Abstract
Many viruses need to stabilize their capsid structure against DNA pressure and for survival in hostile environments. The 9 kDa outer capsid protein (Soc) of bacteriophage T4, which stabilizes the virus, attaches to the capsid during the final stage of maturation. There are 870 Soc molecules that act as “glue” between neighboring hexameric capsomers, forming a “cage” that stabilizes the T4 capsid against extremes of pH and temperature. Here we report a 1.9 Å resolution crystal structure of Soc from the bacteriophage RB69, a close relative of T4. The RB69 crystal structure and a homology model of T4 Soc were fitted into the cryo-electron microscopy reconstruction of the T4 capsid. This established the region of Soc that interacts with the major capsid protein and suggested a mechanism, verified by extensive mutational and biochemical studies, for stabilization of the capsid in which the Soc trimers act as clamps between neighboring capsomers. The results demonstrate the factors involved in stabilizing not only the capsids of T4-like bacteriophages but also many other virus capsids.
Keywords: T4 head structure, bacteriophage capsid assembly, capsid stabilization, virus stabilization proteins, capsid decorative proteins
Introduction
When dsDNA tailed bacteriophages (or phages) assemble their capsid, the assembly of the major capsid protein is nucleated around a core that consists of the dodecameric portal protein and various scaffolding proteins.1,2 Parts of the core are then removed and the capsid expands, initiating DNA packaging.3 Eventually the DNA is packaged to a near crystalline density (~500 mg/ml), exerting ~6 MPa pressure on the capsid shell4. In some viruses, the procapsid expansion exposes sites for binding of outer capsid proteins that stabilize the capsid against the internal pressure of the DNA5. The reinforcements provided by the outer capsid proteins also allow the virus to survive in hostile environments between infections.6 As these proteins are on the outermost surface of the capsid, they have been useful for decorating the capsid with antigens that could be used in biotechnological applications.7-9 However, the principles that govern virus stabilization have remained poorly understood.
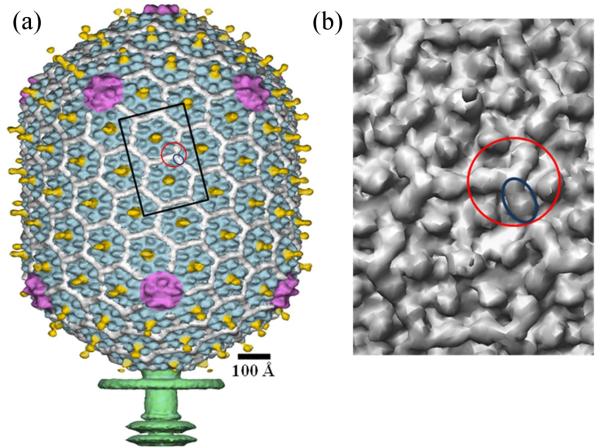
Bacteriophage T4 is a double-stranded DNA virus which belongs to the Myoviridae family and uses E. coli as a host. The T4 virion has a prolate head and a contractile tail, terminating in a baseplate to which are attached six long tail fibers. The ~1200 Å-long and ~860 Å-wide head encapsulates a ~172 kbp DNA genome. It has icosahedral ends and an elongated mid-section (Fig. 1a). The structure of the T4 capsid had been previously determined using cryo-electron microscopy (cryo-EM).10,11 The T4 major capsid protein, the product of gene 23 (gp23), forms a hexagonal lattice characterized by the triangulation Tend = 13 (laevo) for the icosahedral caps and Tmid = 20 for the capsid midsection.10 One vertex of the T4 capsid is the portal for DNA entry, tail attachment and DNA ejection through the tail during the infection process. The portal vertex is formed by a dodecamer of gp20.12 The other 11 capsid vertices are occupied by pentamers of the special vertex protein gp24. The atomic structure of gp24 had been determined using X-ray crystallography.13 A homology model for the major capsid protein gp23 was built, based on the 21% sequence identity between gp23 and gp24.13 Both gp24 and gp23 have a fold similar to the bacteriophage HK97 capsid protein14 with an additional ‘insertion’ (‘I’) domain which forms characteristic protrusions on the T4 capsid surface.13 In the gp24 crystals, the linker regions between the I domain and the rest of the protein structure were disordered. It was proposed that in the virus the insertion domain in gp23 or gp24 molecules interacts with adjacent gp23 or gp24 molecules within the same capsomer, respectively, thus stabilizing the capsomer structure.13
Fig. 1.
Structure of the bacteriophage T4 head (Tend =13 laevo, Tmid = 20). (a) Shaded surface representation of the cryo-EM reconstruction viewed perpendicular to the five-fold axis. gp23 is shown in blue, gp24 is in magenta, Soc is in white, Hoc is in yellow, and the tail is in green10. (b) Close up view of two gp23 capsomers corresponding to the rectangle outlined in black in A. One Soc monomer is outlined by a blue ellipse; one Soc trimer is outlined by a red circle.
Two accessory proteins, Soc (small outer capsid protein with a 9 kDa molecular mass) and Hoc (highly antigenic outer capsid protein with a 40 kDa molecular mass), decorate the T4 capsid surface (Fig. 1). Hoc and Soc are dispensable for capsid formation and bind to the virus only upon capsid expansion in the last stage of capsid maturation6. There are 155 Hoc molecules per capsid. Each Hoc molecule binds to the center of a gp23 hexamer. These Hoc molecules have only a marginal effect on the capsid stability. However, Soc molecules help to stabilize T4 capsids at high temperature and against extremes of pH. This accounts for the almost 104-fold improved survival of wild-type T4 phage compared to Soc-defective phage at pH 10.6.6 However, at neutral pH and room temperature, the T4 particles are stable with or without the presence of Soc. Binding of Soc to T4 polyheads increases their denaturation temperature by 6°C.15 Thus, Soc prevents changes in the capsid structure as a result of unfavorable environments such as pH, temperature and other destabilizing forces.
The T4 capsid contains 870 copies of Soc which attach to the capsid surface at the interfaces between adjacent gp23 hexameric capsomers, thus perhaps serving as a molecular glue.10,11,16 Soc molecules do not bind at the interface between gp24 pentamers and gp23 hexamers nor around the gp20 portal vertex. Ultracentrifugation analysis showed that Soc is a monomer in solution.11 However, the cryoEM reconstruction shows trimeric and dimeric interfaces between Soc molecules on the virus capsid surface (Fig. 1b) and, of these, the trimeric interactions are more extensive (have a greater surface area of contact) than the dimeric interactions.
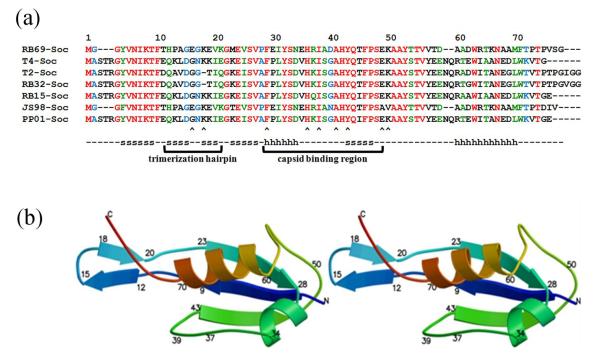
Many T4-like phages have homologous Soc-like molecules that stabilize the capsid (Fig. 2a). Other phages and some eukaryotic viruses such as adenoviruses and herpes viruses also have capsid decorative proteins that form trimers on the viral capsid, possibly involved in capsid stabilization5,17-23 (Table 1), but show no detectable sequence similarity to T4 Soc. In general, these proteins exhibit specificity towards the mature conformational states of their viral capsids.
Fig. 2.
Sequence alignment and structure of Soc. (a) Alignment of Soc sequences from T4-like bacteriophages. Completely conserved residues are colored red, nearly completely conserved residues are colored green and partially conserved residues are colored blue. β-strands and α-helices are marked with an s and h, respectively. Arrow heads indicate residues that have been mutationally characterized. (b) Stereo figure showing the structure of RB69 Soc molecule as a ribbon diagram. The polypeptide is colored with rainbow colors starting with blue at the amino end.
Table 1.
Examples of trimeric capsid stabilization proteins.
Soc might also help to stabilize the capsid against osmotic shock. Some osmotic shock resistant mutations of T4 were mapped to gene 2424 suggesting that the capsid disruption under osmotic stress is initiated at the pentameric vertices where there is no Soc.11 It was hypothesized that the osmotic-shock resistant mutations would allow Soc to bind around the pentameric vertices, thereby stabilizing the virus.11 Alternatively, the mutations in gp24 might cause reinforcement of intermolecular interactions at the capsid vertices without the requirement for Soc. Some T4 gp24-“bypass” mutants also have an osmotic-shock resistant phenotype. These mutants map onto the major capsid protein gene 23 and have amino acid substitutions that permit capsid assembly without the special vertex protein gp24.25,26 In these mutants the pentameric vertices are occupied by gp23, creating five potential binding sites for Soc molecules around each pentameric vertex as verified in one case.27 These additional Soc molecules might stabilize the capsid against osmotic shock.
Hoc and Soc can be fused with different foreign proteins and then assembled on the surface of T4-Hoc−Soc− mutant phage that lacks both Hoc and Soc.8,28-30 This has potential for the development of new vaccines.31 For example, large 90 kDa size anthrax toxin proteins attached to the N-terminus of Soc have been displayed on bacteriophage T4 capsid,7,9 which elicited strong lethal toxin neutralization titers in mice and rabbits31 (Rao, V. unpublished results).
Here we present the crystal structure of Soc protein from bacteriophage RB69, a close relative of T4. As the RB69 Soc has 47% sequence identity to T4 Soc, a reasonably accurate homology model could be built of T4 Soc. The crystal structure and the T4 Soc homology model agree well with the Soc density observed in the cryo-EM reconstruction of the T4 capsid.10 Fitting these atomic models into the cryo-EM reconstruction of the T4 capsid identified the region of the Soc molecule that interacts with two major capsid protein gp23 molecules, thereby gluing together adjacent capsomers. Further reinforcement of the T4 capsid is provided by trimeric interactions of the Soc molecules at the quasi-3-fold axes. Assembly of such triple clamps between hexameric capsomers forms a cage on the T4 capsid surface. Mutant Soc proteins with mutations located in the putative interface with gp23 and in the putative regions controlling the trimeric association between Soc subunits were found to reduce capsid stabilization, confirming the proposed triple clamping reinforcement mechanism.
Results and discussion
Soc binding and stabilization
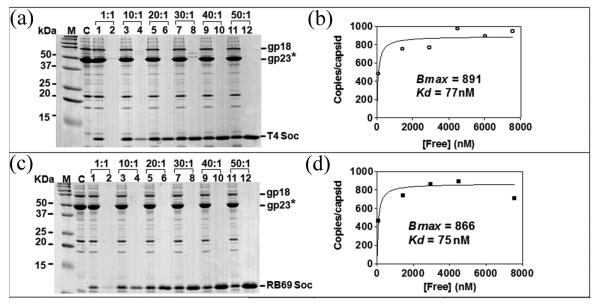
Binding of both T4 Soc and RB69 Soc to the T4 capsid reached a maximum at about 30:1 Soc molecules per binding site (Fig. 3). Both T4 and RB69 Soc proteins saturated the T4 capsid when the expected maximum of 870 copies per capsid had been reached. The binding affinities of Kd = ~75 nM is nearly the same for the T4 and RB69 Soc molecules. The Hoc−Soc− phage completely loses its viability at pH 10.6 (Table 2). On the other hand, as many as about 75% of Hoc−Soc− phage particles retain viability at this pH when saturated with RB69 or T4 Soc (Table 2). Thus, binding of either the T4 Soc or the RB69 Soc to T4 capsid stabilized the capsid. Furthermore, the stabilization properties of the T4 Soc are only slightly better than those of the RB69 Soc. The copy number of capsid-bound Soc remained essentially the same even when the phage lost ~30% viability in the presence of alkaline pH (Table 2; Fig. 4) suggesting that stabilization required more than simple binding of Soc to the capsid.
Fig. 3.
Binding of T4 and RB49 Soc proteins to Hoc−Soc− T4 capsids. Binding of Soc proteins to Hoc−Soc− phage and quantitative analyses were performed as described in Materials and Methods. Panels (a) and (c) lane M, molecular mass standards; C, control Hoc−Soc− phage; lanes 1, 3, 5, 7, 9, and 11 show capsid-bound Soc; lanes 2, 4, 6, 8, 10, and 12 show unbound Soc. The positions of gp18 (tail sheath protein), gp23* (major capsid protein), and Soc are labeled. The saturation binding curves (panels (b) and (d)) were constructed and the apparent Kd (association constant) and Bmax (maximum copy number per phage particle) were determined using the GraphPad PRISM-4 software.
Table 2.
Binding and stabilization properties of T4 and RB69 Soc proteins and their mutants.
| Phage | Saturation binding in vitro with purified recombinant Soc protein |
Kd (nM)a,b |
Bmax (Copies /capsid)a,b |
Phage titer (number of Soc molecules per capsid) |
||
|---|---|---|---|---|---|---|
| pH 7.0 | pH 10.4 | pH 10.6 | ||||
| Controls | ||||||
| T4 WT | - | - | - | 100% (855) | 83.5% (820) | 76.3% (847) |
| T4-Hoc−Soc− | - | - | - | 100% (0) | < 0.3% (0)c | < 0.3% (0)c |
| T4 Soc | 77 | 891 | 100% (813) | 82.3% (831) | 74.8% (816) | |
| RB69 Soc | 75 | 866 | 100% (887) | 78.7% (871) | 66.1% (872) | |
| T4 Soc + RB69 Soc | - | - | 100% (850) | 42% (820) | 30% (805) | |
| N- and C- terminus mutants | ||||||
| T4-Hoc−Soc− | T4 Soc-C3Δ | 75 | 857 | 100% (858) | 84.4% (827) | 74.1% (806) |
| T4Soc-N4Δ | 79 | 847 | 100%( 867 ) | 81.4% (846) | 76.8% (816) | |
| T4Soc-N10Δ | - | 0 | - | - | - | |
| Binding mutants | ||||||
| T4-Hoc−Soc− | RB69Soc-F29A | 963 | 861 | 100% (876) | 23.9% (572) | 2.0% (366) |
| RB69Soc- H36A/I38A |
213 | 876 | 100% (854) | < 0.3%(293)c | < 0.3% (204)c | |
| RB69Soc-I38A | 182 | 863 | 100% (797) | 26.9% (513) | 1.9% (484) | |
| RB69Soc-H36A | 94 | 820 | 100% (853) | 62.2% (707) | 43.4% (603) | |
| RB69Soc-Y43A | 179 | 887 | 100% (819) | 5.9% (534) | < 0.3% (472)c | |
| RB69Soc-A41Y | 343 | 883 | 100% (824) | 16.4% (585) | 10.9% (333) | |
| RB69Soc- E49A/K50A |
92 | 823 | 100% (857) | 83.3% (828) | 69.4% (835) | |
| Trimeric mutants | ||||||
| T4-Hoc−Soc− | RB69Soc-E16R | 80 | 836 | 100% (884) | 24.6% (864) | 6.9% (803) |
| RB69Soc-K18E | 103 | 841 | 100% (886) | 37.1% (878) | 18.1% (822) | |
On average Kd and Bmax values had errors of 35% and 5%, respectively.
The slight variations in Bmax values are within the range of error. Using Bmax = 870 (the actual number of Soc binding sites) produces a change in Kd values of no more than 20%.
No plaques were seen when compared to about 300 plaques in the pH 7 control.
Fig. 4.
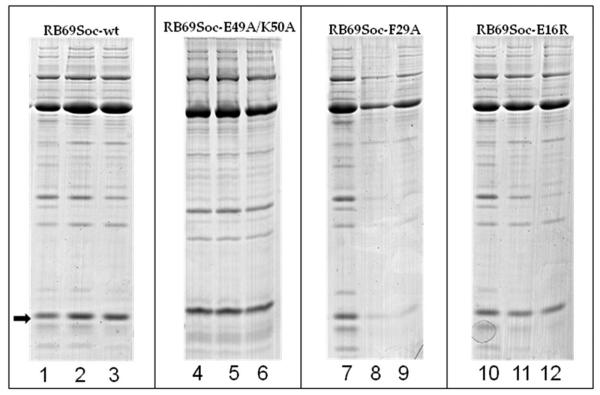
Examples of SDS gels showing the effect of alkaline treatment on the copy number of capsid-bound Soc (Table 2). To determine the copy number of capsid-bound Soc after treatment with alkaline pH (10.4 or 10.6), the purified Soc proteins were bound to T4 Hoc−Soc− phage and processed as described in Materials and Methods. The arrow shows the position of the Soc protein band. RB69Soc-wt is the positive control. RB69Soc-E49A/K50A has a mutation that does not affect capsid binding or stabilization. RB69Soc-F29A has a mutation in the capsid binding surface and binds only weakly to the capsid. Note the loss of capsid-bound Soc after alkaline pH treatment; the RB69Soc-E16R mutation weakens the trimeric interactions between Soc molecules. Note that the copy number of capsid-bound Soc molecules does not change after alkaline pH treatment. In lanes 1, 4, 7, and 10 the phage is at pH 7. In lanes 2, 5, 8, and 11 the phage was exposed to pH 10.4. In lanes 3, 6, 9 and 12, the phage was exposed to pH 10.6.
Structure of the Soc protein
The structure of RB69 Soc was determined to 1.9 Å resolution. The amino acid polypeptide chain of the RB69 Soc molecule could be traced, except for the one N- and the two C-terminal residues. RB69 Soc was found to be an elongated molecule with dimensions of 48 × 28 × 22 Å with an overall tadpole-like shape (Fig. 2b). The head of the tadpole contains a three-stranded β-sheet packed against two α-helices. The presence of helical regions in Soc was not expected based on secondary structural predictions.8 The tail of the tadpole is formed by a β-hairpin consisting of residues Thr 11 - Lys 21 and the extended C-terminus region Thr 72 – Val 78 (Fig. 2b). A structural homology search with the DALI algorithm32 indicated that the Soc protein has no obvious similarity to any other previously determined structure.
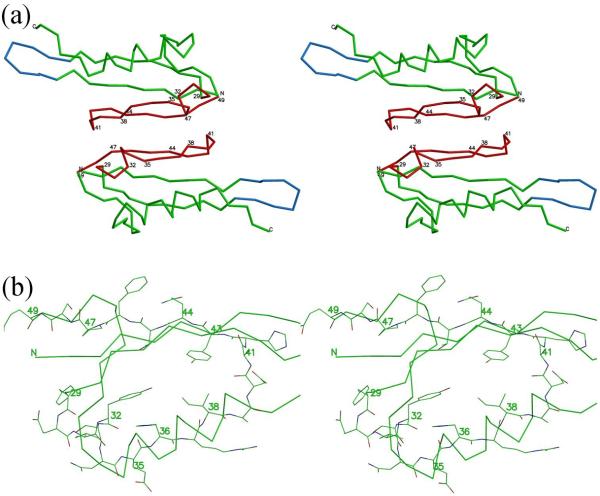
The hexagonal and orthorhombic crystal structures of RB69 Soc (Materials and Methods) have similar dimeric interactions between Soc molecules (Fig. 5). These dimeric interactions are formed by residues 29 – 47 creating a 610 Å2 area of contact.
Fig. 5.
Interface of Soc molecules in the crystal corresponds to the interface between Soc and gp23 in the virus. (a) Stereo diagram showing the Cα backbone trace of two Soc molecules related by two-fold non-crystallographic symmetry. Regions of the Soc molecule that were found to interact with the gp23 capsid shell on the virus are in red. Hairpins involved in the trimerization of the Soc molecules on the capsid surface are in blue. (b) Close view of residues 29-47 involved in interactions with the capsid showing all non-hydrogen atoms. Carbon atoms are green, oxygen atoms are red and nitrogen atoms are blue. Some other parts of the structure are shown as a Cα backbone trace.
Interface between Soc and the T4 capsid
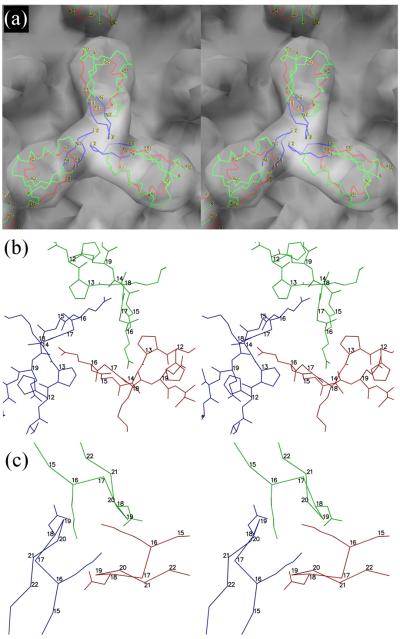
Fitting the RB69 Soc structure into the cryo-EM reconstruction of the T4 capsid showed that the tadpole-like heads of the Soc molecules point towards the quasi-two-fold axes relating adjacent gp23 hexamers, whereas the tails of the Soc molecules are close to the quasi-three-fold axes (Fig. 6). Most of the contacts between the Soc molecules and the T4 capsid are with the tadpole-like head. The RB69 Soc molecule amino acid residues 29 - 49 (the T4 Soc equivalent residues are 32 - 52) form the contact area. Each Soc molecule interacts with two gp23 capsomers, with RB69 residues 29 - 40 contacting one capsomer and residues 44 - 49 interacting with the adjacent capsomer, thus gluing together the adjacent capsomers, presumably leading to the stabilization of the T4 capsid. Residues 41 - 43 may be involved in interactions with either of the adjacent gp23 capsomers. The ~600 Å2 area of interaction of one Soc molecule with a two-fold-related Soc molecule in each of the two crystal structures is roughly the same surface as is used for binding one Soc molecule with gp23 molecules in the T4 capsid. This might be a coincidence but might also indicate that this region is a sticky region capable of easily binding another protein.
Fig. 6.
The Soc trimer. (a) Stereo diagrams of RB69 Soc fitted into the cryo-EM reconstruction of the T4 capsid, viewed down a quasi-three-fold axis. The three RB69 monomers are shown as a Cα backbone colored with the trimerization loop in blue, the gp23 binding region in red and the rest of the structure in green. (b) Stick drawing of the trimerization loop for RB69 with the three different molecules colored red, blue and green. (c) The same as B, but based on the T4 homology model showing the side chains for Lys16 and Asp18. The main chain is shown only as Cα atoms.
The Soc proteins of bacteriophages T4 and RB69 have only 47% sequence identity but the regions interacting with the gp23 protein shell are well conserved (Fig. 2a). Furthermore, unlike the Soc proteins, the major capsid proteins gp23 of T4 and RB69 have a high sequence identity of 94%. Together, this explains the similar affinity of the T4 and RB69 Soc proteins to the T4 capsid.
The correctness of the fitting of the Soc structure into the cryo-EM density (Materials and Methods) was confirmed by analyzing several RB69 Soc mutants with changes in the proposed Soc-capsid interface (Table 2). Each mutant was first examined by circular dichroism to determine that the mutations had no effect on folding (Supplementary Data Fig. S1). Mutations in the clamp region (F29A, H36A/I38A, I38A, A41Y, and Y43A) resulted in a higher binding constant, Kd, of Soc to the capsid (i.e. poorer binding) (Table 2). When these mutant Soc molecules were complexed with the T4-Hoc−Soc− capsid, they could not stabilize the capsid against alkaline pH. The most severe effects were seen with the double mutant H36A/I38A and the single mutant Y43A. These mutants had lost up to 99.7% of phage viability at pH 10.6. Loss of stabilization was due to the dissociation of the weakly bound mutant Soc molecules (Table 2; Fig. 4) and, therefore, loss of reinforcement provided by the Soc clamp. However no strict correlation was observed between Kd or Bmax with phage titer suggesting that different mutations differ in detail as to their impact on the stabilization mechanism.
The E49A/K50A double mutant has mutations on the face of Soc that faces a neighboring, two-fold related Soc molecule on the capsid surface, although there is a 7Å gap between them. As the double mutation has little effect on the stability (Table 2) of the capsid this is consistent with the correctness of the way in which the Soc structure was fitted into the cryoEM density.
The absence of a crystal structure of gp23 and its relatively low homology with gp24, created some uncertainty concerning the precise structure of the T4-Hoc−Soc− pseudo-atomic structure. Nevertheless, given the current homology model of gp23, the regions of gp23 that might be in the Soc binding site would be 267-273 and 463-470 on one hexamer and 257-261 and 493-497 on the adjacent hexamer.
The trimeric interface between Soc molecules bound to the capsid
In RB69 the interfaces between Soc molecules within a trimer on the capsid are formed by the loop containing residues 13 to 18 in the tail of the tadpole-like molecule (Fig. 6). The side chain of Glu-16 points to the neighboring Soc molecule and might be involved in forming a hydrogen bond. Mutating this residue to Arg (Table 2) resulted in 93% loss of phage viability at pH 10.6. Mutating Lys-18 to Glu also resulted in an 82% loss of phage viability at alkaline pH, although the side chain of Lys-18 probably does not interact with the neighboring Soc molecule. Whereas the E16R and K18E affected the stability of the virus, these mutations did not change the affinity of the Soc proteins to the capsid, nor did they cause significant loss of the bound Soc at alkaline pH (Table 2; Fig. 4). Therefore, for the mutants in the Soc trimerization region, unlike for the gp23-binding-site mutants described above, the loss of phage viability at high pH was not due to the dissociation of Soc molecules, but due to the lack of capsid reinforcement at the Soc trimerization sites. These results suggest that merely binding Soc molecules to the capsid is insufficient for capsid stabilization at high pH.
The T4 Soc trimeric interface is formed by loop residues 16 - 21 (Fig. 6) in which the side chain of Lys-16 is located close to the side chain of Asp-18 of the neighboring Soc molecule. Thus, the trimeric interface between T4 Soc molecules might be strengthened by hydrogen bonds or charge attraction. The amino acids that form the Soc trimerization interface, as opposed to the amino acids in the Soc-gp23 interface are poorly conserved between T4 and RB69 (Fig. 2a). Thus the Soc trimerization interactions would be perturbed in heterotrimers formed by RB69 and T4 Soc molecules, thereby reducing the virus stability. The importance of the Soc trimeric interactions for capsid stability could, therefore, be analyzed by binding a mixture of T4 and RB49 Soc to the same capsid. The results showed that these hetero phage particles lost 70% of their viability at alkaline pH, although greater than 90% of Soc molecules were still bound to the capsid (Table 2). Thus the trimeric interactions are important for capsid stabilization, but not for binding Soc to the capsid, when under stress such as alkaline pH.
The N- and C-termini of Soc
The 15 or so C terminal residues are more variable in their amino acid composition than the other residues in Soc. These residues (Figs. 2 and 6a) form in part the exterior surface of the virus which probably permits greater variability. The deletion of a few N- or C-terminal residues of T4 Soc did not affect the binding affinity or the stabilization properties of the protein (Table 2). This is not surprising as these residues are disordered in the RB69 crystal structure and, therefore, are probably flexible when bound to the virus. However, deletion of the 10 N-terminal residues of T4 Soc resulted in improper folding and loss of binding to the T4-Hoc−Soc− capsid (Table 2). The RB69 Soc crystal structure shows that such a deletion would partially destroy the β-sheet located in the tadpole-like head of Soc and, therefore, probably alter the conformation of the Soc binding region.
Mechanisms of T4 capsid stabilization
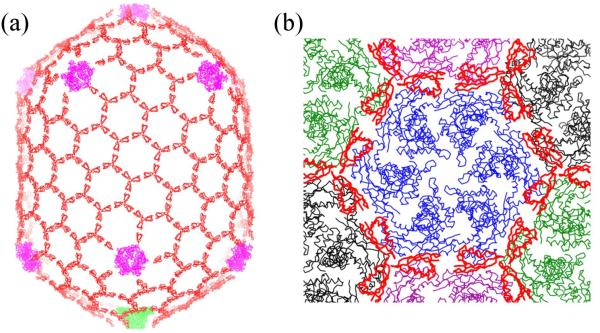
Bacteriophage T4 uses two mechanisms for capsid stabilization. The first mechanism reinforces the structure of the individual hexameric gp23 and pentameric gp24 capsomers. This is accomplished by the presence of an insertion domain, ‘I’, in gp23 and gp24 which interacts with neighboring subunits within the same capsomer.13 The second mechanism involves the Soc protein and is responsible for the reinforcement of the interactions between capsomers. Each Soc molecule binds to two adjacent gp23 molecules, thereby gluing two capsomers together. Trimerization of the Soc molecules results in the formation of a triple clamp (Fig. 7). Assembly of 270 such triple clamps between capsomers leads to the construction of a cage around the T4 capsid, further reinforcing the capsid structure. As Soc is a monomer in solution,11 Soc binding to capsid appears to be a necessary step for trimerization and cage construction. Here we show that both the glue between adjacent gp23 capsomers and the trimerization of Soc molecules (Fig. 7) are important for the T4 capsid stability at alkaline pH. Destabilization of either interaction by mutation resulted in loss of capsid stabilization and loss of phage infectivity after exposure to alkaline pH.
Fig. 7.
The stabilizing Soc cage. (a) The T4 head showing the Soc molecules in red, gp24 in magenta and the portal assembly in green. (b) View of one gp23 hexamer (blue) surrounded by six Soc trimers (red). Also shown are neighboring gp23 hexamers in green, black and magenta.
Evolution
Other bacteriophages that use special mechanisms for capsid stabilization are HK97 and λ. In bacteriophage HK97 inter-capsomer interactions are stabilized by chainmail chemical crosslinking and in bacteriophage λ interactions between capsomers are stabilized by the gpD protein. The location of gpD trimers on the bacteriophage λ capsid surface is close to the location of the chemical cross-links in HK97 capsid. Therefore it was hypothesized33 that gpD in phage λ plays a role analogous to the covalent cross-links in HK97 and that, in the process of evolution, the need for the decorative protein gpD was replaced by the chainmail covalent cross-linking (or vice versa). More probably the two different stabilizing procedures evolved independently from a more general, less stable virus. Bacteriophage T4 particles are stable in normal conditions without Soc, while bacteriophage λ mutants lacking gpD can stably accommodate only ~80% of the genome. In an ancestral T4 bacteriophage, Soc might have played a similar role in stabilizing the capsid as gpD does in present day bacteriophage λ. However, during the evolution of T4 its capsid might have developed the ability to survive at normal conditions without Soc. Probably the T4 major capsid protein interacts more effectively than the major capsid protein in bacteriophage λ with neighboring capsomers, allowing the T4 capsid to sustain the high DNA pressure inside the capsid without further stabilization by decorative proteins. Nevertheless, bacteriophage T4 has retained the dispensable Soc protein in its genome because Soc helps the virus to survive in extreme conditions.
Materials and Methods
Plasmids construction
The wild-type (WT) T4 Soc (T4Soc-wt), N-terminal 4, 10, and 20 amino acid deletion mutants (T4Soc-N4Δ, T4Soc-N10Δ, and Soc-N20Δ), and C-terminal 3, 10, and 20 amino acid deletion mutants (Soc-C3Δ, Soc-C10Δ, and Soc-C20Δ) were constructed by amplifying the respective DNA fragments using specific primers (Supplementary Data Table S1) and phage T4 DNA as template.
The WT RB69 Soc and the RB69 Soc mutants with mutations located in the putative Soc trimerization region, E16R and K18E, were constructed by amplifying the Soc coding sequence using WT or mutant primers and phage RB49 DNA as template (kindly provided by Dr. James Karam, Tulane University) (Supplementary Data Table S1). All other mutants (F29A, H36A, I38A, A41Y, Y43A, H36A/I38A and E49A/K50A) were constructed by the splicing-by-overlap extension (SOE) strategy described previously34 (Supplementary Data Table S1). The end primers contained sites for restriction enzymes NdeI and BamHI required for directional cloning into the phage T7 expression vector, pET28b.
The amplified DNAs were purified by agarose gel electrophoresis, digested with NdeI and BamHI, and ligated with the gel-purified pET-28b DNA digested with the same restriction enzymes. Insertion of Soc DNA results in in-frame fusion with an about 25 amino acid vector sequence containing a hexa-histidine sequence at the N-terminus that is followed by a thrombin cleavage site. The ligated DNA was transformed into E. coli XL10 Gold (Stratagene, La Jolla, CA), mini-prep plasmid DNAs were prepared by alkaline lysis, and the sequence of each clone was confirmed by DNA sequencing of the entire insert (Davis Sequencing, Inc. CA). The clones were then transformed into E. coli codon-plus BL21 (DE3) RIPL (Stratagene), which allowed over-expression of Soc proteins with a hexa-histidine tag at the N-terminus.
Purification of WT and mutant Soc proteins
The BL21 (DE3) RIPL cells harboring Soc clones were induced with 1 mM IPTG for 3 h at 30°C. The cells were harvested by centrifugation (4,000 × g for 15 min at 4°C) and resuspended in 50 ml of HisTrap binding buffer (50 mM Tris-HCl, pH 8.0, 20 mM imidazole, and 300 mM NaCl). The cells were lysed using French-press (Aminco, USA) and the soluble fraction containing the His-tagged fusion protein was separated from the inclusion bodies by centrifugation at 34,000 × g for 20 min. Constructs which expressed Soc in soluble form (all RB69 Soc constructs) were purified from the supernatant and the ones that expressed Soc in the insoluble fraction (T4 WT Soc, N4ΔSoc, and C3ΔSoc) were purified from the inclusion bodies.
To purify Soc from inclusion bodies, the pellet was dissolved in 50 ml of urea buffer (8 M Urea, 50 mM Tris-HCl, pH 8, 20 mM imidazole and 300 mM NaCl) and the sample was centrifuged (34,000 × g) to remove cell debris. The supernatant was loaded onto a HisTrap column pre-equilibrated with the same buffer. The protein was renatured by washing the column with a decreasing urea gradient (8 - 0 M) in binding buffer. The protein was then eluted with 20 - 500 mM linear imidazole gradient in the same buffer. The soluble Soc was purified from the cell-free extract using the same procedure except the loading and wash buffers contained no urea.
The peak fractions containing Soc were pooled and desalted by passing through the Hiprep 10/26 column and concentrated by Amicon Ultra-4 centrifugal filtration (5 kDa cut-off). The protein was then purified by size exclusion chromatography using Hi-Load 16/60 Superdex-200 (prep-grade) gel filtration column (GE Healthcare) in a buffer containing 20 mM Tris-HCl, pH 8 and 100 mM NaCl.
To cleave off the His-tag, the purified protein was incubated with thrombin (EMD Biosciences) (10 units of thrombin per 20 mg of Soc protein) in the cleavage buffer (20 mM Tris-HCl, pH 8.0, 150 mM NaCl, 20 mM imidazole, and 2.5 mM CaCl2) at 8°C for overnight. The cleaved protein was then separated from the His-tag, thrombin, and any uncleaved protein, by His-trap affinity chromatography. The protein recovered in the flow-through fraction was again purified by gel filtration on Superdex-200 column. T4 Soc precipitated at room temperature, even at a concentration of less than about 1 mg/ml. Therefore the protein was kept at a temperature below 8°C although, even at this low temperature, the precipitation was not completely avoided. Thus, T4 Soc samples were flash-frozen in liquid nitrogen for storage. RB69 Soc protein was easier to work with and to crystallize. This protein did not show visible precipitate at 4°C even after a month.
Circular dichroism (CD) spectra of the mutants were compared to the spectrum of the native RB9 Soc protein in order to confirm the proper fold of the RB69 Soc mutants (Supplementary Data Fig. S1). The CD experiments were performed on a Jasco-810 Spectropolarimeter. The spectra in the interval 190 – 260 nm were collected at 4°C with cell path of 0.2 mm. Concentration of the native RB69 sample was determined by its UV absorbance at 280 nm, the spectra of the mutant proteins were scaled to the spectrum of the native RB69 protein.
Crystallization and data collection
Crystallization trials for the RB69 Soc sample with the cleaved His-tag were performed at room temperature and at 4°C. The best crystals were obtained at 4°C, using the hanging drop vapor diffusion method. Drops containing 2 μl of the protein at ~10 mg/ml were mixed with 2 μl of reservoir solution containing 30% PEG 8000, 0.1 M imidazole pH 8.0, 0.2 M NaCl. The drops were equilibrated against 1 ml of the reservoir solution. These conditions produced two different kinds of crystals in the same drop. Their space groups were P61 with the unit cell parameters a = b = 49.12 Å, c = 198.66 Å and C2221 with the unit cell parameters a = 39.22 Å, b = 88.62 Å, c = 81.95 Å. The hexagonal crystals had four molecules in the asymmetric unit whereas the orthorhombic crystals had two molecules in the asymmetric unit. All X-ray datasets were collected at 100 K from crystals flash-frozen in a nitrogen stream. No cryo-protection of the crystals was necessary due to the high concentration of PEG in the mother liquor.
Diffraction datasets from crystals of both P61 and C2221 space groups were collected using a Rigaku R-AXIS-IV++ home diffraction equipment (Table 3). Crystals belonging to the hexagonal space group were soaked for 15 hours in a solution containing 3 mM uranyl acetate, 30% PEG 8000, 0.1 M imidazole pH 8.0, 0.2 M NaCl. A uranium derivative dataset was collected at the home source. Later, a higher resolution dataset was collected of a native hexagonal crystal at the BioCARS beamline 14C of the Advanced Photon Source in Argonne National Laboratory, Argonne, IL (Table 3). Some low resolution reflections in this dataset were overloaded. Measurements of these reflections were obtained from the native dataset collected at the home source by scaling this dataset to the higher resolution dataset collected at the Advanced Photon Source.
Table 3.
Crystal parameters and diffraction data statistics.
| Crystal | RB69 Soc native hexagonal form |
RB69 Soc hexagonal form, uranium derivative |
RB69 Soc native orthorhombic form |
RB69 Soc native hexagonal form Collected at Advanced Photon Source |
|---|---|---|---|---|
| Space group and unit cell parameters, Å |
P61 a=b=49.09 c=198.187 |
P61 a=b=48.85 c=198.21 |
C2221 a=39.22 b=88.62 c=81.95 |
P61 a=b=49.128 c=198.674 |
| Wavelength, Å | 1.5418 | 1.072 | ||
| Detector | Rigaku Raxis IV++ | MAR CCD 165 | ||
| Resolution, Å | 2.30 | 2.30 | 2.26 | 1.9 |
| No. of unique reflections |
11989 | 11827 | 7033 | 21202 |
| Redundancy, ratio | 7.7 | 7.4 | 5.4 | 7.4 |
| Completeness, % | 99.3 | 99.1 | 99.5 | 99.2 |
| Rsym, % | 8.0 | 7.5 | 5.8 | 5.2 |
| Mosaicity (°) | 0.72 | 0.44 | 0.53 | 0.37 |
| I/σ(I) | 35.5 | 40.3 | 21.2 | 33.2 |
| % of reflections with I/σ(I) < 2 |
9.4 | 7.9 | 8.0 | 2.5 |
| Last resolution shell (Å) |
2.38-2.30 | 2.38-2.30 | 2.33-2.26 | 1.97-1.90 |
| Rsym, last shell % | 44.2 | 36.3 | 28.3 | 20.0 |
| I/σ(I), outermost shell |
6.4 | 8.1 | 5.0 | 13.1 |
| % of reflections with I/σ(I) < 2, outermost shell |
23.2 | 18.5 | 18.7 | 8.1 |
The data sets were processed with the program HKL2000.35 The program TRUNCATE36,37 was used to calculate intensity statistics and structure factors.
Structure determination
The structure of the hexagonal crystals was solved with the Single Isomorphous Replacement with Anomalous Scattering (SIRAS) method using the native and uranium derivative datasets. The uranium positions and phases were determined with the program SOLVE38 using data to 2.3 Å resolution. Two heavy atom sites with low occupancies (0.2) were found in the asymmetric unit. Uranium atoms were bound to the Glu-24 side chains of two of the four RB69 Soc molecules in the asymmetric unit. The initial phases were improved by the solvent flattening and histogram matching techniques using the program RESOLVE.39 The resultant map was good enough to distinguish the molecular boundaries and to trace the polypeptide chains of the four molecules independently. The preliminary trace of Cα atoms was used to determine the non-crystallographic symmetry operators. The phases were subsequently improved and extended to 1.9 Å resolution by non-crystallographic symmetry averaging using the program RESOLVE.39 An automatic model building and refinement procedure using the RESOLVE and REFMAC40 programs was used to build a large part of the atomic model. Missing parts of the structure were build manually with the program MIFit41 and COOT.42 The model was subjected to several rounds of crystallographic refinement using the programs CNS43 and PHENIX_REFINE44,45 with manual rebuilding. The crystal structure of the orthorhombic crystals was determined by molecular replacement using the program MolRep,46 and the structure was refined using PHENIX_REFINE.44,45 The quality of the refined structures (Table 4) was examined with the program PROCHECK47 and POLYGON.48 The final atomic model of RB69 Soc contained residues 2 to 77. One N-terminal and two C-terminal residues of the protein were not visible in the electron density maps most probably due to their flexibility.
Table 4.
Crystallographic refinement statistics for RB69 Soc.
| Parameters | Crystal form | ||
|---|---|---|---|
| hexagonal | orthorhombic | ||
| Number of protein non-hydrogen atom | 2349 | 1298 | |
| Number of solvent atoms | 321 | 130 | |
| R-factor (working set) | 0.181 | 0.217 | |
| Rfree (test set, 5% of the data) | 0.229 | 0.263 | |
| Last resolution shell | 1.99 - 1.90 | 2.83 - 2.26 | |
| R-factor in the last resolution shell | 0.207 | 0.256 | |
| R-f Rfree in the last resolution shell | 0.279 | 0.351 | |
| AverageB-factor, (Å2) | 31.09 | 39.63 | |
| RMSD | bond lengths (Å) | 0.005 | 0.002 |
| bond angles (°) | 0.764 | 0.421 | |
| dihedral angles (°) | 15.091 | 11.3 | |
| planarity (°) | 0.004 | 0.003 | |
| chirality (°) | 0.056 | 0.03 | |
| peptide bond ω angle (°) |
5.8 | 5.8 | |
| Percentage of residues in different regions of Ramachandran plot (residues except Gly and Pro) |
most favored regions | 91.8 | 89.8 |
| additional allowed regions |
8.2 | 10.2 | |
| generously allowed regions |
0 | 0 | |
| disallowed regions | 0 | 0 | |
A homology model of the T4 Soc molecule (residues 6 - 79, out of total 80 T4 Soc residues) was built using the program MODELLER.49 The coordinates of RB69 Soc hexagonal and orthorhombic crystal forms have been deposited with the Protein Data Bank.
Fitting into the cryo-EM density
The Cα backbone of the RB69 X-ray structure and the T4 homology model of Soc were fitted into the five- and two-fold symmetry averaged cryo-EM density of the bacteriophage T4 capsid10 (Electron Microscopy Data Bank accession number EMD-1414) using the program COLORES50 and EMFIT.51 Initially the Soc molecules were fitted into the map region closest to the five-fold axis because this region of the reconstruction had the best quality. As these molecules are located close to the capsid vertex they are not involved in trimeric interactions with other Soc molecules. The positions of the centers of the Soc molecules obtained from COLORES were used in the fitting procedure performed using the program EMFIT. The best fit obtained using the EMFIT program agreed, within 1 Å and 5°, of the best fit obtained with the COLORES program. This fit resulted in residues 29 - 49 of RB69 or the T4 Soc equivalent residues 32 - 52, representing part of the head of the tadpole-like structure of Soc, making contact with two neighboring gp23 hexamers that form the capsid shell.
Subsequently, Soc molecules were fitted into the region of the map containing Soc molecules involved in trimeric interactions adjacent to the previously fitted Soc molecules around the capsid vertex. Three Soc molecules were fitted individually using the program COLORES.50 The best fit for two of the three molecules was consistent with the initially fitted Soc molecules in that residues 29 - 49 of RB69 Soc or the equivalent residues 32 - 52 of T4 Soc making contact with gp23 hexamers. However, the best fit obtained for the third molecule was different, probably due to lower quality of the map in this region. As symmetry of the T4 capsid shows that the three Soc molecules must be related by a three-fold axis, the final fit was accomplished by assuming an exact three-fold axis between the molecules while attempting to obtain the best fit to the density (Supplementary Data Table S2).
In vitro assembly of Soc molecules onto the T4 capsid
The purified RB69 Soc remains soluble at room temperature. In contrast, T4 Soc forms a precipitate at room temperature becoming soluble at 8°C. Therefore, all the RB69 and T4 binding experiments were conducted at 8°C.
About 2×1010 T4 Hoc−Soc− phage particles28 were mixed with Soc protein in a ratio varying from 1:1 to 50:1 per Soc binding site on the capsid. These experiments were performed in Protein LoBind tubes (Eppendorf, USA) using the assembly buffer of 50 mM sodium phosphate pH 7.0, 75 mM NaCl, and 1 mM MgSO4. After incubation for 40 min at 8°C, the phage particles were sedimented by centrifugation at 16,000 × g for 40 min. The phage pellet was washed twice with excess assembly buffer and the final pellet was resuspended in 10 μl of assembly buffer and then analyzed by SDS-PAGE. The gels were stained with Coomassie Blue R250 (Bio-Rad) and the protein bands were quantified by laser densitometry (PDSI, GE Healthcare). The density of the Soc, gp23* and gp18 bands were determined for each lane separately and the number of Soc molecules per capsid was calculated using the known copy numbers of gp23* or gp18. A curve relating the number of bound Soc molecules per capsid (Y) and the concentration of unbound protein in the binding reaction (X) (Fig. 3) was not sigmoidal, indicating that there is no cooperativity between neighboring Soc binding sites for either T4 Soc or RB69 Soc to the T4 capsid. The apparent Kd (association constant) and Bmax (maximum copies of Soc bound per capsid) were determined using the equation Y = Bmax*X/(Kd+X) as programmed in the GraphPad PRISM-4 software (GraphPad Software, San Diego California USA, www.graphpad.com).
Stability of phage T4 in alkaline pH
About 108 Hoc−Soc− phage particles were incubated with saturating amounts of Soc protein (~1 μg; Soc protein : Soc binding sites = ~400:1) in phage dilution buffer for 45 min at 8°C to allow binding of Soc to the phage capsid. The total volume of the binding reaction was 10 μl. The samples were then treated with 90 μl of 0.2 M-glycine/NaOH buffer, pH 10.4 or 10.6. As a control, the phage was treated with pH 7.0 assembly buffer. Additional controls in each experiment included WT T4 phage (positive control) and Hoc−Soc− T4 phage (negative control) treated under the same conditions. The samples were then incubated at 37°C for 1 h and neutralized to pH 7.0 by adding 900 μl of phage dilution buffer (26 mM Na2HPO4, 22 mM KH2PO4, 70 mM NaCl, and 1 mM MgSO4, pH 7.0). Small aliquots of these suspensions were used for determining the plaque forming titers of survived phage.
For determining the copy number of Soc remained on the phage following pH treatment, about 6×1010 Hoc−Soc− phages were incubated with Soc at 50:1 ratio for 40 min. After centrifugation and washing with excess phage dilution buffer, the pellet was resuspended in 24 μL of the same buffer and was divided equally into three tubes. Each tube was treated with 92 μl of a different buffer : phage dilution buffer (control); 0.2 M-glycine/NaOH buffer, pH 10.6 (final pH was 10.4); and 0.2 M-glycine/NaOH buffer, pH 10.8 (final pH was 10.6). As above, each experiment contained additional controls; WT T4 phage (positive control) and Hoc−Soc− T4 phage (negative control) treated with the same buffers. After incubation at 37°C for 1 h, phages were sedimented by high speed centrifugation (16,000 × g for 40 min) and subjected to SDS-PAGE (Fig. 4). The copy number of the Soc protein that remained bound to the capsid after each treatment was determined as described above.
Supplementary Material
Acknowledgements
Figures were prepared using the programs MOLSCRIPT,52 Raster3D53 and CHIMERA.54
We thank Tim Schmidt for supporting our home X-ray instrument and the staff of the beam line 14 (BioCars) of the Advanced Photon Source for support of our data collection. We thank Pavel Afonine for advice on the use of the PHENIX_REFINE program. We thank Mike Everly for allowing us to use the CD instrument in Purdue's Department of Chemistry and Stanislav Zakharov for help in the analysis of the CD spectra. We also thank Sheryl Kelly and Carol Greski for help in the preparation of the manuscript.
This work was supported by a National Science Foundation grant (MCB-0443899) to M.G.R. and a National Institutes of Health grant (NIAID, AI056443) to VBR.
Abbreviations used
- cryo-EM
cryo-electron microscopy
- gp
gene product
- ‘I’
‘insertion’
- Soc
small outer capsid protein
- Hoc
highly antigenic outer capsid protein
- LFn
lethal factor N-terminal domain
- WT
wild-type
- SOE
splicing-by-overlap
- CD
circular dichroism
- SIRAS
Single Isomorphous Replacement with Anomalous Scattering
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Accession numbers
The coordinates of RB69 Soc hexagonal and orthorhombic crystal forms have been deposited with the Protein Data Bank and have accession number 3IG9 and 3IGE, respectively.
Supplementary Data
Supplementary data associated with this article can be found, in the online version, at ____________________.
References
- 1.Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. American Society for Microbiology; Washington, D.C.: 1994. pp. 218–258. [Google Scholar]
- 2.Casjens S, Hendrix R. Control mechanisms in dsDNA bacteriophage assembly. In: Calendar R, editor. The Bacteriophages. Vol. 1. Plenum Press; New York: 1988. pp. 15–75. [Google Scholar]
- 3.Rao VB, Feiss M. The bacteriophage DNA packaging motor. Annu. Rev. Genet. 2008;42:647–681. doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 4.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage f29 portal motor can package DNA against a large internal force. Nature (London) 2001;413:748–752. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 5.Sternberg N, Weisberg R. Packaging of coliphage lambda DNA. II. The role of the gene D protein. J. Mol. Biol. 1977;117:733–759. doi: 10.1016/0022-2836(77)90067-5. [DOI] [PubMed] [Google Scholar]
- 6.Ishii T, Yanagida M. The two dispensable structural proteins (soc and hoc) of the T4 phage capsid: their properties, isolation and characterization of defective mutants, and their binding with the defective heads in vitro. J. Mol. Biol. 1977;109:487–514. doi: 10.1016/s0022-2836(77)80088-0. [DOI] [PubMed] [Google Scholar]
- 7.Fokine A, Bowman VD, Battisti AJ, Li Q, Chipman PR, Rao VB, Rossmann MG. Cryo-electron microscopy study of bacteriophage T4 displaying anthrax toxin proteins. Virology. 2007;367:422–427. doi: 10.1016/j.virol.2007.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li Q, Shivachandra SB, Zhang Z, Rao VB. Assembly of the small outer capsid protein, Soc, on bacteriophage T4: a novel system for high density display of multiple large anthrax toxins and foreign proteins on phage capsid. J. Mol. Biol. 2007;370:1006–1019. doi: 10.1016/j.jmb.2007.05.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Shivachandra SB, Leppla SH, Rao VB. Bacteriophage T4 capsid: a unique platform for efficient surface assembly of macromolecular complexes. J. Mol. Biol. 2006;363:577–588. doi: 10.1016/j.jmb.2006.08.049. [DOI] [PubMed] [Google Scholar]
- 10.Fokine A, Chipman PR, Leiman PG, Mesyanzhinov VV, Rao VB, Rossmann MG. Molecular architecture of the prolate head of bacteriophage T4. Proc. Natl. Acad. Sci. U.S.A. 2004;101:6003–6008. doi: 10.1073/pnas.0400444101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Iwasaki K, Trus BL, Wingfield PT, Cheng N, Campusano G, Rao VB, Steven AC. Molecular architecture of bacteriophage T4 capsid: vertex structure and bimodal binding of the stabilizing accessory protein, Soc. Virology. 2000;271:321–333. doi: 10.1006/viro.2000.0321. [DOI] [PubMed] [Google Scholar]
- 12.Driedonks RA, Engel A, ten Heggeler B, van Driel R. Gene 20 product of bacteriophage T4. Its purification and structure. J. Mol. Biol. 1981;152:641–662. doi: 10.1016/0022-2836(81)90121-2. [DOI] [PubMed] [Google Scholar]
- 13.Fokine A, Leiman PG, Shneider MM, Ahvaze B, Boeshans KM, Steven AC, Black LW, Mesyanzhinov VV, Rossmann MG. Structural and functional similarities between the capsid proteins of bacteriophages T4 and HK97 point to a common ancestry. Proc. Natl. Acad. Sci. U.S.A. 2005;102:7163–7168. doi: 10.1073/pnas.0502164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wikoff WR, Liljas L, Duda RL, Tsuruta H, Hendrix RW, Johnson JE. Topologically linked protein rings in the bacteriophage HK97 capsid. Science. 2000;289:2129–2133. doi: 10.1126/science.289.5487.2129. [DOI] [PubMed] [Google Scholar]
- 15.Ross PD, Black LW, Bisher ME, Steven AC. Assembly-dependent conformational changes in a viral capsid protein. Calorimetric comparison of successive conformational states of the gp23 surface lattice of bacteriophage T4. J. Mol. Biol. 1985;183:353–364. doi: 10.1016/0022-2836(85)90006-3. [DOI] [PubMed] [Google Scholar]
- 16.Steven AC, Greenstone HL, Booy FP, Black LW, Ross PD. Conformational changes of a viral capsid protein. Thermodynamic rationale for proteolytic regulation of bacteriophage T4 capsid expansion, cooperativity, and super-stabilization by soc binding. J. Mol. Biol. 1992;228:870–884. doi: 10.1016/0022-2836(92)90871-g. [DOI] [PubMed] [Google Scholar]
- 17.Gilcrease EB, Winn-Stapley DA, Hewitt FC, Joss L, Casjens SR. Nucleotide sequence of the head assembly gene cluster of bacteriophage L and decoration protein characterization. J. Bacteriol. 2005;187:2050–2057. doi: 10.1128/JB.187.6.2050-2057.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang L, Gilcrease EB, Casjens SR, Johnson JE. Highly discriminatory binding of capsid-cementing proteins in bacteriophage L. Structure. 2006;14:837–845. doi: 10.1016/j.str.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 19.Salas M, Vasquez C, Mendez E, Vinuela E. Head fibers of bacteriophage f29. Virology. 1972;50:180–188. doi: 10.1016/0042-6822(72)90358-3. [DOI] [PubMed] [Google Scholar]
- 20.Morais MC, Fisher M, Kanamaru S, Przybyla L, Burgner J, Fane BA, Rossmann MG. Conformational switching by the scaffolding protein D directs the assembly of bacteriophage fX174. Mol. Cell. 2004;15:991–997. doi: 10.1016/j.molcel.2004.08.023. [DOI] [PubMed] [Google Scholar]
- 21.van Oostrum J, Burnett RM. Molecular composition of the adenovirus type 2 virion. J. Virol. 1985;56:439–448. doi: 10.1128/jvi.56.2.439-448.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Parks RJ. Adenovirus protein IX: a new look at an old protein. Mol. Ther. 2005;11:19–25. doi: 10.1016/j.ymthe.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 23.Saad A, Zhou ZH, Jakana J, Chiu W, Rixon FJ. Roles of triplex and scaffolding proteins in herpes simplex virus type 1 capsid formation suggested by structures of recombinant particles. J. Virol. 1999;73:6821–6830. doi: 10.1128/jvi.73.8.6821-6830.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leibo SP, Kellenberger E, Kellenberger-van der Kamp C, Frey TG, Steinberg CM. Gene 24-controlled osmotic shock resistance in bacteriophage T4: probable multiple gene functions. J. Virol. 1979;30:327–338. doi: 10.1128/jvi.30.1.327-338.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson K, Condie B, Mooney DT, Doermann AH. Mutations that eliminate the requirement for the vertex protein in bacteriophage T4 capsid assembly. J. Mol. Biol. 1992;224:601–611. doi: 10.1016/0022-2836(92)90547-w. [DOI] [PubMed] [Google Scholar]
- 26.McNicol LA, Simon LE. A mutation which bypasses the requirement for p24 in bacteriophage T4 capsid morphogenesis. J. Mol. Biol. 1977;116:261–283. doi: 10.1016/0022-2836(77)90216-9. [DOI] [PubMed] [Google Scholar]
- 27.Fokine A, Battisti AJ, Kostyuchenko VA, Black LW, Rossmann MG. Cryo-EM structure of a bacteriophage T4 gp24 bypass mutant: the evolution of pentameric vertex proteins in icosahedral viruses. J. Struct. Biol. 2006;154:255–259. doi: 10.1016/j.jsb.2006.01.008. [DOI] [PubMed] [Google Scholar]
- 28.Shivachandra SB, Rao M, Janosi L, Sathaliyawala T, Matyas GR, Alving CR, Leppla SH, Rao VB. In vitro binding of anthrax protective antigen on bacteriophage T4 capsid surface through Hoc-capsid interactions: a strategy for efficient display of large full-length proteins. Virology. 2006;345:190–198. doi: 10.1016/j.virol.2005.10.037. [DOI] [PubMed] [Google Scholar]
- 29.Ren ZJ, Tian CJ, Zhu QS, Zhao MY, Xin AG, Nie WX, Ling SR, Zhu MW, Wu JY, Lan HY, Cao YC, Bi YZ. Orally delivered foot-and-mouth disease virus capsid protomer vaccine displayed on T4 bacteriophage surface: 100% protection from potency challenge in mice. Vaccine. 2008;26:1471–1481. doi: 10.1016/j.vaccine.2007.12.053. [DOI] [PubMed] [Google Scholar]
- 30.Jiang J, Abu-Shilbayeh L, Rao VB. Display of a PorA peptide from Neisseria meningitidis on the bacteriophage T4 capsid surface. Infect. Immun. 1997;65:4770–4777. doi: 10.1128/iai.65.11.4770-4777.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shivachandra SB, Li Q, Peachman KK, Matyas GR, Leppla SH, Alving CR, Rao M, Rao VB. Multicomponent anthrax toxin display and delivery using bacteriophage T4. Vaccine. 2007;25:1225–1235. doi: 10.1016/j.vaccine.2006.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holm L, Kääriäinen S, Rosenström P, Schenkel A. Searching protein structure databases with DaliLite v.3. Bioinformatics. 2008;24:2780–2781. doi: 10.1093/bioinformatics/btn507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lander GC, Evilevitch A, Jeembaeva M, Potter CS, Carragher B, Johnson JE. Bacteriophage lambda stabilization by auxiliary protein gpD: timing, location, and mechanism of attachment determined by cryo-EM. Structure. 2008;16:1399–1406. doi: 10.1016/j.str.2008.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rao VB, Mitchell MS. The N-terminal ATPase site in the large terminase protein gp17 is critically required for DNA packaging in bacteriophage T4. J. Mol. Biol. 2001;314:401–411. doi: 10.1006/jmbi.2001.5169. [DOI] [PubMed] [Google Scholar]
- 35.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Meth. Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 36.Collaborative Computational Project Number 4 The CCP4 suite: programs for protein crystallography. Acta Crystallogr. sect. D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 37.French S, Wilson K. On the treatment of negative intensity observations. Acta Crystallogr. sect. A. 1978;34:517–525. [Google Scholar]
- 38.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallogr. sect. D. 1999;55:849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Terwilliger TC. Maximum-likelihood density modification. Acta Crystallogr. sect. D. 2000;56:965–972. doi: 10.1107/S0907444900005072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr. D Biol. Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 41.McRee DE. Differential evolution for protein crystallographic optimizations. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2276–2279. doi: 10.1107/S0907444904025491. [DOI] [PubMed] [Google Scholar]
- 42.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 43.Brünger AT, Adams PD, Clore GM, DeLano WL, Gros P, GrosseKunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, Read RJ, Rice LM, Simonson T, Warren GL. Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. sect. D. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 44.Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D Biol. Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- 45.Afonine PV, Grosse-Kunstleve RW, Adams PD. The Phenix refinement framework. CCP4 Newsletter. 2005 July;42 Contribution 8. [Google Scholar]
- 46.Vagin A, Teplyakov A. MOLREP: an automated program for molecular replacement. J. Appl. Crystallogr. 1997;30:1022–1025. [Google Scholar]
- 47.Laskowski RA, Moss DS, Thornton JM. Main-chain bond lengths and bond angles in protein structures. J. Mol. Biol. 1993;231:1049–1067. doi: 10.1006/jmbi.1993.1351. [DOI] [PubMed] [Google Scholar]
- 48.Buehler A, Urzhumtseva L, Lunin VY, Urzhumtsev A. Cluster analysis for phasing with molecular replacement: a feasibility study. Acta Crystallogr. D Biol Crystallogr. 2009;65:644–650. doi: 10.1107/S090744490900969X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sali A, Potterton L, Yuan F, van Vlijmen H, Karplus M. Evaluation of comparative protein modeling by MODELLER. Proteins. 1995;23:318–326. doi: 10.1002/prot.340230306. [DOI] [PubMed] [Google Scholar]
- 50.Wriggers W, Milligan RA, McCammon JA. Situs: a package for docking crystal structures into low-resolution maps from electron microscopy. J. Struct. Biol. 1999;125:185–189. doi: 10.1006/jsbi.1998.4080. [DOI] [PubMed] [Google Scholar]
- 51.Rossmann MG. Fitting atomic models into electron microscopy maps. Acta Crystallogr. sect. D. 2000;56:1341–1349. doi: 10.1107/s0907444900009562. [DOI] [PubMed] [Google Scholar]
- 52.Kraulis P. MOLSCRIPT: a program to produce both detailed and schematic plots of protein structures. J. Appl. Crystallogr. 1991;24:946–950. [Google Scholar]
- 53.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphics. Meth. Enzymol. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 54.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.