutely conserved Arg1096; TL-helix1 residues β′Pro1232 and Leu1236; and TL residues β′Thr1237, Thr1243, and Val1246 10; 25; 26. Likewise, many residues within βa15-a16 are important for the eRNAP II-specific inhibitor α-amanitin. These include Sce RNAP II subunit A Ile756, Ser769, and Gly772 (corresponding to Thermus β′Gln1033, Gln1046, and Ser1049 in β′a15), a large number of residues in the BH, and residues in the TL itself, including absolutely conserved Sce RNAP II A-His1085 (corresponding to Thermus β′His1242) 13; 65. The inhibition mechanism of streptolydigin and α-amanitin appears to be linked to their interaction with the BH and TL elements, and the ability of the inhibitors to alter/influence the conformational equilibria of these structural elements 10; 13; 25; 26; 64, again supporting the notion that concerted conformational rearrangements of the BH and TL elements are a central part of the RNAP catalytic cycle 66.
The N-terminal part of β′a15 (roughly residues β′1021–1036), and the middle of β′a16 (roughly residues β′1231–1247) help form the secondary channel (Fig. 9B). Shared among many bRNAPs is a β-β′ Module 2 domain (BBM2)31, which is inserted between β′a15 and β′a16 (Figs. 9A, 9B). Immediately following TL helix2 is the bRNAP jaw, another SBHM domain that is shared among bRNAPs but not aRNAP or eRNAPs 31.The eRNAP II ‘jaw’ is inserted at precisely this position of the largest subunit, but is not related in structure to the bRNAP jaw 8. The bRNAP jaw likely interacts with Eco RNAP β′In6 69, and is the site of interaction for bacteriophage T7 gp2, a potent inhibitor of Eco RNAPσ70-dependent open promoter complex formation 70; 71.
β′a17-a20; clamp, Sw5 (Fig. 10)
Following the jaw, β′a17-a19 form the core of a structural motif (Fig. 10). β′a17-a19 are linked by segments that are conserved among bRNAP but diverge among aRNAP and eRNAPs.
β′a20 enters into the clamp, where absolutely conserved β′Leu1447 participates in the clamp hydrophobic core. β′a20 then leaves the clamp and forms Sw5, which serves as a hinge mediating clamp movement 11. Most of β′a20 participates in interactions with βa15-a16 as a part of the clamp. Absolutely conserved β′Gly1475 makes van der Waals interactions with absolutely conserved βGly1044 from βa15. Several residues in β′a20 are important for interactions with the antibiotic Myx, including β′Gly1461 and highly conserved β′Gly1469 within Sw5 41; 42.
Discussion
Here, we discuss some general features and observations that arose from this analysis.
High proportion of conserved glycine residues
Nearly half of the absolutely conserved positions in β correspond to Glycines (16 out of 34 residues, 47%). In β′, 8 out of 38 conserved positions correspond to Glycine (21%). Fully 1/3 of the total conserved residues in both large subunits (24 out of 72) are Glycines. For a handful of these positions, the requirement for a Gly residue can be rationalized from the structure. Some of these Gly residues line channels or paths for the nucleic acids (RNA exit channel, βGly1011; path for DNA template, βGly1023/Gly1028/Gly1029/Gly1033, Fig. 4C). Most conspicuously, β′Gly1092 appears to be required to make room for the template DNA sliding past the BH at the +1 position (Fig. 9). Other Gly residues make van der Waals interactions with other conserved residues, and there does not appear to be sufficient room for a side chain (βGly1044, Fig. 4C; β′Gly1080, Fig. 9; β′Gly1475, Fig. 10). The majority of the conserved Gly residues play no obvious functional role, and likely play important structural roles.
Double-psi β-barrels in the RNAP active center
The core catalytic center of the multi-subunit cellular DNA-dependent RNAPs (DDRPs) shares the dual, double-psi β-barrel (DPBB) 72 domain architecture (Fig. S4) with the eukaryotic RNA-dependent RNAPs (RDRPs) 31; 73. Two β-strands from βa11, three from βa14, and one from βa15 make up the six β-strands of the β subunit DPBB motif (Figs. 3A, 4A, S5). Intervening sequences, including the entire flap and βa12 and βa13, constitute large insertions within the DPBB core fold 31. β′a11-a12 make up the β′-DPBB 31 (Figs. 7, S6).
In the RDRPs, the two DPBB domains occur within the same polypeptide chain (Fig. S7), with one DPBB domain contributing the signature DbDGD (‘b’ is a bulky residue) metal-coordinating motif 73. In the DDRPs, the β′ subunit contributes one DPBB with the signature metal-coordinating NADFDGD motif (Fig. 7). The β subunit contributes the second, highly diverged DPBB that lacks the metal-coordinating motif.
In addition to the conservation of the dual-DPBB architecture, the disposition of the two DPBB domains, and the DbDGD metal-coordinating motif, the RDRPs and DDRPs share a number of other universally conserved residues (Fig. S7). In the DDRPs, where structural information on functional complexes is available, these shared residues interact with the nucleotide substrate (DDRP-βR557/RDRP-R671), the RNA transcript (βK846/K743, β′R704/R962), and the template DNA (β′R622/R913). Other shared residues may play critical structural roles (βG847/G744, β′P706/G964). Another DDRP universally conserved residue, βR879, is universally conserved in the RDRPs as K767 (not shown in Fig. S7). In the DDRPs, this residue also interacts with the nucleotide substrate.
As noted by Salgado et al. 73, structure-based alignment of the shared DDRP folds reveals conservation of additional structural features surrounding the enzyme active center, including the BH and TL-helices (Fig. S7). In the RDRP, the two DPPB domains and additional conserved structural features colinearly arranged within a central catalytic domain, while these same features in the DDRPs are shared between two separate subunits (β/β′) and are widely separated in the sequences due to large insertions (Fig. S7), supporting the hypothesis that the DDRPs and RDRPs diverged from a common ancestor, with the accumulation of independent domains in the case of the DDRPs 31.
Switch regions
The Switch (Sw) regions were identified in the first eRNAP II TEC structure as regions that become ordered or change conformation compared with the free RNAP, and were proposed to mediate a conformational ‘switch’ that marks the entry of the enzyme into the stable elongation phase 11. Five Sw regions were originally defined 11, but our analysis suggests that only three of these, Sw2, Sw3, and Sw5, are critical for RNAP function (Table S2). Sw2, contained within β′a11, is highly conserved overall (Table S2), and contains three absolutely conserved residues that make important interactions with the template DNA near the active site (Fig. 7). Substitution of absolutely conserved β′Arg615 to Ala in Sw2 renders aRNAP totally inactive in elongation 27. A number of residues within Sw2 form contacts with the antibiotic Myx 41; 42.
Sw3, contained within βa15, is also highly conserved overall (Table S2), and contains five absolutely conserved residues that line the path of the template DNA around –3/-4 (Fig. 4). Sw5, contained within β′a20 (Fig. 10), is moderately conserved overall (Table S2).
Sw1 begins in β′a18 but continues into an unshared region between β′a18 and β′a19, so much of Sw1 is not shared among all RNAPs. Sw4 is contained within βa15, but the overall sequence is not highly conserved (Table S2).
Interactions with the α subunits
The α subunits of bRNAP initiate RNAP assembly by dimerizing into a platform with which the β and β′ subunits interact 74. One of the α subunits, αI, interacts almost exclusively with the β subunit, while the other subunit, αII, interacts with β′ 6. A heterodimer of the aRNAP subunits D/L or of the eRNAP subunits Rpb3/Rpb11 play the role of the bRNAP αI/αII dimer 75.
An extensive biochemical analysis identified determinants within the Eco bRNAP β subunit involved in the obligatory interaction with the α dimer 29. βa11, βa12, βR14 and βa15 were found to be important for the interaction of β with the α dimer, with βa14 and βa15 being essential. These biochemical findings are completely consistent with our structural analysis of evolutionarily conserved RNAP intersubunit interactions involving β/β′ shared regions. We observed interactions between βa11, βa12, βa14, and βa15 with the αI subunit (Figs. 3A, 4A).
Surprisingly, we do not observe interactions between any β/β′ shared regions and the II subunit. Assembly of core bRNAP is thought to proceed through the pathway 76:
The formation of the α dimer appears to be obligatory for complete core bRNAP assembly. Mutants of α defective in homodimerization are also defective in core bRNAP assembly 77–80. In at least one case, an α mutant defective in homodimerization forms an α1β complex, but β′ is unable to assemble into this complex 79; 80, suggesting that β′ interactions with αII are required for core bRNAP assembly. Nevertheless, the αII/β′ interactions are not evolutionarily conserved. In Thermus RNAP, the interaction between αII and β′ occurs primarily through a domain of β′ located between β′a12 and β′a13 (Thermus β′ residues 794 – 877), a region that is not even shared among bRNAPs and, for example, is poorly conserved between Thermus and Eco bRNAPs. Thus, the incorporation of αII into the RNAP complex appears to be due to it’s ability to dimerize with αI, combined with interactions with β′ that are highly idiosyncratic.
Materials and Methods
Figures were constructed using PyMOL (http://www.pymol.org). Structural analyses utilized the programs O 81, COOT 82, and programs from the CCP4 package 83.
Supplementary Material
Acknowledgments
W.J.L. was supported by National Institutes of Health MSTP grant GM07739 and The W.M. Keck Foundation Medical Scientist Fellowship. We thank Lars Westblade, Chris Lima, and Tom Muir for helpful discussions and advice. This work was supported by NIH GM061898 and GM053759 to S.A.D.
Abbreviations
- aRNAP
archaeal RNA polymerase
- BBM2
β-β′ Module 2
- BH
Bridge Helix
- bRNAP
bacterial RNA polymerase
- DDRP
DNA-dependent RNA Polymerase
- DPBB
Double-psi β-barrel
- Eco
Escherichia coli
- eRNAP
eukaryotic RNA polymerase
- FL2
Fork Loop 2
- MSA
Multiple sequence alignment
- Myx
Myxopyronin
- pRNAP
plastid RNA polymerase
- RDRP
RNA-dependent RNA Polymerase
- RNAP
RNA polymerase
- SBHM
Sandwich barrel hybrid motif
- Sce
Saccharomyces cerevisiae
- Sw2
Switch2
- Sw3
Switch 3
- Sw5
Switch 5
- TEC
Ternary elongation complex
- TL
Trigger Loop
- vRNAP
viral RNA polymerase
- ZNR
Zn-ribbon
Footnotes
Please visit http://darstlab.org/supp/RNAP_MSA_2009 to download the BlaFA and other custom programs, RNAP BlaFA pattern files, sequence files, alignments, annotation files, phylogenetic trees, intergenic gap analysis, shared sequence region positions, and the lineage-specific insertions details.
References
- 1.Cramer P. Multisubunit RNA polymerases. Curr Opinion Struct Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 2.Archambault J, Friesen JD. Genetics of RNA polymerases I, II, and III. Microbiol Rev. 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jokerst RS, Weeks JR, Zehring WA, Greenleaf AL. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol Gen Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 4.Lane WJ, Darst SA. Molecular evolution of multi-subunit RNA polymerases: sequence analysis. 2009 doi: 10.1016/j.jmb.2009.10.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sweetser D, Nonet M, Young RA. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc Natl Acad Sci USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 7.Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008;451:851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 9.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 10.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 11.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 12.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–54. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brueckner F, Cramer P. Structural basis of transcription inhibition by alpha-amanitin and implications for RNA polymerase II translocation. Nat Struct Mol Biol. 2008;15:811–818. doi: 10.1038/nsmb.1458. [DOI] [PubMed] [Google Scholar]
- 14.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: Separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 15.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–9. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 16.Kettenberger H, Armache KJ, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 17.Caffrey DR, Dana PH, Mathur V, Ocano M, Hong EJ, Wang YE, Somaroo S, Caffrey BE, Potluri S, Huang ES. PFAAT version 2.0: a tool for editing, annotating, and analyzing multiple sequence alignments. BMC Bioinformat. 2007;8:381. doi: 10.1186/1471-2105-8-381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 19.Deaconescu AM, Chambers AL, Smith AJ, Nickels BE, Hochschild A, Savery NJ, Darst SA. Structural basis for bacterial transcription-coupled DNA repair. Cell. 2006;124:507–520. doi: 10.1016/j.cell.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 20.Smith AJ, Savery NJ. RNA polymerase mutants defective in the initiation of transcription-coupled DNA repair. Nucleic Acids Res. 2005;33:755–64. doi: 10.1093/nar/gki225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Severinov K, Darst SA. A mutant RNA polymerase that forms unusual open promoter complexes. Proc Natl Acad Sci USA. 1997 doi: 10.1073/pnas.94.25.13481. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Campbell EA, Korzheva N, Mustaev A, Murakami K, Nair S, Goldfarb A, Darst SA. Structural mechanism for rifampicin inhibition of bacterial RNA polymerase. Cell. 2001;104:901–912. doi: 10.1016/s0092-8674(01)00286-0. [DOI] [PubMed] [Google Scholar]
- 23.McClure WR, Cech CL. On the mechanism of rifampicin inhibition of RNA synthesis. J Biol Chem. 1978;253:8949–8956. [PubMed] [Google Scholar]
- 24.Feklistov A, Mekler V, Jiang Q, Westblade LF, Irschik H, Jansen R, Mustaev A, Darst SA, Ebright RH. Rifamycins do not function by allosteric modulation of binding of Mg2+ to the RNA polymerase active center. Proc Natl Acad Sci USA. 2008;105:14820–14825. doi: 10.1073/pnas.0802822105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Temiakov D, Zenkin N, Vassylyeva MN, Perederina A, Tahirov TH, Kashkina E, Savkina M, Zorov S, Nikiforov V, Igarashi N, Matsugaki N, Wakatsuki S, Severinov K, Vassylyev DG. Structural basis of transcription inhibition by antibiotic streptolydigin. Mol Cell. 2005;19:655–666. doi: 10.1016/j.molcel.2005.07.020. [DOI] [PubMed] [Google Scholar]
- 26.Tuske S, Sarafianos SG, Wang X, Hudson B, Sineva E, Mukhopadhyay J, Birktoft JJ, Leroy O, Ismail S, Clark ADJ, Dharia C, Napoli A, Laptenko O, lee J, Borukhov S, Ebright RH, Arnold E. Inhibition of bacterial RNA polymerase by streptolydigin: Stabilization of a straight-bridge-helix active-center conformation. Cell. 2005;122:541–552. doi: 10.1016/j.cell.2005.07.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Naji S, Bertero MG, Spitalny P, Cramer P, Thomm M. Structure-function analysis of the RNA polymerase cleft loops elucidates initial transcription, DNA unwinding and RNA displacement. Nucleic Acids Res. 2008;36:676–687. doi: 10.1093/nar/gkm1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Thomm M, Reich C, Grunberg S, Naji S. Mutational studies of archaeal RNA polymerase and analysis of hybrid RNA polymerases. Biochem Soc Trans. 2009;37:18–22. doi: 10.1042/BST0370018. [DOI] [PubMed] [Google Scholar]
- 29.Wang Y, Severinov K, Loizos N, Fenyö D, Heyduk E, Heyduk T, Chait BT, Darst SA. Determinants for Escherichia coli RNA polymerase assembly within the β subunit. J Mol Biol. 1997;270:648–662. doi: 10.1006/jmbi.1997.1139. [DOI] [PubMed] [Google Scholar]
- 30.Anantharaman V, Koonin EV, Aravind L. Regulatory potential, phyletic distribution and evolution of ancient, intracellular small-molecule-binding domains. J Mol Biol. 2001;307:1271–1292. doi: 10.1006/jmbi.2001.4508. [DOI] [PubMed] [Google Scholar]
- 31.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct Biol. 2003;3:1–23. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gross CA, Chan C, Dombroski A, Gruber T, Sharp M, Tupy J, Young B. The functional and regulatory roles of sigma factors in transcription. Cold Spring Harbor Symp Quant Biol. 1998;63:141–155. doi: 10.1101/sqb.1998.63.141. [DOI] [PubMed] [Google Scholar]
- 33.Kuznedelov K, Minakhin L, Niedziela-Majka A, Dove SL, Rogulja D, Nickels BE, Hochschild A, Heyduk T, Severinov K. A role for interaction of the RNA polymerase flap domain with the sigma subunit in promoter recognition. Science. 2002;295:855–857. doi: 10.1126/science.1066303. [DOI] [PubMed] [Google Scholar]
- 34.Murakami K, Masuda S, Campbell EA, Muzzin O, Darst SA. Structural basis of transcription initiation: An RNA polymerase holoenzyme/DNA complex. Science. 2002;296:1285–1290. doi: 10.1126/science.1069595. [DOI] [PubMed] [Google Scholar]
- 35.Mooney RA, Artsimovitch I, Landick R. Information processing by RNA polymerase: Recognition of regulatory signals during RNA chain elongation. J Bacteriol. 1998;180:3265–3275. doi: 10.1128/jb.180.13.3265-3275.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Toulokhonov I, Artsimovitch I, Landick R. Allosteric control of RNA polymerase by a site that contacts nascent RNA hairpins. Science. 2001;292:730–733. doi: 10.1126/science.1057738. [DOI] [PubMed] [Google Scholar]
- 37.Toulokhonov I, Landick R. The flap domain is required for pause RNA hairpin inhibition of catalysis by RNA polymerase and can modulate intrinsic termination. Mol Cell. 2003;12:1125–1136. doi: 10.1016/s1097-2765(03)00439-8. [DOI] [PubMed] [Google Scholar]
- 38.Gregory BD, Nickels BE, Garrity SJ, Severinova E, Minakhin L, Urbauer RJ, Urbauer JL, Heyduk T, Severinov K, Hochschild A. A regulator that inhibits transcription by targeting an intersubunit interaction of the RNA polymerase holoenzyme. Proc Natl Acad Sci USA. 2004;101:4554–4559. doi: 10.1073/pnas.0400923101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nechaev S, Kamali-Moghaddam M, Andre E, Leonetti JP, Geiduschek EP. The bacteriophage T4 late-transcription coactivator gp33 binds the flap domain of Escherichia coli RNA polymerase. Proc Natl Acad Sci USA. 2004;101:17365–17370. doi: 10.1073/pnas.0408028101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mustaev A, Kashlev M, Zaychikov E, Grachev M, Goldfarb A. Active center rearrangement in RNA polymerase initiation complex. J Biol Chem. 1993;268:19185–19187. [PubMed] [Google Scholar]
- 41.Belogurov GA, Vassylyeva MN, Sevostyanova A, Appleman JR, Xiang AX, Lira R, Webber SE, Klyuyev S, Nudler E, Artsimovitch I, Vassylyev DG. Transcription inactivation through local refolding of the RNA polymerase structure. Nature. 2009;457:332–335. doi: 10.1038/nature07510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Mukhopadhyay J, Das K, Ismail S, Koppstein D, Jang M, Hudson B, Sarafianos S, Tuske S, Patel J, Jansen R, Irschik H, Arnold E, Ebright RH. The RNA polymerase ‘switch region’ is a target for inhibitors. Cell. 2008;135:295–307. doi: 10.1016/j.cell.2008.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: Implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 44.Murakami K, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 Å resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 45.Vassylyev DG, Sekine S, Laptenko O, Lee J, Vassylyeva MN, Borukhov S, Yokoyama S. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 Å resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 46.Naryshkina T, Kuznedelov K, Severinov K. The role of the largest RNA polymerase subunit lid element in preventing the formation of extended RNA-DNA hybrid. J Mol Biol. 2006;361:634–643. doi: 10.1016/j.jmb.2006.05.034. [DOI] [PubMed] [Google Scholar]
- 47.Toulokhonov I, Landick R. The role of the lid element in transcription by E. coli RNA polymerase. J Mol Biol. 2006;361:644–658. doi: 10.1016/j.jmb.2006.06.071. [DOI] [PubMed] [Google Scholar]
- 48.Arthur TM, Burgess RR. Localization of a sigma70 binding site on the N terminus of the Escherichia coli RNA polymerase beta′ subunit. J Biol Chem. 1998;273:31381–7. doi: 10.1074/jbc.273.47.31381. [DOI] [PubMed] [Google Scholar]
- 49.Arthur TM, Anthony LC, Burgess RR. Mutational analysis of beta β′260–309, a sigma 70 binding site located on Escherichia coli core RNA polymerase. J Biol Chem. 2000;275:23113–9. doi: 10.1074/jbc.M002040200. [DOI] [PubMed] [Google Scholar]
- 50.Belogurov GA, Vassylyeva MN, Svetlov V, Klyuyev S, Grishin NV, Vassylyev DG, Artsimovich I. Structural basis for converting a general transcription factor into an operon-specific virulence regulator. Mol Cell. 2007;26:117–129. doi: 10.1016/j.molcel.2007.02.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kuznedelov K, Korzheva N, Mustaev A, Severinov K. Structure-based analysis of RNA polymerase function: the largest subunit’s rudder contributes critically to elongation complex stability and is not involved in the maintenance of RNA-DNA hybrid length. EMBO J. 2002;21:1369–1378. doi: 10.1093/emboj/21.6.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Sosunov V, Zorov S, Sosunova E, Nikolaev A, Zakeyeva I, Bass I, Goldfarb A, Nikiforov V, Severinov K, Mustaev A. The involvement of the aspartate triad of the active center in all catalytic activieis of multisubunit RNA polymerase. Nucleic Acids Res. 2005;33:4202–4211. doi: 10.1093/nar/gki688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zaychikov E, Martin E, Denissova L, Kozlov M, Markovtsov V, Kashlev M, Heumann H, Nikiforov V, Goldfarb A, Mustaev A. Mapping of catalytic residues in the RNA polymerase active center. Science. 1996;273:107–109. doi: 10.1126/science.273.5271.107. [DOI] [PubMed] [Google Scholar]
- 54.Minakhin L, Bhagat S, Brunning A, Campbell EA, Darst SA, Ebright RH, Severinov K. Bacterial RNA polymerase subunit ω and eukaryotic RNA polymerase subunit RPB6 are sequence, structural, and functional homologs and promote RNA polymerase assembly. Proc Natl Acad Sci USA. 2001;98:892–897. doi: 10.1073/pnas.98.3.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 57.Reeder TC, Hawley DK. Promoter proximal sequences modulate RNA polymerase II elongation by a novel mechanism. Cell. 1996;87:767–777. doi: 10.1016/s0092-8674(00)81395-1. [DOI] [PubMed] [Google Scholar]
- 58.Nickels BE, Hochschild A. Regulation of RNA polymerase through the secondary channel. Cell. 2004;118:281–284. doi: 10.1016/j.cell.2004.07.021. [DOI] [PubMed] [Google Scholar]
- 59.Kettenberger H, Armache KJ, Cramer P. Architecture of the RNA polymerase II-TFIIS complex and implications for mRNA cleavage. Cell. 2003;114:347–357. doi: 10.1016/s0092-8674(03)00598-1. [DOI] [PubMed] [Google Scholar]
- 60.Laptenko O, Lee J, Lomakin I, Borukhov S. Transcript cleavage factors GreA and GreB act as transient catalytic components of RNA polymerase. EMBO J. 2003;23:6322–6334. doi: 10.1093/emboj/cdg610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Opalka N, Chlenov M, Chacon P, Rice WJ, Wriggers W, Darst SA. Structure and function of the transcription elongation factor GreB bound to bacterial RNA polymerase. Cell. 2003;114:335–345. doi: 10.1016/s0092-8674(03)00600-7. [DOI] [PubMed] [Google Scholar]
- 62.Sosunova E, Sosunov V, Kozlov M, Nikiforov V, Goldfarb A, Mustaev A. Donation of catalytic residues to RNA polymerase active center by transcription factor Gre. Proc Natl Acad Sci USA. 2003 doi: 10.1073/pnas.2536698100. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Perederina A, Svetlov V, Vassylyeva M, Tahirov T, Yokoyama S, Artsimovitch I, Vassylyev D. Regulation through the secondary channel - structural framework for ppGpp-DksA synergism during transcription. Cell. 2004;118:297–309. doi: 10.1016/j.cell.2004.06.030. [DOI] [PubMed] [Google Scholar]
- 64.Bushnell DA, Cramer P, Kornberg RD. Structural basis of transcription: Alpha-amanitin-RNA polymerase II cocrystal at 2.8 Å resolution. Proc Natl Acad Sci USA. 2002;99:1218–1222. doi: 10.1073/pnas.251664698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kaplan CD, Larsson KM, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Tan L, Wiesler S, Trzaska D, Carney H, Weinzierl R. Bridge helix and trigger loop perturbations generate superactive RNA polymerases. J Biol. 2008;7:40. doi: 10.1186/jbiol98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bar-Nahum G, Epshtein V, Ruckenstein AE, Rafikov R, Mustaev A, Nudler E. A ratchet mechanism of transcription elongation and its control. Cell. 2005;120:183–193. doi: 10.1016/j.cell.2004.11.045. [DOI] [PubMed] [Google Scholar]
- 68.Kireeva ML, Nedialkov YA, Cremona GH, Purtov YA, Lubkowska L, Malagon F, Burton ZF, Strathern JN, Kashlev M. Transient reversal of RNA polymerase II active site closing controls fidelity of transcription elongation. Mol Cell. 2008;30:557–566. doi: 10.1016/j.molcel.2008.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Chlenov M, Masuda S, Murakami KS, Nikiforov V, Darst SA, Mustaev A. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase β′ subunit. J Mol Biol. 2005;353:138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 70.Nechaev S, Severinov K. Inhibition of Escherichia coli RNA polymerase by bacteriophage T7 gene 2 protein. J Mol Biol. 1999;289:815–26. doi: 10.1006/jmbi.1999.2782. [DOI] [PubMed] [Google Scholar]
- 71.Nechaev S, Yuzenkova J, Niedziela-Majka A, Heyduk T, Severinov K. A novel bacteriophage-encoded RNA polymerase binding protein inhibits transcription initiation and abolishes transcription termination by host RNA polymerase. J Mol Biol. 2002;320:11–22. doi: 10.1016/S0022-2836(02)00420-5. [DOI] [PubMed] [Google Scholar]
- 72.Castillo RM, Mizuguchi K, Dhanaraj V, Albert A, Blundell TL, Murzin AG. A six-stranded double-psi beta barrel is shared by several protein superfamilies. Structure. 1999;7:227–236. doi: 10.1016/s0969-2126(99)80028-8. [DOI] [PubMed] [Google Scholar]
- 73.Salgado PS, Koivunen MRL, Makeyev EV, Bamford DH, Stuart DI, Grimes JM. The structure of an RNAi polymerase links RNA silencing and transcription. PLoS Biol. 2006;4:2274–2281. doi: 10.1371/journal.pbio.0040434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Zillig W, Palm P, Heil A. Function and reassembly of subunits of DNA-dependent RNA polymerase. In: Losick R, Chamberlin M, editors. RNA Polymerase. Cold Spring Harbor Laboratory; Cold Spring Harbor, NY: 1976. pp. 101–125. [Google Scholar]
- 75.Zhang G, Darst SA. Structure of the Escherichia coli RNA polymerase α subunit amino-terminal domain. Science. 1998;281:262–266. doi: 10.1126/science.281.5374.262. [DOI] [PubMed] [Google Scholar]
- 76.Ishihama A. Subunit assembly of Escherichia coli RNA polymerase. Adv Biophys. 1981;14:1–35. [PubMed] [Google Scholar]
- 77.Kimura M, Fujita N, Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase. Deletion analysis of the amino-terminal assembly domain. J Mol Biol. 1994;242:107–115. doi: 10.1006/jmbi.1994.1562. [DOI] [PubMed] [Google Scholar]
- 78.Kimura M, Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase: Amino acid substitution within the amino-terminal assembly domain. J Mol Biol. 1995;254:342–349. doi: 10.1006/jmbi.1995.0621. [DOI] [PubMed] [Google Scholar]
- 79.Kimura M, Ishihama A. Functional map of the alpha subunit of Escherichia coli RNA polymerase: Insertion analysis of the amino-terminal assembly domain. J Mol Biol. 1995;248:756–767. doi: 10.1006/jmbi.1995.0258. [DOI] [PubMed] [Google Scholar]
- 80.Kimura M, Ishihama A. Subunit assembly in vivo of Escherichia coli RNA polymerase: role of the amino -terminal assembly domain of alpha subunit. Genes to Cells. 1996;1:517–528. doi: 10.1046/j.1365-2443.1996.d01-258.x. [DOI] [PubMed] [Google Scholar]
- 81.Jones TA, Zou J-Y, Cowan S, Kjeldgaard M. Improved methods for building protein models in electron denstiy maps and the location of errors in these models. Acta crystallographica. 1991;A47:110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 82.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 83.Collaborative Computational Project. The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.