Abstract
The explosion of protein sequences deduced from genetic code has led to both a problem and a potential resource: Efficient data use requires interpreting the functional impact of sequence change without experimentally characterizing each protein variant. Several groups have hypothesized that interpretation could be aided by analyzing the sequences of naturally-occurring homologues. To that end, myriad sequence/function analyses have been developed to predict which conserved, semi-, and non-conserved positions are functionally important. These positions must be discriminated from the non-conserved positions that are functionally silent. However, the assumptions that underlie sequence analyses are based on experimental results that are sparse and usually designed to address different questions. Here, we use three homologues from a test family common to bioinformatics – the LacI/GalR transcription repressors – to test a common assumption: If a position is functionally important for one family member, it has similar importance in all homologues. We generated experimental sequence/function information for each non-conserved position in the 18 amino acids that link the DNA-binding and regulatory domains of three LacI/GalR homologues. We find that the functional importance of each position is preserved among the three linkers, albeit to different degrees. We also find that every linker position contributes to function, which has two-fold implications. (1) Since the linker positions range from highly to semi- to non-conserved, and contribute to affinity, selectivity, and allosteric response, we assert that sequence/function analyses must identify positions in the LacI/GalR linkers to be qualified as “successful”. Many analyses overlook this region, since most of the residues do not directly contact ligand. (2) No position in the LacI/GalR linker is functionally silent. This finding is inconsistent with another underlying principle of many analyses: Using sequence sets to discriminate important from non-contributing positions obligates silent positions, which denotes that most homologues tolerate a variety of amino acid substitutions at the position without functional change. Instead, additional combinatorial mutants in the LacI/GalR linkers show that particular substitutions can be silent in a context dependent manner. Thus, specific permutations of sequence change (rather than change at silent positions) would facilitate neutral drift during evolution. Finally, the combinatorial mutants also reveal functional synergy between semi- and non-conserved positions. Such functional relationships would be missed by analyses that rely primarily upon co-evolution.
Keywords: LacI/GalR repressor proteins, specificity determinants, protein engineering, bioinformatics, protein evolution
Introduction
Protein sequence alignments are omnipresent in biological research. A common use is for identification of conserved positions (e.g. 1; 2; 3; 4; 5; 6); if an uncharacterized protein is homologous to known proteins, one might infer a similar gross structure or function (reviewed in 7). Recently, an increasing number of sequence analyses (e.g. 5; 8; 9; 10; 11; 12; 13; 14; 15; 16; 17) attempt to identify which nonconserved positions are important to protein function; these sites are locationsa where amino acids may be substituted to derive unique variations of the overall function of the protein family. Successful recognition of these positions has great potential for guiding protein engineering and will be key to deducing the biological effects of polymorphisms.
Identifying the locations of important nonconserved sites requires more than a sequence alignment: The relevant positions must be discriminated from functionally-silent nonconserved positions, where substitution allows neutral drift during evolution18. To separate the two classes of nonconserved positions, analyses like those referenced above employ various statistical methods, in combination with various structural or functional information. In following the development and proliferation of these analyses, we have noted several features in their design and application that impede their usefulness. We recently discussed the importance of carefully defining subfamilies of protein sequences before carrying out these analyses19. Here, we address two additional concerns: First, many analyses assume that each sequence position is either functional or silent for all proteins within a family. We are not aware that this has been demonstrated systematically using multiple homologues. Thus, we are experimentally determining whether functional importance is preserved for given positions in the LacI/GalR transcription regulators, a family commonly used to benchmark prediction algorithms. Second, the performance of sequence-based algorithms is often evaluated by comparing the locations of predicted sites to known binding sites. This criterion neglects the fact that other regions – such as sequences that link domains – are vital to full protein function.
Here, we report the results of experiments that directly address the first concern – whether the functional importance of nonconserved positions is preserved in the LacI/GalR family. To highlight our second concern, we focus experiments on ~18 amino acids which link the DNA-binding and regulatory domains of the LacI/GalR proteins (Figure 1A, B). We previously found that amino acid substitutions at nonconserved linker positions can alter DNA binding affinity, selectivity, and allosteric response to a signaling molecule20.
Figure 1. Repressor structures.
(A, B) The panels show two views of the LacI dimer. Figures were made with Chimera 64 using the pdb file 1efa 42 The close-up view in (B) is rotated ~90 degrees relative to that in (A). DNA is at the top of each structure (blue sticks) and is bound to the DNA-binding domain of each protein. Magenta highlights the N-linker (LacI 45–49); green, the hinge helix (LacI 50–58); and yellow, the C-linker (LacI 59–62). The side chains of all linker residues are shown in wire frame; note the contacts formed between two linker helices of a dimer and between the linker and the large regulatory domains. The LacI anti-inducer ligand is depicted in black space-filling representation in the ligand-binding pockets between the two subdomains of the regulatory domains. (C) Cartoons showing the domain structures of LacI, LLhP, LPhP, LLhG, and LGhG. Ovals represent the DNA-binding domains, rectangles portray the linkers, and the large shapes depict the regulatory domains. Cyan indicates the sequence is from LacI; dotted pink indicates the sequence is from PurR, and striped green indicates that the sequence is from GalR. Chimera nomenclature used the following scheme: The first letter in the name indicates the source of the DNA binding domain (here always LacI), the second letter (followed by “h”) indicates the source of the linker, and the last capital letter indicates the source of the regulatory domain.
In designing the previous and current experiments, we wished to compare the functional outcomes of analogous mutations in several homologues. Such studies could be carried out using a set of natural LacI/GalR proteins, but interpretation would be complicated by the fact that each protein binds a different DNA sequence. To standardize comparisons, we have engineered a series of transcription repressors that comprise identical DNA binding domains but different regulatory domains (Figure 1C). The intervening linker that joins the 2 domains can be derived from various homologues. Using these chimeras, we can directly compare the effect of linker mutations on the function of the common DNA-binding domain.
Previously, we reported experimental results for two chimeras comprising the LacI DNA-binding domain, the LacI linker, and either the PurR or the GalR regulatory domain (respectively named LLhP and LLhG 19; 21, Figure 1C). Using a “host-guest” strategy, we substituted up to thirteen amino acids into several nonconserved linker positions, which resulted in a range of functional changes. Although their regulatory domains differ, LLhP and LLhG have the same linker sequence – that of LacI. We have now created LPhP (with the PurR linker and regulatory domain) and LGhG (with the GalR linker and regulatory domain), which provide two additional contexts for examining the functional roles of nonconserved positions (Figure 1C). In the current work, we use the strategy of exchanging single and combinatorial substitutions between the PurR or GalR and LacI linker sequences of the LPhP/LLhP and LGhG/LLhG pairs. This approach allows us to assess both (1) the functional importance of various positions in different linker sequences, and (2) whether a particular amino acid substitution results in the same type of functional change when other positions in the linker are varied.
RESULTS
The common function of the LacI/GalR transcription regulators is to bind operator DNA and thereby modulate transcription of downstream genes. Many family members – including the lactose repressor protein (LacI), the purine repressor protein (PurR) and the galactose repressor protein (GalR) – repress transcription (reviewed in 22). DNA binding, and thus repression, is modulated when the repressor protein binds a small metabolite. For both LacI and GalR, repression is relieved by binding inducer molecules, which are respectively allolactose (or the gratuitous inducer isopropyl-β-D-thiogalactoside [IPTG]) 23; 24 and galactose (or fucose) 25. For PurR, repression is enhanced by binding guanine or hypoxanthine 26.
When we previously created the chimeric repressors from the LacI DNA-binding domain and homologous regulatory domains, we determined that the regulatory domain dictates the direction of response to binding effector. For example, LLhP (a chimera with the LacI DNA-binding domain and linker and the PurR regulatory domain), is co-repressed; whereas the LLhG chimera with the GalR regulatory domain is induced, each by their respective ligands 19; 21. In this study, we expected and observed similar effector responses for LPhP and LGhG chimeras.
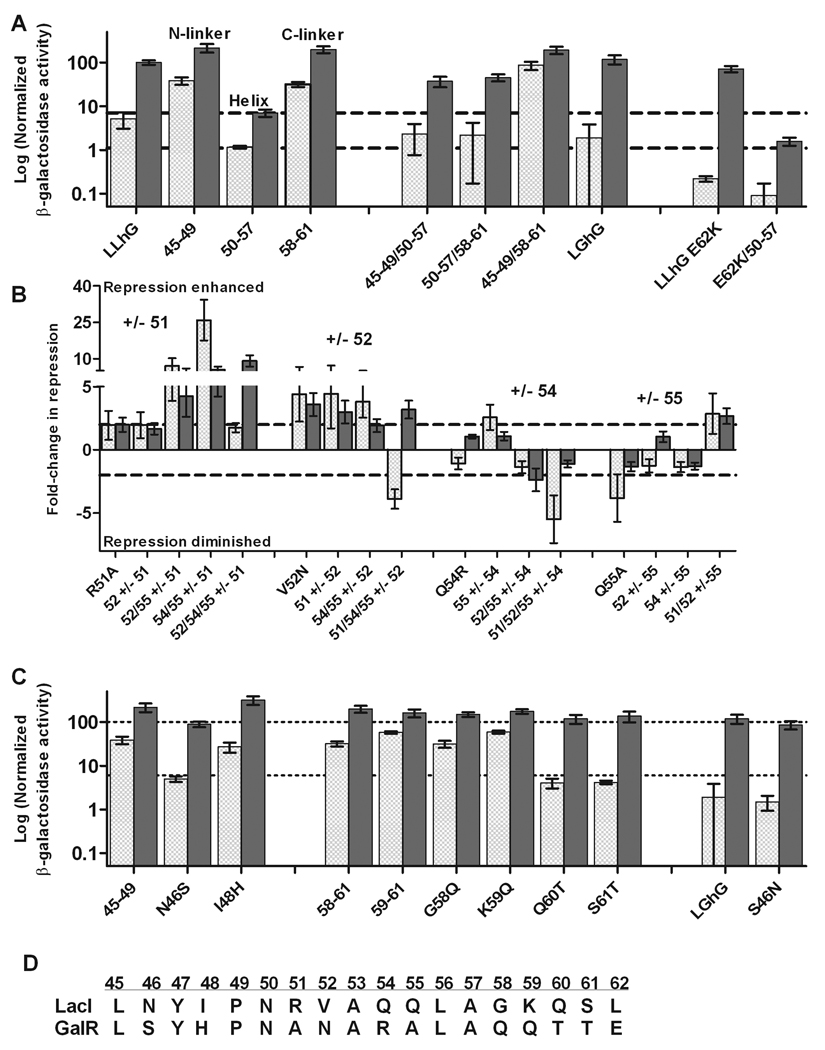
LPhP and LGhG were constructed in order to compare the functional contributions of PurR and GalR linker positions to those in the LacI linkers in LLhP and LLhG19; 21. In Materials and Methods, we present our criteria for designing the chimeric repressors and for interpreting in vivo repression resultsb. We found that the chimera LPhP was a very poor repressor, about 100-fold weaker than was previously shown for LLhP21, with reporter activities that were about the same as “empty” plasmid pHG165 (Figure 2A). In contrast, we found that LGhG was a stronger repressor than LLhG19. This result can be better seen in the picture of a plate assay (lighter blue colonies) than in the liquid culture assay, because LGhG has high error in the latter (Figure 2B and C). Differences between the plate and liquid culture assays (as well as high error) for LGhG are reminiscent of, although not as severe as, the toxicity previously seen for several LLhG variants19.
Figure 2. Repression by LLhP, LPhP, LLhG, and LGhG.
(A, B) In liquid culture assays, low values of β-galactosidase activity correlate with increased repression. Light gray hatched bars represent values determined in the absence of effector; checkered bars represent values determined from cells grown in the presence of corepresssor; and solid gray bars represent values determined from cells grown in the present of inducer. In order to facilitate comparison to previous works, the PurR and GalR chimeras were normalized to different standards. The two normalization scales differ ~50-fold. Average values shown are from 3–6 independent assays (each with duplicate samples from 1× and 2× cell culture); error bars represent one standard deviation. (A) LLhP and LPhP were normalized to LacI + IPTG, the value of which was set to 100 (heavy dashed line). pHG165 is the plasmid without a repressor gene. To aid visual inspection, dotted lines indicate the levels of “no repression” by pHG165 and strong repression by LLhP+co-repressor. (B) LLhG and LGhG variants were normalized to LLhG+inducer fucose, which was set to 100 (heavy dashed line). pHG165a is the plasmid with a knocked-out lacO1 site without a repressor gene (See Methods and footnote f.) To aid visual inspection, dotted lines indicate the level repression by LLhG. (C) In plate assays, lighter colonies correspond to stronger repression; darker blue colonies correspond to weaker repression. The images were created by scanning the plates to create a tif file. The picture was processed by varying the contrast and brightness of the whole file so that it matched visual inspection. Adobe Photoshop was used to hide hand-written labels around the circumference and to add typed labels.
Functional differences between pairs of chimeras that differ only in their linker sequences (LLhP/LPhP or LLhG/LGhG) must arise from sequence differences in linker positions. Previously, we found that at least 4 nonconserved positions contribute to function in LLhP 21 and at least 9 contribute to LLhG function 19. We now asked whether the same and/or additional nonconserved positions lead to functional differences between the LPhP/LLhP and LGhG/LLhG pairs. Our initial strategy was to determine which residues of the LPhP or LGhG linkers must be exchanged with the LacI sequence in order to recapitulate LLhP or LLhG function. Our expectation was that exchanging important nonconserved residues would impact in vivo repression. In contrast, exchanging amino acid residues at a “silent” position would have no effect on repression.
Results for the interconversion of LLhP/LPhP and LLhG/LGhG are presented below. During the course of this work, we created various combinatorial substitutions of linker residues, which allowed us to monitor whether the outcome of a particular substitution is the same (independent) or different (synergistic) in multiple sequence contexts. If independent, one would expect both of the following to hold: (1) the substitution would have very similar outcomes in different combinations of linker sequences; and (2) mirrored exchanges between chimera pairs would have opposite effects - for example LLhG V52N would have the opposite effect of LGhG N52V. If a substitution is synergistic with other linker positions, one or both of the above patterns will be absent. Data in Figure 3 and Figure 4 are plotted to facilitate comparing the effects of a particular substitution in multiple linker contexts.
Figure 3. Repression by LPhP variants.
The numbers in the X-axis names indicate which positions have been exchanged from the PurR to the LacI residue (Table 1). The complete linker amino acid sequence for each LPhP variant is given in Supplemental Table 1. (A) Results from liquid culture assays of LPhP variants. The normalization scale is that of Figure 2A. Dotted lines are to aid visual inspection by indicating the levels of LLhP repression +/− corepressor. (B, C) Fold-change in repression for given substitutions (indicated on the plots), calculated by comparing repression for the variant without the substitution. For example, the fold-change of “57–61+/− 48” is calculated from the ratio of LPhP/48/57–61 and LPhP/57–61 repression. Positive values indicate enhanced repression; negative values indicate diminished repression; data used to calculate these values are in Supplemental Table 2. Dashed lines indicate 2-fold change, below which differences cannot be reliably discriminated. All repressor variants are based on LPhP/57–61, except the indicated LLhP “mirror” substitutions.
Figure 4. Repression by LLhG/LGhG variants.
The numbers in the X-axis names indicate which positions have been exchanged between the GalR and LacI sequences (Table 1). The complete linker amino acid sequence for each LLhG/LGhG variant is given in Supplemental Table 3. (A) Results from liquid culture assays of LLhG variants with LGhG linker segments. The normalization scale is that of Figure 2B. Dotted lines are to aid visual inspection and indicate the levels of LLhG/50–57 repression +/− inducer. (B) Fold-change in repression for LLhG helix combinatorial substitutions, calculated by comparing repression for the variant without the substitution. For example, the fold-change of “52+/−51” is calculated for site 51 from the ratio of LLhG/51/52 vs LLhG/52 repression; in the second group, the same variant is designated as “51+/−52” and the fold-change is calculated for site 52 from LLhG/51/52 vs LLhG51. Positive values indicate enhanced repression; negative values indicate diminished repression; data used to calculate these values are in Supplemental Table 4. Dashed lines indicate 2-fold change, below which differences cannot be reliably discriminated. (C) Results from liquid culture assays of LLhG N- and C-linker variants; repression by LGhG and LGhG S46N, which is one of the few LGhG variants that could be reliably quantitated in the liquid culture assay. Dashed lines indicate the repression levels of “wild-type” LLhG, +/− inducer.
Interconversion of LPhP and LLhP
In addition to the conserved YxPxxxAxxL motif (described Materials and Methods), the LacI and PurR linker sequences contain identical residues at sites 45 and 52; position 62 is the same in LLhP and LPhP. Thus, 11 positions might contribute to the functional differences between LLhP and LPhP.
Exchanging LPhP positions 57–61 allows measurable lacO1 repression
To determine the functional contributions from nonconserved LPhP linker positions, we would ideally detect either enhanced or diminished repression as a consequence of amino acid substitution. However, if unmodified LPhP contained a “lethal” substitution that always precluded repression, we would never be able to detect the contributions from other positions. Indeed, the amino acid difference at position 57 (Table 1) might be lethal: This nonconserved residue directly contacts DNA and the nature of this side chain is important for DNA-binding in both LacI and PurR 27; 28. Thus, PurR-derived K57 in LPhP is probably not appropriate for binding lacO1. However, the K57A mutation did not restore any lacO1 repression to LPhP (Supplementary Figure 1), suggesting that this position alone is insufficient to account for differences with LLhP.
Table 1.
Linker sequences and sequence entropya
| 45 | 46 | 47 | 48 | 49 | 50 | 51 | 52 | 53 | 54 | 55 | 56 | 57 | 58 | 59 | 60 | 61 | 62 | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| LacI | ℒ | 𝒩 | 𝒴 | ℐ | 𝒫 | N | R | V | A | Q | Q | L | A | G | K | Q | S | L |
| PurR | ℒ | ℋ | 𝒴 | 𝒮 | 𝒫 | S | A | V | A | R | S | L | K | V | N | H | T | K |
| GalR | ℒ | 𝒮 | 𝒴 | ℋ | 𝒫 | N | A | N | A | R | A | L | A | Q | Q | T | T | E |
| SEbs | 0.29 | 0.70 | 0.00 | 0.82 | 0.00 | 0.25 | 0.60 | 0.66 | 0.00 | 0.34 | 0.69 | 0.00 | 0.65 | 0.98 | 0.68 | 0.93 | 0.37 | 0.94 |
Different font styles are used to indicate the N-linker (italic font, dotted underlined numbers), hinge helix (normal font, solid underlined numbers), and C-linker (bold font, dashed underlined numbers). Note that the hinge helix in PurR is one residue longer than the helix in LacI; we presume this is true for LLhP and LPhP; the structure is unknown for the GalR linker. Gray shading indicates positions that are conserved. In addition to the YxPxxxAxxL motif, position 45 is conserved as L in the three proteins.
SE is sequence entropy of the position calculated from 1082 nonredundant sequences in the YxPxxxAxxL subfamily of the LacI/GalR proteins. This parameter describes the frequency and randomness for the occurrence of different amino acids at each position. Calculations are described in Methods; a value of 0.0 is perfectly conserved; a value of 1.3 would indicate equal representation of all 20 amino acids.
We devised several additional strategies to identify an LPhP construct that was at least partially-functional. Three were unsuccessful (Supplementary Figure 1 and Supplementary Table 1): (1) exchanging combinations of the three semi- and non-conserved positions that directly contact DNA (50, 54, and 57); (2) exchanging combinations of the 4 nonconserved positions previously known to diminish repression in LLhP (48, 55, 58, and 61) 21; and (3) testing whether substitutions at the end of the C-linker (position 62) would enhance LPhP/K57A repression, as it did in LLhG 19. Finally, noting the extreme impact of C-linker substitutions on LLhP 20; 21, we exchanged the entirety of positions 58–61 plus K57A (hereafter referred to as “LPhP/57–61”). Intermediate repression was gained, 10-fold more than LPhP but 10-fold less than LLhP (Figure 3A). Like “wild-type” LLhP, the LPhP/57–61 allosteric response in the presence of adenine was about 2-fold. Repression from lacO1 appears to require that position 57 be exchanged to the LacI sequence; exchanging the C-linker alone (LPhP/58–61) does not result in measurable repression (Figure 3A).
Substitutions of N-linker and helix positions
We next used LPhP/57–61 as a background for exploring the functional contributions of nonconserved linker positions in the N-linker (46, 48) and helix (50, 51, 54, and 55). Our strategy was to exchange multiple combinations of residues between the LacI and PurR sequences and to look for consistent effects (or lack thereof) on repression, as well as to compare results to the “mirror” substitution in LLhP.
Since the I48S and Q55S substitutions had a big impact on LLhP function21, we started with opposite substitutions of S48I and S55Q in LPhP/57–61, alone and in combination with other nonconserved helix positions. In Figure 3B–C, results are reported as “fold-change” relative to the construct without the substitution (i.e. the ratio in repression for LPhP/48/57–61 to LPhP/57–61; values used to calculate fold-change are in Supplementary Table 2). The S48I substitution enhanced repression relative to LPhP/57–61 and three additional variants (Figure 3B), albeit to different extents (1.5 to 15-fold). These results mirrored the effects previously seen for LLhP I48S, which showed ~5-fold diminished repression (Figure 3B 21). For S55Q substitutions, most LPhP constructs showed little change in repression (within two-fold of the parent construct; Figure 3C, dashed lines). The exception was LPhP/57–61 (+) co-repressor, where adding S55Q enhanced repression by 5-fold. The change in the “mirror” LLhP substitution Q55S was more pronounced, with repression diminished ~10-fold (21, Figure 3C).
The LPhP/48/55/57–61 variant has repression characteristics that are very similar to LLhP (Figure 3A), and the two proteins differ by only 4 amino acids. This raised the question as to whether any of the four remaining nonconserved positions (46, 50, 51, 54) are functionally silent, and can be substituted without altering function. Position 54 directly contacts DNA. Adding the R54Q mutations to LPhP/48/55/57–61 moves the chimera one residue closer to the sequence of LLhP and should restore a “native” interaction between the linker and lacO1. Thus, we expected that this substitution would have little functional effect or enhanced repression. Paradoxically, R54Q diminished repression relative to LPhP/48/55/57–61, as well as in two other linker backgrounds (Figure 3C). Therefore, at least one of the three remaining nonconserved positions (46, 50, and 51) must also be exchanged to facilitate the good repression of LLhP. We exchanged each of the three remaining positions, again expecting enhanced repression as the chimera became more like the LLhP sequence. On the contrary, individually substituting positions 46, 50c, and 51 weakened repression relative to LPhP 48/54/55/57–61, by about 3-, 30- and 3-fold, respectively (Figure 3 B, C).
These results lead us to conclude that the functional contributions of helix positions 51 and 54 are synergistic with each other. First, their respective substitutions diminished repression in LPhP 48/54/55/57–61, even though each makes the chimera more similar to the sequence of the strong repressor LLhP. Second, the mirror substitutions in LLhP for sites 51 and 54 also diminished repression (Figure 3 B and C). We cannot ascertain whether position 50 also participates in the synergy, since LPhP 48/50/54/55/57–61 does not show protein in a DNA-pulldown assay (see footnote c). Positions 51 and 54 flank two residues that are conserved as V and A in LacI and PurR. Either the PurR sequence (A51V52A53R54) or the LacI sequence (R51V52A53Q54) allows strong repression, but intermediate sequences diminish repression.
The H46N substitution had slightly diminished repression (perhaps slightly more than the 2-fold limit of detection for the assay) in two LPhP variants, and the mirror N46H substitution in LLhP slightly enhanced repression (Figure 3C). Therefore, this N-linker position might alter function independently of the linker helix sequence.
Substitutions of C-linker positions
We assessed the contributions of the nonconserved C-linker positions 59 and 60 using both LPhP57–61 and the tightly repressing LPhP 48/55/57–61 contexts. Positions 59 and 60 have high sequence entropy (Table 1),dicating that other homologues utilize a variety of amino acids at these positions. However, the presence of PurR residues at positions 59 and 60 in LPhP 57/58/61 precludes measurable repression (Supplementary Figure 1). Further, when either position was changed from the LacI to the PurR residue in LPhP48/55/57–61, repression was diminished 30- to 100-foldd (Supplementary Table 2). Thus, we conclude that in vivo repression of the LacI:PurR chimeras is abolished if any C-linker position contains a PurR residue. It will be interesting to determine (1) whether the C-linker variants bind DNA weakly and nonspecifically, like LLhP G58L and G58T 20 or (2) whether the variants behave like LLhP S61C and S61M, which in vitro bind DNA with reasonably high affinity in the presence of co-repressor, but cannot repress in vivo because they require more corepressor than is apparently available 20.
In summary, all 11 positions that differ between LPhP and LLhP alter function when the amino acid is exchanged between PurR and LacI sequences. No functionally silent linker positions were identified in the LacI:PurR chimeras.
Interconversion of LGhG and LLhG
Beyond the conserved YxPxxxAxxL motif, the LacI and GalR linker sequences contain three identical residues at sites 45, 50, and 57; LLhG and LGhG have identical residues at position 62. Thus, LLhG and LGhG differ at 10 positions; of these, only position 54 directly contacts DNA.
We first planned to parallel the LPhP/LLhP experiments, exchanging LGhG positions so that it became more like LLhG sequence. However, most LGhG variants exhibited mismatched results between plate assays and liquid culture values and/or very slow growth rates in liquid cultures. For example, a variant that resulted in a white colony in the plate assay showed very high values in the liquid culture assay. As discussed in Methods, we consider the plate results to be more reliable in this case. In order to obtain variants with reliable liquid culture values, we adopted the opposite strategy and exchanged the N-linker, C-linker, and hinge helix from LGhG into LLhG (Figure 4, Supplementary Table 3).
Functional effects from exchanging the helix were striking (Figure 4A). Relative to LLhG, the helix swap of GalR residues 50–57 enhanced repression 5-fold in the absence of inducer and by more than 10-fold in presence of inducer. Together, these resulted in a two to three-fold decrease in response to inducer. Exchanging the hinge helix also increased repression in the LLhG/E62K context (a strongly repressing C-linker variant19), by 2-fold in the absence and >30-fold in the presence of inducer (Figure 4 A).
Conversely, exchanging either the N- or C-linker of LLhG diminished repression alone (LLhG/45–49 or 58–61; ~6-fold each) or in combination (LLhG/45–49/58–61; ~18-fold) (Figure 4A). However, when the N- or C-linker was exchanged simultaneously with helix residues 50–57, repression in the absence of inducer was diminished by at most 2-fold relative to LLhG/50–57. A greater functional change occurred in the presence of inducer, with a 5- to 6-fold loss of repression compared to 50–57 alone. Thus, the N- and C-linker sequences in LLhG/LGhG may modify functional contributions from the helix, but they do not dominate the functional outcome in the way that the C-linker dominates LLhP/LPhP function.
Substitutions of helix positions
In the linker helix, 4 positions are conserved between LacI and GalR, whereas 4 are not (51, 52, 54, and 55). Using LLhG and LLhG/50–57, we exchanged the 4 nonconserved residues singly and in various combinations. In Figure 4B, we present the fold-change in repression for each variant, which is calculated as a ratio relative to the repression of the parent construct. For example, the repression of LLhG/51/52 is used to calculate two values of “fold-change” (Figure 4B) – once relative to LLhG/R51A, which quantitates the effect of exchanging position 52; and once relative to LLhG V52N, which quantitates the effect of exchanging position 51. Values used to calculate fold-change are in Supplementary Table 4.
No subset of substitutions in the LLhG helix recapitulated LLhG/50–57 function (Supplementary Table 4), and no position showed a consistent outcome for all contexts (Figure 4B). (1) Some exchanges at position 51 showed very little effect (2-fold is at the limit of reliably detected change), whereas other showed 25-fold enhanced repression. (2) Exchanging position 52 in the 51/54/55 background to create the fully exchanged 50–57 variant showed dramatic differences in the presence and absence of inducer, with ~4-fold loss in the absence and 3-fold enhancement in the presence of inducer. In all other contexts tested, V52N enhanced repression 3 to 5-fold. The mirror substitution of LGhG N52V also showed enhanced repression in plate assays relative to LGhG (Figure 2C). (3) Addition of Q54R to LLhG/51/52/55 diminished repression 5-fold in the absence of inducer, whereas the same substitution in LLhG and two other combinations had little effect. (4) The Q55A substitution diminished repression in LLhG nearly 4-fold (19 , Figure 4B). However, this substitution is essentially neutral in the context of V52N and Q54R; and repression is enhanced 2–3 fold when Q55A is used to form the triple substitution 51/52/55. Thus, several LLhG/LGhG helix positions show significant context dependence (synergy) with the other helix positions.
N-linker positions that contribute to differences between LLhG and LGhG
Subregion exchange studies showed that exchanging the N-linker (45–49) worsened repression of LLhG (Figure 4A). Because three N-linker positions are conserved between LacI and GalR, LLhG/45–49 only differs from LLhG by 2 amino acids, at positions 46 and 48. The I48H substitution was previously shown to diminish repression of LLhG (Figure 4C)19. We constructed and characterized the LLhG N46S substitution, and found it had no effect on repression (Figure 4C). Thus, all effects in LLhG/45–49 appear to derive from position 48; indeed, the measured liquid culture values of the two constructs are statistically equivalent. Further, the reverse LGhG substitutions appear to mirror these effects: S46N has very little effect (Figure 4C) and H48I appears to enhance LGhG repression in the plate assay (Figure 2C).
Although N46S and S46N had little or no effect on repression of LLhG and LGhG, respectively, we have become acutely aware of the distinction between a silent substitution and a silent position. For the latter to occur, all amino acids must be tolerated at the position without causing a change in function (with the exception of residues that might disrupt the structure, such as proline or glycine). Therefore, we further tested the functional importance of site 46 using codon-randomizing mutagenesis in the LLhG, LLhG/E62K, and LGhG backgrounds. Data are shown in Table 2 for repression in either the plate or liquid culture assay (depending upon whether the latter could be reliably quantitated). Combined with exchange data (Figure 4), a total of 12 amino acid substitutions were obtained at position 46. Of these, the variants with H, R, D, I, N, and S showed little change in repression relative to the parent protein; those with P or E had diminished repression; those with C or F had enhanced repression; we did not detect protein or repression for W variants; and a variant with V had measurable repression in spite of having undetectable protein levels by SDS-PAGE. Thus, amino acid changes at position 46 can impact repression, and, although the LacI-to-GalR exchange substitutions were silent, position 46 is not functionally silent.
Table 2.
Repression by Random Variants of LLhG and LGhG-like proteins
| Repression Phenotypea | ||||||
|---|---|---|---|---|---|---|
| Repressor Variant | no inducer | (+) inducer | Overall effectb | |||
| LLhG | LB/B | B | ||||
| 5.2 | (2.1) | 100 | (11) | |||
| LLhG E62K | VLB | B | ||||
| 0.22 | (0.03) | 71 | (12) | |||
| LLhG/50–57 | LB | LB | ||||
| 1.2 | (0.1) | 7.0 | (1.4) | |||
| LLhG/50–57/58–61 | LB/B | B | ||||
| 2.2 | (2.0) | 45 | (8) | |||
| LGhG | LB | B | ||||
| 1.9 | (1.9) | 118 | (28) | |||
| 46 H | LGhG | 22 | (8) | 81 | (8) | D |
| 46 P | LGhG | 140 | (11) | 120 | (17) | D |
| 46 R | LGhG | LB/B | LB | |||
| 46 W | LGhG | no protein in pull-down | ||||
| 46 E | LLhG E62K | 3.2 | (0.5) | 91 | (11) | D |
| 46 H | LLhG E62K | 0.27 | (0.05) | 57 | (7) | = |
| 46 W | LLhG E62K | no protein in pull-down | ||||
| 46 V | LLhG E62K | 20 | (6.5) despite no protein | |||
| 46 C | LGhG | W | B | |||
| 46 D | LGhG | 5.8 | (2.7) | 150 | (25) | D |
| 46 F | LGhG | 0.54 | (0.12) | 41 | (8) | E |
| 46 I | LGhG | 7.4 | (1.7) | 95 | (5) | D |
| 46 R | LGhG | Spkld | ||||
| 58 C | LGhG | W | B | E? | ||
| 58 G | LGhG | Rich = LB/B | ||||
| Min = B | B | |||||
| 160 | (40) | 120 | (16) | |||
| 58 P | LGhG | 330 | (50) | 180 | (13) | D |
| 58 S | LGhG | W | B | E? | ||
| 58 V | LGhG | W | B | E? | ||
| 59 L | LLhG/50–57/58–61 | 4.3 | (2.8) | 150 | (60) | D |
| 59 P | LLhG/50–57/58–61 | B | B | D | ||
| 59 F | LGhG | 300 | (74) | 250 | (50) | D |
| 59 H | LGhG | 16 | (3) | 180 | (40) | D |
| 59 L | LGhG | 130 | (80) | 150 | (30) | D |
| 59 N | LGhG | 7.7 | (3.2) | 160 | (20) | D |
| 59 V | LGhG | 220 | (70) | 200 | (45) | D |
| 60 A | LLhG/50–57 | W | W | E | ||
| 60 I | LLhG/50–57 | 0.70 | (0.36) | W | E | |
| 60 S | LLhG/50–57 | 0.44 | (0.07) | 45 | (4) | E |
| 60 T | LLhG/50–57 | W | W | E | ||
| 60 A | LLhG/50–57/58–61 | W | LB/B | E | ||
| 60 E | LLhG/50–57/58–61 | 59 | (4) | 109 | (8) | D |
| 60 M | LLhG/50–57/58–61 | W | LB/B | E | ||
| 60 P | LLhG/50–57/58–61 | 130 | (30) | 150 | (30) | D |
| 60 S | LLhG/50–57/58–61 | W | LB | |||
| 60 V | LLhG/50–57/58–61 | W 2.4 | (0.9) | 40 | (4) | E? |
| 60 D | LGhG | 9.7 | (1.3) | 110 | (7) | D |
| 60 E | LGhG | 12 | (2) | 140 | (20) | D |
| 60 K | LGhG | W | LB | E | ||
| 60 R | LGhG | W | W | E | ||
| 60 S | LGhG | 5.5 | (3.2) | 110 | (30) | D |
| 60 V | LGhG | VLB | B | = | ||
| 61 L | LLhG/50–57 | 0.23 | (0.05) | 54 | (6.7) | E |
| 61 M | LLhG/50–57 | 0.62 | (0.42) | 79 | (12) | E? |
| 61 P | LLhG/50–57 | W | W | |||
| 61 T | LLhG/50–57 | 0.15 | (0.09) | 22 | (11) | E |
| 61 I | LLhG/50–57/58–61 | 14 | (3) | 108 | (6.9) | D |
| 61 K | LLhG/50–57/58–61 | 3.4 | (0.8) | 79 | (10) | = |
| 61 R | LLhG/50–57/58–61 | 4.3 | (0.1) | 90 | (10) | D |
| 61 S | LLhG/50–57/58–61 | 1.9 | (0.7) | 21 | (4) | = |
| 61 P | LGhG | LB | B | D | ||
| 61 Q | LGhG | 150 | (60) | 135 | (16) | D |
Results from both plate and liquid culture assays are shown for the parent variants in the top 5 rows. For the variants obtained after random mutagenesis, results from liquid culture β-galactosidase assays are shown when values were in reasonable agreement with plate assays. Values reported are the average of 3 to 8 determinations; numbers in parentheses are the standard deviation; inducer was fucose. Most assays were performed in the high-throughput format, but some were also measured using the original protocol to confirm agreement between the two techniques. If plate assays indicated stronger repression, those values are shown instead of liquid culture results (see Methods); if the match is ambiguous, both results are shown. “W”, white (strong repression); “LB”, light blue, moderate repession; “B”, blue, poor repression; “Spkld” shows a mixture of white and blue colonies, similar to that seen for some LLhP variants14.
Substitutions either enhance (E), diminish (D), or do not change (=) repression relative to repression of the parent protein. “E?” indicates that enhanced repression is near the detection limit of the assay.
C-linker positions that contribute to differences between LLhG and LGhG
Subregion exchange studies showed that exchanging the C-linker of LLhG with that of GalR worsened repression (Figure 4A). None of the four C-linker positions are conserved between LacI and GalR. In LLhG, two of the single exchange substitutions – G58Q and K59Q – diminished repression, whereas Q60T and S61T have essentially no impact on function (19 and this work, Figure 4C). The little change for sites 60 and 61 could indicate silent substitutions or silent positions. Earlier “host-guest” random mutagenesis experiments in LLhG show convincing functional contributions from varied amino acids at sites 58, 59, and 60, but little impact from substitution at site 61. At the time of the latter publication, we presumed this position was functionally important because of its contribution in LLhP. Here, we wished to determine the functional importance in alternative linkers. Thus, we repeated the random mutagenesis of sites 58, 59, 60, and 61 in three chimeras that contain large sections of the GalR linker (LLhG/50–57, LLhG/50–57/58–61, and LGhG). Results of functional assays are presented in Table 2.
At position 58 of LGhG, repression was effectively abolished by changing the GalR glutamine to the LacI glycine. Three other variants (V, C, and S) show possibly enhanced repression in plate assays (and enhanced repression appears to correlate with unreliable liquid culture values). For position 59, repression was diminished in 7 variants ranging from 4-fold, 15-fold, to 100-fold. For position 60, 12 substitutions were isolated in various contexts. S, T, A, V, M and I had little effect; D, E, and P diminished repression more than 2-fold and up to 70-fold; K and R enhanced repression. Of the nine substitutions isolated at position 61, M and S showed little functional change; Q, I, K, and R diminished repression; L and T enhanced repression. Notably, the proline 61 substitution had very little effect on the strong repression of LLhG/50–57, despite its potential to alter backbone structure. All but a few C-linker repressors showed strong protein bands in pull-down assays; exceptions are noted in Table 2.
In summary, all 10 positions that are not conserved between the LacI and GalR linker regions can be substituted to alter function in the GalR-like linker sequence.
Rugged sequence/function landscapes
In the process of exchanging residues between the pairs LLhP:LPhP/57–61 and LLhG:LLhG/50–57, we created a number of “evolutionary” trajectories between the two endpoints. In Figure 5 we present a landscape for each of the chimera pairs in the tightest repression conditions (LacI:PurR chimeras +co-repressor, LacI:GalR chimeras without inducer). Both landscapes are rugged and resemble landscapes that have been reconstructed for naturally-evolved proteins 29. Further, in both chimera landscapes, one can trace a smooth trajectory between endpoints that avoids frequent, violent changes in function. However, our two chimera landscapes differ significantly in the relationship of the smooth path to the rest of the landscape.
Figure 5. “Evolutionary” landscapes between chimera pairs.
Repression assay values for combinatorial variants are plotted as columns. (A) Conversion of LPhP/57–61 to LLhP. (B) Conversion of LLhG to LLhG/50–57. In both panels, the legends of the X axes indicate how many substitutions have been made in the variants of that row. Connecting lines are drawn in the baseplane to show “evolutionary” paths of consecutive substitutions. In (A), the dashed lines show two constrained pathways which avoid variants with poor repression. In (B), the dashed and dotted lines indicate two nearly neutral pathways, during which improved repression occurs primarily at a single step.
Conversion of LPhP/57–61 to the tighter repressor LLhP shows at least two trajectories that are constrained to a trough (Figure 5A, dashed curved lines). Along this pathway, repression is mostly enhanced and avoids variants with greatly diminished repression. Constrained evolution has been invoked to describe the acquisition of five amino acid substitutions that lead to a penicillin-resistant β-lactamase; only a few permutations of the possible 120 allow constant positive selection of penicillin-resistance 30.
Conversion of LLhG into the tighter repressor LLhG/50–57 shows two nearly neutral trajectories in the absence of inducer (Figure 5B, dotted curved lines): Only one mutation in each series has a dramatic effect on function, whereas repression at other steps is within 2-fold of the preceding value. These trajectories avoid both extremely weak and extremely strong repression, allowing many “neutral” changes. Notably, the same substitutions are not neutral in other linker contexts. In a natural process, the existence of neutral pathways would allow complex new sequences to evolve with minimal impact on the organism. A similar process has been proposed for the evolution of influenza A hemagluttinin, to explain how constant genetic drift of multiple positions in the viral epitope results in punctuated antigenic change 31.
DISCUSSION
Functional contributions of positions in the YxPxxxAxxL LacI/GalR linkers
Substitution of 11 analogous, nonconserved positions alters repressor function in the LacI, PurR, and GalR linkers. Thus, the functional importance of these positions is likely to be preserved in the YxPxxxAxxL subfamily of the LacI/GalR transcription regulators. Furthermore, the assumption behind many sequence analyses appears to be valid, if the proper subfamily of sequences is utilized. We do not yet know if the same linker positions contribute to functions of LacI/GalR proteins that contain alternative linker sequences, such as CytR (see 19 for discussion).
Another similarity among the linkers studied is the functional synergy of positions at the start of the helix. In the LacI: PurR chimeras, positions 51/54 are clearly synergistic; position 52 is conserved; and results for position 50 are inconclusive. The effects of substitutions at position 55 might also be context-dependent, as indicated by differences in the magnitude of functional change. In the LacI:GalR chimeras, the clearly synergistic positions are 52, 54, and 55; 51 substitution exhibits different magnitudes in various contexts; and position 50 is conserved. Synergy within the helix is not surprising, given the known hydrogen bonding and side chain interactions of helices. Nonetheless, the linker helix does not appear to function like “normal” helices, since our previous “host/guest” studies show no correlation with position-specific helical propensities 19; 21; 32; 33.
Implications for current sequence/function analyses
The LacI/GalR homologues are among the most commonly used to develop algorithms that predict functionally important sequences (e.g. 8; 11; 12; 14; 34; 35; 36; 37; 38). Predictions about the functional importance of linker positions vary widely; some studies identify several linker positions as functionally important, others do not identify any. Experimentally, we find that, when combined with the conserved positions, every YxPxxxAxxL linker position contributes to repressor function. Thus, this region is clearly a functional “hotspot”. Any successful identification algorithm should highlight the functional importance of the LacI/GalR linkers.
Although a successful program should highlight the linker, a particular analysis need not detect all positions in the linker: As we noted before 19, different algorithms appear to identify different categories of positions. Nonetheless, the LacI/GalR linker positions represent all possibilities (conserved, semi-conserved, and nonconserved), with a range of functional contributions (substitutions alter affinity, selectivity, and allostery 20). Therefore, we reiterate our statement that a successful analysis should identify at least a few positions of this region. Analyses that do not implicate the linker might be misdirected by their reliance upon ligand binding positions to assess algorithm success. This requirement could place inappropriate selective pressure on algorithm development, driving the analysis away from the behavior of natural proteins.
Finally, the rugged evolutionary landscapes (Figure 5) illustrate why experiments with single proteins or single substitutions should not be used to benchmark functional importance of the position for all homologues in the family. For example, in the context of LL G, the Q54R substitution had no effect and would not be identified as functionally important. however, substitution of this position clearly impacts other chimeric variants; therefore, substitution of this position might impact other homologues.
Evolution of the linker sequence and functional correlations
Several attempts have been made to extract functional interactions from sequence by finding which positions co-evolve (e.g. 12; 14; 16; 39; 40; 41). Co-evolving positions should have similar sequence entropies. However, we note that functionally important, nonconserved positions are not constrained to interact only with other nonconserved, co-evolving positions. In the interconversion of the LacI:GalR chimeras, synergy is seen between positions with high (51, 55) and moderate/low (50, 54) sequence entropies. A nonconserved position might also functionally interact with a conserved residue. Such might be the case for position 48. In representative LacI/GalR structures (e.g. 42; 43), the side chain of position 48 interacts with a region of the regulatory domain that is fairly well conserved between LacI, PurR, and GalR. Position 48 was not detected as functionally important by methods that incorporate co-evolution, although our current and previous 19; 21 experiments clearly show its functional contributions. Thus, we believe that co-evolution analyses of functionally important positions will be incomplete, at best.
Indeed, our data compel us to re-visit an even older assumption about protein evolution: that some nonconserved positions are silent – not important for either structure or function. The LacI/GalR YxPxxxAxxL linkers have no silent positions. We now discriminate between a neutral substitution, which we do detect in some chimeric variants, and a neutral position, which should remain insensitive to substitution in all homologue contexts. Sequence/function analyses that attempt to discriminate functionally important from silent nonconserved positions obligate the assumption of silent positions. Although the lack of silent positions in the linker might be due to the unique importance of this region, we now wonder whether neutral positions ever exist in other regions of the homologues. Alternatively, evolution might rely upon neutral pathways like the ones observed in the landscapes shown in Figure 5.
Conclusion
At least three avenues for progress in sequence/function analyses are suggested by the current LacI/GalR studies: (1) Although the functional importances are preserved for the YxPxxxAxxL linkers of the LacI/GalR proteins, we noted previously that other LacI/GalR linkers lack this motif and thus proposed that family/subfamily delineation might greatly impact predictions by various algorithms19. This is a subject of our ongoing research with the LacI/GalR proteins and should be the first consideration in undertaking studies of other protein families. If the proper subfamilies are identified, the current work supports the common assumption –functional importance is preserved for a given position in multiple homologues. (2) We observe that while single substitutions in the LacI/GalR linkers are sometimes “silent”, no linker position was truly functionally neutral. Since neutrality of some nonconserved positions is an underlying premise of many sequence/function analyses, we must determine whether the LacI/GalR linker result is the exception or the rule for the protein universe. If protein families do not commonly have silent positions, we must identify alternative relationships to serve as the bases of useful sequence/function analyses. (3) We observe that linker residues with different sequence entropies show functional synergy. Similar interactions between conserved, semi-conserved, and nonconserved positions will likely occur in other proteins; algorithms based primarily upon co-evolution (and thus similar sequence entropy) will miss the functional importance of these positions and relationships.
The current work clearly illustrates that nuanced and complex relationships exist between residue positions to convey function to each protein. Current sequence/function analyses show promise, especially if multiple algorithms are applied to each protein family13; 19; 44. However, future progress will be greatly enhanced by the design and implementation of experiments that explicitly identify underlying principles, and the use of these principles to improve (not necessarily simplify) bioinformatics algorithms.
MATERIALS AND METHODS
Features of the linker structure important to the design of chimeric variants
As stated above, the central problem to be addressed is “Which nonconserved positions can be varied to modify the common function and confer unique attributes to each homologue?” In designing experimental studies, this question cannot be divorced from the question of “how” functional change happens: We make the assumption that functional modification occurs by changing side chain interactions or dynamics; effects on backbone structure should be subtle, leaving the conserved fold intact. Therefore, we must consider whether a particular substitution dramatically disrupts structure. Since we cannot directly determine structure for the hundreds of variants in this and related studies, we consider the following structural aspects:
The LacI/GalR linker region contains two unstructured regions that flank a helix (Figure 1; 42; 43). Many lines of evidence, including recent analysis of small angle X-ray scattering data, indicate that the LacI helix undergoes a critical coil-helix transition upon binding DNA (42; 43; 45; 46; 47 and references therein). Since the helix-coil transition might occur in other homologues, we must consider whether helix formation is precluded by substitution of a nonconserved position. However, results from LacI, LLhP, and LLhG “host-guest” substitutions (with multiple amino acids at each site) showed no correlation between function of the variant repressors and position-specific helical propensity 19; 21; 32; 33. Thus, the ability to form a helix does not appear to be abolished by most substitutions.
The flanking regions, which we denote as N- and C-linkers, have no regular secondary structure and thus are not likely to be disrupted by most substitutions. These regions encompass positions 45–49 and 58/59–61/62, respectively. The ends of the C-linker are difficult to delineate: The start of the C-linker can be varied by extending the helix, which is one position longer in PurR than in LacI 48. The end of the C-linker was originally defined at position 61 21, since position 62 is not sensitive to mutagenesis in LacI 27. However, changes at position 62 have a large impact on LLhG function 19, which led us to re-consider the boundary. In LPhP and LGhG, this issue is not relevant, since the linker is continuous with the PurR or GalR regulatory domain. Position 63 is considered to be in the regulatory domain, as it participates in the central -sheet of that region. Supplementary Table 1 and 3 show precisely which positions are exchanged in the chimeric variants of the current study.
Calculation of sequence entropy for linker positions
The linkers of natural homologues intersperse conserved and nonconserved positions (Table 1). Many family members have a conserved linker motif – YxPxxxAxxL – which corresponds to LacI positions 47, 49, 53, and 56. Homologues lacking this motif have been proposed to comprise a separate subfamily 19; 38e. We collected more than 1000 natural sequences of the LacI/GalR family using a BLAST search of Swiss-Prot (data not shown; 49; 50). Using the subfamily of 1082 nonredundant sequences that conserve this motif, we calculated sequence entropy values 51 for all linker positions, with the modification from the referenced work that we placed each of the 20 amino acids in its own “class”. The rationale for this modification was based on the observation that conserved position 56 only tolerates L or M in LacI and PurR 27; 52, and thus grouping side chains by chemical “similarities” is an oversimplification. Sequence entropy values are presented in Table 1; a value 0.0 represents perfect conservation, whereas a value of 1.3 represents a perfectly random distribution of amino acids.
Construction of LPhP, LGhG, and variants
Chimeric protein LPhP comprises LacI positions 1–45 and PurR 44–341. LGhG comprises LacI 1–45 and GalR 44–343. Note that the numbering of wild-type PurR and GalR linkers each differ from LacI sequence by 2, because of differences in the length of the DNA-binding domains. The amino acid sequences of the linkers are shown in Table 1.
All primers for mutagenesis and sequencing were purchased from Integrated DNA Technology (Coralville, IA) or the Biotechnology Support Center at KUMC. Primer sequences are listed in Supplementary Table 5. DNA sequencing was carried out by the KUMC Biotechnology Support Center or Northwoods, DNA Inc. (Solway, MN).
The lacI gene and promoter regions were derived from the pLS1 plasmid 53, which has an engineered SacI site at LacI codon 45. pLS1 also contains a Bsu36I site downstream of the gene encoding LacI. PurR codons for amino acids 44–340 were amplified from E. coli DH5α (Invitrogen, Carlsbad, CA) using colony PCR (PCR Mastermix, Promega, Madison, WI) using primers that added (1) a SacI site at position 45, (2) an additional stop codon after the original stop codon and (3) a Bsu36I site following the second stop codon. PCR products were ligated into pGemT vector (Promega, Madison, WI). White colonies were grown overnight in 3ml cultures of 2xYT and plasmid DNA purified with QIAprep Spin Miniprep Kit (Qiagen, Valencia, CA) or Quantum Prep Plasmid Miniprep Kit (Bio-rad, Chicago, IL). Plasmids were screened with restriction digestion for the appropriate insert and sequenced using primers to the SP6 and T7 sites on the pGemT vector. Any mutations that arose during amplification of the PurR gene from E. coli, which used a low-fidelity polymerase, were corrected using Quikchange site-directed mutagenesis (Stratagene, La Jolla, CA) so that the final sequence matched the published Swiss-PROT sequence 50.
For ligation of the final LPhP construct, pLSI and the pGemT plasmids containing the appropriate inserts were cut with SacI and Bsu36I. Fragments were separated by gel electrophoresis and gel purified using a Montage Ultra Free column (Millipore, Billerica, MA). The vector was dephosphorylated using Calf Alkaline Phosphatase. Fragments were ligated at 16 °C overnight, and transformed to DH5α Max or High Efficiency cells (Invitrogen, Carlsbad, CA). To increase repeatability in repression assays, the gene for LPhP was subcloned onto plasmid pHG165f 21; in typically nonlinear experimental chronology, subcloning was carried out first with the LPhP/K57A variant and site directed mutagenesis was then used to revert to the original LPhP sequence.
LGhG was created by mutating the linker of LLhG on the plasmid pHG165af. Like LLhG19, LGhG was toxic unless inducer fucose was added to the media or unless the “E230K” substitution was added to the GalR regulatory domain. As before19, all LLhG and LGhG variants reported in this manuscript contain E230K; for simplicity in the tables and figures, this mutation is not explicitly noted.
Variants of LPhP, LGhG, and LLhG were created with either site-directed or codon-randomizing mutagenesis using Quikchange, as reported previously 19; 21 and using the primers listed in Supplementary Table 5. The coding regions of all variants were fully sequenced to ascertain that no additional mutations were introduced.
Verification of protein expression and activity
Expression of active chimeric variants in E. coli. 3.300 cells was verified in crude cell extracts as described before 19. These assays detect DNA-binding events with affinities as low as 10−7 M and differentiate binding to operator from binding to nonspecific DNA sequences. Briefly, short synthetic stretches of DNA containing either the natural operator 54 lacO1, a tight-binding variant 55 lacOsym, or a nonspecific sequence 56 Onon were 5’-biotinylated (Integrated DNA Technologies, Coralville, IA) and coupled to streptavidin magnetic beads (New England Biolabs, Ipswich, MA). Cells expressing the chimeric variants were grown overnight in LB media, pelleted, and lysed by freeze-thaw in the presence of lysozyme; the resulting supernatant was incubated with the DNA-beads. After magnetic separation and washing, the proteins associated with the beads were assayed by SDS-PAGE. Detection was first with Coomassie followed by silver stain. For all but a few variants (noted in Tables, Figures, and Supplementary Tables and Figures), the bands corresponding to the chimeric variants were clearly evident when compared to “empty” control cells that did not express a repressor. Bands for LPhP and LGhG ran at positions equivalent to LLhP and LLhG, which have been purified and verified by mass spectrometry and other techniques (20 and unpublished data). The detection limits of the gel stains were used to estimate that chimera concentrations were at least three to four orders of magnitude higher than the single genomic lacO1 binding site employed in the phenotypic repression assays described below 19. Even for most variants with low affinity/repression for lacO1, we detected significant protein using lacOsym, which indicates that lost repression of lacO1 does not necessarily equate with total loss of DNA binding.
Measurement of in vivo β-galactosidase activity
Repression assays were carried out as described previously for LLhP and LLhG 19; 21. Briefly, we utilized E. coli 3.300 cells (E. coli Genetic Stock Center, Yale University) 57, which have an interrupted lacI gene but an intact genomic lacZYA operon controlled by the operator sequence lacO1. Expression of the lac operon was assessed on both plates and in liquid culture by monitoring β-galactosidase activity. Plate assays 53; 58; 59 utilized standard LB media with the blue-white indicator 5-bromo-4-chloro-3-indolyl β-D-galactopyranoside (Xgal)59 and 100 ug/ml ampicillin. White colonies express repressor variants that are capable of repressing the reporter β-galactosidase gene. Blue colonies express repressor variants that are incapable of repression, and light blue colonies arise from chimeric variants with partial function.
Liquid culture assays 59; 60; 61 utilized minimal media with 2% glycerol as a carbon source. Low values of reporter activity correlate with strong repression of lacO1 in the natural E. coli lac operon; high values indicate diminished repression. All LPhP and many LGhG variants were assayed using the procedure outlined in 21. LGhG random variants and others presented in Table 2 were assayed using the high-throughput, 96-well assay method described in 19. The latter method was adapted for tight repressors by concentrating the cells after the final growth in a smaller volume of either media or enzyme assay buffer prior to initiating the assay. In switching assays to the high-throughput format, we repeated numerous controls and re-assayed previous LLhG variants, to demonstrate that the two techniques resulted in equivalent values (data not shown).
To facilitate comparison of liquid culture repression values with previous works, we continued the separate normalization protocols developed for LLhP and LLhG 19; 21. To normalize measurements of LPhP variants, we assayed wild-type LacI −/+ inducer IPTG in parallel with each day’s assay. The number of β-galactosidase units measured for LacI in the presence of IPTG was set to 100 (Figure 2a, thick dashed line) and used to normalize the LPhP samples (values for LLhP with no mutations were too low to serve as a calibration point). For LGhG studies, a sample of LLhG was always assayed in parallel +/− inducer fucose; the value in the presence of fucose was set to 100 (Figure 2b, thick dashed line) and used to normalize results from other LLhG and LGhG constructs. Experiments were performed three to six separate times for each variant, always from a fresh transformation (LPhP variants) or from a plate that was less than a week old (LGhG/LLhG variants). Reported values are averages, with error bars representing one standard deviation.
In vivo repression assays were performed in the absence and presence of the relevant effectors: Both PurR and LLhP show enhanced DNA-binding and repression in the presence of guanine or hypoxanthine, which bind to the regulatory domain 20; 21; 62; 63. In the in vivo assays, the effector is adenine (25 µg/ml )26, which is metabolized to hypoxanthine. We expected and observed similar behavior for LPhP variants that are capable of repression. GalR and LLhG are induced by galactose or fucose 19; 25; LGhG variants show analogous behaviors. For in vivo assays of chimeras with a GalR regulatory domain, inducer was 20 mM fucose.
We also verified that liquid culture values were consistent with plate results. Although the plate assays do not have a concrete cut-off point between “blue”, “light blue”, and other shades, a survey of over 100 LLhG variants 19 indicated the plate colors correlate with the ranges of liquid culture values indicated in Supplementary Table 3. Several LGhG variants showed discrepancies between the two assays, always appearing to be worse repressors in liquid culture than on the plates. We repeated the plate assays using minimal MOPS media to better match the conditions of the two assays; results were consistent for the two plate media and still showed tighter repression than in liquid culture. Since we cannot imagine artifactual means for acquiring repression in plate assays, and we can imagine many scenarios for losing repression in the liquid culture assays (which have longer growth times and might be more sensitive to slightly toxic chimeric variants), we report the plate assay results in cases where the two techniques do not match.
Interpretation of in vivo repression assays
In vivo repression levels are impacted by the repressor’s DNA binding affinity, allosteric response, and nonspecific binding to other genomic DNA. In the current results, we anticipate that changes in repression will most often correlate with changes in DNA binding affinity, as was seen for several variants of LLhP 20. Although repressor protein concentration could also contribute to in vivo results, we have ruled that out for most of the chimeric variants (see above). In addition, in vivo assays also have potential to be affected by unexpected events, such as interactions with other E. coli proteins. Therefore, repression assays are uniquely suited to capture myriad functional changes that might occur upon amino acid substitution.
Effector concentrations can also impact in vivo repression results. This appears to be a factor for the LLhP S61C/M variants, which did not repress in vivo even though the purified proteins did bind lacO1 DNA in the presence of co-repressor 20. However, these LLhP variants required higher co-repressor concentrations to bind lacO1, and we hypothesized that the LLhP in vivo co-repressor concentrations were not high enough to allow repression. Nonetheless, the “no repression” phenotype reflects a significant functional change in these repressor variants, in this case due to altered co-repressor allosteric response.
In addition to repression levels, an in vivo allosteric response can be calculated from the ratio of repression in the absence and presence of co-repressor or inducer. LLhP has a small but reproducible allosteric ratio in vivo: co-repressor increases repression ~2-fold for “wild-type” and up to 10-fold for some variants at positions 48 and 55 21. Similar values are obtained for LPhP variants (Figure 2A and Figure 3). In vitro, the allosteric ratio for “wild-type” LLhP is 100 to 200-fold, and substitutions at positions 48 and 55 did not significantly alter this value 20. One confounding feature may again be that the metabolites which serve as co-repressors are restricted to a narrow range in vivo – even when exogenous co-repressor is added – so that the in vivo allosteric response is too small to accurately detect differences arising from amino acid substitutions.
The in vivo allosteric change of LLhG is 20-fold (Figure 2B), which is consistent with a preliminary value determined for purified LLhG of ~30-fold (unpublished data). The in vivo allosteric response of LLhG/E62K is >300-fold (Figure 4A). Inducer fucose is not metabolized by E. coli 3.300 cells and thus its concentration is regulated only by uptake levels. Most LLhG variants have induced levels of LacZ activity that are comparable to the negative control plasmid, pHG165a (Figure 2). A few variants retain significant repression in the presence of inducer fucose (19 and this work). Possible sources for changes in the in vivo allosteric response are: (1) a loss of allosteric response at the protein level, so that repressors remain bound to operator in the presence of inducer or (2) a dramatic loss of non-specific binding affinity for other areas of the genome, so that more repressor is available to bind the operator in vivo. Either of these scenarios represents a significant functional change.
Supplementary Material
Liquid culture results for LPhP exchange substitutions that do not restore lacO1 repression. The successful exception is 57–61. The numbers in the X-axis names indicate which positions have been exchanged between the PurR and LacI sequences (Table 1). The complete linker amino acid sequence for each variant is given in Supplemental Table 1. All variants express protein, as seen in the pull-down assay, except LPhP 48/55/57/58/59/61. For this protein, we cannot discriminate between total loss of DNA binding for lacO1 and lacOsym, undetectable protein expression, or enhanced protein degradation. The normalization scale is that for Figure 2A. (Top panel) Note that positions 50 and 54 directly contact DNA in both LacI and PurR 48 and are moderately conserved across the family. PurR is one of the few family members that lacks 50N or 54Q. However, creating the LPhP R54Q mutation alone and in combination with K57A did not restore any lacO1 repression. Surprisingly, the construct with exchanges at all three positions 50/54/57 could not be created, despite extensive mutagenesis efforts. Another construct that included these three exchanges did not result in detectable protein (Supplementary Table 2). We speculate that this combination of residues could result in a repressor that is toxic to E. coli. (Middle panel). The substitutions in these variants are of the 4 nonconserved positions that were previously shown to impact LLhP repression21. (Lower panel) Substitutions in the LPhP C-linker.
Acknowledgements
This work was supported by grants from the NIH: GM079423 to LSK and P20 RR17708 from the Institutional Development Award program of the National Center for Research Resources. We thank Drs. Sarah Bondos (Texas A&M Health Science Center), Susan Egan (KU-Lawrence) and Aron Fenton (KUMC) for comments on the manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Sometimes called “specificity determinants”.
Previous LLhP and LLhG studies utilized different normalization standards for values of in vivo reporter activity (described in Materials and Methods). To facilitate comparisons with earlier works, we have kept the separate normalization standards for LPhP and LGhG. Data for LacI repression, as well as reporter activity in the absence of repressor, are shown on both normalization scales in Figure 2.
The S50N substitution inserts a LacI side chain that directly contacts lacO1 DNA; all three direct contacts (50, 54, and 57) are LacI sequence in LPhP 48/50/54/55/57–61; and the sequence only differs from LLhP by two positions. However, we cannot detect protein for this variant in the DNA pulldown assay. Thus, we cannot discriminate between greatly diminished DNA binding and the absence of protein in vivo.
One of these two variants – LPhP/48/55/57/58/59/61 – did not show protein in DNA pull-down assays. This variant may have a structural disruption (although the C-linker does not have regular secondary structure), enhanced proteolysis, or might have completely lost ability to bind both lacO1 and lacOsym DNA.
In addition to the linker motif, the two subfamilies have different types of DNA-binding sites.
Note that LPhP is on plasmid pHG165 whereas LGhG is on pHG165a. The difference in the two plasmids is that pHG165 contains a lacO1 binding site that was extensively mutated in pHG165a to preclude potential competition with the genomic lacO1 site of the 3.300 cells that is utilized in the liquid culture assay. However, direct comparison of LLhG in the two different plasmids showed no functional difference, probably because of the vast excess of expressed protein 19. Likewise, LLhP function is statistically indistinguishable when expressed from either plasmid (data not shown).
References
- 1.Armon A, Graur D, Ben-Tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J Mol Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- 2.Landau M, Mayrose I, Rosenberg Y, Glaser F, Martz E, Pupko T, Ben-Tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–W302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Valdar WS. Scoring residue conservation. Proteins. 2002;48:227–241. doi: 10.1002/prot.10146. [DOI] [PubMed] [Google Scholar]
- 4.Capra JA, Singh M. Predicting functionally important residues from sequence conservation. Bioinformatics. 2007;23:1875–1882. doi: 10.1093/bioinformatics/btm270. [DOI] [PubMed] [Google Scholar]
- 5.Manning J, Jefferson E, Barton G. The contrasting properties of conservation and correlated phylogeny in protein functional residue prediction. BMC Bioinformatics. 2008;9:51. doi: 10.1186/1471-2105-9-51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lichtarge O, Bourne HR, Cohen FE. An Evolutionary Trace Method Defines Binding Surfaces Common to Protein Families. Journal of Molecular Biology. 1996;257:342. doi: 10.1006/jmbi.1996.0167. [DOI] [PubMed] [Google Scholar]
- 7.Watson JD, Laskowski RA, Thornton JM. Predicting protein function from sequence and structural data. Current Opinion in Structural Biology. 2005;15:275–284. doi: 10.1016/j.sbi.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 8.Ye K, Vriend G, Ijzerman AP. Tracing evolutionary pressure. Bioinformatics. 2008;24:908–915. doi: 10.1093/bioinformatics/btn057. [DOI] [PubMed] [Google Scholar]
- 9.Fischer JD, Mayer CE, Soding J. Prediction of protein functional residues from sequence by probability density estimation. Bioinformatics. 2008;24:613–620. doi: 10.1093/bioinformatics/btm626. [DOI] [PubMed] [Google Scholar]
- 10.La D, Sutch B, Livesay DR. Predicting protein functional sites with phylogenetic motifs. Proteins: Structure, Function, and Bioinformatics. 2005;58:309–320. doi: 10.1002/prot.20321. [DOI] [PubMed] [Google Scholar]
- 11.Dukka Bahadur KC, Livesay DR. Improving position-specific predictions of protein functional sites using phylogenetic motifs. Bioinformatics. 2008;24:2308–2316. doi: 10.1093/bioinformatics/btn454. [DOI] [PubMed] [Google Scholar]
- 12.Pei J, Cai W, Kinch LN, Grishin NV. Prediction of functional specificity determinants from protein sequences using log-likelihood ratios. Bioinformatics. 2006;22:164–171. doi: 10.1093/bioinformatics/bti766. [DOI] [PubMed] [Google Scholar]
- 13.Pazos F, Rausell A, Valencia A. Phylogeny-independent detection of functional residues. Bioinformatics. 2006;22:1440–1448. doi: 10.1093/bioinformatics/btl104. [DOI] [PubMed] [Google Scholar]
- 14.Kalinina OV, Novichkov PS, Mironov AA, Gelfand MS, Rakhmaninova AB. SDPpred: a tool for prediction of amino acid residues that determine differences in functional specificity of homologous proteins. Nucleic Acids Res. 2004;32:W424–W428. doi: 10.1093/nar/gkh391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sankararaman S, Sjolander K. INTREPID - INformation-theoretic TREe traversal for Protein functional site IDentification. Bioinformatics. 2008:btn474. doi: 10.1093/bioinformatics/btn474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lee B, K P, D K. Analysis of the residue-residue coevolution network and the functionally important residues in proteins. Proteins: Structure, Function, and Bioinformatics. 2008;72:863–872. doi: 10.1002/prot.21972. [DOI] [PubMed] [Google Scholar]
- 17.Bromberg Y, Yachdav G, Rost B. SNAP predicts effect of mutations on protein function. Bioinformatics. 2008;24:2397–2398. doi: 10.1093/bioinformatics/btn435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kimura M. Evolutionary rate at the molecular level. Nature. 1968;217:624–626. doi: 10.1038/217624a0. [DOI] [PubMed] [Google Scholar]
- 19.Meinhardt S, Swint-Kruse L. Experimental identification of specificity determinants in the domain linker of a LacI/GalR protein: Bioinformatics-based predictions generate true positives and false negatives. Proteins: Structure, Function, and Bioinformatics. 2008;73:941–957. doi: 10.1002/prot.22121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhan H, Taraban M, Trewhella J, Swint-Kruse L. Subdividing repressor function: DNA binding affinity, selectivity, and allostery can be altered by amino acid substitution of nonconserved residues in a LacI/GalR homologue. Biochemistry. 2008;47:8058–8069. doi: 10.1021/bi800443k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tungtur S, Egan SM, Swint-Kruse L. Functional consequences of exchanging domains between LacI and PurR are mediated by the intervening linker sequence. Proteins. 2007;68:375–388. doi: 10.1002/prot.21412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Swint-Kruse L, Matthews KS. Allostery in the LacI/GalR family: variations on a theme. Curr Opin Microbiol. 2009;12:129–137. doi: 10.1016/j.mib.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jobe A, Bourgeois S. lac Repressor-operator interaction. VI. The natural inducer of the lac operon. J Mol Biol. 1972;69:397–408. doi: 10.1016/0022-2836(72)90253-7. [DOI] [PubMed] [Google Scholar]
- 24.Barkley MD, Riggs AD, Jobe A, Burgeois S. Interaction of effecting ligands with lac repressor and repressor-operator complex. Biochemistry. 1975;14:1700–1712. doi: 10.1021/bi00679a024. [DOI] [PubMed] [Google Scholar]
- 25.Majumdar A, Rudikoff S, Adhya S. Purification and properties of Gal repressor:pL-galR fusion in pKC31 plasmid vector. J Biol Chem. 1987;262:2326–2331. [PubMed] [Google Scholar]
- 26.Makaroff CA, Zalkin H. Regulation of Escherichia coli purF. Analysis of the control region of a pur regulon gene. J Biol Chem. 1985;260:10378–10387. [PubMed] [Google Scholar]
- 27.Suckow J, Markiewicz P, Kleina LG, Miller J, Kisters-Woike B, Müller-Hill B. Genetic studies of the Lac repressor. XV: 4000 single amino acid substitutions and analysis of the resulting phenotypes on the basis of the protein structure. J Mol Biol. 1996;261:509–523. doi: 10.1006/jmbi.1996.0479. [DOI] [PubMed] [Google Scholar]
- 28.Glasfeld A, Koehler AN, Schumacher MA, Brennan RG. The role of lysine 55 in determining the specificity of the purine repressor for its operators through minor groove interactions. J Mol Biol. 1999;291:347–361. doi: 10.1006/jmbi.1999.2946. [DOI] [PubMed] [Google Scholar]
- 29.Poelwijk FJ, Kiviet DJ, Weinreich DM, Tans SJ. Empirical fitness landscapes reveal accessible evolutionary paths. Nature. 2007;445:383–386. doi: 10.1038/nature05451. [DOI] [PubMed] [Google Scholar]
- 30.Weinreich DM, Delaney NF, DePristo MA, Hartl DL. Darwinian Evolution Can Follow Only Very Few Mutational Paths to Fitter Proteins. Science. 2006;312:111–114. doi: 10.1126/science.1123539. [DOI] [PubMed] [Google Scholar]
- 31.Koelle K, Cobey S, Grenfell B, Pascual M. Epochal evolution shapes the phylodynamics of interpandemic influenza A (H3N2) in humans. Science. 2006;314:1898–1903. doi: 10.1126/science.1132745. [DOI] [PubMed] [Google Scholar]
- 32.Zhan H, Swint-Kruse L, Matthews KS. Extrinsic interactions dominate helical propensity in coupled binding and folding of the lactose repressor protein hinge helix. Biochemistry. 2006;45:5896–5906. doi: 10.1021/bi052619p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kumar S, Bansal M. Dissecting alpha-helices: position-specific analysis of alpha-helices in globular proteins. Proteins. 1998;31:460–476. doi: 10.1002/(sici)1097-0134(19980601)31:4<460::aid-prot12>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 34.Chakrabarti S, Panchenko AR. Coevolution in defining the functional specificity. Proteins: Structure, Function, and Bioinformatics. 2009;75:231–240. doi: 10.1002/prot.22239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mirny LA, Gelfand MS. Using orthologous and paralogous proteins to identify specificity-determining residues in bacterial transcription factors. J Mol Biol. 2002;321:7–20. doi: 10.1016/s0022-2836(02)00587-9. [DOI] [PubMed] [Google Scholar]
- 36.Chakrabarti S, Bryant SH, Panchenko AR. Functional Specificity Lies within the Properties and Evolutionary Changes of Amino Acids. J Mol Biol. 2007;373:801–810. doi: 10.1016/j.jmb.2007.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye K, Anton Feenstra K, Heringa J, Ijzerman AP, Marchiori E. Multi-RELIEF: a method to recognize specificity determining residues from multiple sequence alignments using a Machine-Learning approach for feature weighting. Bioinformatics. 2008;24:18–25. doi: 10.1093/bioinformatics/btm537. [DOI] [PubMed] [Google Scholar]
- 38.Francke C, Kerkhoven R, Wels M, Siezen RJ. A generic approach to identify Transcription Factor-specific operator motifs; Inferences for LacI-family mediated regulation in Lactobacillus plantarum WCFS1. BMC Genomics. 2008;9:145. doi: 10.1186/1471-2164-9-145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 40.Yip KY, Patel P, Kim PM, Engelman DM, McDermott D, Gerstein M. An integrated system for studying residue coevolution in proteins. Bioinformatics. 2008;24:290–292. doi: 10.1093/bioinformatics/btm584. [DOI] [PubMed] [Google Scholar]
- 41.Dunn SD, Wahl LM, Gloor GB. Mutual information without the influence of phylogeny or entropy dramatically improves residue contact prediction. Bioinformatics. 2008;24:333–340. doi: 10.1093/bioinformatics/btm604. [DOI] [PubMed] [Google Scholar]
- 42.Bell CE, Lewis M. A closer view of the conformation of the Lac repressor bound to operator. Nat Struct Biol. 2000;7:209–214. doi: 10.1038/73317. [DOI] [PubMed] [Google Scholar]
- 43.Schumacher MA, Glasfeld A, Zalkin H, Brennan RG. The X-ray structure of the PurR-guanine-purF operator complex reveals the contributions of complementary electrostatic surfaces and a water-mediated hydrogen bond to corepressor specificity and binding affinity. J Biol Chem. 1997;272:22648–22653. doi: 10.1074/jbc.272.36.22648. [DOI] [PubMed] [Google Scholar]
- 44.Kalinina O, Gelfand M, Russell R. Combining specificity determining and conserved residues improves functional site prediction. BMC Bioinformatics. 2009;10:174. doi: 10.1186/1471-2105-10-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Files JG, Weber K. Limited proteolytic digestion of lac repressor by trypsin. Chemical nature of the resulting trypsin-resistant core. J Biol Chem. 1976;251:3386–3391. [PubMed] [Google Scholar]
- 46.Geisler N, Weber K. Isolation of amino-terminal fragment of lactose repressor necessary for DNA binding. Biochemistry. 1977;16:938–943. doi: 10.1021/bi00624a020. [DOI] [PubMed] [Google Scholar]
- 47.Taraban M, Zhan H, Whitten AE, Langley DB, Matthews KS, Swint-Kruse L, Trewhella J. Ligand-induced conformational changes and conformational dynamics in the solution structure of the lactose repressor protein. J Mol Biol. 2008;376:466–481. doi: 10.1016/j.jmb.2007.11.067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Swint-Kruse L, Larson C, Pettitt BM, Matthews KS. Fine-tuning function: correlation of hinge domain interactions with functional distinctions between LacI and PurR. Protein Sci. 2002;11:778–794. doi: 10.1110/ps.4050102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 50.Bairoch A, Apweiler R. The SWISS-PROT protein sequence database and its supplement TrEMBL in 2000. Nucleic Acids Research. 2000;28:45–48. doi: 10.1093/nar/28.1.45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mirny L, Shakhnovich E. Evolutionary conservation of the folding nucleus. Journal of Molecular Biology. 2001;308:123–129. doi: 10.1006/jmbi.2001.4602. [DOI] [PubMed] [Google Scholar]
- 52.Arvidson DN, Lu F, Faber C, Zalkin H, Brennan RG. The structure of PurR mutant L54M shows an alternative route to DNA kinking. Nat Struct Biol. 1998;5:436–441. doi: 10.1038/nsb0698-436. [DOI] [PubMed] [Google Scholar]
- 53.Swint-Kruse L, Zhan H, Fairbanks BM, Maheshwari A, Matthews KS. Perturbation from a distance: mutations that alter LacI function through long-range effects. Biochemistry. 2003;42:14004–14016. doi: 10.1021/bi035116x. [DOI] [PubMed] [Google Scholar]
- 54.Gilbert W, Maxam A. The nucleotide sequence of the lac operator. Proc Natl Acad Sci U S A. 1973;70:3581–3584. doi: 10.1073/pnas.70.12.3581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sadler JR, Sasmor H, Betz JL. A perfectly symmetric lac operator binds the lac repressor very tightly. Proc Natl Acad Sci U S A. 1983;80:6785–6789. doi: 10.1073/pnas.80.22.6785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Falcon CM, Matthews KS. Operator DNA sequence variation enhances high affinity binding by hinge helix mutants of lactose repressor protein. Biochemistry. 2000;39:11074–11083. doi: 10.1021/bi000924z. [DOI] [PubMed] [Google Scholar]
- 57.Luria SE, Adams JN, Ting RC. Transduction of lactose-utilizing ability among strains of E. coli and S. dysenteriae and the properties of the transducing phage particles. Virology. 1960;12:348–390. doi: 10.1016/0042-6822(60)90161-6. [DOI] [PubMed] [Google Scholar]
- 58.Swint-Kruse L, Elam CR, Lin JW, Wycuff DR, Shive Matthews K. Plasticity of quaternary structure: twenty-two ways to form a LacI dimer. Protein Sci. 2001;10:262–276. doi: 10.1110/ps.35801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Miller JH. A Short Course in Bacterial Genetics: A Laboratory Handbook for Escherichia coli and Related Bacteria. Plainview, N.Y: Cold Spring Laboratory Press; 1992. [Google Scholar]
- 60.Neidhardt FC, Bloch PL, Smith DF. Culture medium for enterobacteria. J Bacteriol. 1974;119:736–747. doi: 10.1128/jb.119.3.736-747.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bhende PM, Egan SM. Amino acid-DNA contacts by RhaS: an AraC family transcription activator. J Bacteriol. 1999;181:5185–5192. doi: 10.1128/jb.181.17.5185-5192.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Choi KY, Zalkin H. Structural characterization and corepressor binding of the Escherichia coli purine repressor. J Bacteriol. 1992;174:6207–6214. doi: 10.1128/jb.174.19.6207-6214.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Meng LM, Nygaard P. Identification of hypoxanthine and guanine as the co-repressors for the purine regulon genes of Escherichia coli. Mol Microbiol. 1990;4:2187–2192. doi: 10.1111/j.1365-2958.1990.tb00580.x. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Liquid culture results for LPhP exchange substitutions that do not restore lacO1 repression. The successful exception is 57–61. The numbers in the X-axis names indicate which positions have been exchanged between the PurR and LacI sequences (Table 1). The complete linker amino acid sequence for each variant is given in Supplemental Table 1. All variants express protein, as seen in the pull-down assay, except LPhP 48/55/57/58/59/61. For this protein, we cannot discriminate between total loss of DNA binding for lacO1 and lacOsym, undetectable protein expression, or enhanced protein degradation. The normalization scale is that for Figure 2A. (Top panel) Note that positions 50 and 54 directly contact DNA in both LacI and PurR 48 and are moderately conserved across the family. PurR is one of the few family members that lacks 50N or 54Q. However, creating the LPhP R54Q mutation alone and in combination with K57A did not restore any lacO1 repression. Surprisingly, the construct with exchanges at all three positions 50/54/57 could not be created, despite extensive mutagenesis efforts. Another construct that included these three exchanges did not result in detectable protein (Supplementary Table 2). We speculate that this combination of residues could result in a repressor that is toxic to E. coli. (Middle panel). The substitutions in these variants are of the 4 nonconserved positions that were previously shown to impact LLhP repression21. (Lower panel) Substitutions in the LPhP C-linker.