Abstract
Background & Aims
NPSR1, the receptor for neuropeptide S (NPS), is expressed by gastrointestinal (GI) enteroendocrine (EE) cells, and is involved in inflammation, anxiety and nociception. NPSR1 polymorphisms are associated with asthma and inflammatory bowel disease. We aimed to determine whether NPS induces expression of GI neuropeptides; and to associate NPSR1 single nucleotide polymorphisms (SNPs) with symptom phenotype and GI functions in health and functional GI disorders (FGID).
Methods
The effect of NPS on mRNA expression of neuropeptides was assessed using real-time PCR in NPSR1-tranfected HEK293 cells. Seventeen NPSR1 SNPs were successfully genotyped in 699 subjects from a regional cohort of 466 FGID patients and 233 healthy controls. Associations were sought using sex-adjusted regression analysis and false discovery rate (FDR) correction.
Results
NPS-NPSR1 signaling induced increased expression of CCK, VIP, PYY, and somatostatin. There were no significant associations with phenotypes of FGID symptoms. There were several NPSR1 SNPs associated with individual motor or sensory functions; the associations of SNPs rs2609234, rs6972158 and rs1379928 with colonic transit rate remained significant after FDR correction. The rs1379928 polymorphism was also associated with pain, gas and urgency sensory ratings at 36 mm Hg distension, the level pre-specified for formal testing. Associations with rectal sensory ratings were not significant after FDR correction.
Conclusions
Expression of several neuropeptides is induced upon NPS-NPSR1 signaling; NPSR1 variants are associated with colonic transit in FGID. The role of the NPS system in FGID deserves further study.
INTRODUCTION
Irritable bowel syndrome (IBS) is the most common gastrointestinal disorder, of largely unknown etiology and pathobiology. Diet, stressful life experiences, prior gastrointestinal infections, altered mucosal immunity, intestinal dysmotility, and abnormal gut-brain interactions have all been proposed as pathogenetic factors. [1] IBS is associated with altered motor, sensory, psychological, mucosal dysfunctions including enterochromaffin cell hyperplasia, a low mucosal inflammatory state and increased intestinal mucosal permeability. [2–5] There is increasing evidence that IBS might be associated with impaired gut epithelial barrier, and a dysregulated immune response to intestinal bacteria [6]; however, the magnitude and the nature of the inflammatory infiltrate is different from that observed in Crohn’s disease (CD) and ulcerative colitis (UC). Inflammatory barrier diseases, including also asthma, atopic dermatitis and psoriasis, affect organs that are at the interface with the environment (such as the lungs and skin), and show genetic predisposition. Despite several studies of candidate genes related to diverse mechanisms (detailed in reference 7), a genetic component in IBS has not been convincingly proven; in contrast, several loci of genetic linkage have been identified in asthma, atopic dermatitis and inflammatory bowel disease (IBD). [8]
The neuropeptide S (NPS) is a recently identified bioactive 20 amino acid peptide whose primary sequence is highly conserved in different species. [9,10] NPS selectively binds and activates an orphan G-protein coupled receptor (GPCR), named neuropeptide S receptor (NPSR1). [9,10] NPS is expressed in discrete nuclei in the brainstem, such as the locus coeruleus (LC) area and the parabrachial nucleus. NPS activates its G protein-coupled receptor at low nanomolar agonist concentrations and induces elevation of intracellular Ca2+ and cAMP, thereby acting as an excitatory transmitter. The NPS system can modulate stress responses, increase arousal and wakefulness, and is involved in anxiety behavior and extinction, possibly by selectively inhibiting the evoked release of serotonin (5-HT) and norepinephrine (NE) in the frontal cortex, by acting directly on 5-HT and NE nerve terminals. [9–12]
NPSR1 (also called GPR154 or G-protein coupled receptor for asthma susceptibility [GPRA]) is a 7-transmembrane G protein-coupled receptor, whose function is poorly characterized. Like its ligand NPS, NPSR1 is mainly expressed in the brain, in regions mediating anxiety and stress responses such as the amygdaloid complex and the paraventricular hypothalamic nucleus, and in the hippocampus. [13] NPSR1 mRNA was also found in the area related to descending control system of pain, such as the periaqueductal gray (PAG), raphe nuclei, and lateral parabrachial nucleus (PBN); antinociception induced by intracerebroventricular administration of NPS suggests a possible role of the NPS-NPSR system in the regulation of pain transmission. [14] This is not mediated through opiate receptors.
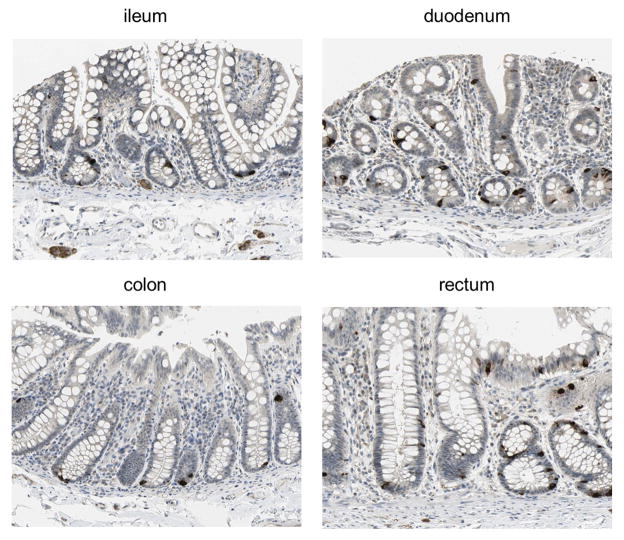
Previous studies have reported weak NPSR1 expression in epithelial cells of several organs and tissues (including small and large intestine), and an increase of this expression during inflammation, such as IBD and asthma. [15,16] Immunohistochemistry (IHC) data on human normal gut tissue recently made publicly available (Figure 1) were generated with an affinity-purified (monospecific) NPSR1-antibody. [17] Thus, the ileum, duodenum, colon and rectum clearly revealed strong and specific staining of enteroendocrine cells. Moreover, positive NPSR1-staining was detected in submucosal neurons (see ileum in Figure 1). The NPSR1 pattern of expression in the enteroendocrine cells and enteric nervous system (ENS), and the second messengers (e.g. Ca ++ and cAMP) activated by NPS-NPSR1 interaction suggest this system may modulate gastrointestinal motor and sensory functions.
Figure 1. NPSR1 is expressed in intestinal enteroendocrine cells.
Immunohistochemistry stainings performed with an affinity purified NPSR1-specific antibody (HPA007489) on human normal intestinal tissue, reported in the Human Protein Atlas portal (http://www.proteinatlas.org). Full size images and additional information are publicly available at the website, hosted by the Swedish Human Proteome Resource (HPR) program.
The neuropeptide S receptor gene (NPSR1) has received much attention as a susceptibility locus for asthma and related traits, and inflammatory bowel disease (IBD). [15,16,18–22] While no individual single nucleotide polymorphism (SNP) was found to be responsible for the observed associations, in most of these studies the genetic information was contained within a haplotype block formed by 7 SNPs (rs323917, rs323922, rs324377, hopo546333, rs324384, rs324396, and rs740347), mapping within a 70-kb region between exons 2 and 3 of the NPSR1 gene. In addition, there is some evidence of an association of a functional NPSR1 SNP (Asn107Ile, dbSNP rs324981) with airway hyperresponsiveness (a hallmark of asthma) and Crohn’s disease. [15,21] In these and related studies, the expression of both major isoforms of NPSR1 (NPSR1-A and NPSR1-B, which derive from alternative splicing) has been detected in blood leukocytes and in the epithelium of several organs including the lung and the intestine, and has been shown to increase during inflammation in asthma and inflammatory bowel diseases. [15,16,23,24]
Thus, based on the expression and potential role of NPS in enteroendocrine cell function, inflammation and nociception, our aims were: first, to determine the direct effects of NPS on expression of GI neuropeptides in a cell line; second, to test associations of NPSR1 polymorphisms with symptom phenotype and GI functions in health and functional GI disorders (FGID). Our results provide evidence that NPS can induce mRNA expression of several neuropeptides that affect GI functions, and also point to the involvement of NPSR1 in the genetic susceptibility to the intermediate phenotypes of altered colonic transit and to a lesser extent, satiation and rectal sensation, in humans.
MATERIALS AND METHODS
Overall Design
Phenotypes of human participants were studied at two levels: symptom phenotype using consensus criteria by means of validated questionnaires of gastrointestinal symptoms [25] and somatic symptoms in a majority (63%) of the participants; second, intermediate phenotypes of gastrointestinal functions associated with FGID by means of validated motor and sensory studies.
The physiological measurements have been used extensively to characterize motor and sensory functions in patients with FGID and to document effects of pharmacological agents in health and disease states. These methods are described in the Supplementary material. These were: satiation volume (maximum tolerated volume, MTV) and symptoms after a nutrient drink test (n=116, of which 37 were healthy volunteers [HV]); gastrointestinal (n=268, with 37 HV), small bowel transit (colonic filling at 6 h [CF6], n=219, with 30 HV), and colonic (n=172, with 30 HV) transit of solid food and residue by dual isotope scintigraphy; gastric volume (fasting and post-meal accommodation) by 99mTc-SPECT (n=228 with 32 HV), and rectal compliance and sensation by barostat (n=116, with 30 HV). These are intermediate phenotypes that have been described extensively in patients with FGID, particularly, IBS, functional constipation and diarrhea and functional dyspepsia.
Participants
The participants have been extensively described in prior publication, [26] and consisted of 466 patients with Rome II positive FGIDs [19 chronic abdominal pain (14 F, 5 M, mean age 47 y), 175 diarrhea-predominant IBS (IBS-D or functional diarrhea [144 F, 21 M, mean age 45 y)], 155 constipation-predominant IBS (IBS-C) or functional constipation [148 F, 7 M, mean age 45 y], 84 IBS-mixed or alternating (IBS-Alt, [77 F and 7 M, mean age 41 y]), and 33 dyspepsia ([13 F and 20 M, mean age 53 y]] and 233 HV [165 F, and 68 M, mean age 37 y]. All were recruited to studies of symptom phenotype and genotype from 2000–2007. [26–29] All resided within 150 miles of Rochester, Minnesota. Participants had been recruited for the original studies by means of letters or public advertisements and had signed informed consent for the respective studies; all patients fulfilled Rome II criteria, and in addition, functional dyspepsia patients were identified by upper abdominal pain and discomfort related to food ingestion. [30] The study was approved by the Mayo Clinic Institutional Review Board; all participants had provided written permission for research studies based on their medical records and DNA samples. An experienced coordinator used the validated bowel symptom questionnaire, electronic medical record (SM), and information from direct physician interview and examination (MC) to characterize the subtype of FGID.
Gene Expression Analysis
HEK-293 cells (http://www.atcc.org) were transfected with Fugene 6 reagent (Roche, Indianapolis, IN) and 0.5 μg of either NPSR1-A expression plasmid [24] or an empty (control) vector, according to manufacturer’s instructions. Twenty-four hours after transfection, cells were stimulated for six hours with NPS at a final concentration of 1 μM in the culture medium. Total RNA was then isolated from HEK293 cells with RNeasy Plus Mini Kit (Qiagen GmbH, Hilden, Germany), and reverse-transcribed with the SuperScript™ First-Strand Synthesis System for RT-PCR (Invitrogen, Carlsbad, CA). Real-time PCR reactions were carried out in duplicate for each gene on total RNAs from three independent experiments in an ABI Prism 7500 Fast Real-time PCR System (Applied Biosystems, Foster City, CA), according to manufacturer’s instructions. Primer sequences were designed across exons for each gene with the Primer ExpressTM 2.0 software (Applied Biosystems), and are reported in Supplementary Table 1. Analysis of cholecystokinin (CCK), calcitonin-related polypeptide alpha (CALCA), ghrelin (GHRL), motilin (MLN), peptide YY (PYY), pancreatic polypeptide (PPY), somatostatin (SST) and vasoactive intestinal peptide (VIP) mRNA expression was performed using glyceraldehyde 3-phosphate dehydrogenase (GAPDH) as internal housekeeping gene for normalization, and the comparative ΔΔCT method for relative quantification. [31] Gene expression was calculated and expressed in fold changes (arbitrary units), relative to the values obtained for the cells transfected with the empty vector (control).
NPSR1 Genotyping
Selection of SNPs
Twenty SNPs were tested in the NPSR1 gene. These SNPs were selected based on the following criteria: 11 SNPs (rs323917, rs323922, rs324377, hopo546333, rs324396, rs740347, rs2609234, rs714588, rs1379928, rs10278663, rs324384) had been previously associated with asthma and/or inflammatory bowel diseases, either individually or as part of informative haplotype blocks;[15,16,18–22] 4 SNPs (rs324981, Ile107Asn; rs34705969, Phe197Cys; rs727162, Arg241Ser; rs6972158, Arg344Gln) represent all NPSR1 coding variations known to occur in Caucasian populations (www.hapmap.org); 5 SNPs (rs2168890, rs1963499, rs887020, rs2530547, rs1419793) map in a region upstream the ATG start codon corresponding to NPSR1 predicted promoter (Mauro D’Amato, unpublished).
Method
Genotyping was carried out using TaqMan (Applied Biosystems, Foster City, CA) according to the manufacturer’s instructions, using 10–20ng of genomic DNA for each sample. Primers and probes were Assay-by Design except hopo546333 which was a custom assay (Applied Biosystems). Following PCR amplification, end reactions are read on the ABI Prism 7900HT using Sequence Detection Software (Applied Biosystems). TaqMan assays were performed at the Mayo Clinic Genotyping Shared Resource, Mayo Clinic Rochester. Hardy-Weinberg Equilibrium (HWE) calculations were performed to verify that each marker was within allelic equilibrium in controls.
Statistical Analyses
Single-marker analyses were performed with the SNPassoc 1.5–8 package implemented in R (http://www.r-project.org and http://www.creal.cat/jrgonzalez/software.htm). [32] The overall association of SNPs with FGIDs was assessed using logistic regression, combining all disorders into one group and, separately, using the individual FGID subtypes. Separate analyses with available individual physiological measurements or quantitative variables (somatic symptom score and BMI) were carried out with linear regression and, when needed, variables were log transformed in order to ensure normality. Due to small sample size, carriership of the rare alleles was used in all tests of association. Analyses were adjusted by including gender as covariate in the model, and the potential effect of BMI was also evaluated by testing whether it represented a confounder when appropriate. Linkage disequilibrium (LD) and haplotype block structure (solid spine of LD) were investigated in Haploview 4.1 (http://www.broad.mit.edu/mpg/haploview). Haplotypes in the identified LD blocks were tested with the haplo.stats 1.3.0 package implemented in R (http://mayoresearch.mayo.edu/mayo/research/schaid_lab/software.cfm), in a two step process in which they were first inferred by using an Expectation Maximization method, and later tested with either logistic or linear regression adjusted for gender. False discovery rate (FDR) correction was applied to assess whether associations were still significant after correction for multiple comparisons. For the calculation of the effect size of individual SNPs on quantitative physiological measurements, mean differences between carriers and non-carriers of the tested allele were calculated, and expressed as percent over the mean of the entire sample for the specific variable.
RESULTS
Effect of NPS-NPSR1 Signaling on Neuropeptides Expression
Human epithelial HEK293 cells were transfected with NPSR1 cDNA and, after stimulation with NPS for six hours, neuropeptides mRNA levels were quantified by Real-Time PCR. As shown in Figure 2, the expression of NPSR1 in HEK293 cells induced up-regulation of several neuropeptides upon NPS stimulation, compared to parental HEK293 cells not expressing the receptor. In particular, while NPS induced higher expression of CCK, VIP, PYY and to some extent GHRL, SST showed the strongest induction with up to > 40 fold increase in mRNA levels compared to the control cells. This experiment therefore demonstrates that NPS-NPSR1 signaling can influence the expression of several neuropeptides that are involved in the modulation of gut function(s).
Figure 2. Effect of NPS on neuropeptides expression.
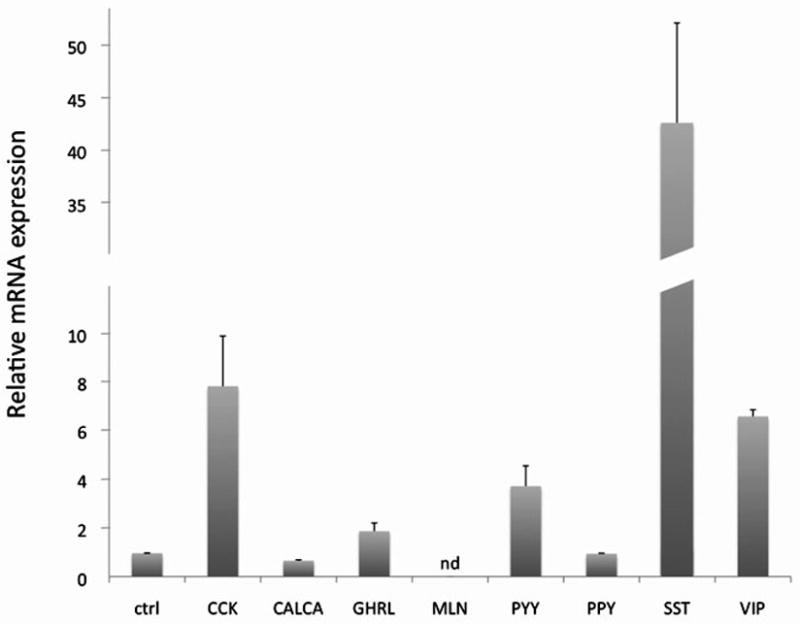
HEK293 cells were transfected in duplicate experiments with NPSR1 cDNA or empty vector (ctrl) and stimulated with 1 μM NPS for six hours. mRNA levels for each gene were measured by quantitative Real-Time PCR and are expressed as relative fold induction (+SD), compared to control cells. CCK = cholecystokinin; CALCA = calcitonin-related polypeptide alpha; GHRL = ghrelin; MLN = motilin; PYY = peptide YY; PPY = pancreatic polypeptide; SST = somatostatin; VIP = vasoactive intestinal peptide. nd = not detected.
Genotyping
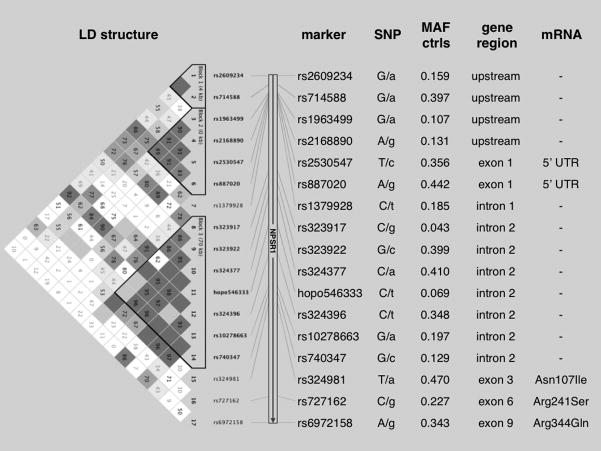
Twenty SNPs (dbSNP rs323917, rs323922, rs324377, hopo546333, rs324396, rs740347, rs2609234, rs714588, rs1379928, rs10278663, rs324384, rs324981, rs34705969, rs727162, rs6972158, rs2168890, rs1963499, rs887020, rs2530547, rs1419793) spanning approximately 200 kb of Chromosome 7p and including the entire NPSR1 coding region (Figure 3), were genotyped in all 699 study participants. Eighteen SNPs showed 100% success rate; two markers (rs34705969 and rs324384) failed to cluster in the TaqMan assay, and were therefore excluded from downstream analyses. Allelic frequencies for the SNP rs1419793 showed significant deviation from HWE in the control group (p < 0.001); therefore, this marker was also excluded. Allelic frequencies at each marker, linkage disequilibrium (LD), and haplotype block structure in the region were similar to those previously shown in other studies of populations of European Caucasian origin (Figure 3). [15,16,18–20,22]
Figure 3. LD map and characteristics of the studied NPSR1 SNPs.
Left: LD and haplotype block structure (LD structure) obtained from Haploview 4 analysis of genotyping data. The numbers in each box correspond to LD coefficient D’ between respective SNPs (only values for LD <100% are reported). Center: SNPs (marker) are listed with alleles at each locus (SNP - minor allele in lower case) and minor allele frequency in controls (MAF ctrl). Right: position of each SNP within then NPSR1 coding region (gene region), and corresponding effect on mRNA (mRNA).
The 17 NPSR1 SNPs passing all quality controls were tested for association with susceptibility to FGID symptom phenotypes and intermediate (physiological) phenotypes.
Association with Symptom Phenotype
There were no significant associations of NPSR1 genotypes with symptom phenotype of FGIDs, either when tested as different sub-phenotypes (IBS-C, IBS-D, IBD-Alt, dyspepsia, compared to healthy controls) or when grouped together (FGID compared to healthy controls, data not shown). Two NPSR1 SNPs, rs2530547 and rs324377, appeared to influence somatic symptom scores (Supplementary Table 2), but the association was not significant after FDR correction for the multiple tests performed. In addition, they were not significantly associated with any gastrointestinal physiological measurements (see below).
Association with Intermediate Phenotypes: GI Motor and Sensory Functions
Several associations were detected between individual gastrointestinal functions and NPSR1 polymorphisms, which are reported in Table 1.
Table 1.
Associations (P values) of NPSR1 SNPs with motor function phenotypes in all study participants.
| N | rs2609234 | rs714588 | rs1963499 | rs2168890 | rs2530547 | rs887020 | rs1379928 | rs323917 | rs323922 | rs324377 | hopo546333 | rs324396 | rs10278663 | rs740347 | rs324981 | rs727162 | rs6972158 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele tested | A | A | A | G | C | G | T | G | C | A | T | T | A | C | A | G | G | |
|
Gastrointestinal transit | ||||||||||||||||||
| Log(GE T ½) | 268 | .675 | .928 | .254 | .200 | .733 | .713 | .282 | nt | .770 | .780 | .565 | .122 | .885 | .119 | .884 | .719 | .512 |
| CF6 | 219 | .877 | .528 | .818 | .996 | .630 | .827 | .912 | nt | .057 | .080 | .303 | .846 | .004 | .052 | .104 | .219 | .322 |
| Log(GC24) | 172 | .055 | .469 | .486 | .423 | .922 | .947 | .002 | nt | .196 | .239 | .104 | .970 | .783 | .007 | .480 | .475 | .035 |
| Log(GC48) | 171 | .0002 | .071 | .220 | .068 | .678 | .493 | .006 | nt | .395 | .344 | .337 | .777 | .485 | .026 | .728 | .072 | .009 |
|
Satiation | ||||||||||||||||||
| Log(MT Vol) | 116 | .175 | .826 | .821 | .430 | .831 | .374 | .839 | nt | .838 | .838 | nt | .943 | .648 | .928 | .538 | .839 | .971 |
| Aggregate Score | 116 | .392 | .245 | .830 | .094 | .198 | .182 | .108 | nt | .442 | .274 | nt | .935 | .953 | .780 | .011 | .456 | .018 |
|
Gastric Volume and Accommodation | ||||||||||||||||||
| Log(Fast GVol) | 228 | .269 | .316 | .449 | .375 | .132 | .693 | .559 | nt | .290 | .162 | .881 | .956 | .637 | .873 | .087 | .979 | .990 |
| Log(Av Fed GVol) | 228 | .892 | .410 | .881 | .879 | .361 | .519 | .865 | nt | .305 | .384 | .344 | .330 | .420 | .135 | .026 | .303 | .944 |
| Delta Vol | 228 | .685 | .783 | .457 | .615 | .954 | .663 | .709 | nt | .777 | .906 | .160 | .277 | .283 | .090 | .220 | .195 | .935 |
NOTE: GE T ½ = gastric half emptying time; CF6 = colonic filling at 6 hours; GC24/GC48 = colonic geometric center at 24/48 hours; MT Vol = maximum tolerated volume; Aggregate Score = aggregate score of postprandial symptoms (nausea, fullness, bloating, pain); Fast GVol = fasting gastric volume; Av Fed GVol = average feeding gastric volume; Delta Vol = Fast GVol - Av Fed GVol; nt = not tested (allele frequency below 0.05). Significant P values (< 0.05) are reported in bold, and are underlined when significant even after False Discovery Rate (FDR) correction for the multiple tests performed.
Two SNPs, rs324981 and rs6972158, were associated with the aggregate symptom score 30 minutes after a fully satiating meal. Of these, the polymorphism rs324981 (which corresponds to the Asn107Ile coding SNP) was also associated with postprandial gastric volume.
Five NPSR1 polymorphisms showed association with individual lower gastrointestinal motor function measurements. Specifically, the SNPs rs1379928, rs740347 and rs6972158 (corresponding to the Arg344Gln coding variant) were all associated with colonic transit rate at both 24 and 48 hours; the SNP rs2609234 was associated with GC48, while the SNP rs10278663 showed association with colonic filling at 6 hours (surrogate of small bowel transit). The associations of rs1379928 with both GC24 and GC48, and of rs2609234 and rs6972158 with GC48 remained significant after FDR correction for multiple tests.
The results obtained from the statistical analysis of NPSR1 SNPs and sensory functions are summarized in Table 2. Interestingly, among others, the SNP rs1379928 (which showed strong association with colonic transit at both 24 and 48 hours) also was associated with rectal sensory ratings of gas, urgency and pain. This polymorphism is, therefore, of particular interest, since it shows consistent associations with both motor and sensory physiological endpoints.
Table 2.
Associations (P values) of NPSR1 SNPs with rectal compliance, and sensory ratings.
| N | rs2609234 | rs714588 | rs1963499 | rs2168890 | rs2530547 | rs887020 | rs1379928 | rs323917 | rs323922 | rs324377 | hopo546333 | rs324396 | rs10278663 | rs740347 | rs324981 | rs727162 | rs6972158 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Allele tested | A | A | A | G | C | G | T | G | C | A | T | T | A | C | A | G | G | |
| Compliance Pr50 | 112 | .920 | .977 | .911 | .516 | .652 | .633 | .118 | nt | .202 | .316 | nt | .956 | .893 | .889 | .225 | .598 | .880 |
| Gas at 36 mm Hg | 102 | .434 | .900 | .314 | .652 | .192 | .676 | .012 | nt | .494 | .436 | nt | .087 | .014 | .926 | .960 | .918 | .662 |
| Urgency at 36 mm Hg | 101 | .220 | .679 | .616 | .852 | .546 | .479 | .041 | nt | .539 | .334 | nt | .308 | .277 | .497 | .905 | .996 | .243 |
| Pain at 36 mm Hg | 102 | .044 | .013 | .694 | .942 | .886 | .864 | .041 | nt | .545 | .303 | nt | .921 | .098 | .588 | .980 | .229 | .727 |
NOTE: Sensation ratings of gas/urgency/pain at 36 mm Hg distension (VAS score); nt = not tested (allelic frequency below 0.05). Significant P values (< 0.05) are reported in bold. No P values remained significant after correction for the multiple tests performed.
No additional findings, beyond those from individual SNPs, were identified when NPSR1 haplotypes at LD blocks 1, 2 and 3 (Figure 1) were tested for association with either FGID symptom or intermediate physiological phenotypes (data not shown). Thus, the only haplotypic association identified was entirely due to linkage disequilibrium with rs2609234, and identical to the association at the single marker level. However, haplotype associations may have been hampered by low power due to the small sample size.
Inclusion as covariate in the statistical analyses of BMI, (which affects colonic functions, [33] and which is associated with some NPSR1 SNPs in our population [Supplementary Table 2]), did not significantly modify the estimated genetic associations (data not shown).
Quantitative Differences in Motor and Sensory Endpoints in Association with NPSR1 SNPs
To provide an estimate of the potential effect of NPSR1 polymorphism on gastrointestinal functions, the SNPs showing significant associations in the initial analyses in tables 1 and 2, were selected to quantify their associations and the direction of variation in motor and sensory ratings. In this exploratory analysis, several SNPs showed effects of substantial size, expressed in Table 3 as percent increase or decrease over the mean value of physiological endpoints in each of the NPSR1 genotypes of interest. In particular, the SNPs rs2609234, rs1379928 and rs6972158 (whose associations withstood correction for multiple tests) were associated with > 10% accelerations over average colonic transit (GC24 and/or GC48).
Table 3.
Quantitative effect of NPSR1 SNPs of interest on physiological endpoints.
| rs2609234 | rs714588 | rs1379928 | rs10278663 | rs740347 | rs324981 | rs6972158 | |
|---|---|---|---|---|---|---|---|
| Allele tested | A | A | T | A | C | A | G |
| GE T ½ | |||||||
| CF6 | −27.1% | ||||||
| GC24 | +15.5% | +16.2% | +10.1% | ||||
| GC48 | +16.0% | +11.1% | +8.1% | +10.0% | |||
| MT Vol | |||||||
| Aggregate Score | +23.1% | +18.6% | |||||
| Fast GVol | |||||||
| Av Fed GVol | −5.7% | ||||||
| Delta Vol | |||||||
| Compliance Pr50 | |||||||
| Gas at 36 mm Hg | +22.5% | ||||||
| Urgency at 36 mm Hg | +9.9% | −5.5% | |||||
| Pain at 36 mm Hg | +20.7% | +24.7% | +19.9% |
NOTE: The direction of the relative contribution of tested alleles to each variable is expressed as mean percent increase (+) or decrease (−), calculated from the difference between carriers and non-carriers over the entire sample. Only effects for significant (p < 0.05) associations are reported, and those withstanding FDR correction for multiple tests (as reported in Table 1) are underlined. Abbreviations used to define variables are the same as from previous tables.
Functional Analysis of NPSR1 SNP rs1379928
Since the SNP rs1379928 was associated both with colonic transit and rectal sensory functions, we pursued preliminary studies to assess the functional effect(s) of this polymorphism. SNP rs1379928 is intronic and therefore is unlikely to have an effect on the corresponding protein.
To evaluate whether the SNP rs1379928 is of direct functional relevance, we tested its potential to affect DNA-protein interactions in electrophoretic mobility shift assays (EMSAs) with nuclear extracts from three different cell lines, SH-SY5Y (neuroblastoma), Colo205 (colon adenocarcinoma), and HEK293 (embrionic kidney epithelial cells). As shown in Supplementary Figure 1, however, this analysis did not disclose allelic qualitative or quantitative differences in DNA-protein complex formation. Although cell line-specific patterns were observed (with Colo205 cells differing from SH-SY5Y and HEK293), the two variants (T and C) at the rs1379928 SNP gave rise to identical DNA-protein complex profiles in the 3 cell lines tested. Although functional effects cannot be excluded, this experiment did not provide direct evidence that the rs1379928 polymorphism impacts protein binding to DNA.
DISCUSSION
This study shows that the expression of neuropeptides typically produced by enteroendocrine cells increases upon NPS-NPSR1 signaling in an in vitro model system. In addition, we have studied 17 NPSR1 SNPs in an ethnically homogeneous group of FGID patients and healthy controls from one geographical region of the United States. Our results provide evidence of involvement of NPSR1 in the genetic susceptibility to intermediate phenotypes of gastrointestinal function. We have shown that discrete NPSR1 polymorphisms are associated with colonic transit and, to a lesser extent, with rectal sensations and postprandial satiation symptoms that may predispose to perturbed gastrointestinal functions in patients with FGIDs.
Few candidate genes have hitherto been tested for association with genetic susceptibility to FGID and/or related phenotypes, and there is no compelling evidence coming from replications in different populations of any one marker of susceptibility. [7,26–29,34–36]. The NPSR1 gene encompasses 220 kb of genomic DNA on chromosome 7p14, and, in this region, there are hundreds of SNPs, which have been identified through sequencing of individuals from different ethnic groups (http://www.hapmap.org). To increase the likelihood of detecting association, our candidate gene strategy was based on the selection of NPSR1 SNPs either previously associated with disease (asthma and IBD), [15,16,18–22] or of highest potential functional relevance (promoter and coding SNPs). Most of the association signals identified in previous studies come from haplotypes tagged by 7 intronic SNPs (rs323917, rs323922, rs324377, hopo546333, rs324384, rs324396 and rs740347) that are part of an LD block between exon 2 and 3 of the NPSR1 gene. [15,16,18–22] Whereas these SNPs were included in our study, the multi-marker (haplotype) inference analysis was not sufficiently powered to confirm their relevance in the relatively small sample beyond the detected single-marker associations.
Several NPSR1 polymorphisms located in different regions of the gene provided evidence of association. The most significant associations, which withstood correction for multiple comparisons, involved 3 SNPs associated with colonic transit, namely rs2609234 and rs6972158 (associated with GC24), and rs1379928 (associated with both GC24 and GC48). These SNPs had a considerable effect on colonic transit, corresponding to a 10–16% mean acceleration of colonic transit in carriers of the tested alleles. The rs2609234 polymorphism maps 9 kb upstream the NPSR1 coding region, and is thus likely to represent a marker in LD with as-yet unidentified functional variation(s). The rs6972158 SNP corresponds to an Arg344Gln amino acid change in the cytoplasmic tail of NPSR1, the receptor domain predicted to convey transduction signals intracellularly, and hence it may be functionally relevant. However, this variant has not been functionally characterized.
We were particularly interested in the SNP rs1379928, which affects colonic transit and rectal sensory ratings of gas, urgency and pain, and provided the strongest signal of association with asthma in two different populations (European-American and Costa Rican Hispanic). [22] This polymorphism is intronic (intron 1), does not impact on the functional properties of the receptor, and it may relate to intragenic regulatory elements or represent an LD proxy for other causative polymorphisms. In an electrophoretic mobility shift assays on 3 different cell lines, we failed to provide evidence of differential binding of nuclear proteins to DNA sequences from the two rs1379928 alleles, and therefore further studies are needed to assess whether this SNP is truly functionally relevant.
Several associations of nominal significance (p<0.05) were also detected in this study; their relevance will require confirmation in larger samples. The association of the SNP rs324981 with postprandial aggregate symptoms score and gastric volumes is of potential interest as it was associated with IBD, and with bronchial hyperresponsiveness to metacholine in Chinese asthmatics. [15,21] This polymorphism also corresponds to an NPSR1 Asn107Ile amino acid change that affects both the level of expression of the protein on the cell membrane, and the potency of the ligand NPS. [37–39] Carriage of the A allele, (Asn 107, characterized by lower NPSR1 biological activity), was associated with a 23.1% increase in postprandial aggregate score following a full satiating meal, and a 5.7% reduction of postprandial gastric volume following a full satiating meal; both would be expected to induce earlier satiation, as might occur in functional dyspepsia. Because the current study involved only 33 small patients with dyspepsia, this association with symptom and intermediate phenotypes should be tested in the future.
The biological function of the NPSR1-NPS system in the gut is still poorly understood; in an in vitro model system we have provided here indirect evidence that signaling through NPSR1 can induce increased expression of somatostatin, CCK, VIP, PYY and, possibly, ghrelin. Although their induction remains to be verified in the intestinal enteroendocrine cell type(s) expressing the receptor, many of these and other peptides and hormones up-regulated by NPS (such as substance P, neurotensin and the alpha subunit of chorionic gonadotropin, LH, FSH and TSH, [40]) are involved in the control of physiological motor and sensory functions in the gastrointestinal tract. [41–44] It is therefore conceivable that genetic alterations affecting NPSR1 expression or function might result in excessive or diminished neuropeptides induction leading to perturbed gut function, and thus represent at least one potential mechanism to explain the genetic associations reported here. Of note, inhibition of distal colonic transit rate has been recently shown in mice upon intracerebroventricular administration of NPS. [45] Similar to other neuropeptides and their receptors, it is plausible that NPS-NPSR1 signaling plays a role in the complex gut-brain axis, participating in modulation of inflammatory responses, anxiety and gut functions including nociception. Further investigations should thus use the data from this study to explore the associations of specific NPSR1 genetic variants with different FGIDs, bowel dysfunction, and the IBS pain, gas or urgency.
As with such candidate gene approaches, we may not have identified the specific variation(s) responsible for the intermediate phenotypes of FGIDs. The NPSR1 is a complex locus with several LD blocks and many polymorphisms. Our targeted analysis takes us at least one step closer to narrowing down the number of SNPs to be evaluated in functional studies or to identifying risk alleles that may be in linkage disequilibrium with the detected SNPs of interest. In addition, our study did not focus on symptom phenotypes but on intermediate physiological phenotypes or biomarkers, and therefore direct clinical relevance is unclear.
The strengths of the study include the use of intermediate phenotypes that have been extensively studied, are associated with symptoms of FGID, and their coefficient of variation is thoroughly characterized providing a measurable effect of the associated SNPs. We used false discovery rate (FDR) correction to provide a conservative interpretation of the observed associations, and analyzed the data adjusting for gender. In contrast to the significant findings for multiple physiological functions, we did not observe any significant associations with symptom phenotypes of FGID. We believe this simply reflects the greater power of genotype-intermediate phenotype association studies compared to genetic epidemiology association studies, which require much greater sample sizes. [46]
In conclusion, our study provides the first evidence of an association of NPSR1 polymorphisms and gastrointestinal motor and sensory functions that are relevant to IBS and functional dyspepsia. Replication studies will be important to confirm these findings in independent populations, although these may be difficult to accomplish as we are unaware that an IBS cohort with the same intermediate phenotypes measured is available. Further analyses of NPSR1 function(s) are encouraged to elucidate its role in FGIDs and the potential associations with epithelial barrier functions, inflammation, sensation, transit and satiation in health and disease.
Supplementary Material
Acknowledgments
Dr. Camilleri is supported in part by RO1 grant DK54681 from National Institutes of Health. Dr. D’Amato is supported by the Groschinskys Minnesfond, the Ruth and Richard Juhlin’s Foundation, and the Prof. Nanna Svartz Fund.
Abbreviations
- CALCA
calcitonin-related polypeptide alpha
- CCK
cholecystokinin
- CI
confidence interval
- FDR
false discovery rate
- GHRL
ghrelin
- IBS
irritable bowel syndrome
- IBS-D
diarrhea-predominant irritable bowel syndrome
- IBS-C
constipation-predominant irritable bowel syndrome
- IBS-Alt
irritable bowel syndrome with alternating bowel function
- MLN
motilin
- NPS
neuropeptide S
- NPSR1
neuropeptide S receptor
- OR
odds ratio
- PCR
polymerase chain reaction
- PPY
pancreatic polypeptide
- PYY
peptide YY
- SNP
single nucleotide polymorphism
- SST
Somatostatin
- VIP
vasoactive intestinal peptide
Footnotes
Conflict of Interest: No conflicts of interest exist.
Authors’ contributions: M. Camilleri: study conceptualization, writing protocol and paper; P. Carlson: NPSR1 genotyping; A.R. Zinsmeister: database management and statistical analysis; S. McKinzie: patient recruitment and classification through questionnaires and medical record review; I. Busciglio: rectal sensation studies; D. Burton: scintigraphic transit and SPECT imaging; M. Zucchelli: statistical/association analysis; M. D’Amato: expertise on genotyping, writing manuscript, EMSA studies, neuropeptide expression studies in HEK293 cells.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Camilleri M. Mechanisms in IBS: something old, something new, something borrowed. Neurogastroenterol Motil. 2005;17:311–316. doi: 10.1111/j.1365-2982.2004.00632.x. [DOI] [PubMed] [Google Scholar]
- 2.Camilleri M, McKinzie S, Busciglio I, et al. Prospective study of motor, sensory, psychological and autonomic functions in patients with irritable bowel syndrome. Clin Gastroenterol Hepatol. 2008;6:772–781. doi: 10.1016/j.cgh.2008.02.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dorn SD, Palsson OS, Thiwan SI, et al. Increased colonic pain sensitivity in irritable bowel syndrome is the result of an increased tendency to report pain rather than increased neurosensory sensitivity. Gut. 2007;56:1202–1209. doi: 10.1136/gut.2006.117390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Spiller RC, Jenkins D, Thornley JP, et al. Increased rectal mucosal enteroendocrine cells, T lymphocytes, and increased gut permeability following acute Campylobacter enteritis and in post-dysenteric irritable bowel syndrome. Gut. 2000;47:804–811. doi: 10.1136/gut.47.6.804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dunlop SP, Hebden J, Campbell E, et al. Abnormal intestinal permeability in subgroups of diarrhea-predominant irritable bowel syndromes. Am J Gastroenterol. 2006;101:1288–1294. doi: 10.1111/j.1572-0241.2006.00672.x. [DOI] [PubMed] [Google Scholar]
- 6.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 7.Saito YA. Genes and irritable bowel syndrome: is there a link? Curr Gastroenterol Rep. 2008;10:355–362. doi: 10.1007/s11894-008-0069-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schreiber S, Rosenstiel P, Albrecht M, et al. Genetics of Crohn disease, an archetypal inflammatory barrier disease. Nat Rev Genet. 2005;6:376–388. doi: 10.1038/nrg1607. [DOI] [PubMed] [Google Scholar]
- 9.Xu YL, Reinscheid RK, Huitron-Resendiz S, et al. Neuropeptide S: a neuropeptide promoting arousal and anxiolytic-like effects. Neuron. 2004;43:487–497. doi: 10.1016/j.neuron.2004.08.005. [DOI] [PubMed] [Google Scholar]
- 10.Okamura N, Reinscheid RK. Neuropeptide S: a novel modulator of stress and arousal. Stress. 2007;10:221–226. doi: 10.1080/10253890701248673. [DOI] [PubMed] [Google Scholar]
- 11.Raiteri L, Luccini E, Romei C, et al. Neuropeptide S selectively inhibits the release of 5-HT and noradrenaline from mouse frontal cortex nerve endings. Br J Pharmacol. 2009 Apr 3; doi: 10.1111/j.1476-5381.2009.00163.x. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jüngling K, Seidenbecher T, Sosulina L, et al. Neuropeptide S-mediated control of fear expression and extinction: role of intercalated GABAergic neurons in the amygdala. Neuron. 2008;59:298–310. doi: 10.1016/j.neuron.2008.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu YL, Gall CM, Jackson VR, et al. Distribution of neuropeptide S receptor mRNA and neurochemical characteristics of neuropeptide S-expressing neurons in the rat brain. J Comp Neurol. 2007;500:84–102. doi: 10.1002/cne.21159. [DOI] [PubMed] [Google Scholar]
- 14.Li W, Chang M, Peng YL, et al. Neuropeptide S produces antinociceptive effect at the supraspinal level in mice. Regul Pept. 2009 Apr 2; doi: 10.1016/j.regpep.2009.03.013. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 15.D’Amato M, Bruce S, Bresso F, et al. Neuropeptide S receptor 1 gene polymorphism is associated with susceptibility to inflammatory bowel disease. Gastroenterology. 2007;133:808–817. doi: 10.1053/j.gastro.2007.06.012. [DOI] [PubMed] [Google Scholar]
- 16.Laitinen T, Polvi A, Rydman P, et al. Characterization of a common susceptibility locus for asthma-related traits. Science. 2004;304:300–304. doi: 10.1126/science.1090010. [DOI] [PubMed] [Google Scholar]
- 17. [accessed May 14, 2009]; www.proteinatlas.org, hosted by the Swedish Human Proteome Reseource Program.
- 18.Kormann MS, Carr D, Klopp N, et al. G-protein-coupled receptor polymorphisms are associated with asthma in a large German population. Am J Respir Crit Care Med. 2005;171:1358–1362. doi: 10.1164/rccm.200410-1312OC. [DOI] [PubMed] [Google Scholar]
- 19.Melen E, Bruce S, Doekes G, et al. Haplotypes of G protein-coupled receptor 154 are associated with childhood allergy and asthma. Am J Respir Crit Care Med. 2005;171:1089–1095. doi: 10.1164/rccm.200410-1317OC. [DOI] [PubMed] [Google Scholar]
- 20.Malerba G, Lindgren CM, Xumerle L, et al. Chromosome 7p linkage and GPR154 gene association in Italian families with allergic asthma. Clin Exp Allergy. 2007;37:83–89. doi: 10.1111/j.1365-2222.2006.02615.x. [DOI] [PubMed] [Google Scholar]
- 21.Feng Y, Hong X, Wang L, et al. G protein-coupled receptor 154 gene polymorphism is associated with airway hyperresponsiveness to methacholine in a Chinese population. J Allergy Clin Immunol. 2006;117:612–617. doi: 10.1016/j.jaci.2005.11.045. [DOI] [PubMed] [Google Scholar]
- 22.Hersh CP, Raby BA, Soto-Quirós ME, et al. Comprehensive testing of positionally cloned asthma genes in two populations. Am J Respir Crit Care Med. 2007;176:849–857. doi: 10.1164/rccm.200704-592OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pulkkinen V, Majuri ML, Wang G, et al. Neuropeptide S and G protein-coupled receptor 154 modulate macrophage immune responses. Hum Mol Genet. 2006;15:1667–1679. doi: 10.1093/hmg/ddl090. [DOI] [PubMed] [Google Scholar]
- 24.Vendelin J, Pulkkinen V, Rehn M, et al. Characterization of GPRA, a novel G protein-coupled receptor related to asthma. Am J Respir Cell Mol Biol. 2005;33:262–270. doi: 10.1165/rcmb.2004-0405OC. [DOI] [PubMed] [Google Scholar]
- 25.Talley NJ, Phillips SF, Wiltgen CM, et al. Assessment of functional gastrointestinal disease: the bowel disease questionnaire. Mayo Clin Proc. 1990;65:1456–1479. doi: 10.1016/s0025-6196(12)62169-7. [DOI] [PubMed] [Google Scholar]
- 26.Camilleri M, Carlson P, Zinsmeister AR, et al. Mitochondrial DNA and gastrointestinal motor and sensory functions in health and functional gastro-intestinal disorders. Am J Physiol Gastrointest Liver Physiol. 2009;296:G510–516. doi: 10.1152/ajpgi.90650.2008. [DOI] [PubMed] [Google Scholar]
- 27.Kim HJ, Camilleri M, Carlson PJ, et al. Association of distinct α2 adrenoceptor and serotonin-transporter polymorphisms associated with constipation and somatic symptoms in functional gastrointestinal disorders. Gut. 2004;53:829–837. doi: 10.1136/gut.2003.030882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Andresen V, Camilleri M, Kim HJ, et al. Is there an association between GNβ3 C825T genotype and lower functional gastrointestinal disorders? Gastroenterology. 2006;130:1985–1994. doi: 10.1053/j.gastro.2006.03.017. [DOI] [PubMed] [Google Scholar]
- 29.Camilleri CE, Carlson PJ, Camilleri M, et al. A study of candidate genotypes associated with dyspepsia in a U.S. community. Am J Gastroenterol. 2006;101:581–592. doi: 10.1111/j.1572-0241.2006.00481.x. [DOI] [PubMed] [Google Scholar]
- 30.Castillo EJ, Camilleri M, Locke GR, III, et al. A community based, controlled study of the epidemiology and pathophysiology of dyspepsia. Clin Gastroenterol Hepatol. 2004;2:985–996. doi: 10.1016/s1542-3565(04)00454-9. [DOI] [PubMed] [Google Scholar]
- 31.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 32.Gonzalez JR, Armengol L, Sole X, et al. SNPassoc: an R package to perform whole genome association studies. Bioinformatics. 2007;23:644–645. doi: 10.1093/bioinformatics/btm025. [DOI] [PubMed] [Google Scholar]
- 33.Delgado-Aros S, Camilleri M, Garcia MA, et al. High body mass alters colonic sensory-motor function and transit in humans. Am J Physiol Gastrointest Liver Physiol. 2008;295:G382–388. doi: 10.1152/ajpgi.90286.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeo A, Boyd P, Lumsden S, et al. Association between a functional polymorphism in the serotonin transporter gene and diarrhoea predominant irritable bowel syndrome in women. Gut. 2004;53:1452–1458. doi: 10.1136/gut.2003.035451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Kerkhoven LA, Laheij RJ, Jansen JB. Meta-analysis: a functional polymorphism in the gene encoding for activity of the serotonin transporter protein is not associated with the irritable bowel syndrome. Aliment Pharmacol Ther. 2007;26:979–986. doi: 10.1111/j.1365-2036.2007.03453.x. [DOI] [PubMed] [Google Scholar]
- 36.Camilleri M, Busciglio I, Carlson P, et al. Candidate genes and sensory functions in health and irritable bowel syndrome. Am J Physiol Gastrointest Liver Physiol. 2008;295:G219–225. doi: 10.1152/ajpgi.90202.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Reinscheid RK, Xu YL, Okamura N, et al. Pharmacological characterization of human and murine neuropeptide S receptor variants. J Pharmacol Exp Ther. 2005;315:1338–1345. doi: 10.1124/jpet.105.093427. [DOI] [PubMed] [Google Scholar]
- 38.Roth AL, Marzola E, Rizzi A, et al. Structure-activity studies on neuropeptide S: identification of the amino acid residues crucial for receptor activation. J Biol Chem. 2006;281:20809–20816. doi: 10.1074/jbc.M601846200. [DOI] [PubMed] [Google Scholar]
- 39.Bernier V, Stocco R, Bogusky MJ, et al. Structure/function relationships in the neuropeptide s receptor: molecular consequences of the asthma-associated mutation N107I. J Biol Chem. 2006;281:24704–24712. doi: 10.1074/jbc.M603691200. [DOI] [PubMed] [Google Scholar]
- 40.Vendelin J, Bruce S, Holopainen P, et al. Downstream target genes of the neuropeptide S-NPSR1 pathway. Hum Mol Genet. 2006;15:2923–2935. doi: 10.1093/hmg/ddl234. [DOI] [PubMed] [Google Scholar]
- 41.Shimizu Y, Matsuyama H, Shiina T, et al. Tachykinins and their functions in the gastrointestinal tract. Cell Mol Life Sci. 2008;65:295–311. doi: 10.1007/s00018-007-7148-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bornstein JC, Costa M, Grider JR. Enteric motor and interneuronal circuits controlling motility. Neurogastroenterol Motil. 2004;16(Suppl 1):34–38. doi: 10.1111/j.1743-3150.2004.00472.x. [DOI] [PubMed] [Google Scholar]
- 43.Olsson C, Holmgren S. The control of gut motility. Comp Biochem Physiol A Mol Integr Physiol. 2001;128:481–503. doi: 10.1016/s1095-6433(00)00330-5. [DOI] [PubMed] [Google Scholar]
- 44.Camilleri M, Grudell AB. Appetite and obesity: a gastroenterologist’s perspective. Neurogastroenterol Motil. 2007;19:333–341. doi: 10.1111/j.1365-2982.2006.00864.x. [DOI] [PubMed] [Google Scholar]
- 45.Han RW, Chang M, Peng YL, et al. Central Neuropeptide S inhibits distal colonic transit through activation of central Neuropeptide S receptor in mice. Peptides. 2009;30:1313–1317. doi: 10.1016/j.peptides.2009.03.012. [DOI] [PubMed] [Google Scholar]
- 46.Mayer EA. The challenge of studying the biology of complex, symptom-based GI disorders. Gastroenterology. 2008;134:1826–1827. doi: 10.1053/j.gastro.2008.05.003. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.