Abstract
Werner Syndrome is an autosomal inherited disease that is characterized by premature aging. The gene mutated in Werner Syndrome (WS), WRN, encodes both a 3'→5' DNA helicase and a 3'→5' DNA exonuclease. Amongst the WS phenotypes is an exceptionally high incidence of sarcomas. We asked whether spontaneous sarcomas, not known to be associated with WS, also harbor mutations or unreported single nucleotide polymorphisms (SNPs) in WRN. We analyzed RNA or DNA sequences within the helicase and exonuclease domains from 51 and 69 matched sarcoma and adjacent normal tissues, respectively. Among a total of 13 nucleotide polymorphisms detected, we identified three novel non-synonymous polymorphisms: c.611C>T, c.809_810insT, and c.1882C>G. We further characterized one, c.611C>T, which results in substitution of an evolutionarily conserved proline at amino acid 204 in the exonuclease domain with leucine. We show that P204L WRN exhibits a reduction of WRN exonuclease activity; the specific activity is ~10-fold lower than that of wild-type WRN. In contrast, the helicase activity of P204L WRN is reduced less than 2-fold.
INTRODUCTION
Werner syndrome (WS) is an uncommon, autosomal recessive disorder. To date, approximately 1300 cases have been reported ([1], OMIM #277700). WS patients exhibit many phenotypic changes associated with aging; the symptoms are typically manifested during the post-adolescent years. These changes include the premature development of osteoporosis, cataracts, arthritis, alopecia, diabetes, and heart disease [2]. Consistent with this, primary cells from WS patients exhibit reduced lifespan in vitro. In addition to aging phenotypes, WS patients also have an increased propensity to develop specific neoplasms, including sarcomas, thyroid carcinomas, and meningiomas. Consistent with a cancer-prone phenotype, WS cells are genetically unstable and exhibit enhanced sensitivity to multiple DNA-damaging agents [3–5].
The gene mutated in WS, WRN, encodes a helicase that belongs to the evolutionarily conserved RecQ-family of DNA helicases [6]. It is located on chromosome 8p12 and encodes a 1432-amino-acid protein, WRN [7]. In humans, the RecQ-family of DNA helicases also includes RecQ1, RecQ5, the Bloom protein (BLM), and Rothmund-Thomson protein (RTS). Mutations in BLM and RTS are responsible for Bloom syndrome (BS), and Rothmund-Thomson syndrome (RTS), respectively. Similar to WS mutations, BS and RTS mutations are associated with genetic instability and individuals with these syndromes have an increased risk of developing malignancies, although the spectrum of cancers appears to be different in these diseases [8].
All characterized RecQ helicases exhibit an ATP-dependent 3'→5' DNA unwinding activity. WRN is unique amongst human RecQ helicases in that it also encodes a 3'→5' DNA exonuclease at the N-terminus [9]. The sequence of the WRN exonuclease is partially homologous to the 3'→5' proofreading exonuclease domains of E.coli DNA polymerase I and RNaseD [10]. Biochemical and molecular studies suggest that WRN is involved in multiple DNA metabolic pathways, including replication, recombination, and repair [11]. An emerging body of evidence increasingly supports the hypothesis that WRN is involved in resolving replication forks stalled at DNA lesions or at alternate DNA structures [12]. The function of WRN in multiple DNA repair pathways, including: base-excision repair, single- and double-strand break repair, mismatch repair, and telomere maintenance (reviewed in [13]) implies that WRN is required for genomic maintanence. Loss of WRN activity could be associated with increased mutagenesis and tumor progression. However, the precise roles of the WRN helicase and exonuclease activities in these processes have yet to be established.
Currently, more than 50 different disease-associated WS mutations have been reported. All except one of these mutations are single-base substitutions, splice variants, insertions, or deletions causing frameshifts that lead to premature termination of the WRN protein [14–17]. Loss of functional WRN is believed to occur through nonsense-mediated mRNA decay (NMD), accelerated degradation of the truncated protein, or failed nuclear translocation of the WRN protein [18]. One WS patient has been reported to be homozygous for two missense mutations in WRN that reduce protein stability in vitro and result in loss of detectable WRN in vivo [17]. Thus, regardless of the type of mutation, all disease-associated genetic changes result in the elimination of functional WRN within the nucleus.
Many non-synonymous single nucleotide polymorphisms (SNPs) have been reported in WRN and some of these have been associated with breast cancer susceptibility, bone density loss, diabetes, and cardiovascular health [19–22]. We have previously analyzed the effects of several SNPs on WRN enzymatic activities. Although many exhibit only subtle reductions of WRN helicase and exonuclease activities, we identified an infrequent SNP, R834C, that decreases helicase and helicase-coupled exonuclease activity by > 95% [23]. This polymorphism maps to the central helicase core of WRN and interferes with ATP binding and hydrolysis, quintessential for DNA unwinding. Importantly, unlike WS-associated mutations that result in loss of WRN protein, R834C has little, or no effect on protein expression [23]. The clinical manifestations of R834C are currently being evaluated.
Diseases caused by mutations in the RecQ-family DNA helicases, such as WS, BS, and RTS, all share a marked propensity for developing specific neoplasms. The association between mutations in WRN and increased incidence of mesenchymal cancers distinguishes WS from both BS and RTS. The ratio of non-epithelial to epithelial malignancies is approximately 10-fold greater in WS patients than in the general population. The propensity for sarcomas is especially marked; soft tissue sarcomas, which comprise less than 1% of all neoplasms worldwide, constitute over 15% of WS-related malignancies [24]. Therefore, we hypothesize that nucleotide sequence alterations in WRN can be a major factor in the development of human sarcomas, and the prevalence of specific SNPs in WRN may be greater in individuals with spontaneous sarcomas. We analyzed WRN genomic DNA and mRNA extracted from soft tissue sarcomas of non-WS individuals in search of an elevated frequency of SNPs and somatic mutations in WRN. Due to the unique presence of an exonuclease domain and the evolutionary conservation of the helicase domain in WRN, we targeted our search for mutations and SNPs specifically to these regions.
MATERIALS AND METHODS
Tissue Samples
Soft tissue sarcomas (various types and grades, each contained >80% malignant cells upon histological examination) and adjacent normal tissues were obtained at the time of surgery, immediately frozen, and stored at −80°C. Tumor samples were predominantly composed of malignant cells, and no tumor cells were observed in adjacent normal tissue upon histological examination. No additional patient information was available.
RNA Extraction, Amplification, and Sequence Analysis
RNA was extracted from 56 matched tumor and normal tissues using RNA-STAT-60 according to manufacturer's recommendations (Tel-Test, Friendswood, TX). The WRN exonuclease and helicase domains (exons 2–21) were reverse transcribed and the cDNA was amplified as a single amplicon using the Access Reverse Transcription PCR System (Promega, Madison, WI). All primers used are listed in Supplemental Table 1. The cDNA was used as a template for the amplification of the exonuclease domain (exons 2–10) and the helicase domain (exons 12–21) separately. The Expand High Fidelity PCR System and protocol were used for all routine amplification reactions (Roche, Indianapolis, IN). Products were purified with a Montage PCR96 Clean-up Plate Kit (Millipore, Billerica, MA) and sequenced (DNA Sequencing Facility, University of Washington). Sequence data was analyzed using Sequencher 4.1.4 (Gene Codes Corporation, Ann Arbor, MI). Tumor and normal sequence data were compared with WRN cDNA reference sequences obtained from http://www.ensembl.org (gene ID ENSG00000165392) at 25% secondary peak sensitivity threshold. Each chromatogram was inspected individually. All nucleotide substitutions were confirmed by reamplification and resequencing of the RNA and corresponding genomic DNA.
DNA Extraction, Amplification, and Sequence Analysis
Genomic DNA was prepared from matched tumor and normal tissues using DNA-STAT-60 according to manufacturer's protocol (Tel-Test, Friendswood, TX). The exonuclease domain (exons 4–5, 6–7) was amplified from genomic DNA extracted from 36 sarcomas (18 were the same as above) by nested PCR. Exons 4 and 5 were amplified together, and exons 6 and 7 were amplified together using nested primers. Purification, sequencing, sequence analysis, and confirmation were performed as described above.
Detection of c.611C>T Substitutions in Genomic DNA
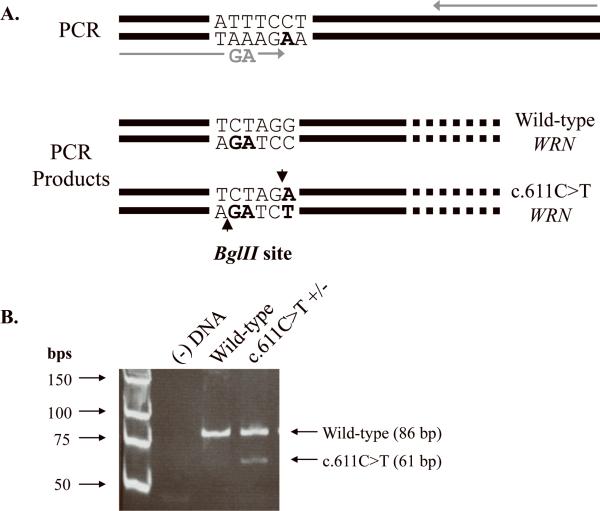
A high-sensitivity screen for the c.611C>T substitution was performed on genomic DNA from 199 additional sarcomas using a PCR-based assay. Exons 6–7 were amplified as described above. Nested primers were designed such that a BglII restriction enzyme site would be created if the c.611C>T substitution were present (Figure 3). BglII cleavage of wild-type DNA results in two fragments, 1249 bp and 86 bp in length, whereas that of a product, heterozygous for c.611C>T, results in four fragments, 1249 bp, 86 bp, 61 bp, and 25 bp in length. Genomic DNA from the tumor sample from which c.611C>T was initially identified was used as a positive control. 10 μ-L final amplified product was digested with 5 U BglII (New England Biolabs, Ipswich, MA) at 37°C for 90 min. Reaction aliquots were electrophoresed through a 12% polyacrylamide gel to resolve the smaller sized fragments.
Fig. 3. Screen for the c.611C>T mutation in 199 additional sarcomas.
(A) Schematic of the screen for the c.611C>T mutation in WRN. PCR amplification with mutagenic primers (grey arrows) introduces two nucleotide changes that create a BglII restriction site only if the c.611C>T mutation is present. Amplification of a wild-type sequence with these same primers does not result in an additional cleavable BglII site. (B) PCR amplification results in a 1335 bp product. BglII cleavage of a wild-type product results in two fragments, 1249 bp (not shown) and 86 bp in length (lane 3: wild-type WRN). BglII digestion of amplified DNA, heterozygous for c.611C>T WRN, generates four fragments, 1249 bp (not shown), 86 bp, 61 bp, and 25 bp (not shown), in length (lane 4: c.611C>T WRN). The digestion products were visualized following electrophoresis through a 12% polyacrylamide gel. In order to better resolve the 86 bp and 61 bp products, the smallest 25 bp fragment was electrophoresed off the gel. Lane 1: low molecular weight ladder and lane 2: (−) DNA control.
Growth of 293T Cells
293T kidney epithelial tumor cells were grown in Dulbecco's modified minimal essential medium (Cellgro, Manassas, VA) supplemented with 10% fetal bovine serum (HyClone, Logan, UT), 2 mM L-glutamine, and 100 U/mL penicillin G sulfate and 100 μg/mL streptomycin sulfate (Invitrogen, Carlsbad, CA) in a humidified, 5% CO2 incubator at 37°C. Cultures were propagated by dilution into fresh medium, and harvested at densities of 2×106 cells/mL.
WRN DNA Constructs
The pCS2+MT construct was used as the vector-only control. pMM229 encodes mycepitope-tagged WRN cDNA whose expression is driven by the CMV promoter. The pMM229 construct was used for mutagenesis, cloning, and expression in human cells [23]. The c.611C>T substitution and a silent PstI restriction site were introduced into wild-type WRN cDNA using Pfu Ultra Taq DNA polymerase and the QuikChange XL site-directed mutagenesis kit (Stratagene, La Jolla, CA). Mutagenized clones were identified by diagnostic PstI (New England Biolabs, Ipswich, MA) digests, verified by sequencing, and subcloned into a nonmutagenized pMM229 backbone to create the pMM229-c.611C>T construct.
Transfection of WRN DNA
pCS2+MT, pMM229, and pMM229-c.611C>T DNA constructs were purified using a QIAfilter Plasmid Maxi Kit (Qiagen, Valencia, CA). 293T cells, seeded at a density of 1×106 cells/60 mm dish, were transiently transfected with 4 μg of plasmid DNA by calcium phosphate precipitation in HEPES 24 hr after seeding. The medium was exchanged 12 hr following transfection, and cells were harvested 38 hr after media change. Harvested cells were washed with phosphate buffered saline, immediately flash frozen in liquid nitrogen, and stored at −80°C [23]. Transfection efficiency was monitored using standard GFP and lacZ transfection efficiency assays.
Cell Lysis and Immunoprecipitation
Total protein lysates were obtained by high-salt lysis of 293T cells expressing the mycepitope tag alone, myc-wild-type WRN, or myc-P204L WRN. WRN was immunoprecipitated from 500 μg total protein using a myc epitope-specific monoclonal antibody, as previously described [25]. WRN immune precipitates (IPs) were immediately assayed for helicase and/or exonuclease activity.
Measurement of WRN Protein Levels
Wild-type and P204L WRN levels were measured by semi-quantitative Western blot analysis, using a mouse anti-WRN monoclonal antibody (BD Transduction Laboratories, San Jose, CA), and a sheep anti-mouse horseradish peroxidase-conjugated antibody (GE Healthcare Life Sciences, Piscataway, NJ). Proteins were detected with an ECL system (GE Healthcare Life Sciences, Piscataway, NJ) and visualized on a STORM Phosphorimager (GE Healthcare Life Sciences, Piscataway, NJ). The relative levels of WRN protein were quantified with ImageQuant 5.2 software (GE Healthcare Life Sciences, Piscataway, NJ).
Assays for WRN Enzymatic Activity
Three distinct assays were performed to measure WRN helicase and exonuclease activities. All activity assays utilized a 5' 32P-labeled 20-nt oligonucleotide centrally annealed to a 46-nt oligonucleotide (*20/46), as previously described [23]. The 13-nt overhangs on either side of the *20-mer enable WRN helicase and WRN exonuclease activities to be assayed simultaneously. Helicase-only activity was measured by using a 3'-blocked *20-mer that is not degraded by WRN exonuclease (*20X), while the exonuclease-only activity was measured in the absence of unwinding by inclusion of the non-hydrolyzable ATP analog, ATPγS. Helicase activity was monitored by the separation of the *20-mer/*20X-mer from the 46-mer. Exonuclease activity was detected by the appearance of products shorter than the *20-mer.
Fresh immune precipitates were prepared for each assay. Immune precipitates containing wild-type WRN were assayed at dilutions of 1:30 and 1:10; both helicase and exonuclease activities were within the linear range of detection at these concentrations. Immune precipitates containing P204L WRN were assayed at 10-fold higher concentrations compared to wild-type WRN in order to compensate for differences in protein expression. Reaction products were resolved on a native 12% polyacrylamide gel (coupled helicase/exonuclease and helicase) or through a urea-14% polyacrylamide gel (exonuclease), visualized with a STORM Phosphorimager (GE Healthcare Life Sciences, Piscataway, NJ), and quantified with ImageQuant 5.2 software (GE Healthcare Life Sciences, Piscataway, NJ). Helicase activity was quantified as the ratio of unwound product (*20-mer) to the sum of intact substrate and unwound product (*20 + *20/46). Likewise, exonuclease activity was quantified by the ratio of degradation products to non-degraded substrate plus exonucleolytic products.
The substrate utilized in the coupled exonuclease/helicase assay affords the advantage of examining simultaneous unwinding and degradation of DNA. The sequence of the *20-mer is such that exonucleolytic loss of a single nucleotide migrates anomalously in native polyacrylamide gels. Thus, the lower band represents the unwound *20-mer, whereas the upper band represents exonucleolytic products of the unwound *20-mer. This was independently confirmed from the positions of migration of heat-denatured *19/46 and *20/46 DNA substrates, wherein the 19-mer and 20-mer were 5'-radiolabeled [23].
RESULTS
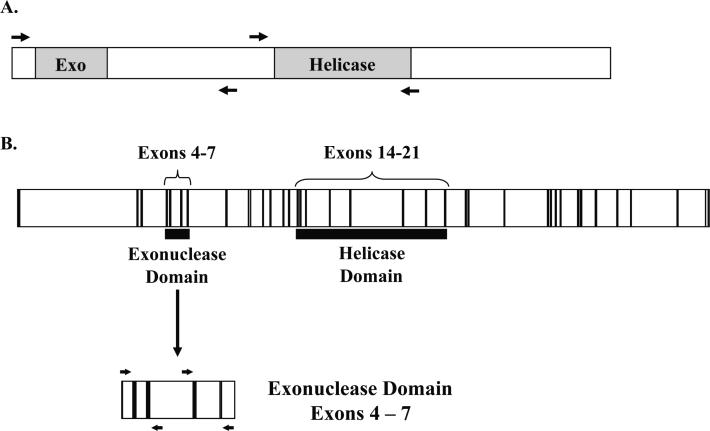
To determine the prevalence of WRN DNA sequence changes in sarcomas, we amplified and sequenced functionally important regions of genomic WRN and its transcript. Due to the evolutionary conservation of the helicase domain within the RecQ-family helicases and the uniqueness of the exonuclease domain to WRN, we focused our analyses on these two regions (Figure 1).
Fig. 1. Schematic of the WRN transcript and genomic WRN DNA.
(A) Regions of the WRN transcript containing the exonuclease and helicase domains (shown in grey) were amplified and sequenced, as indicated by the flanking arrows. (B) The region encoding the WRN exonuclease domain (exons 4–7) was amplified and sequenced from genomic DNA (exons are shown in black, introns shown in white). Due to large intronic regions, exons 4 and 5 were amplified together, and exons 6 and 7 were amplified together as indicated by the flanking arrows. Each exon was sequenced individually.
Substitutions in the Helicase Domain
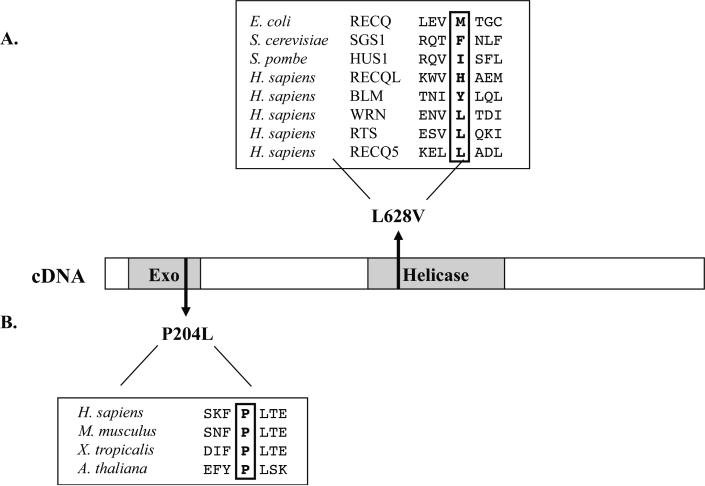
The helicase domain of WRN was amplified and sequenced using mRNA from 51 sarcomas and matched adjacent normal tissues. The results are summarized in Table 1. A heterozygous substitution in exon 16, c.1882C>G, was detected in one tumor sample, as well as in the corresponding adjacent normal tissue. This nucleotide change has not been previously reported; it results in a non-synonymous L628V amino acid substitution within the WRN helicase domain. Amino acid sequence alignments of E.coli RecQ, S. cerevisiae SGS1, S. pombe HUS2, and the five human RecQ-family helicases show that leucine is not conserved at this position (Figure 2a). We also identified c.2361G>T (L787) in exon 20, a known synonymous SNP, as either a homozygous or heterozygous substitution in 18 samples at an allelic frequency of 0.25. The frequency of this polymorphism is 0.4–0.5 in the general population, according to the GeneSNPs SNP database (http://www.genome.utah.edu/genesnps/).
Table 1.
WRN helicase region SNPs.
| Mutation | Location | Nucleotide Position | Amino Acid Substitution | Frequency | Reported Frequency |
|---|---|---|---|---|---|
| C to G | Exon 16 | 1882 | L628V | 0.01 | - |
| G to T | Exon 20 | 2361 | - | 0.25 | 0.4–0.5 |
Reported frequencies were obtained from the GeneSNPs SNP database (http://www.genome.utah.edu/genesnps/).
Fig. 2. Location of two novel non-synonymous polymorphisms identified in WRN.
(A) L628V results from a C to G transversion in exon 16 of the WRN helicase domain. Amino acid sequence comparisons of the helicase regions of other RecQ-family DNA helicases indicate that L628 is a poorly conserved residue. (B) P204L results from a C to T transition in exon 6 of the WRN exonuclease domain. Amino acid sequence comparisons of WRN exonuclease homologues show that the proline at this position is strictly conserved.
Substitutions in the Exonuclease Domain
The WRN exonuclease domain was amplified and sequenced using mRNA from 51 sarcoma and adjacent normal tissues (same samples as above), and using genomic DNA from 18 additional sarcoma and adjacent normal tissues. The results are summarized in Table 2. Eleven different sequence changes were observed within and adjacent to the exonuclease domain, all of which were present in both tumor and adjacent normal tissue. Eight of the eleven substitutions are previously reported polymorphisms present in the general population: four in noncoding regions, as well as two synonymous and two non-synonymous polymorphisms in the coding region. The two synonymous SNPs are c.503T>C (C171) in exon 6 and c.1155G>A (E385) in exon 9, which we found at allelic frequencies of 0.54 (reported 0.13–0.46) and 0.01 (reported 0.02–0.09), respectively. Non-synonymous SNPs c.340G>A (V114I) and c.1162G>A (M387I) were found at allelic frequencies of 0.06 (reported 0.02–0.17) and 0.03 (reported 0.02–0.07), respectively.
Table 2.
WRN exonuclease region SNPs.
| Mutation | Location | Nucleotide Position | Amino Acid Substitution | Frequency | Reported Frequency |
|---|---|---|---|---|---|
| G to A | Exon 4 | 340 | V114I | 0.06 | 0.02–0.17 |
| G to C | Intron 4/5 | +4/−477 | - | 0.03 | - |
| A to T | Intron 4/5 | +20/−461 | - | 0.06 | 0.02–0.06 |
| A to G | Intron 4/5 | +176/−305 | - | 0.06 | 0.05–0.06 |
| T to C | Exon 6 | 503 | - | 0.54 | 0.13–0.46 |
| C to T | Exon 6 | 611 | P204L | 0.01 | - |
| C to A | Intron 6/7 | +135/−941 | - | 0.03 | 0.01 |
| T to A | Intron 6/7 | +1053/−23 | - | 0.06 | 0.04–0.05 |
| Insert T* | Exon 8 | 809 | - | 0.01 | - |
| G to A* | Exon 9 | 1155 | - | 0.01 | 0.02–0.09 |
| G to A* | Exon 9 | 1162 | M387I | 0.03 | 0.02–0.07 |
Indicate mutations that occur just outside the WRN exonuclease domain.
Reported frequencies were obtained from the GeneSNPs SNP database (http://www.genome.utah.edu/genesnps/).
Interestingly, we identified three previously unreported nucleotide substitutions; only the two coding region substitutions are discussed here. One tumor sample was heterozygous for an insertion of a single thymidine at position 809 in exon 8, immediately downstream of the exonuclease domain. The resulting frameshift creates a premature stop codon three amino acids following the insertion. This insertion was also observed in the mRNA, suggesting that the mutant message was not subjected to nonsense-mediated decay. The predicted protein product of this transcript is a truncated WRN protein containing only the exonuclease domain. Although previous characterization of WRN indicates that expression of the exonuclease domain alone is sufficient for exonucleolytic activity in vitro, this truncated form of WRN may be functionally null due to the loss of its nuclear localization signal [26]. In addition to the insertion mutation, another tumor contained a heterozygous nucleotide substitution, c.611C>T, within exon 6 of the exonuclease domain. c.611C>T results in an amino acid substitution of proline with leucine at residue 204 (P204L). Amino acid sequence alignments of the WRN exonuclease homologues from Mus musculus, Xenopus tropicalis, and Arabidopsis thaliana, show strict conservation of proline at this residue (Figure 2b). We focused on the c.611C>T because it was an unreported polymorphism and because the proline to leucine substitution is likely affect protein function. Thus, c.611C>T could represent a novel non-synonymous polymorphism with structural and/or functional consequences for the WRN protein.
Prevalence of c.611C>T WRN in Sarcomas
To determine if c.611C>T WRN found in 1 of 69 sarcomas is commonly present in sarcomas, an additional 199 sarcomas were analyzed using high-throughput nested PCR and restriction enzyme digestion (see Materials and Methods). As shown in Figure 3, amplification of genomic DNA using mutagenic primers creates a BglII recognition site only when the c.611C>T substitution is present. Thus, in addition to a 1249 bp and a 86 bp product, two smaller fragments, 25 bp and 61 bp in size, are expected, and were observed, upon BglII digestion of PCR products containing the c.611C>T WRN substitution. The smaller 25 bp fragment is not shown here in order to better visualize the difference between the 86 bp (wild-type) and 61 bp (c.611C>T) fragments (Figure 3, lane 4). None of the additional 199 sarcoma-derived PCR products displayed these two fragments indicating the absence of this WRN SNP.
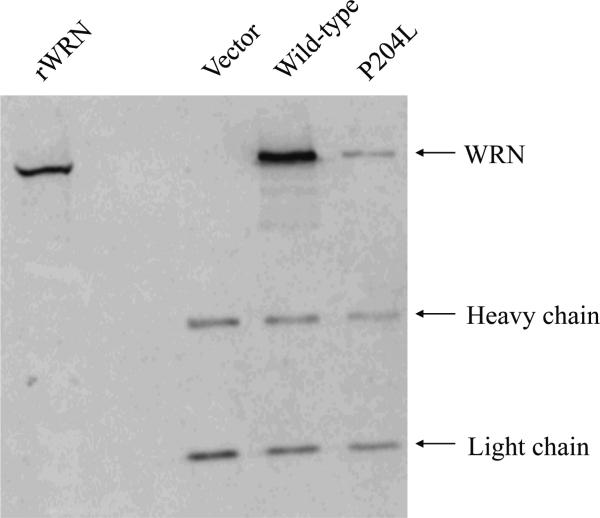
Levels of P204L WRN
Semi-quantitative Western blot analyses of IPs were performed to determine expression levels of P204L WRN from several independent transfections of 293T cells. The levels of P204L WRN were consistently reduced 10-fold relative to that of wild-type WRN (Figure 4). In order to determine if this decrease in WRN protein is caused by reduced mRNA levels, we compared wild-type WRN and c.611C>T WRN mRNA levels using quantitative RT-PCR and Northern blot analysis. Both assessments showed that c.611C>T WRN mRNA was present at levels comparable to that of wild-type WRN (data not shown), suggesting that reduction of protein levels occurs by post-transcriptional mechanisms.
Fig. 4. Western blot analysis of P204L WRN.
Immune precipitates from vector alone, wild-type WRN, and c.611C>T WRN transfections were resolved by electrophoresis through an 8.5% polyacrylamide gel. The gel was transferred to PVDF, probed with a monoclonal anti-WRN antibody, and visualized on a Phosphorimager following ECL detection. Bands corresponding to WRN, and immunoglobulin heavy and light chains are indicated. Lane 1: (rWRN) purified recombinant WRN; lane 2: vector only control; lane 3: wild-type WRN; lane 4: P204L WRN. P204L WRN levels were 10-fold lower relative to wild-type WRN.
Enzymatic Characterization of P204L WRN
The c.611C>T substitution results in a change from an evolutionarily conserved proline to a leucine in the exonuclease domain of WRN. To determine if this substitution affects WRN function, we analyzed enzymatic activity in immune precipitated samples prepared from transfected 293T cells. Three assays were performed to measure helicase and exonuclease activity. A coupled helicase/exonuclease assay was carried out initially to evaluate both activities simultaneously. These assays reproducibly demonstrated that P204L WRN had a <2-fold reduction in helicase activity when compared to equal amounts of wild-type WRN (Figure 5). Because the exonuclease activity detected in this assay is dependent on helicase function, exonuclease activity cannot be accurately quantified using this assay. However, it is important to note that the helicase-coupled exonuclease activity of P204L WRN was diminished to a greater extent than the helicase activity.
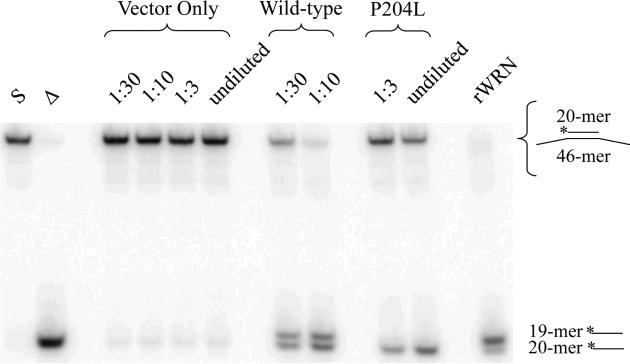
Fig. 5. Coupled helicase/exonuclease activity of P204L WRN.
Equivalent amounts of wild-type and P204L WRN were assayed for coupled helicase and exonuclease activities; reaction aliquots were electrophoresed through a 12% native polyacrylamide gel and visualized by PhopshorImager analysis, as described in Materials and Methods. Lane 1: (S) *20/46 substrate; lane 2: (Δ) heat-denatured *20/46 substrate; lanes 3-6: vector only control; lanes 7-8: wild-type WRN; lanes 9-10: P204L WRN; lane 11: (rWRN) purified recombinant WRN. The 20-mer represents the unwound product while the 19-mer represents exonucleolytic products. The sequence of the 20-mer affects its migration through a native polyacrylamide gel, therefore, the 19-mer appears to migrate more slowly than the 20-mer (see Materials and Methods and [23]).
To more precisely quantify the effects of P204L on WRN helicase and WRN exonuclease, we evaluated each activity independently. Helicase activity was measured in the absence of exonuclease activity by using a non-degradable substrate. Consistent with the results obtained in the coupled helicase/exonuclease assay, P204L WRN exhibited <2-fold reduction of unwinding activity compared to equal amounts of wild-type WRN (Figure 6). We also measured WRN exonuclease activity in the absence of DNA unwinding by inhibiting the ATP-dependent helicase with non-hydrolyzable ATPγS. In agreement with the aforementioned observation that the exonuclease activity of P204L WRN appeared more compromised relative to its helicase activity, the exonuclease-only assay revealed that P204L WRN has >8-fold lower exonuclease activity than wild-type WRN (Figure 7).
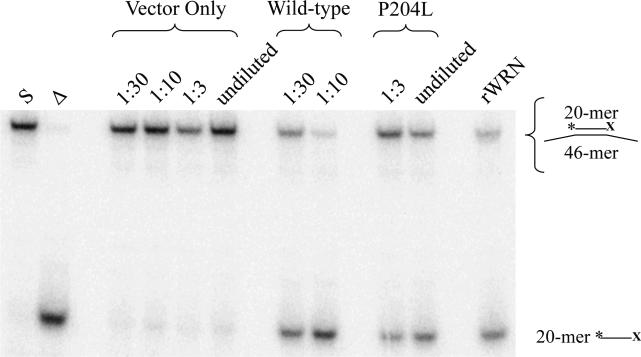
Fig. 6. Helicase activity of P204L WRN in the absence of exonuclease function.
Wild-type and P204L immune precipitates were assayed for helicase activities at the indicated dilutions. Equivalent amounts of WRN protein, as determined by Western blot analysis were assayed. Helicase reactions were performed with a non-degradable substrate in which the 3'end of the *20-mer is blocked (*20X) to prevent the action of WRN exonuclease. Lane 1: (S) *20X/46 substrate; lane 2: (Δ) heat-denatured *20X/46 substrate; lanes 3-6: vector only control; lanes 7-8: wild-type WRN; lanes 9-10: P204L WRN; lane 11: (rWRN) purified recombinant WRN. Helicase activity was determined by comparing the intensity of the lower band, *20X, to the sum of upper and lower band intensities, and found to be reduced 1.6-fold in P204L WRN relative to wild-type.
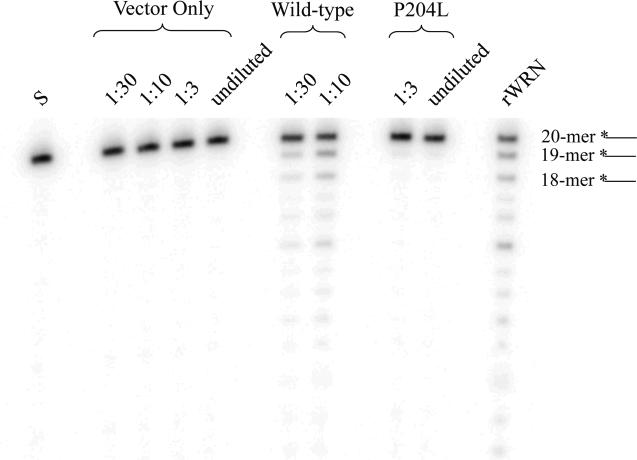
Fig. 7. Exonuclease activity of P204L WRN in the absence of helicase function.
Wild-type and P204L immune precipitates were assayed for exonuclease activity at the indicated dilutions, determined by Western blot analysis to represent equivalent protein amounts. Exonuclease reactions were performed in the presence of the non-hydrolyzable ATP analog, ATPγS, to abolish helicase activity. Lane 1: (S) *20/46 substrate; lanes 2-5: vector only control; lanes 6-7: wild-type WRN; lanes 8-9: P204L WRN, lane 10: (rWRN) purified recombinant WRN. The uppermost band represents the *20-mer, while the lower bands represent exonuclease degradation products. Relative exonuclease activity is determined by comparing the sum intensities of exonuclease degradation products to the total intensity of the reaction. Helicase-independent exonuclease activity was reduced 8.1-fold in P204L WRN relative to wild-type WRN.
These results together suggest that the c.611C>T substitution does not affect the expression of WRN at the transcriptional level. However, this nucleotide substitution results in a non-synonymous P204L amino acid alteration that a) reduces protein levels 10-fold, b) reduces helicase activity <2-fold, and c) markedly, >8-fold, decreases exonuclease activity.
DISCUSSION
The unusually high prevalence of sarcomas in WS prompted us to ask whether spontaneous sarcomas harbor mutations in the WRN gene. Spontaneous sarcomas may occur in individuals that bear one mutant WRN allele and yet do not manifest other signs of premature aging. While WS is an uncommon inherited disease and biallelic mutations in WRN have been documented in most cases of WS, heterozygous carriers of single pathologic mutant WRN alleles are much more prevalent in the general population [27]. In addition, a large number of SNPs have been documented in both the coding and noncoding sequences of WRN and these could be associated with spontaneous sarcomas.
To study sarcoma-specific mutations in WRN, we sequenced the conserved exonuclease and helicase domains of WRN in human sarcomas. No changes at the nucleotide level were detected in WRN amplified from sarcomas that were not also present in tissue adjacent to the surgical margins. These “normal” adjacent tissues were free of tumor cells upon microscopic examination. Taken together, our results suggest that mutations in the exonuclease and helicase domains of WRN are not characteristic of spontaneous sarcomas. However, there are several caveats to consider. We only sequenced exons that encode the helicase and exonuclease domains, and did not interrogate other coding and noncoding segments of WRN. In addition, we did not enrich for tumor cells, and thus, our analysis is limited by the percentage of tumor cells compared to non-cancerous stromal cells that may be present in each sample. Using DNA sequencing, we can only detect WRN mutations if they are present at a frequency greater than 15% in any given population of cells. Thus, most current methods for DNA sequencing only detect clonal mutations in tumors; i.e. mutations that drive tumor progression or those that “piggy-back” during clonal proliferation. In order to detect subclonal or random mutations one either needs to start with single cells or with single DNA molecules. Finally, large deletions, deletions present in a single allele, and epigenetic changes cannot be detected by the methods we employed.
In addition to SNPs previously reported in the GeneSNPs database, four new SNPs were identified in WRN. The possibility that one or more of these SNPs is associated with an increase in sarcoma incidence would require large epidemiological studies. Two of the four newly identified nucleotide changes are unlikely to affect the expression or function of WRN. The G to C intronic transversion found in the splice donor site following exon 4 in the exonuclease domain could potentially result in aberrant splicing, however, sequencing of mRNA showed only evidence of the wild-type transcript. The second mutation, a C to G transversion in exon 16, produces a non-synonymous amino acid change from a leucine to a valine. Although it is found within the helicase domain, the conservative nature of the amino acid substitution makes it less likely to affect WRN helicase activity.
One of the SNPs encoding a proline to leucine substitution is likely to represent a new class of amino acid substitutions that alters WRN activity. WS-associated mutations invariably result in loss of functional WRN protein. Whereas most known WRN polymorphisms are neutral, some result in reduced helicase and/or exonuclease activities without any change in WRN protein levels. For example, the WRN R834C polymorphism yields a protein that has less than 1% of helicase activity compared to wild-type WRN [23]. The c.611C>T substitution we detected is a unique WRN polymorphism; it not only reduces the level of WRN protein in the cell, but it also decreases the exonuclease activity while leaving the DNA unwinding activity largely intact. This decrease in WRN protein may be due to a negative feedback mechanism that is responsive to the mutant protein, or due to loss of protein stability. Our results suggest that the P204L variant of WRN has the potential to be clinically relevant, especially if expression of the wild-type allele becomes compromised.
Molecular analyses have revealed multiple functions of WRN in DNA metabolism and the maintenance of genome integrity. Genetic changes that compromise WRN activities may result in genomic instability, which could contribute to cancers. Furthermore, the rarity of sarcomas in the general population and the propensity for sarcomas in individuals lacking functional WRN may suggest a role of WRN in the development of sarcomas. Here, we identified several novel constitutional variants in WRN within a small population of sarcoma patients. Our characterization of one of these genetic changes, c.611C>T WRN, shows that these single nucleotide changes have the potential to compromise WRN enzymatic functions.
Supplementary Material
Acknowledgements
We thank Drs. Kiersten Henderson and Adam Hughes for critical reading of the manuscript and members of the Loeb laboratory for helpful discussions. This research was funded by NCI grant, PO1- CA77852.
REFERENCES
- 1.Shimaoka Y, Hatamochi A, Hamasaki Y, et al. Case of Werner's syndrome with pancreatic carcinoma. J Dermatol. 2007;34(9):674–676. doi: 10.1111/j.1346-8138.2007.00357.x. [DOI] [PubMed] [Google Scholar]
- 2.Epstein CJ, Martin GM, Schultz AL, Motulsky AG. Werner's syndrome a review of its symptomatology, natural history, pathologic features, genetics and relationship to the natural aging process. Medicine (Baltimore) 1966;45(3):177–221. doi: 10.1097/00005792-196605000-00001. [DOI] [PubMed] [Google Scholar]
- 3.Fukuchi K, Martin GM, Monnat RJ., Jr Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc Natl Acad Sci U S A. 1989;86(15):5893–5897. doi: 10.1073/pnas.86.15.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin GM, Sprague CA, Epstein CJ. Replicative life-span of cultivated human cells. Effects of donor's age, tissue, and genotype. Lab Invest. 1970;23(1):86–92. [PubMed] [Google Scholar]
- 5.Salk D, Au K, Hoehn H, Martin GM. Cytogenetics of Werner's syndrome cultured skin fibroblasts: variegated translocation mosaicism. Cytogenet Cell Genet. 1981;30(2):92–107. doi: 10.1159/000131596. [DOI] [PubMed] [Google Scholar]
- 6.Soultanas P, Wigley DB. Unwinding the `Gordian knot' of helicase action. Trends Biochem Sci. 2001;26(1):47–54. doi: 10.1016/s0968-0004(00)01734-5. [DOI] [PubMed] [Google Scholar]
- 7.Yu CE, Oshima J, Fu YH, et al. Positional cloning of the Werner's syndrome gene. Science. 1996;272(5259):258–262. doi: 10.1126/science.272.5259.258. [DOI] [PubMed] [Google Scholar]
- 8.Bachrati CZ, Hickson ID. RecQ helicases: suppressors of tumorigenesis and premature aging. Biochem J. 2003;374(Pt 3):577–606. doi: 10.1042/BJ20030491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Opresko PL, Cheng WH, von Kobbe C, Harrigan JA, Bohr VA. Werner syndrome and the function of the Werner protein; what they can teach us about the molecular aging process. Carcinogenesis. 2003;24(5):791–802. doi: 10.1093/carcin/bgg034. [DOI] [PubMed] [Google Scholar]
- 10.Moser MJ, Holley WR, Chatterjee A, Mian IS. The proofreading domain of Escherichia coli DNA polymerase I and other DNA and/or RNA exonuclease domains. Nucleic Acids Res. 1997;25(24):5110–5118. doi: 10.1093/nar/25.24.5110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fry M, Loeb LA. The three faces of the WS helicase. Nat Genet. 1998;19(4):308–309. doi: 10.1038/1188. [DOI] [PubMed] [Google Scholar]
- 12.Fry M, Loeb LA. Human werner syndrome DNA helicase unwinds tetrahelical structures of the fragile X syndrome repeat sequence d(CGG)n. J Biol Chem. 1999;274(18):12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 13.Bohr VA. Rising from the RecQ-age: the role of human RecQ helicases in genome maintenance. Trends Biochem Sci. 2008;33(12):609–620. doi: 10.1016/j.tibs.2008.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moser MJ, Oshima J, Monnat RJ., Jr WRN mutations in Werner syndrome. Hum Mutat. 1999;13(4):271–279. doi: 10.1002/(SICI)1098-1004(1999)13:4<271::AID-HUMU2>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 15.Oshima J, Yu CE, Piussan C, et al. Homozygous and compound heterozygous mutations at the Werner syndrome locus. Hum Mol Genet. 1996;5(12):1909–1913. doi: 10.1093/hmg/5.12.1909. [DOI] [PubMed] [Google Scholar]
- 16.Yu CE, Oshima J, Wijsman EM, et al. Mutations in the consensus helicase domains of the Werner syndrome gene. Werner's Syndrome Collaborative Group. Am J Hum Genet. 1997;60(2):330–341. [PMC free article] [PubMed] [Google Scholar]
- 17.Huang S, Lee L, Hanson NB, et al. The spectrum of WRN mutations in Werner syndrome patients. Hum Mutat. 2006;27(6):558–567. doi: 10.1002/humu.20337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsumoto T, Imamura O, Yamabe Y, et al. Mutation and haplotype analyses of the Werner's syndrome gene based on its genomic structure: genetic epidemiology in the Japanese population. Hum Genet. 1997;100(1):123–130. doi: 10.1007/s004390050477. [DOI] [PubMed] [Google Scholar]
- 19.Wang Z, Xu Y, Tang J, et al. A polymorphism in Werner syndrome gene is associated with breast cancer susceptibility in Chinese women. Breast Cancer Res Treat. 2009 doi: 10.1007/s10549-009-0327-z. [DOI] [PubMed] [Google Scholar]
- 20.Ogata N, Shiraki M, Hosoi T, Koshizuka Y, Nakamura K, Kawaguchi H. A polymorphic variant at the Werner helicase (WRN) gene is associated with bone density, but not spondylosis, in postmenopausal women. J Bone Miner Metab. 2001;19(5):296–301. doi: 10.1007/s007740170013. [DOI] [PubMed] [Google Scholar]
- 21.Hirai M, Suzuki S, Hinokio Y, et al. WRN gene 1367 Arg allele protects against development of type 2 diabetes mellitus. Diabetes Res Clin Pract. 2005;69(3):287–292. doi: 10.1016/j.diabres.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 22.Kuningas M, Slagboom PE, Westendorp RG, van Heemst D. Impact of genetic variations in the WRN gene on age related pathologies and mortality. Mech Ageing Dev. 2006;127(3):307–313. doi: 10.1016/j.mad.2005.11.005. [DOI] [PubMed] [Google Scholar]
- 23.Kamath-Loeb AS, Welcsh P, Waite M, Adman ET, Loeb LA. The enzymatic activities of the Werner syndrome protein are disabled by the amino acid polymorphism R834C. J Biol Chem. 2004;279(53):55499–55505. doi: 10.1074/jbc.M407128200. [DOI] [PubMed] [Google Scholar]
- 24.Goto M, Miller RW, Ishikawa Y, Sugano H. Excess of rare cancers in Werner syndrome (adult progeria) Cancer Epidemiol Biomarkers Prev. 1996;5(4):239–246. [PubMed] [Google Scholar]
- 25.Shen JC, Gray MD, Oshima J, Kamath-Loeb AS, Fry M, Loeb LA. Werner syndrome protein. I. DNA helicase and dna exonuclease reside on the same polypeptide. J Biol Chem. 1998;273(51):34139–34144. doi: 10.1074/jbc.273.51.34139. [DOI] [PubMed] [Google Scholar]
- 26.Huang S, Beresten S, Li B, Oshima J, Ellis NA, Campisi J. Characterization of the human and mouse WRN 3'-->5' exonuclease. Nucleic Acids Res. 2000;28(12):2396–2405. doi: 10.1093/nar/28.12.2396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Moser MJ, Bigbee WL, Grant SG, et al. Genetic instability and hematologic disease risk in Werner syndrome patients and heterozygotes. Cancer Res. 2000;60(9):2492–2496. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.