Abstract
Notch signaling is a highly conserved pathway important for normal embryonic development and as well as cancer. We previously demonstrated a role for Notch3 in lung cancer pathogenesis. Notch3 inhibition resulted in tumor apoptosis and growth suppression. In vitro, these effects were enhanced when the EGFR pathway was also inhibited, suggesting significant crosstalk between these two pathways. How Notch3 and EGFR/MAPK pathways cooperate in modulating apoptosis is not yet known. In this study, we provide evidence that Notch3 regulates Bim, a BH-3-only protein via MAPK signaling. Furthermore, loss of Bim expression prevents tumor apoptosis induced by Notch3 inhibition. Using γ-secretase inhibitor and erlotinib in a xenograft model, Bim induction and tumor inhibition were enhanced compared to either agent alone, consistent with our previous observation of significant synergism between Notch and EGFR/ras/MAPK signaling. Thus, our data support the hypothesis that Notch3 not only plays a crucial role in lung cancer through regulating apoptosis but also cooperates with the EGFR/MAPK pathway in modulating Bim.
Keywords: Notch3, Bim, Apoptosis, Lung Cancer
INTRODUCTION
Notch3 belongs to a family of proteins essential for cellular differentiation in a variety of developing tissues. In mammals, there are four Notch receptors (Notch1 to Notch4) and two families of ligands, Jagged (Jagged1, 2) and Delta-like (Dll1, −3, −4). Ligand binding results in two successive proteolytic cleavages by an ADAM-type metalloprotease and by a γ-secretase, respectively (for a recent review see (Roy et al., 2007)). The released Notch intracellular domain (NICD) translocates to the nucleus, binds to transcription factor CSL (CBF1, Sel, Lag-1), and induces expression of target genes, such as the Hairy-enhancer of Split (HES) and hairy and Enhancer-of-split related with YRPW motif (Hey) gene families.
Aberrant activation of Notch proteins is associated with cancer phenotypes (Das et al., 2004; Curry et al., 2005; Duechler et al., 2005; Reedijk et al., 2005). Notch3 is overexpressed in about 40% of non-small cell lung cancers (NSCLC), and suppression of Notch3 results in the loss of the malignant phenotype both in vitro and in vivo models (Haruki et al., 2005; Konishi et al., 2007). Tumor suppression is enhanced when Notch inhibition is combined with epidermal growth factor (EGF) pathway inhibitors in vitro. These observations support the findings in both the development and the cancer literature that Notch and EGFR/ras/MAPK pathways are interdependent. Whether ras enhances or antagonizes Notch signaling in development appears to be context-dependent. In cancer, however, many studies have suggested that the ability of ras or Notch to transform cells depends on the cooperative relationship between them (Dievart et al., 1999; Fitzgerald et al., 2000; Weijzen et al., 2002; Haruki et al., 2005).
The role of the Notch pathway in tumorigenesis also involves the regulation of the apoptotic pathway. The Bcl-2-related proteins are key regulators of apoptosis. There are three subfamilies, the pro-survival Bcl-2 like proteins, such as Bcl-2, Bcl-xL and Mcl-1, the pro-apoptotic proteins Bax and Bak, and the BH3-only proteins, which share homology with the Bcl-2 family only in the BH3 region (Heiser and Hebrok, 2004). The BH3-only proteins function mainly as initiators of apoptosis. They are activated by cellular stress, such as DNA damage in the case of Noxa and Puma, or by growth factor deprivation in the case of Hrk and Bim. Other BH3-only proteins such as Bid are activated by death receptors via caspase-8 (Borner, 2003). Recently, it has been reported that Bim is required to induce apoptosis in several cancer types, including lung cancer (Gomez-Bougie et al., 2005; Gong et al., 2007). Little is known about the relationship between the Notch3 pathway and Bim regulation. Since Bim is induced by growth factor deprivation, we hypothesize that the crosstalk between EGFR/ras and Notch3 modulates apoptosis through the regulation of Bim.
In this study, we demonstrate that Notch3 modulates apoptosis through the Bcl-2 protein family. We also provide evidence that the Notch3 and MAPK pathways modulate Bim to regulate apoptosis. Although Notch3 is known to regulate apoptosis through other pathways such as the NF- κB pathway, this is the first study linking Bim to Notch3-dependent apoptosis in lung cancer.
RESULTS
Notch3 modulates apoptosis through the regulation of pERK and Bcl-2-related proteins but has no effect on the Akt/PI3K pathway
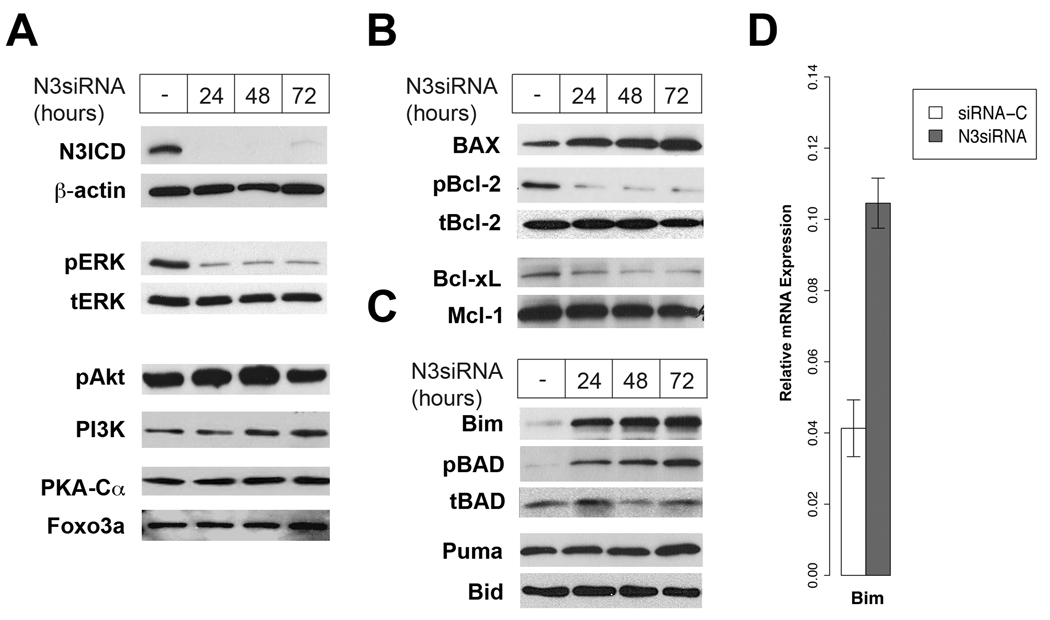
Consistent with our earlier work, loss of Notch3 activity resulted in the downregulation of pERK, a member of the MAPK family. Interestingly, while Notch3 modulated the MAPK pathway, no effect on phospho-Akt, PI3K, PKC-α and Foxo3a was observed (Figure 1A). Notch3 affects the PI3K-Akt pathway in some experimental systems but not others, suggesting that, unlike the MAPK pathway, cellular context is more important for the Notch3 modulation of the PI3K-pAkt pathway (Campos et al., 2002; Wang et al., 2007). Using Notch3 siRNA, we also observed that Notch regulates both pro-apoptotic and pro-survival proteins (Figure 1B). Although no change was detected in Mcl-1, phospho-Bcl-2 (pBCl2) and Bcl-xL were downregulated in HCC2429 when the cells were transfected with Notch3 siRNA, compared to control siRNA. Conversely, the pro-apoptotic protein Bax was upregulated. Taken together, these observations are consistent with the hypothesis that Notch3 is oncogenic in lung cancer.
Figure 1.
Notch3 regulates apoptosis through the modulation of pERK and Bcl-2 proteins in lung cancer cell line HCC2429. (A) Consistent with previous studies, Notch3 siRNA downregulates pERK expression after 48 hours of treatment with Notch3 siRNA. Expressions of pAkt, Foxo3a, PKA-Cα and PI3K are unchanged, suggesting that in lung cancer the MAPK pathway controls Notch3-dependent apoptosis. (B) Inhibition of Notch3 with siRNA upregulates pro-apoptotic Bax and downregulates expressions of the Bcl-2 pro-survival proteins pBcl-2, Bcl-xL and Mcl-1. (C) Among the BH3-only proteins, induction of Bim can be observed. By contrast, no change in the expression of Puma or Bid is seen. Note that pBad is upregulated in Notch3 siRNA treated cells compared to siRNA control. Similar to Bim, cytokines and growth factors regulate pBad, further supporting previous observations of significant crosstalk between Notch and EGFR pathway. (D) Compared to siRNA control, an increase of Bim mRNA was observed when lung cancer cell line HCC2429 was transfected with Notch3 siRNA, suggesting that Notch3 regulated Bim transcriptionally. In all siRNA experiments, non-silencing siRNA was used as control.
BH3-only proteins is a subset of the Bcl-2 family. They function as damage sensors and respond to distinct forms of cellular stress or DNA damage (Borner, 2003). Since the Notch pathway is known to crosstalk with the ras/MAPK pathway, and pro-apoptotic Bim is induced by growth factor deprivation, we hypothesized that Bim is the mediator in Notch3 regulation of apoptosis. This hypothesis is consistent with our earlier observation that Notch3-mediated apoptosis is enhanced in the presence of growth factor deprivation (Haruki et al., 2005). In the present study, Bim expression was increased after transfection with Notch3 siRNA (Figure 1C). Phospho-BAD, a pro-apoptotic protein that is modulated by cytokines/growth factors withdrawal, was similarly affected. Puma functions as sensor for DNA damage and has been shown to be a direct target of p53-mediated apoptosis. On the other hand, Bid is a target of the death receptor ligands. No change in the levels of Puma and Bid was observed when tumor cells were transfected with Notch3 siRNA, suggesting that in some lung cancers Notch3-dependent apoptosis is not mediated by p53 or the death receptors, but by the modulation of growth factors and cytokines. We detected significantly higher mRNA expression of Bim in tumor cells transfected with Notch3 siRNA, compared to siRNA control (Figure 1D). It is known that the Bim promoter possesses binding sites for Foxo3a, Mybs, and c-Jun, and mutation of any one of these sites abolishes Bim transcription in response to growth factor deprivation (Biswas et al., 2007). Since we have shown that Notch3 has no effect on Foxo3a, transcriptional regulation of Bim by Notch3 is indirect and more likely through c-jun and the ras/MAPK pathway.
Inhibition of Notch3 with siRNA induces apoptosis
Inhibition of the Notch pathway by either a dominant-negative receptor (DN) or by the γ-secretase-inhibitor MRK-003 results in tumor apoptosis (Haruki et al., 2005). Since both the DN receptor and γ-secretase-inhibitor potentially target all of the Notch family receptors, we used the small interference RNA (siRNA) system to determine whether these observations are specific to Notch3. Apoptosis markers such as cleaved PARP and cytochrome-c are induced by Notch3 siRNA (Figure 2A). Furthermore, caspase-3 activity was enhanced in lung cancer cells treated with Notch3 siRNA (Fig. 2B). While alterations in apoptotic markers PARP and cytochrome-c were observed, no change in apoptosis was detected with annexin V staining when cells were maintained in 10% FCS (Figure 2C). However, in the presence of growth deprivation, the percentage of apoptotic cells was significantly increased in Notch3 siRNA-treated cells compared with vector control. This observation is consistent with our previous studies with the γ-secretase inhibitor MRK-003 and Notch3 DN, in which apoptosis was seen in serum-free conditions (Haruki et al., 2005; Konishi et al., 2007). These results demonstrate that while Notch3 can modulate many components of apoptosis such as Bcl-xL and Bax in full-serum conditions, a noticeable effect on apoptosis requires growth factor deprivation.
Figure 2.
The loss of Notch3 expression induces apoptosis. (A) Inhibition of Notch3 induces expression of cleaved PARP and cytochrome c in both lung cancer cell lines HCC2429 and H460. The Notch3 overexpressing cell line HCC2429 was transfected with Notch3 siRNA or control siRNA, and lysate was collected after 24 hours and 48 hours. (B) In addition, induction of caspase-3 activity was also observed in HCC2429 after transfection with Notch3 siRNA at 24 and 48 hours. The induction of caspase-3 by Notch3 siRNA was reversed by the addition of caspase-3 inhibitor. (C) The increased apoptotic fraction induced with Notch3 siRNA transfection in cell lines HCC2429 and H460 was dependent on serum-free conditions. While we observed induction of PARP, cytochrome c and increased caspase-3 activity, apoptosis measured using Annexin V/PI was observed only in the presence of low serum. Asterisks (*) denotes statistical significance with p < 0.05.
Bim is necessary for induction of apoptosis in Notch3 knockout cells
To determine whether Bim is necessary for Notch3-dependent apoptosis, we used Bim siRNA to suppress Bim in HCC2429 cells transfected with Notch3 siRNA. The loss of Bim abrogated the induction of PARP by Notch3 siRNA (Figure 3A). In further support of our hypothesis, we observed that Bim knockdown abolished Notch3 siRNA induced apoptosis in the absence of serum compared with vector control suggesting that the effect of Notch3 on apoptosis is dependent on Bim (Figure 3B). A small increase in apoptosis was detected in Bim negative cells in both serum-free and 10% FCS conditions. Interestingly, Bim downregulation also resulted in downregulation of Notch3 suggesting a feedback loop existed between the two pathways.
Figure 3.
Notch3-mediated apoptosis is dependent on Bim in vitro. (A) Twenty-four hours after Notch3 siRNA transfection, lung cancer cell line HCC2429 was transfected with Bim siRNA. Cells were harvested after an additional 24 hours. Reduction of Bim expression was observed after 24 hours as compared control siRNA. The loss of Bim prevented induction of PARP by Notch3 siRNA. (B) In serum free media, the loss of Bim abrogates apoptosis, suggesting that Bim is necessary for Notch3-dependent apoptosis. No significant change is observed in 10% FCS condition. In all siRNA experiments, non-silencing siRNA was used as control. The difference between SiRNA-C and Bim SiRNA in N3SiRNA treated cells were statistical significance with p < 0.05.
Effect of Notch3 on Bim depends on intact MAPK signaling
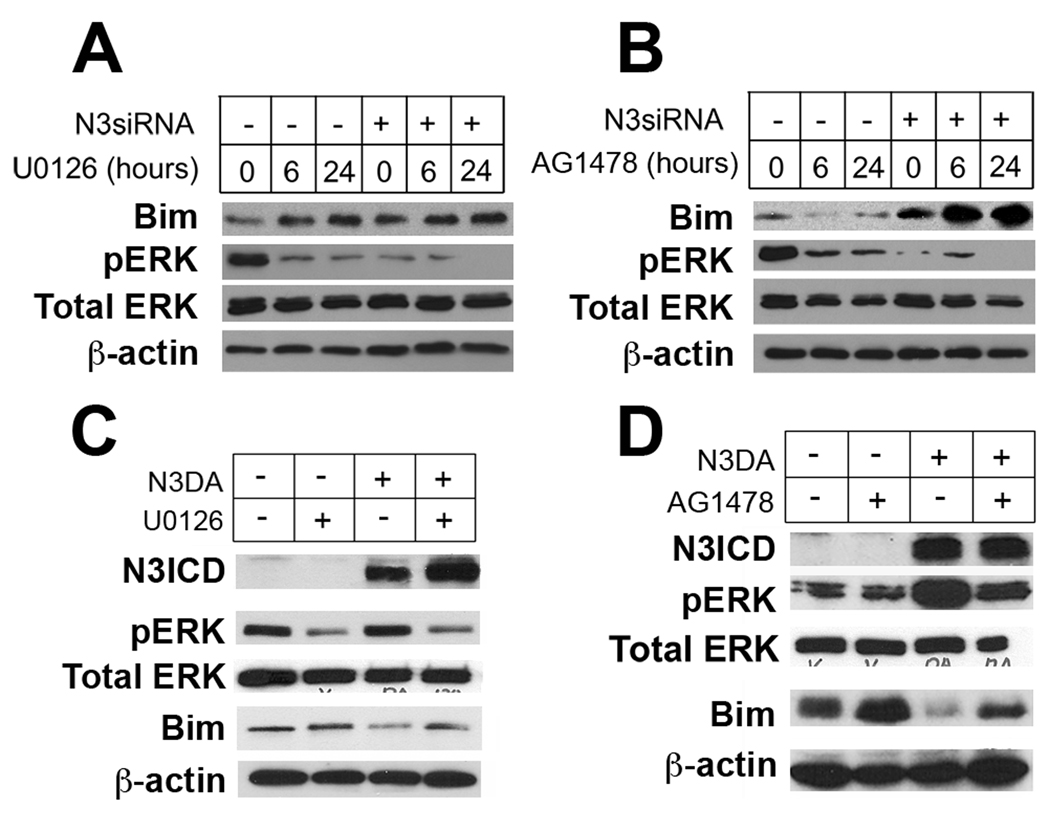
To better define the interaction between Notch3 and EGFR/ras/MAPK pathways in Bim modulation, we examined the effect of the MEK inhibitor U0126 and the EGFR inhibitor AG1478 on lung cancer cells treated with Notch3 siRNA. Interestingly, Bim expression is induced with the MEK inhibitor U0126, while the added loss of Notch3 signaling did not augment Bim expression (Figure 4A). One explanation for this observation is that the effect Notch3 has on Bim is dependent on intact MAPK signaling and that inhibiting Notch3 by siRNA is not of added benefit. On the other hand, when HCC2429 cells were treated with both Notch3 siRNA and AG1478 at 5 µM, Bim expression was induced significantly compared to Notch3 siRNA, U0126 or AG1478 alone (Figure 4B). The Akt-PI3K pathway regulates Bim through phosphorylation of the transcription factor Foxo3a (Urbich et al., 2005). Thus, inhibiting both Notch3 and EGFR synergistically affects Bim through both Foxo3a and MAPK signaling.
Figure 4.
Notch3 and EGFR/ras/MAPK pathways cooperate in regulating Bim expression. (A) Bim is upregulated when HCC2429 was transfected with Notch3 siRNA as compared to control at 0 times, whereas inhibition of Notch3 signaling does not increase Bim expression over treatment with U0126 alone at both 6 and 24 hours, suggesting that Notch3 regulates Bim expression mainly through pERK. (B) AG1478 alone slightly diminishes Bim expression. However, treatment with both AG1478 and N3SiRNA resultes in the synergistic induction of Bim, compared to either treatment alone. Inhibition of pERK and EGFR with U0126 (C) and AG1478 (D), respectively, preventing Notch3-dependent suppression of Bim, supporting the hypothesis that modulation of Bim and apoptosis in lung cancer by Notch3 is dependent on intact EGFR/ras/MAPK signaling.
To confirm that Notch3 regulates Bim is through MAPK, we transfected activated Notch3 (N3DA) into COS cells treated with either U0126 or AG1478. As expected, U0126 prevented Bim suppression by activated Notch3, further supporting our contention that the anti-apoptotic effect of Notch3 is MAPK-dependent (Figure 4C). On the other hand, inhibition of EGFR with AG1478 could only partially reverse the effect of activated Notch3 on Bim expression despite near suppression of pERK, suggesting that Bim is also modulated by a MAPK-independent pathway (Figure 4D).
Gamma-secretase and EGFR tyrosine kinase inhibition enhance anti-tumor activity in vivo and Bim expression
We previously observed that in vitro the combination of γ-secretase inhibitor MRK-003 and the EGFR tyrosine kinase inhibitor (AG1478) was more effective in inhibiting tumor growth than either agent alone (Konishi et al., 2007). To determine whether the combination of γ-secretase inhibitor MRK-003 and the EGFR tyrosine kinase inhibitor erlotinib also enhances tumor activity in vivo, we utilized a xenograft model. The combination of MRK-003 and erlotinib significantly reduced tumor size compared with either single agent alone (Fig. 5A). We also noted that the combination of MRK-003 and erlotinib resulted in greater expression of Bim in the mouse tumors, compared to either agent alone (Figure 5B). Given the role of Notch pathway in angiogenesis, we tumor measured microvessels density (MVD) (data not shown). No change in tumor MVD was observed, supporting the hypothesis that mechanism of tumor inhibition in vivo involves Bim modulation. Finally, whether this effect is due to Notch inhibition is uncertain due to the non-specificity of GSI. However, we previously have demonstrated that loss of Notch3 rendered the GSI ineffective, suggesting that at least our lung cancer model, the antitumor effect may be Notch3 dependent (Konishi et al., 2007).
Figure 5.
Combination of Notch inhibition with MRK-003 and EGFR inhibition with erlotinib has greater anti-tumor effect in vivo and enhances Bim expression. (A) H460 cells were inoculated subcutaneously into nude mice and treatment was initiated when tumors were palpable. Treatment with the combination of erlotinib and MRK-003 resulted in the lowest rate of tumor growth, compared to treatment with either alone. The asterisk (*) denotes statistical significance, p < 0.05 when comparing treatment with the combination vs. MRK-003 alone. The difference between erlotinib alone and the combination is statistically significant across all time points except for Day 1. (B) Bim expression was markedly higher in tumor treated with MRK-003 and erlotinib than either MRK-003 or erlotinib alone.
DISCUSSION
Like Notch, the Bcl-2 protein family members play central roles in both development and cancer, facilitating strict organ morphogenesis during embryonic development and maintenance of tissue homeostasis. These proteins are regulators of programmed cell death through the integration of diverse extra- and intracellular death signals. In this study, we demonstrated that loss of Notch3 resulted in downregulation of the pro-survival proteins, Bcl-2 and Bcl-xL and upregulation of the pro-apoptotic protein Bax, as well as the BH3-only proteins Bim and Bad. In tumorigenesis, activated Notch3 has been shown to induce T-cell leukemia through the constitutive activation of NF-κB (Bellavia et al., 2000). Activated Notch1 has been shown to inhibit p53-mediated apoptosis (Bocchetta et al., 2003; Beverly et al., 2005). In this study, we examined the effect of Notch3 on both the NF-κB pathway and p53 using Notch3 siRNA. While our findings indicate that Bim is a target of Notch3 signaling, we were unable to discern appreciable changes in the levels of p53 or NF-κB-related proteins (data not shown), suggesting that the role of Notch3 in apoptosis is distinct, and context dependent.
Bim is a BH3-only member of the Bcl-2-like family of proteins. Loss or withdrawal of cytokines and growth factors specifically induce its expression. Once Bim is activated, it binds and inactivates Bcl-2-like pro-survival proteins, leading to cytochrome c release from mitochondria and caspase activation. Furthermore, Bim is required to mediate EGFR inhibitor-induced apoptosis in lung cancer cells, also supporting a significant interaction between Bim and the EGFR/ras/MAPK pathway (Costa et al., 2007; Gong et al., 2007; Wang et al., 2007). In this paper, we demonstrate not only that the loss of Notch3 results in upregulation of Bim, but moreover that Bim expression is further enhanced when both the EGFR and Notch3 pathways were inhibited. This finding provides additional evidence for the crosstalk between the EGFR and Notch3 in modulating apoptosis.
While Notch signaling regulates apoptosis through the NF-κB and p53 pathways in some cells, Bim appears to be necessary for the induction of Notch3-dependent apoptosis in lung cancer. This finding is supported by the recent observation that Notch inhibition with a γ-secretase inhibitor upregulates Bim in malignant melanomas (Qin et al., 2004). While it is possible that this effect is unrelated to Notch inhibition due to the potential lack of specificity of the γ-secretase inhibitors, our siRNA data suggest that Bim upregulation results from specific Notch3 knockdown, and that Notch-induced apoptosis is indeed dependent on this upregulation of Bim.
Withdrawal of growth factors results in induction of Bim through either of the two major EGFR-dependent pathways: the Raf/MAPK and Akt/PI3K (Shinjyo et al., 2001). The Akt/PI3K pathway is known to negatively affect Bim through downregulation of Foxo3a. In our study, no change in pAkt or pFoxoa3 was noted in the lung cancer cell lines transfected with Notch3 siRNA, indicating that Notch3 regulates Bim expression through the MAPK pathway and not the Akt pathway. Activated Notch3 was unable to rescue U0126 induced apoptosis, providing further evidence that Notch3 modulation may be entirely dependent on MAPK pathway. This contention is consistent with our previous findings that Notch3 regulates the MAPK activation through modulating MAPK phosphatase.
In summary, we have characterized one mechanism by which Notch3 plays an important role in the biology of cancer through the modulation of apoptosis. While the crucial role of Notch signaling in apoptosis has been well-established in the context of cancer as well as development, the mechanism by which Notch3 affects apoptosis in lung cancer remained obscure to date. In this study, we provided evidence that Bim is necessary for Notch3-dependent apoptosis and that the effect of Notch3 on Bim is through MAPK regulation.
Finally, we previously observed a cooperative relationship between the Notch3 pathway and the EGFR pathway in maintaining the tumor phenotype. Our data have shown that this cooperative relationship also involves the modulation of Bim. Interestingly, Bim expression is upregulated in lung cancer cell lines sensitive to inhibitors of EGFR signaling, but not in resistant cell lines (Gong et al., 2007). Taken together, these observations suggest that the concomitant inhibition of Notch and EGFR pathways represents a rational strategy for promoting apoptosis in lung cancer and potentially overcoming treatment resistance.
MATERIALS AND METHODS
Cell lines and inhibitors
The Notch3 expressing lung cancer cell line, HCC2429, was established as previously described (Dang et al., 2000). The NSCLC cell line H460 and COS cell line were obtained from American Type Culture Collection (ATCC) and maintained in RPMI with 10% FCS. Activated Notch3 was created previously and cloned into pBabe retroviral vector. AG1478 and U0126 were obtained from Calbiochem (La Jolla, CA), and erlotinib was obtained from Genetech, Inc (San Francisco, CA). The formulation and the in vivo dosing of γ-sectretase inhibitor, MRK-003, was provided by Merck & Co., Inc and was described previously (Lewis et al., 2007).
Notch3 and Bim Small interfering RNAs
Notch3-overexpressing cell lines HC2429 and H460 were seeded at 1.5 × 105 cells per 6-well plate the day before transfection. The Notch3 siRNA sequence is 5’-CACCUAUAACUGCCAGUGC-3’ and was synthesized by Qiagen. Cells were transfected with 100 nmol/L siRNA in Opti-MEM medium (Invitrogen, Calsbad, CA) using LipofectAMINE 2000 (Invitrogen, Calsbad, CA) according to the manufacturer’s recommendation. The efficiency of siRNA transfection was measured with Western blot analysis. Bim siRNA (catalog number M-004383-01-0010) was purchased from Dharmacon Research (Lafayette, CO). An unspecific (non-silencing) siRNA against the target sequence 5′ AAT TCT CCG AAC GTG TCA CGT TT 3′ (cat. no. 80-11310, Xeragon) was used as controls.
Caspase-3 Cellular Activity
HCC2429 cells was transfected with Notch3 siRNA or control as described above. After 24 or 48 hours, the cells were harvested in lysis buffer, and the cell extracts were used to determine caspase-3 activity, using the Caspase-3 Cellular Activity Assay Kit (Calbiochem, La Jolla, CA). Caspase-3 activity was measured in cell lysates in the presence and absence of Caspase-3 inhibitor at 0.1 µM final concentration.
Apoptosis Analysis
Cells transfected with Notch3 siRNA for 48 hours were maintained in 10% FCS-RPMI or serum-free medium. These cells were stained with FITC-conjugated annexin V and propidium iodide using Annexin V-FITC Apoptosis Detection kit (Calbiochem, La Jolla, CA). The percentage of apoptotic cells was determined with a Beckman Coulter FACS Calibur Flow Cytometer. In dual Notch3 and Bim siRNA transfection experiments, the cells were transfected with Bim siRNA twenty-four hours after Notch3 siRNA transfection. Twenty-four hours after Bim siRNA transfection, the cells were harvested and analyzed for apoptosis, as described above. Caspase-3 activity was measured using the Caspase-3 Cellular Activity Assay Kit (Calbiochem, La Jolla, CA) according to manufacture’s recommendation.
RNA Extraction and Real-Time PCR
Total RNA was extracted with Trizol (Invitrogen, Carlsbad, CA) from HCC2429 cells 24 hrs after transfection of either Notch3 siRNA or control siRNA. RNA was reverse transcribed with the iScript cDNA synthesis kit (Bio-RAD, Hercules, CA). Real-time PCR was performed with the iQ5 multicolor Real-Time PCR detection system (Bio-RAD). Fifty µl mixture was used for reaction, which contains 5 µl cDNA sample (0.5–1 µg/µl), 300 nM forward primers for Bim (GCAGATATGCGCCCAGAGAT) or β-actin (ATGGCTCCGGTATGTGCAA), 300 nM reverse primers for Bim (AAGCGTTAAACTCGTCTCCGATA) or β-actin (TGTCTTTCTGGCCCATACCAA), and 25 µl SYBR Green Supermix (Bio-RAD). After incubation at 50°C for 2 min followed by 95°C for 10 min, the reaction was carried out for 40 cycles of the following: 95°C for 15 sec and 60°C for 1 min. The threshold cycle value (Ct) was obtained using the iCycler Optical system interface software. Mean Ct of Bim was calculated from triplicate measurements and normalized with the mean Ct of the control gene for β-actin.
Antibodies and Western Blot Analysis
Notch3 was detected with a rabbit polyclonal antibody at 1:500 dilution (Orbigen, Inc., San Diego, CA). The rabbit antibodies to PARP, Bcl-xL, phosho-Bcl-2, Puma, BAX, phospho-extracellular signal-regulated kinase (pERK), ERK, phospho-Akt, Akt, phosphoinositide 3-kinase (PI3K), and PKA C-α, were purchased from Cell signaling Technology (Danvers, MA). The rabbit antibodies to Mcl-1 and Bim were purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Calbiochem (La Jolla, CA), respectively. The rabbit antibodies to phospho-Foxo3a and Foxo3a were from Millipore (Billerica, MA). Monoclonal cytochrome C antibody was obtained from BD Biosciences (San Jose, CA). Proteins were stained with Ponceau S to confirm equal loading of in each analysis. For Western blot analysis of mitogen-activated protein kinase (MAPK), Akt, PI3K and PKA-Cα activation, the cells were transfected with Notch3 siRNA before serum deprivation and maintained in serum-free medium for 24 hours before the addition of serum. The cells were harvested after designated time intervals.
In vivo tumorigenicity
Athymic 4- to 6-week-old female nude mice (nu+/nu+) were used for the tumor xenograft models. H460 (1 × 106 cells in the volume of 200 µl of PBS) was inoculated subcutaneously (s.c.) into the right posterior legs of the mice. Treatment was initiated when tumors were palpable. Erlotinib (100 mg/kg) was administered orally every other day to the mice alone or in combination with MRK-003 (150 mg/kg) given daily for 3 days followed by 4 days off. Erlotinib was diluted in 1% methylcellulose/0.1%Tween 80. MRK-003 was diluted in 0.5% methylcellulose. Tumors were measured every 2 days with a caliper. Tumor Volume (TV) was calculated with the formula: TV = (Length) × (Width)2 / 2. Percentage tumor volume (% TV) on day X was calculated as: %TV = (tumor volume on day X / tumor volume on day 1) × 100.
Statistical analyses
The size of implanted tumors at different time points after treatment was compared with that of control groups. Unless specifically stated, statistical inference in comparative experiments both in vivo and in vitro was obtained using unpaired, two-sided Student’s t test. For all determinations, the differences were considered significant when P value is < 0.05.
Acknowledgments
Grant Support: This study was supported by NCI 1R01 CA115707
REFERENCES
- 1.Roy M, Pear WS, Aster JC. Curr Opin Genet Dev. 2007;17:52–59. doi: 10.1016/j.gde.2006.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Curry CL, Reed LL, Golde TE, Miele L, Nickoloff BJ, Foreman KE. Oncogene. 2005;24:6333–6344. doi: 10.1038/sj.onc.1208783. [DOI] [PubMed] [Google Scholar]
- 3.Das I, Craig C, Funahashi Y, Jung KM, Kim TW, Byers R, Weng AP, Kutok JL, Aster JC, Kitajewski J. J Biol Chem. 2004;279:30771–30780. doi: 10.1074/jbc.M309252200. [DOI] [PubMed] [Google Scholar]
- 4.Duechler M, Shehata M, Schwarzmeier JD, Hoelbl A, Hilgarth M, Hubmann R. Leukemia. 2005;19:260–267. doi: 10.1038/sj.leu.2403592. [DOI] [PubMed] [Google Scholar]
- 5.Reedijk M, Odorcic S, Chang L, Zhang H, Miller N, McCready DR, Lockwood G, Egan SE. Cancer Res. 2005;65:8530–8537. doi: 10.1158/0008-5472.CAN-05-1069. [DOI] [PubMed] [Google Scholar]
- 6.Haruki N, Kawaguchi KS, Eichenberger S, Massion PP, Olson S, Gonzalez A, Carbone DP, Dang TP. Cancer Res. 2005;65:3555–3561. doi: 10.1158/0008-5472.CAN-04-3132. [DOI] [PubMed] [Google Scholar]
- 7.Konishi J, Kawaguchi K, Vo H, Haruki N, Gonzalez A, Carbone DP, Dang TP. Cancer Res. 2007 doi: 10.1158/0008-5472.CAN-07-1022. [DOI] [PubMed] [Google Scholar]
- 8.Dievart A, Beaulieu N, Jolicoeur P. Oncogene. 1999;18:5973–5981. doi: 10.1038/sj.onc.1202991. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald K, Harrington A, Leder P. Oncogene. 2000;19:4191–4198. doi: 10.1038/sj.onc.1203766. [DOI] [PubMed] [Google Scholar]
- 10.Weijzen S, Rizzo P, Braid M, Vaishnav R, Jonkheer SM, Zlobin A, Osborne BA, Gottipati S, Aster JC, Hahn WC, Rudolf M, Siziopikou K, Kast WM, Miele L. Nat Med. 2002;8:979–986. doi: 10.1038/nm754. [DOI] [PubMed] [Google Scholar]
- 11.Heiser PW, Hebrok M. Cell Cycle. 2004;3:270–272. [PubMed] [Google Scholar]
- 12.Borner C. Mol Immunol. 2003;39:615–647. doi: 10.1016/s0161-5890(02)00252-3. [DOI] [PubMed] [Google Scholar]
- 13.Gomez-Bougie P, Oliver L, Le Gouill S, Bataille R, Amiot M. Oncogene. 2005;24:8076–8079. doi: 10.1038/sj.onc.1208949. [DOI] [PubMed] [Google Scholar]
- 14.Gong Y, Somwar R, Politi K, Balak M, Chmielecki J, Jiang X, Pao W. PLoS Med. 2007;4:e294. doi: 10.1371/journal.pmed.0040294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Campos AH, Wang W, Pollman MJ, Gibbons GH. Circ Res. 2002;91:999–1006. doi: 10.1161/01.res.0000044944.99984.25. [DOI] [PubMed] [Google Scholar]
- 16.Wang T, Holt CM, Xu C, Ridley C, P OJR, Baron M, Trump D. Cell Signal. 2007;19:2458–2467. doi: 10.1016/j.cellsig.2007.07.019. [DOI] [PubMed] [Google Scholar]
- 17.Biswas SC, Shi Y, Sproul A, Greene LA. J Biol Chem. 2007;282:29368–29374. doi: 10.1074/jbc.M702634200. [DOI] [PubMed] [Google Scholar]
- 18.Urbich C, Knau A, Fichtlscherer S, Walter DH, Bruhl T, Potente M, Hofmann WK, de Vos S, Zeiher AM, Dimmeler S. FASEB J. 2005;19:974–976. doi: 10.1096/fj.04-2727fje. [DOI] [PubMed] [Google Scholar]
- 19.Bellavia D, Campese AF, Alesse E, Vacca A, Felli MP, Balestri A, Stoppacciaro A, Tiveron C, Tatangelo L, Giovarelli M, Gaetano C, Ruco L, Hoffman ES, Hayday AC, Lendahl U, Frati L, Gulino A, Screpanti I. Embo J. 2000;19:3337–3348. doi: 10.1093/emboj/19.13.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bocchetta M, Miele L, Pass HI, Carbone M. Oncogene. 2003;22:81–89. doi: 10.1038/sj.onc.1206097. [DOI] [PubMed] [Google Scholar]
- 21.Beverly LJ, Felsher DW, Capobianco AJ. Cancer Res. 2005;65:7159–7168. doi: 10.1158/0008-5472.CAN-05-1664. [DOI] [PubMed] [Google Scholar]
- 22.Costa DB, Halmos B, Kumar A, Schumer ST, Huberman MS, Boggon TJ, Tenen DG, Kobayashi S. PLoS Med. 2007;4:1669–1679. doi: 10.1371/journal.pmed.0040315. discussion 1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang YF, Jiang CC, Kiejda KA, Gillespie S, Zhang XD, Hersey P. Clin Cancer Res. 2007;13:4934–4942. doi: 10.1158/1078-0432.CCR-07-0665. [DOI] [PubMed] [Google Scholar]
- 24.Qin JZ, Stennett L, Bacon P, Bodner B, Hendrix MJ, Seftor RE, Seftor EA, Margaryan NV, Pollock PM, Curtis A, Trent JM, Bennett F, Miele L, Nickoloff BJ. Mol Cancer Ther. 2004;3:895–902. [PubMed] [Google Scholar]
- 25.Shinjyo T, Kuribara R, Inukai T, Hosoi H, Kinoshita T, Miyajima A, Houghton PJ, Look AT, Ozawa K, Inaba T. Mol Cell Biol. 2001;21:854–864. doi: 10.1128/MCB.21.3.854-864.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dang TP, Gazdar AF, Virmani AK, Sepetavec T, Hande KR, Minna JD, Roberts JR, Carbone DP. J Natl Cancer Inst. 2000;92:1355–1357. doi: 10.1093/jnci/92.16.1355. [DOI] [PubMed] [Google Scholar]
- 27.Lewis HD, Leveridge M, Strack PR, Haldon CD, O'Neil J, Kim H, Madin A, Hannam JC, Look AT, Kohl N, Draetta G, Harrison T, Kerby JA, Shearman MS, Beher D. Chem Biol. 2007;14:209–219. doi: 10.1016/j.chembiol.2006.12.010. [DOI] [PubMed] [Google Scholar]