Abstract
The cell cycle regulator cyclin E1 is aberrantly expressed in a variety of human cancers. In breast cancer, elevated cyclin E1 correlates with poor outcome, as do high cytoplasmic levels of the stress-induced RNA binding protein Human antigen R (HuR). We showed previously that increased cytoplasmic HuR elevates cyclin E1 in MCF-7 breast cancer cells by stabilizing its mRNA. We show here that cold-inducible RNA binding protein (CIRP) co-regulates cyclin E1 with HuR in breast cancer cells. CIRP had been shown to interact with HuR in Xenopus laevis oocytes and to be decreased in endometrial cancer. To investigate if human CIRP and HuR co-regulate cyclin E1, HuR and CIRP levels were altered in MCF-7 cells and effects on cyclin E1 assessed. Altering HuR expression resulted in a reciprocal change in CIRP expression, while altering CIRP expression resulted in corresponding changes in HuR and cyclin E1 expression. CIRP and HuR co-precipitated in the presence of RNA and CIRP enhanced HuR binding to the cyclin E1 mRNA and increased cyclin E1 mRNA stability. CIRP co-localized with HuR predominantly in the nucleus, but also in discrete cytoplasmic foci identified as stress granules. CIRP overexpression increased the number of HuR-containing stress granules, while its knockdown decreased them. Our results suggest that CIRP positively regulates HuR, ultimately resulting in increased protein synthesis of at least one of its targets.
Keywords: mRNA stability, MCF-7, cell cycle
INTRODUCTION
Several independent studies have demonstrated that the cell cycle regulator cyclin E1 and its low molecular weight isoforms are aberrantly expressed in a variety of human tumor types including breast cancer [1]. Cyclin E1 is a positive regulator of the G1/S phase transition and is essential for cell cycle re-entry from G0 and oncogenic transformation [2]. Elevated cyclin E1 is a strong prognostic indicator due to significant correlation with poor outcome in patients with breast cancer [3,4]. Similar to aberrant expression of cyclin E1, high cytoplasmic levels of the RNA binding protein (RBP) HuR are closely associated with high histologic grade, large primary tumor size, and poor survival in breast cancer, as well as in colon, lung, and ovarian cancers [5–9]. HuR is a stress-induced RBP that shuttles from the nucleus to the cytoplasm to stabilize and promote translation of its target mRNAs [10,11]. These findings established both cyclin E1 and HuR as important molecular markers for tumor aggressiveness.
In a recent study, we demonstrated that elevated cytoplasmic HuR in MCF-7 breast cancer cells increased cyclin E1 mRNA stability and growth-promotion, defining a new mechanism of cyclin E1 deregulation in breast cancer [12]. Since HuR, like any RBP, has many potential target mRNAs, we performed microarray analysis of MCF-7 cells treated with HuR siRNA to identify other targets. Our screen resulted in the identification of another stress-induced RBP, cold-inducible RBP (CIRP).
CIRP belongs to the glycine-rich RBP family, which possesses an RNA recognition motif (RRM), and a carboxyl-terminal domain containing several RGG motifs [13]. CIRP is expressed in a wide variety of tissues and cells and can be induced by cellular stresses such as cold shock, UV irradiation and hypoxia [14–16]. Upon stress induction, CIRP shuttles from the nucleus to the cytoplasm to stabilize target mRNAs [17,18]. It mediates suppression of cell growth with prolongation of G1 phase [14], and contributes to the suppression of apoptosis induced by tumor necrosis factor-α [19]. CIRP also contributes to the maintenance of normal cellular function [20,21]. CIRP is suggested to participate in cell cycle regulation of normal human endometrium and loss of its expression may be involved in endometrial carcinogenesis [22]. Of particular relevance to our studies was the report that in Xenopus laevis oocytes, CIRP stabilizes AU-rich element (ARE)-containing mRNAs via interaction with the Xenopus homolog of HuR [23].
In the present study, we tested the hypothesis that human CIRP and HuR cooperatively regulate cyclin E1, elevating its expression in breast cancer cells. We show that CIRP is upregulated in breast cancer cells as compared to nontumorigenic breast epithelial cells. CIRP positively modulates the expression of both cyclin E1 and HuR in MCF-7 breast cancer cells, enhancing HuR binding to cyclin E1 mRNA as well as cyclin E1 mRNA half-life. Thus, CIRP contributes to cyclin E1 overexpression by positively regulating HuR. This is one of the first studies to show that CIRP expression is increased in breast cancer cells, and that it upregulates two proteins implicated in the etiology of breast cancer that also serve as prognostic markers.
MATERIAL AND METHODS
Cell culture
MCF-7 and T47D cells (American Type Culture Collection (ATCC), Manassas, VA) were cultured in DMEM (Invitrogen, Carlsbad, CA) containing 10% fetal bovine serum (FBS). MDA-MB-231 cells (ATCC) were grown in DMEM/F12 (Sigma, St. Louis, MO) supplemented with 10% FBS. MCF10A cells (ATCC) were grown in DMEM/F12 supplemented with 5% FBS, 20 ng/ml epidermal growth factor (Sigma), 0.01 mg/ml insulin (Sigma), 500 ng/ml hydrocortisone (Sigma). AG11132 and AG11137 normal human mammary epithelial cells (NIA Cell Culture Repository, Coriell Institute, NJ) were grown in MEGM (Lonza Group Ltd. Basel, Switzerland). All media contained 100 units/ml penicillin, and 100 µg/ml streptomycin.
Plasmid constructs
pGEM-T-easy-cyclin E1 3' untranslated region (3'UTR) and pGEM-T-easy-cyclin E1CR378 were constructed as described previously [12]. FLAG-tagged CIRP was generated as described [24] except MCF10A cDNA was used as template with primers (FLAG-tag is underlined): forward, 5’-GAGCGGGACACCATGGCATCAGATGAAGGC-3’; reverse, 5’-GCAAGCTTTTCACTTGTCATCGTCGTCCTTGTAGTCCTCGTTGTGTGTAGTA-3’ (Genbank accession No. BC000403). FLAG-CIRP was cloned into pGEM-T-easy (Promega, Madison, WI) and then subcloned into pTracer-CMV2 (Invitrogen). For generation of glutathione S-transferase- (GST) CIRP, a cDNA encoding residues 2-172 of CIRP was generated by PCR with a BamHI-linked forward primer 5’-CGGGATCCGCATCAGATGAAGGCAAAC-3’ and an EcoRI-linked reverse primer 5’-CGGAATTCCTCGTTGTGTGTAGCGTAACTG-3’ using MCF10A cDNA as template. The PCR product was digested with BamHI and EcoRI and ligated into pGEX-2T digested with the same enzymes. The resulting pGEX-CIRP was used to produce GST-CIRP in BL21DE3 pLysS E. coli as recommended (Amersham Biosciences, Uppsala, Sweden). Protein concentration was determined using Bio-Rad Protein Assay (Bio-Rad, Hercules, CA). All constructs were sequenced.
Cell transfections
CIRP Silencer siRNA was constructed using Silencer siRNA Construction Kit (Ambion, Austin, TX). The control siRNA and four CIRP siRNAs were transfected into MCF-7 cells and 72 hr later CIRP silencing assayed by immunoblotting. CIRP siRNA #2 (sense 5'-AATGTCTTTCACAACCACCACCCTGTCTC-3' and anti-sense 5'-AAGTGGTGGTTGTGAAAGACACCTGTCTC-3') was used as it decreased CIRP expression by 80%, while the expression of actin did not change. MCF-7 cells at 70% to 80% confluence in 6-well plates were transfected with 4 µg pTracer-CMV2 vector, pTracer-CMV2-CIRP, or 40 nM control or CIRP siRNA using lipofectamine 2000 (Invitrogen). 72 hr later, total cell lysates, or cytoplasmic and nuclear fractions were immunoblotted. Transfection with HuR and siHuR constructs was as previously described [12]. Transfection efficiencies were between 60–70%.
Immunoblot analysis
Primary antibodies against CIRP (Proteintech Group, Inc., Chicago, IL), cyclin E1 and HuR (Santa Cruz Biotechnology, Santa Cruz, CA) and HRP-conjugated anti-rabbit or anti-mouse secondary antibodies (Santa Cruz) were used. The blots were developed using SuperSignal West Pico Chemiluminescent Substrate (Pierce, Rockford, IL). Membranes were stripped and reprobed for β-actin (Sigma) or β-tubulin (Santa Cruz) as loading controls for total and cytoplasmic extracts or with α-histone deacetylase (HDAC1) for nuclear fractions (Santa Cruz). The density of protein bands was measured using Kodak Molecular Imaging Software. The relative quantity of protein was calculated after normalization to β-actin.
Real-time RT-PCR
To assay mRNA level, total RNA was isolated from cells using Trizol reagent (Invitrogen). For mRNA stability assays, 24 h after transfection with control siRNA, CIRP siRNA, pTracer-CMV2, or pTracer-CMV2-CIRP, cells were synchronized at late G1 by serum deprivation for 24 hr followed by 24 hr incubation with aphidicolin (5 µg/ml). Total RNA was extracted at the indicated time points following addition of actinomycin D (5 µg/ml) to inhibit new transcription. mRNA was reverse transcribed using random hexamers. The resulting cDNAs were amplified by real-time PCR using SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA). Primers were: CIRP forward 5’-TTTGGGTTTGTCACCTTTGA-3’ and reverse 5’-ACTTGCCTGCCTGGTCTACT-3’; HuR forward 5’-GACATCGGGAGAACGAATTT-3’ and reverse 5’-TGCTGAACAGGCTTCGTAAC-3’; cyclin E1 forward 5'-CGGCTCGCTCCAGGAA-3' and reverse 5'-TCATCTGGATCCTGCAAAAAAA-3'; glyceraldehyde-3-phosphate dehydrogenase (GAPDH; Genbank accession No. XM-006959), forward 5'-ATGGAAATCCCATCACCATCTT-3' and reverse 5'-CGCCCCACTTGATTTTGG-3'. Threshold cycles (Ct) were normalized to GAPDH and data expressed as relative mRNA level.
GST-pull down and immunoprecipitation assays
For GST-pull downs, 10 µg GST or GST-CIRP was incubated with 100 µl cell lysate (10 µg/µl) supplemented with 900 µl lysis buffer (1% Triton X-100, 150 mM NaCl, 2 mM MgCl2, 2 mM CaCl2, 10 mM HEPES, pH 7.4; 1 µg/ml each of leupeptin, pepstatin, chymotrypsin, and PMSF) at 4°C with agitation overnight. 80 µl Glutathione-Sepharose (50% slurry) was added and incubated for an additional 3–4 hr at 4°C with agitation, washed extensively, resuspended in 2× SDS sample buffer and the supernatants used for HuR immunoblotting. For immunoprecipitation assays, 1 mg of cell lysate with or without RNase A treatment was pre-cleared with 20 µl protein A-agarose (Santa Cruz) and then incubated with 1 µg normal rabbit IgG or anti-CIRP antibody at 4°C with agitation overnight. 20 µl protein A-agarose was added and lysates incubated for 3–4 hr at 4°C with agitation and then pelleted. Supernatants were used for β-actin immunoblotting to monitor protein loading. Washed beads were resuspended in 2× SDS sample buffer and supernatants used for CIRP and HuR immunoblotting.
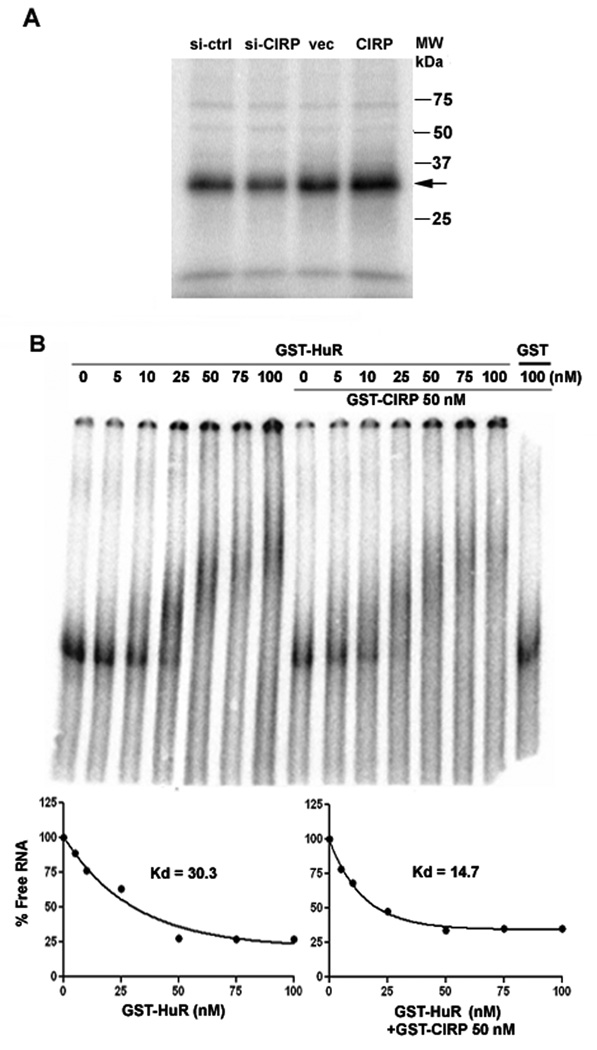
Gel shift and UV-crosslink analyses
32P-UTP-labeled or unlabeled cyclin E1 3'UTR or cyclin E1 CR378 (partial cyclin E1 coding region) were generated by in vitro transcription. For UV-crosslink assays, 500 ng of GST-CIRP was pre-incubated for 15 min with 100 molar excess of unlabeled cyclin E1 3'UTR or cyclin E1 CR378 before addition of 32P-labeled cyclin E1 3'UTR. RNA-protein complexes were UV-crosslinked, resolved by SDS-PAGE, and dried gels analyzed on a Storm phosphorimager (Molecular Dynamics, Sunnyvale, CA). For UV-crosslink immunoprecipitation, 100 µg cell lysate was incubated with 32P-labeled cyclin E1 3'UTR or cyclin E1 CR378, UV-crosslinked and digested with RNase T1 and RNase A. CIRP or HuR were immunoprecipitated as described and dried gels analyzed by phosphorimaging. For gel shift assays, an increasing amount of GST-HuR (0, 10, 25, 50, 75, 100 nM) alone or with 50 nM GST-CIRP was incubated for 20 min with 1 fmol 32P-labeled cyclin E1 3'UTR. The complexes were resolved by 5% native PAGE and phosphorimaged. Free vs. bound RNA was quantitated and Kd determined using one-phase exponential decay (Graphpad Prism software).
Immunofluorescence microscopy
MCF-7 cells plated on coverslips were transfected with control siRNA, CIRP siRNA, pTracer-CMV2, or pTracer-CMV2-CIRP. After 72 hr, cells were fixed, permeabilized, and nonspecific binding sites blocked in PBS with 0.1% Tween-20, 1% BSA and 5% normal serum. Cells were incubated at 4°C overnight with mouse anti-HuR (1:1,000 in PBS with 0.1% Tween-20 and 0.5% BSA). Co-immunostaining was performed similarly with mouse anti-HuR and rabbit anti-CIRP (1:200) or goat anti-TIA-1 (1:200, Santa Cruz). Secondary antibodies were labeled with Cyanine 3 (CY3) for HuR or AlexaFluor 488 for CIRP and TIA-1. Mounting solution contained DAPI and anti-fade agent (Vector laboratories, Inc, Burlingame, CA). Cells were visualized using an Olympus BH2-RFCA inverted microscope (Olympus Optical Co. LTD, Japan) equipped for epifluorescence. Images were collected using standard filter sets for AlexaFluor, CY3 and DAPI.
Statistics
Values were expressed as mean ± SD and assessed by Student's t test. P-values < 0.05 were considered to be statistically significant. All experiments were repeated at least three times.
RESULTS
HuR knockdown increased CIRP expression in MCF-7 cells
We identified CIRP as a gene upregulated in response to HuR knockdown in MCF-7 cells. We focused on CIRP as it co-regulates mRNAs with the Xenopus ortholog of HuR [23]. CIRP prolongs G1 phase in mammalian cells [14] as does HuR knockdown [12] and is decreased in endometrial cancer [22]. Based on these observations, we hypothesized that human CIRP and HuR may cooperatively regulate cyclin E1 expression in human breast cancer cells.
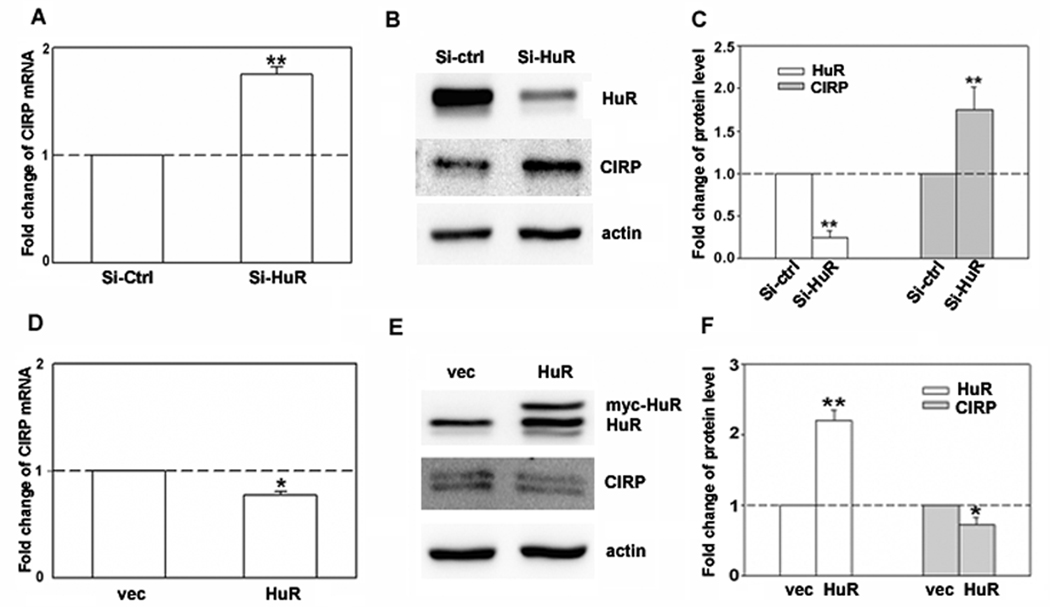
To confirm our microarray results showing CIRP upregulation in response to HuR knockdown, we performed real-time PCR and immunoblot analysis for CIRP in MCF-7 cells treated with HuR siRNA. As shown in Figure 1, HuR knockdown increased both CIRP mRNA (A) and protein (B and C) by 2-fold. This increase was consistent with the 2-fold increase in CIRP mRNA seen by microarray analysis (not shown). To test if HuR overexpression would have the opposite effect, we overexpressed HuR by transfecting pcDNA3.1mycHuR into MCF-7 cells and assessed CIRP mRNA and protein levels. Myc-HuR expression (Fig. 1E, upper band on HuR blot) slightly but significantly decreased both CIRP mRNA (Fig. 1D) and protein levels (Fig. 1E and F). A larger decrease in CIRP upon HuR overexpression may not have been seen because HuR is already elevated in MCF-7 cells as compared to non-transformed mammary epithelial cells ([12] and Fig. 2D). These data suggested that HuR regulates CIRP or the CIRP mRNA, either directly or indirectly. In this and following experiments actin was used as a loading control. Since actin was recently shown to be a target of HuR in HeLa cells [25], we also used tubulin as a loading control with identical results (Supplemental Fig. S1).
Fig. 1.
Regulation of CIRP expression by HuR knockdown and overexpression in MCF-7 cells. A and D, Real-time RT-PCR was used to analyze CIRP mRNA level. Total RNA was harvested 72 hr after transfection of HuR siRNA (Si-HuR), control siRNA (Si-ctrl), pcDNA3.1mycHuR (HuR), or pcDNA3.1 vector (vec). Data were normalized to GAPDH and expressed as relative mRNA levels. B and E, The protein levels of HuR and CIRP in MCF-7 cells were analyzed by sequential western blotting 72 hr after transfection. Equal loading of protein was determined by re-probing of blots for β-actin. C and F, The relative quantity of protein was calculated after normalization to β-actin. n = 4, * p < 0.05, **, p < 0.01 versus control siRNA or pcDNA3.1 vector.
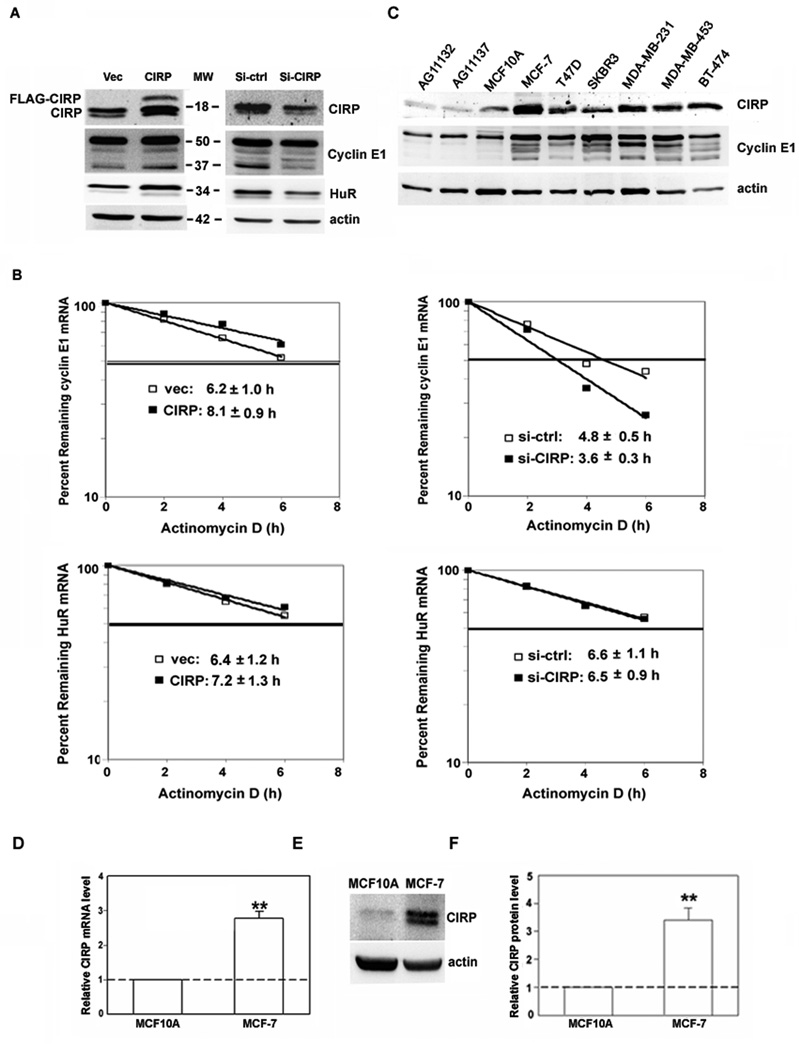
Fig. 2.
Regulation of cyclin E1 and HuR expression by CIRP knockdown and overexpression. A, MCF-7 cells were transfected with CIRP siRNA (Si-CIRP), control siRNA (Si-ctrl), pTracer-CMV2 (Vec), or pTracer-CMV2-CIRP (CIRP). Total protein was extracted 72 hr after transfection and the levels of CIRP, cyclin E1 and HuR were analyzed by western blotting. Equal loading of protein was determined by re-probing for β-actin. B, MCF-7 cells were synchronized at late G1 phase 24h after transfection. Total RNA was extracted at the indicated time after the addition of actinomycin D. Real time RT-PCR was used to analyze HuR and cyclin E1 mRNA level. Data were normalized to GAPDH mRNA and plotted on semi logarithmic scales. p < 0.05 for cyclin E1 mRNA in CIRP/siCIRP cells vs. controls. C, Western blotting for CIRP and cyclin E1 in total protein extracts from normal mammary epithelial cells (lanes 1–2: AG11132, AG11137), immortalized nontumorigenic mammary epithelial cells (lane 3: MCF10A) and 6 breast carcinoma cell lines (lanes 4–9: MCF-7, T47D, SKBR3, MDA-MB-231, MDA-MB-453, BT-474). The blot was stripped and re-probed for β-actin to control for protein loading. D, CIRP mRNA level was analyzed by real-time RT-PCR and expressed as relative level after normalization to GAPDH. E, CIRP protein level was analyzed by western blotting and blots stripped and re-probed for β-actin. F, The relative quantity of CIRP protein was calculated from E after normalization to β-actin. For all experiments, n = 3, **, p < 0.01 versus MCF10A.
CIRP contributed to cyclin E1 upregulation in MCF-7 cells
We showed previously that one target mRNA stabilized by HuR in MCF-7 cells was that encoding cyclin E1. To test if CIRP can also target cyclin E1 mRNA, we manipulated CIRP expression and assessed cyclin E1 protein level. CIRP was overexpressed by transfecting MCF-7 cells with pTracer-CMV2-CIRP or silenced by transfecting with CIRP siRNA. As shown in the left panel of Figure 2A, compared to vector alone, overexpression of CIRP increased cyclin E1 protein level. In contrast, CIRP silencing decreased cyclin E1 (Fig. 2A, right). Identical to results seen upon altering HuR level [12], a 37-kDa low molecular weight (LMW) form of cyclin E1 was preferentially affected by changes in CIRP expression. When control levels are set to 1, full length and LMW forms are increased to 1.2 and 1.6, respectively, upon CIRP overexpression and decreased to 0.7 and 0.4 upon CIRP silencing. Interestingly, when CIRP was overexpressed, HuR protein level increased, and when CIRP was silenced, HuR protein level decreased (Fig. 2A). These results showed that CIRP positively regulates HuR expression and suggested that CIRP might regulate cyclin E1 expression via its effects on HuR.
To determine if a change in mRNA stability accompanies the change in cyclin E1 and HuR protein levels upon CIRP alteration, we analyzed mRNA half-life in CIRP altered cells. Figure 2B (top) shows that the half-life of cyclin E1 mRNA increases upon CIRP overexpression and decreases upon CIRP knockdown. In contrast, HuR mRNA half-life is not significantly affected by changes in CIRP expression (Fig. 2B, bottom). These results show that CIRP regulates cyclin E1 but not HuR mRNA stability.
CIRP was increased in breast cancer cells
We next assessed endogenous CIRP level in several different breast carcinoma cell lines. As shown in Figure 2C, when compared to nontransformed human mammary epithelial cells (lanes 1–3) and normalized to actin to control for protein loading, CIRP expression was higher in 6 breast cancer cell lines (lanes 4–9) that also overexpress cyclin E1 and its LMW forms. When nontumorigenic MCF10A and tumorigenic MCF-7 cells were directly compared, endogenous CIRP mRNA (Fig. 2D) and protein (Fig. 2E and F) were higher in MCF-7 cells, with a 2.8-fold difference in mRNA and 3.4-fold difference in protein. The difference in CIRP level was not an indirect effect of maintaining MCF10A and MCF-7 cells in different media, as we obtained identical results with cells grown in the same media (data not shown). These data show that CIRP was increased in breast cancer cells that overexpressed cyclin E1. When combined with the CIRP overexpression and knockdown studies, results suggest that CIRP positively regulates cyclin E1, perhaps via HuR, to contribute to its overexpression in breast cancer.
CIRP binds the Cyclin E1 mRNA
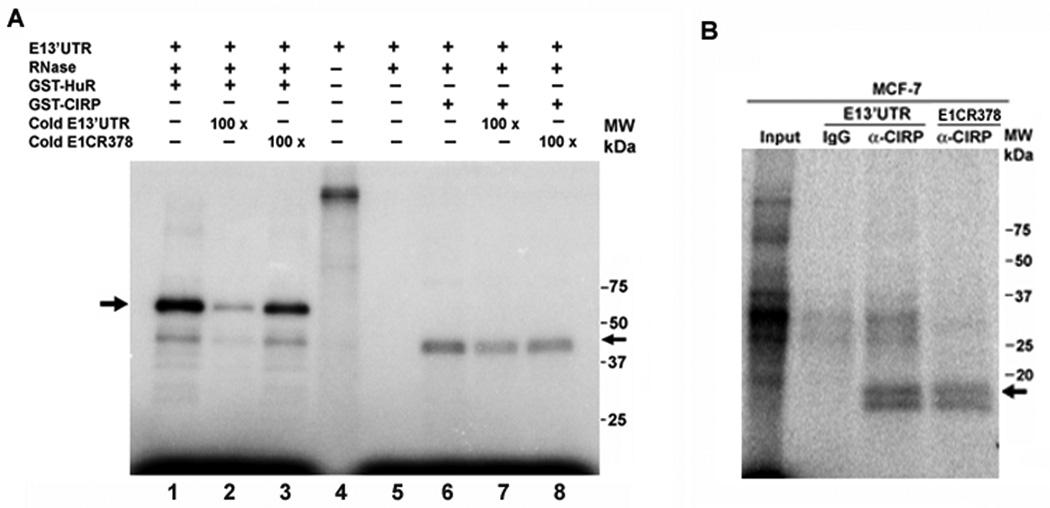
We carried out UV-crosslink and immunoprecipitation assays to ask whether cyclin E1 mRNA was bound by CIRP. As shown in Figure 3A, UV-crosslink competition analysis showed binding of GST-CIRP to the 3' untranslated region (3'UTR) of cyclin E1 mRNA (lane 6). Binding was partially competed by unlabeled cyclin E1 3'UTR at 100 molar excess (E13'UTR, lane 7), but also by a competitor consisting of part of the cyclin E1 coding region (E1CR378, lane 8). In contrast, GST-HuR binding was well competed by E13'UTR (lane 2) but not E1CR378 (lane 3) competitor RNA. Immunoprecipitation of UV-crosslinked cytoplasmic extract of MCF-7 cells verified that endogenous CIRP binds the 3'UTR of cyclin E1 mRNA (Fig. 3B). An 18-kDa protein was precipitated by CIRP-specific antibodies (α-CIRP) with both cyclin E1 3'UTR and E1CR378, but not by nonspecific IgG. A 35-kDa nonspecific complex binding the cyclin E1 3’UTR was precipitated with both IgG and CIRP antibody. These results indicate that endogenous CIRP binds to the cyclin E1 3’UTR, but also binds the cyclin E1 coding region.
Fig. 3.
CIRP and HuR bind the cyclin E1 mRNA. A, UV-crosslink competition analysis. An equal amount of GST-CIRP or GST-HuR was incubated with 32P-labeled cyclin E1 mRNA 3’UTR (E13’UTR) in the presence of 0 (lanes 1 and 6) or 100 molar excess unlabeled specific (cold E13’UTR, lanes 2 and 7) or nonspecific (cold E1CR378, lanes 3 and 8) competitor. Arrows indicate protein binding to the cyclin E1 3’UTR. Lane 4, radiolabeled RNA alone; lane 5, radiolabeled RNA treated with RNase in the absence of added protein. B, UV-crosslink immunoprecipitation analysis. MCF-7 cell extract was incubated with 32P-labeled cyclin E1 mRNA 3’UTR (E13’UTR) or cyclin E1CR378 mRNA. After crosslinking and RNase treatment, RNA-protein complexes were precipitated with CIRP antibody (α-CIRP) or normal IgG followed by protein A/G agarose beads and resolved on a 12% SDS-polyacrylamide gel. Input lane is the UV-crosslinked 32P-labeled cyclin E1 3’UTR with MCF-7 cell extract. Arrow indicates the 18-kDa protein immunoprecipitated by CIRP antibody. Experiments were repeated at least 3 times.
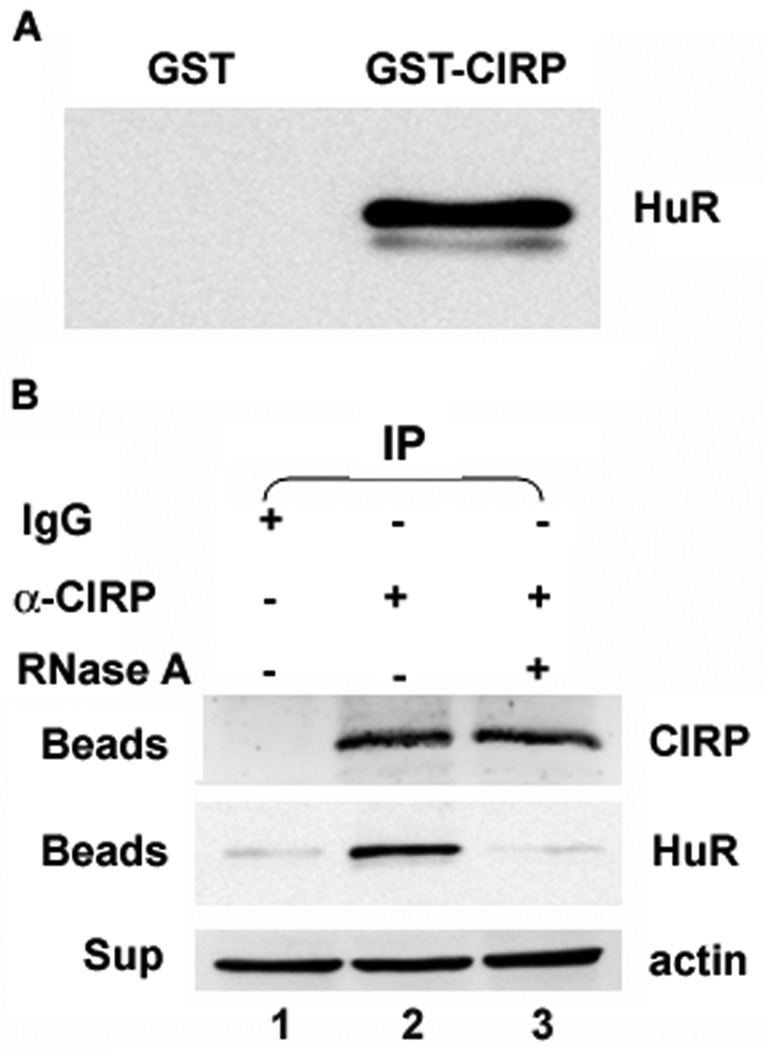
CIRP co-precipitated with HuR dependent on the presence of RNA
We next asked whether CIRP could be regulating cyclin E1 via effects on HuR. Based on published studies showing that the Xenopus orthologs of CIRP and HuR interact, we examined whether human CIRP could directly interact with HuR. As shown in the HuR immunoblot in Figure 4A, GST-CIRP but not GST pulled down endogenous HuR from MCF-7 cell extracts. To ask if the endogenous proteins associate, we immunoprecipitated CIRP from MCF-7 cells and performed western analysis for HuR. Figure 4B shows that HuR co-precipitated with CIRP (lane 2), but not with normal IgG (lane 1). Co-precipitation of HuR and CIRP was dependent on the presence of RNA as RNase A treatment abolished co-precipitation (lane 3). Thus, CIRP and HuR appear to bind to an overlapping pool of RNAs.
Fig. 4.
CIRP co-precipitates with HuR. A, GST pull-down assay: GST or GST-CIRP was incubated with MCF-7 cell extract. GST-CIRP was pulled down with Glutathione-Sepharose and eluted proteins immunoblotted for HuR. B, MCF-7 cell extract treated with or without RNase A was incubated with CIRP antibody (α-CIRP) or normal IgG followed by protein A/G agarose beads. The co-precipitated proteins eluted from the beads (Beads) were resolved on a 10% SDS-polyacrylamide gel and immunoblotted for CIRP or HuR. The supernatants (Sup) were immunoblotted for β-actin to monitor equal amounts of starting protein. Experiments were repeated at least 3 times.
CIRP increased HuR binding to the cyclin E1 3'UTR
The RNA-dependent association of CIRP and HuR prompted us to test whether CIRP could affect the binding of HuR to the cyclin E1 3'UTR. To test this, we performed UV-crosslink analysis with extracts of MCF-7 cells whose CIRP level was altered, followed by immunoprecipitation of HuR. Figure 5A shows that HuR binding to the cyclin E1 3'UTR decreased upon CIRP knockdown (si-CIRP) and increased upon CIRP overexpression (CIRP). These changes could be due to the altered HuR level, and/or to CIRP facilitating HuR binding to the cyclin E1 3’UTR. To ask if CIRP influences HuR binding to the cyclin E1 3’UTR, native gel shifts were performed. Figure 5B shows that GST-HuR bound to the cyclin E1 3'UTR with a Kd of 30.3 nM. The addition of GST-CIRP but not GST alone decreased the Kd to 14.7 nM. These results were reproducible, as similar results were obtained in three separate experiments, showing that CIRP can influence HuR binding to cyclin E1 mRNA. Despite this, the effect is only two-fold, suggesting that the positive effects of CIRP on cyclin E1 can mostly be explained by the increase in HuR expression.
Fig. 5.
CIRP enhances the binding of HuR to the cyclin E1 mRNA 3’UTR. A, MCF-7 cells were transfected with control siRNA (si-ctrl), CIRP siRNA (si-CIRP), pTracer-CMV2 vector (vec), or pTracer-CMV2-CIRP (CIRP). Equal amounts of cell lysates were incubated with 32P-labeled cyclin E1 mRNA 3’UTR, UV-crosslinked, and treated with RNase. The resulting RNA-protein complexes were precipitated with HuR antibody/protein A/G agarose and resolved on a 10% SDS-polyacrylamide gel. Arrow indicates radiolabeled protein immunoprecipitated by the HuR antibody. B, Gel shift assay. 32P-labeled cyclin E1 mRNA 3’UTR (1 fmol) was incubated with the indicated amounts of GST-HuR +/− 50 nM GST-CIRP. RNA-protein complexes were resolved on a 5% native polyacrylamide gel, phophorimaged and Kd calculated using a one-phase exponential decay. All experiments were repeated at least 3 times.
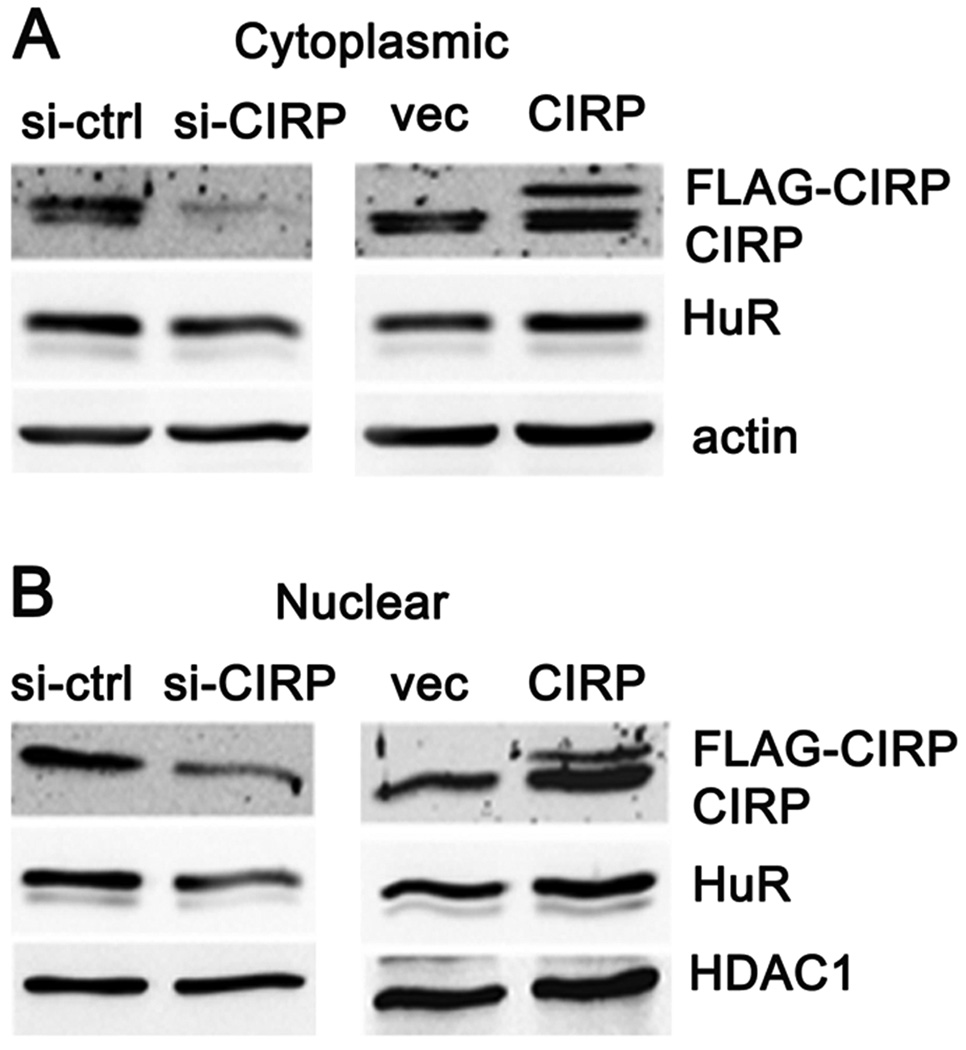
CIRP overexpression increased cytoplasmic foci of HuR
Despite their role in cytoplasmic mRNA stabilization, HuR and CIRP are predominantly nuclear proteins [17,18,26,27]. Upon cellular stress, a fraction of these proteins translocates to the cytoplasm where they bind target mRNAs and prevent their decay, presumably enhancing cell survival in response to stress [9,17,18,26]. Overexpression of CIRP could be increasing the pool of cytoplasmic HuR available for binding cyclin E1 mRNA. To address this possibility, we first compared the level of HuR in cytoplasmic and nuclear fractions from MCF-7 cells in which CIRP was either overexpressed or silenced. Figure 6 shows that CIRP overexpression increased both cytoplasmic (A) and nuclear (B) HuR, while CIRP silencing decreased both. Quantitation of HuR (and CIRP) level showed that there was no shift in nuclear/cytoplasmic ratio, just an overall change in protein level (not shown). Interestingly, CIRP blots of cytoplasmic fractions displayed two bands, while those of nuclear fractions displayed one band. These bands may result from post-translational modification of CIRP and their presence varied in immunoblots of whole cell extracts (compare Fig. 1B and E, Fig. 2, and S1), suggesting that CIRP post-translational modification may vary with stress and/or subcellular localization.
Fig. 6.
CIRP increases both cytoplasmic and nuclear HuR. MCF-7 cells were transfected with control siRNA (si-ctrl), CIRP siRNA (si-CIRP), pTracer-CMV2 vector (vec), or pTracer-CMV2-CIRP (CIRP). A, Cytoplasmic (cyto) and B, nuclear (Nuclear) protein was extracted 72 hr after transfection and the levels of CIRP and HuR analyzed by western blotting. Equal protein loading was determined by re-probing for β-actin (cyto) or HDAC1 (Nuclear).
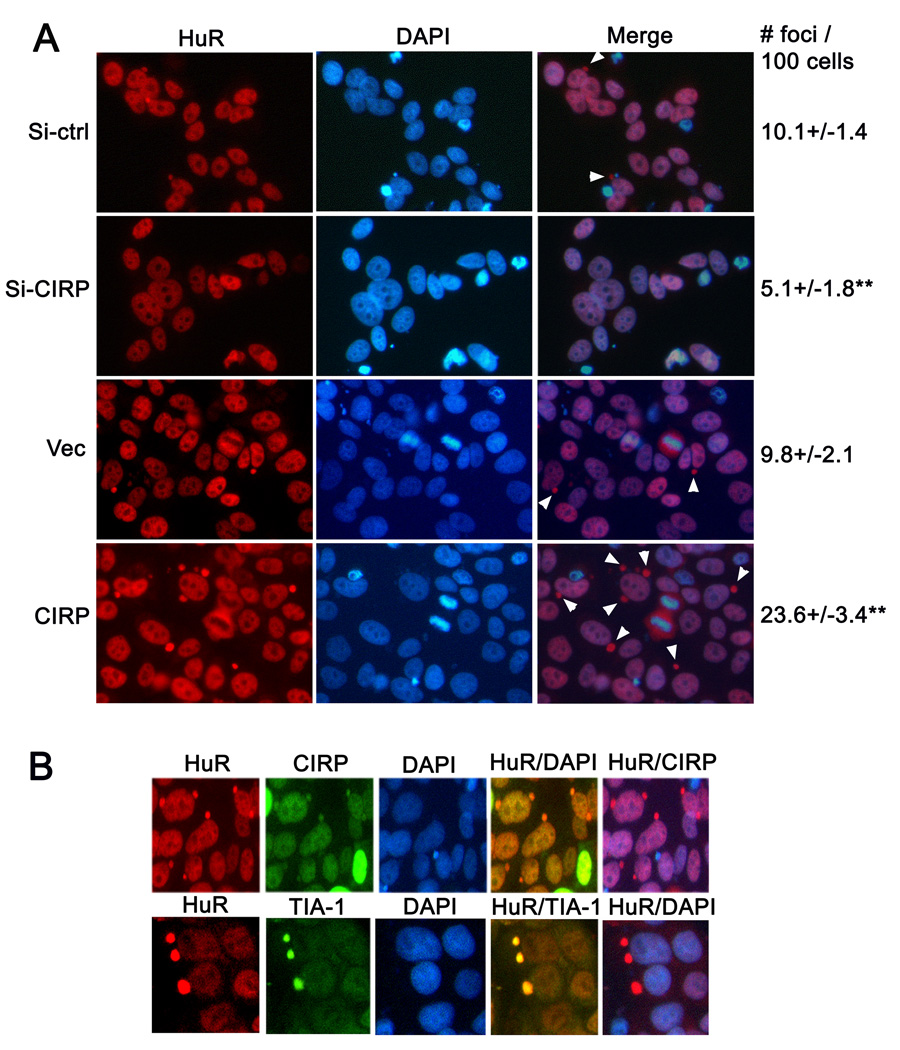
Next, we used immunofluorescence microscopy to verify that altering CIRP does not affect the subcellular localization of HuR. Figure 7A shows that HuR was primarily nuclear when visualized by microscopy. HuR was also present in the cytoplasm localized to small round structures (white arrowheads in Merge panels). These structures or foci were often closely associated with the nucleus but did not contain DNA, as could be seen by comparing DAPI and Merge panels. Knockdown of CIRP decreased the number of these structures from 10% to 5% of the total HuR (Si-ctrl vs. Si-CIRP), while CIRP overexpression (CIRP vs. Vec) increased them to about 24%.
Fig. 7.
CIRP increases HuR containing cytoplasmic foci. MCF-7 cells were transfected with control siRNA (si-ctrl), CIRP siRNA (si-CIRP), pTracer-CMV2 vector (vec), or pTracer-CMV2-CIRP (CIRP). A, HuR localization was assessed using immunofluorescence analysis. Representative fluorescence micrographs of cells 72 hr after transfection are shown. HuR staining was visualized with Cy-3 conjugated secondary antibody (red). Nuclei were stained with DAPI (blue). White arrowheads in the merge panel indicate cytoplasmic foci containing HuR but not DAPI. Cytoplasmic foci containing HuR were counted in 500 cells for each condition and shown on the right as foci number per 100 cells. ** p< 0.01 as compared to Si-ctrl or Vec. B, HuR co-localization with CIRP and TIA-1 was assessed in MCF-7 cells transfected with pTracer-CMV2-CIRP using immunofluorescence analysis. Representative fluorescence micrographs of cells 72 hr after transfection are shown. HuR staining was visualized with Cy-3 conjugated secondary antibody (red). CIRP and TIA-1 were visualized with Alexa Fluor 488 (green) conjugated secondary antibody). Nuclei were stained with DAPI (blue). All experiments were repeated at least 3 times.
We used co-immunofluorescence analysis to ask if CIRP and HuR co-localized and to query the identity of the cytoplasmic foci. As previously reported by others [17] we found that CIRP, like HuR, was predominantly nuclear (Figure 7B, top). Upon overexpression, CIRP accumulated in the same cytoplasmic foci as HuR (Figure 7B, top). We next queried the identity of the foci, specifically asking if they correspond to stress granules (SGs), cytoplasmic foci that form in response to cytoplasmic or endoplasmic reticulum stresses. CIRP was recently shown to be recruited to SGs in response to stress or upon its overexpression, which induces SG formation [17]. To determine if the foci correspond to SGs, we immunostained for TIA-1, an RBP that is considered a SG marker [28,29] As shown in Figure 7B (second), TIA-1 was present in the foci containing HuR and CIRP, identifying them as SGs. Taken together, these data are consistent with a model in which CIRP upregulation increases overall HuR level but does not affect its nuclear to cytoplasmic distribution. The number of cytoplasmic stress granules containing HuR and CIRP increased in response to CIRP upregulation, probably due to increased overall protein levels.
DISCUSSION
In this study, we show that expression of the stress-induced RBP CIRP is increased in breast cancer cells, where it contributes to increased levels of the RBP HuR and the G1-S regulator cyclin E1. We previously showed that higher cytoplasmic HuR in MCF-7 breast cancer cells contributed to upregulation of cyclin E1 via stabilization of its mRNA [12]. We next observed that HuR knockdown increased CIRP level, concomitant with a decrease in cyclin E1 and G1-phase cell cycle arrest. CIRP had been shown to cause G1-phase arrest [14] in mammalian cells and to co-regulate mRNAs with HuR in a Xenopus model system [23]. Based on these collective data, we hypothesized that CIRP and HuR may also co-regulate target mRNAs in human cells. To test this, we overexpressed and knocked down CIRP in MCF-7 cells and monitored cyclin E1 level. Cyclin E1 protein and its mRNA stability increased upon CIRP overexpression, and decreased upon CIRP knockdown, suggesting positive regulation of cyclin E1 by CIRP. A greater effect was seen on the 37-kDa LMW form of cyclin E1 as compared to full length cyclin E1. LMW isoforms arise from proteolytic processing of full length cyclin E1, appear to be cancer specific, and may be a function of the total amount of cyclin E1 present in a cell [30]. Thus the preferential change in LMW cyclin E1 may be secondary to the change in full-length protein. Regardless of origin, LMW cyclin E1 forms hyperactive kinase complexes with Cdk2 that are resistant to cdk inhibitors, is associated with genetic instability and is predictive of poor clinical outcome [31–33].
CIRP bound to both the cyclin E1 3'UTR and coding region, suggesting that regulation may be via multiple binding sites. It is also possible that CIRP binds nonspecifically to the cyclin E1 mRNA and that CIRP regulates cyclin E1 only via its regulation of HuR, a possibility that is currently under investigation. CIRP regulated HuR protein level but did not affect HuR mRNA stability, suggesting an alternate mode of increasing HuR protein. CIRP co-precipitated with HuR in the presence of RNA, showing that they bound an overlapping pool of RNAs. Consistent with this, CIRP enhanced HuR binding to the cyclin E1 3’UTR. Together, these data suggest that CIRP positively regulates cyclin E1, at least in part via its regulation of HuR.
RBPs are key components in the post-transcriptional regulation of gene expression. These proteins contain well-conserved RNA binding domains mediating RNA contact but also auxiliary domains involved in protein-protein interactions and subcellular targeting [34]. The complement of interacting RBPs determines the stability and translation of mRNAs [35]. Xenopus orthologs of CIRP and HuR stabilize ARE-containing mRNAs cooperatively, but at distinct steps [23]. Another member of the glycine rich RBP family, RBM3, was recently shown to co-stabilize mRNAs with HuR, to be overexpressed in several different cancers, and to promote oncogenesis [36]. Our results are consistent with these prior studies. Comparison of CIRP level in 3 different nontumorigenic breast epithelial cells with 6 breast carcinoma cell lines showed that CIRP protein was higher in the breast cancer cells, correlating with higher cyclin E1 protein. As well as increasing HuR overall protein level, CIRP enhanced HuR binding to the cyclin E1 3'UTR, suggesting that CIRP stabilizes cyclin E1 mRNA and increases cyclin E1 protein level at least in part via its effects on HuR.
Both CIRP and HuR shuttle from the nucleus to the cytoplasm in response to stress [10,37,38]. We found that altering CIRP expression did not change the nuclear to cytoplasmic distribution of HuR or itself. When assessed by immunofluorescence microscopy, HuR was primarily nuclear but was also present in distinct cytoplasmic foci. The number of these foci increased in response to CIRP overexpression, and decreased upon CIRP knockdown. These cytoplasmic structures contained CIRP as well as TIA-1, distinguishing them as SGs. Untranslated mRNAs and several RBPs, including HuR [29,39], are found in SGs, which appear to act as triage centers that sort, remodel and export specific mRNPs for translational re-initiation, decay, or storage [40]. Consistent with our results, HuR is a known component of SGs [29,39], CIRP is recruited to SGs in response to stress, and CIRP overexpression induces SG formation [17]. As one proposed function of SGs is marking mRNPs for translational re-initiation and the presence of HuR is thought to be one of the “marks” [41,42], the accumulation of HuR containing SGs upon CIRP overexpression could ultimately increase cyclin E1 expression by marking its mRNP for translational re-initiation upon export. Cyclin E1 mRNA stabilization upon CIRP overexpression is also consistent with this possibility. It is also possible that the increase in SGs is due to sequestration of excess CIRP/HuR and does not affect cyclin E1 level.
Our observation that CIRP is elevated in breast cancer contrasts with a reported decrease in CIRP in endometrial cancer [22]. CIRP has been shown to play roles in maintenance of normal cellular functions and morphogenesis, biological rhythms, suppression of cell growth induced by cold stress, and protection against apoptosis [14,16,19–22]. Based on this, one explanation for the apparent differences in CIRP levels in breast and endometrial cancers is that CIRP may play different roles depending on the types, stress responses, and physiological state of tissues and cells. In agreement with this possibility, CIRP has been reported to activate translation in response to UV irradiation [43] and to suppress translation in response to oxidative stress [17]. Although it is possible that cancer cell lines differ from primary human tumors in their CIRP expression level, a recent study reported upregulation of CIRP protein in 28% of 193 tumors studied, including 33% of breast cancers (11 of 33) [44]. This same study showed that CIRP increased proliferation when expressed in mouse embryo fibroblasts, consistent with a role in growth promotion suggested by our studies.
Regardless of whether CIRP regulates cyclin E1 via increasing HuR, or if CIRP and HuR collaborate to regulate cyclin E1, our initial observation was that of CIRP upregulation in response to HuR knockdown. This may result from a compensatory mechanism to maintain either a high level of HuR or cyclin E1. It could also be explained if HuR or one of its targets, destabilized or translationally repressed CIRP mRNA. Since aberrant expression of either HuR or cyclin E1 contributes to malignant transformation and tumorigenesis in breast and other cancers [45–48], CIRP, as a positive regulator of these two proteins, may also contribute to tumorigenesis.
In summary, the current study shows that CIRP positively regulates and interacts with HuR, promoting its presence in SGs and its cyclin E1 mRNA binding activity. Since HuR stabilizes cyclin E1 mRNA via binding its 3'UTR [12], the most likely mechanism by which CIRP contributes to cyclin E1 overexpression is by increasing the levels of HuR and enhancing its actions. Future experiments will address how CIRP functions in this capacity, and how HuR regulates CIRP. These results are the first to show CIRP overexpression in breast cancer cell lines versus breast epithelial cells and suggest that targeting it could reduce cell proliferation and thus tumor growth by decreasing expression of HuR and downstream targets such as cyclin E1.
Supplementary Material
Supp fig-S1. Regulation of CIRP expression by HuR knockdown and overexpression in MCF-7 cells. The protein levels of HuR and CIRP in MCF-7 cells were analyzed by sequential western blotting 72 hr after transfection. Equal loading of protein was determined by re-probing of blots for β-actin or β-tubulin.
ACKNOWLEDGMENTS
We thank Drs. Wenlan Liu and Nora Perrone-Bizzozero (UNM-HSC, Albuquerque, NM) and Dr. David Port (UCHSC, Denver, CO) for critical reading of the manuscript and technical advice. Dr. David Port also generously provided GST-HuR plasmid and pcDNA3.1mycHuR. We thank Dr. Bridget Wilson (UNM-HSC and Cancer Center) for providing breast cancer cell lysates (SKBR3, MDA-MB-453 and BT-474).
This work was funded by a grant from the National Cancer Institute-National Institutes of Health to RSH (R01CA095898).
The abbreviations used are
- ARE
AU-rich element
- ATCC
American Type Culture Collection
- CIRP
cold-inducible RNA binding protein
- CS-RBD
consensus sequence-RNA binding domain
- CY3
cyanine 3
- ElrA
elav-related protein A
- ERK
extracellular signal regulated kinase
- E1CR378
cyclin E1 coding region 378
- FBS
fetal bovine serum
- GST
glutathione S-transferase
- HDAC1
histone deacetylase 1
- hDCP1a
human decapping protein 1a
- HuR
human antigen R
- LMW
low molecular weight
- RBP
RNA binding protein
- RGG
arginine glycine glycine
- RRM
RNA recognition motif
- SG
stress granule
- TIA-1
T-cell intracytoplasmic antigen
- siRNA
small interfering RNA
- 3’UTR
3’ untranslated region
REFERENCES
- 1.Akli S, Keyomarsi K. Cyclin E and its low molecular weight forms in human cancer and as targets for cancer therapy. Cancer Biol Ther. 2003;2:S38–S47. [PubMed] [Google Scholar]
- 2.Hwang HC, Clurman BE. Cyclin E in normal and neoplastic cell cycles. Oncogene. 2005;24:2776–2786. doi: 10.1038/sj.onc.1208613. [DOI] [PubMed] [Google Scholar]
- 3.Keyomarsi K, O'Leary N, Molnar G, Lees E, Fingert HJ, Pardee AB. Cyclin E, a potential prognostic marker for breast cancer. Cancer Res. 1994;54:380–385. [PubMed] [Google Scholar]
- 4.Porter PL, Malone KE, Heagerty PJ, et al. Expression of cell-cycle regulators p27Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med. 1997;3:222–225. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 5.Blaxall BC, Dwyer-Nield LD, Bauer AK, Bohlmeyer TJ, Malkinson AM, Port JD. Differential expression and localization of the mRNA binding proteins, AU-rich element mRNA binding protein (AUF1) and Hu antigen R (HuR), in neoplastic lung tissue. Mol Carcinog. 2000;28:76–83. [PubMed] [Google Scholar]
- 6.Denkert C, Weichert W, Winzer KJ, et al. Expression of the ELAV-like protein HuR is associated with higher tumor grade and increased cyclooxygenase-2 expression in human breast carcinoma. Clin Cancer Res. 2004;10:5580–5586. doi: 10.1158/1078-0432.CCR-04-0070. [DOI] [PubMed] [Google Scholar]
- 7.Denkert C, Weichert W, Pest S, et al. Overexpression of the embryonic-lethal abnormal vision-like protein HuR in ovarian carcinoma is a prognostic factor and is associated with increased cyclooxygenase 2 expression. Cancer Res. 2004;64:189–195. doi: 10.1158/0008-5472.can-03-1987. [DOI] [PubMed] [Google Scholar]
- 8.Heinonen M, Bono P, Narko K, et al. Cytoplasmic HuR expression is a prognostic factor in invasive ductal breast carcinoma. Cancer Res. 2005;65:2157–2161. doi: 10.1158/0008-5472.CAN-04-3765. [DOI] [PubMed] [Google Scholar]
- 9.Lopez de Silanes I, Fan J, Yang X, et al. Role of the RNA-binding protein HuR in colon carcinogenesis. Oncogene. 2003;22:7146–7154. doi: 10.1038/sj.onc.1206862. [DOI] [PubMed] [Google Scholar]
- 10.Brennan CM, Steitz JA. HuR and mRNA stability. Cell Mol Life Sci. 2001;58:266–277. doi: 10.1007/PL00000854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cherry J, Karschner V, Jones H, Pekala PH. HuR, an RNA-binding protein, involved in the control of cellular differentiation. In Vivo. 2006;20:17–23. [PubMed] [Google Scholar]
- 12.Guo X, Hartley RS. HuR Contributes to Cyclin E1 Deregulation in MCF-7 Breast Cancer Cells. Cancer Res. 2006;66:7948–7956. doi: 10.1158/0008-5472.CAN-05-4362. [DOI] [PubMed] [Google Scholar]
- 13.Nishiyama H, Higashitsuji H, Yokoi H, et al. Cloning and characterization of human CIRP (cold-inducible RNA-binding protein) cDNA and chromosomal assignment of the gene. Gene. 1997;204:115–120. doi: 10.1016/s0378-1119(97)00530-1. [DOI] [PubMed] [Google Scholar]
- 14.Nishiyama H, Itoh K, Kaneko Y, Kishishita M, Yoshida O, Fujita J. A glycine-rich RNA-binding protein mediating cold-inducible suppression of mammalian cell growth. J Cell Biol. 1997;137:899–908. doi: 10.1083/jcb.137.4.899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sheikh MS, Carrier F, Papathanasiou MA, et al. Identification of several human homologs of hamster DNA damage-inducible transcripts. Cloning and characterization of a novel UV-inducible cDNA that codes for a putative RNA-binding protein. J Biol Chem. 1997;272:26720–26726. doi: 10.1074/jbc.272.42.26720. [DOI] [PubMed] [Google Scholar]
- 16.Wellmann S, Buhrer C, Moderegger E, et al. Oxygen-regulated expression of the RNA-binding proteins RBM3 and CIRP by a HIF-1-independent mechanism. J Cell Sci. 2004;117:1785–1794. doi: 10.1242/jcs.01026. [DOI] [PubMed] [Google Scholar]
- 17.De Leeuw F, Zhang T, Wauquier C, Huez G, Kruys V, Gueydan C. The cold-inducible RNA-binding protein migrates from the nucleus to cytoplasmic stress granules by a methylation-dependent mechanism and acts as a translational repressor. Exp Cell Res. 2007;313:4130–4144. doi: 10.1016/j.yexcr.2007.09.017. [DOI] [PubMed] [Google Scholar]
- 18.Yang C, Carrier F. The UV-inducible RNA-binding protein A18 (A18 hnRNP) plays a protective role in the genotoxic stress response. J Biol Chem. 2001;276:47277–47284. doi: 10.1074/jbc.M105396200. [DOI] [PubMed] [Google Scholar]
- 19.Sakurai T, Itoh K, Higashitsuji H, et al. Cirp protects against tumor necrosis factor-alpha-induced apoptosis via activation of extracellular signal-regulated kinase. Biochim Biophys Acta. 2006;1763:290–295. doi: 10.1016/j.bbamcr.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 20.Nishiyama H, Xue JH, Sato T, et al. Diurnal change of the cold-inducible RNA-binding protein (Cirp) expression in mouse brain. Biochem Biophys Res Commun. 1998;245:534–538. doi: 10.1006/bbrc.1998.8482. [DOI] [PubMed] [Google Scholar]
- 21.Peng Y, Yang PH, Tanner JA, et al. Cold-inducible RNA binding protein is required for the expression of adhesion molecules and embryonic cell movement in Xenopus laevis. Biochem Biophys Res Commun. 2006;344:416–424. doi: 10.1016/j.bbrc.2006.03.086. [DOI] [PubMed] [Google Scholar]
- 22.Hamid AA, Mandai M, Fujita J, et al. Expression of cold-inducible RNA-binding protein in the normal endometrium, endometrial hyperplasia, and endometrial carcinoma. Int J Gynecol Pathol. 2003;22:240–247. doi: 10.1097/01.PGP.0000070851.25718.EC. [DOI] [PubMed] [Google Scholar]
- 23.Aoki K, Matsumoto K, Tsujimoto M. Xenopus cold-inducible RNA-binding protein 2 interacts with ElrA, the Xenopus homolog of HuR, and inhibits deadenylation of specific mRNAs. J Biol Chem. 2003;278:48491–48497. doi: 10.1074/jbc.M308328200. [DOI] [PubMed] [Google Scholar]
- 24.Harwell RM, Porter DC, Danes C, Keyomarsi K. Processing of cyclin E differs between normal and tumor breast cells. Cancer Res. 2000;60:481–489. [PubMed] [Google Scholar]
- 25.Dormoy-Raclet V, Menard I, Clair E, et al. The RNA-binding protein HuR promotes cell migration and cell invasion by stabilizing the beta-actin mRNA in a U-rich-element-dependent manner. Mol Cell Biol. 2007;27:5365–5380. doi: 10.1128/MCB.00113-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Atasoy U, Watson J, Patel D, Keene JD. ELAV protein HuA (HuR) can redistribute between nucleus and cytoplasm and is upregulated during serum stimulation and T cell activation. J Cell Sci. 1998;111(Pt 21):3145–3156. doi: 10.1242/jcs.111.21.3145. [DOI] [PubMed] [Google Scholar]
- 27.Wang W, Caldwell MC, Lin S, Furneaux H, Gorospe M. HuR regulates cyclin A and cyclin B1 mRNA stability during cell proliferation. Embo J. 2000;19:2340–2350. doi: 10.1093/emboj/19.10.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jamison JT, Kayali F, Rudolph J, Marshall M, Kimball SR, DeGracia DJ. Persistent redistribution of poly-adenylated mRNAs correlates with translation arrest and cell death following global brain ischemia and reperfusion. Neuroscience. 2008;154:504–520. doi: 10.1016/j.neuroscience.2008.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kawai T, Lal A, Yang X, Galban S, Mazan-Mamczarz K, Gorospe M. Translational control of cytochrome c by RNA-binding proteins TIA-1 and HuR. Mol Cell Biol. 2006;26:3295–3307. doi: 10.1128/MCB.26.8.3295-3307.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spruck C, Sun D, Fiegl H, et al. Detection of low molecular weight derivatives of cyclin E1 is a function of cyclin E1 protein levels in breast cancer. Cancer Res. 2006;66:7355–7360. doi: 10.1158/0008-5472.CAN-05-3240. [DOI] [PubMed] [Google Scholar]
- 31.Akli S, Keyomarsi K. Low-molecular-weight cyclin E: the missing link between biology and clinical outcome. Breast Cancer Res. 2004;6:188–191. doi: 10.1186/bcr905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Akli S, Zheng PJ, Multani AS, et al. Tumor-specific low molecular weight forms of cyclin E induce genomic instability and resistance to p21, p27, and antiestrogens in breast cancer. Cancer Res. 2007;64:3198–3208. doi: 10.1158/0008-5472.can-03-3672. [DOI] [PubMed] [Google Scholar]
- 33.Hunt KK, Keyomarsi K. Cyclin E as a prognostic and predictive marker in breast cancer. Semin Cancer Biol. 2005;15:319–326. doi: 10.1016/j.semcancer.2005.04.007. [DOI] [PubMed] [Google Scholar]
- 34.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Linker K, Pautz A, Fechir M, Hubrich T, Greeve J, Kleinert H. Involvement of KSRP in the post-transcriptional regulation of human iNOS expression-complex interplay of KSRP with TTP and HuR. Nucleic Acids Res. 2005;33:4813–4827. doi: 10.1093/nar/gki797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sureban SM, Ramalingam S, Natarajan G, et al. Translation regulatory factor RBM3 is a proto-oncogene that prevents mitotic catastrophe. Oncogene. 2008 doi: 10.1038/onc.2008.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gallouzi IE, Brennan CM, Stenberg MG, et al. HuR binding to cytoplasmic mRNA is perturbed by heat shock. Proc Natl Acad Sci U S A. 2000;97:3073–3078. doi: 10.1073/pnas.97.7.3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Furneaux H, Cheng H, et al. HuR regulates p21 mRNA stabilization by UV light. Mol Cell Biol. 2000;20:760–769. doi: 10.1128/mcb.20.3.760-769.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gallouzi IE, Brennan CM, Steitz JA. Protein ligands mediate the CRM1-dependent export of HuR in response to heat shock. Rna. 2001;7:1348–1361. doi: 10.1017/s1355838201016089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Anderson P, Kedersha N. RNA granules. J Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 42.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Stress-induced reversal of microRNA repression and mRNA P-body localization in human cells. Cold Spring Harb Symp Quant Biol. 2006;71:513–521. doi: 10.1101/sqb.2006.71.038. [DOI] [PubMed] [Google Scholar]
- 43.Yang R, Weber DJ, Carrier F. Post-transcriptional regulation of thioredoxin by the stress inducible heterogenous ribonucleoprotein A18. Nucleic Acids Res. 2006;34:1224–1236. doi: 10.1093/nar/gkj519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Artero-Castro A, Callejas FB, Castellvi J, et al. Cold-inducible RNA-binding protein bypasses replicative senescence in primary cells through extracellular signal-regulated kinase 1 and 2 activation. Mol Cell Biol. 2009;29:1855–1868. doi: 10.1128/MCB.01386-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ahn MJ, Kim BH, Jang SJ, et al. Expression of cyclin D1 and cyclin E in human gastric carcinoma and its clinicopathologic significance. J Korean Med Sci. 1998;13:513–518. doi: 10.3346/jkms.1998.13.5.513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Cam WR, Masaki T, Shiratori TY, et al. Activation of cyclin E-dependent kinase activity in colorectal cancer. Dig Dis Sci. 2001;46:2187–2198. doi: 10.1023/a:1011962915280. [DOI] [PubMed] [Google Scholar]
- 47.Keyomarsi K, Pardee AB. Redundant cyclin overexpression and gene amplification in breast cancer cells. Proc Natl Acad Sci U S A. 1993;90:1112–1116. doi: 10.1073/pnas.90.3.1112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sui L, Dong Y, Ohno M, et al. Implication of malignancy and prognosis of p27(kip1), Cyclin E, and Cdk2 expression in epithelial ovarian tumors. Gynecol Oncol. 2001;83:56–63. doi: 10.1006/gyno.2001.6308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supp fig-S1. Regulation of CIRP expression by HuR knockdown and overexpression in MCF-7 cells. The protein levels of HuR and CIRP in MCF-7 cells were analyzed by sequential western blotting 72 hr after transfection. Equal loading of protein was determined by re-probing of blots for β-actin or β-tubulin.