Abstract
Transcription in all cellular organisms is performed by multi-subunit, DNA-dependent RNA polymerases that synthesize RNA from DNA templates. Previous sequence and structural studies have elucidated the importance of shared regions common to all multi-subunit RNA polymerases. In addition RNA polymerases contain multiple lineage-specific domain insertions involved in protein-protein and protein-nucleic acid interactions. We have created comprehensive multiple sequence alignments using all available sequence data for the multi-subunit RNA polymerase large subunits, including the bacterial β and β′ subunits and their homologues from archaebacterial RNA polymerases, the eukaryotic RNA polymerases I, II, and III, the nuclear-cytoplasmic large double-stranded DNA Virus RNA polymerases, and plant plastid RNA polymerases. In order to overcome technical difficulties inherent to the large subunit sequences, including large sequence length, small and large lineage-specific insertions, split subunits, and fused proteins, we created an automated and customizable sequence retrieval and processing system. In addition, we used our alignments to create a more expansive set of shared sequence regions and bacterial lineage-specific domain insertions. We also analyzed the intergenic gap between the bacterial β and β′ genes.
Keywords: Evolution, RNA polymerase, Sequence analysis
Introduction
In all cellular organisms the process of transcription is driven by a large multi-subunit molecular machine, the DNA-dependent RNA Polymerase (RNAP) 1. Bacteria contain a single DNA-dependent multi-subunit RNAP (bRNAP) comprising five core subunits: β′ (usually around 150 kDa), β (150 kDa), two copies of α (40 kDa each), and ω (10 kDa) 2. Eukaryotes contain three DNA-dependent multi-subunit cellular RNAPs (eRNAP I, II, and III) comprising 10 common subunits (Rpb1-3, Rpb5-6, Rpb8-12) plus an additional 4, 2, and 5 subunits, respectively 1. In addition to eRNAP I, II, and III, plants contain two additional multi-subunit RNAPs: i) cellular eRNAP IV 3, and ii) an organelle plastid (chloroplast) RNAP (pRNAP) 4; 5 closely related to cyanobacterial RNAP. Archaea contain only one RNAP (aRNAP) composed of 12 subunits, 11 of which are similar to eRNAP II subunits. In general, DNA viruses contain single subunit DNA-dependent RNAPs unrelated in sequence and structure to the multi-subunit RNAPs found in the other branches of life. However, the Nuclear-Cytoplasmic Large double-stranded DNA Viruses (NCLDVs) contain an eRNAP-like enzyme presumably acquired from their eukaryotic hosts (vRNAP) 6; 7.
With the availability of the first large subunit (bRNAP β and β′ homologs) sequences it became apparent that bRNAP and eRNAPs shared several regions of sequence similarity connected by intervening segments of divergent sequence. Sweetser et al. 8 defined conserved sequence regions (A-I) for the bacterial β subunit and its homologs by aligning the sequences of the Escherichia coli (Eco) β subunit and its eRNAP II homolog from Saccharomyces cerevisiae (Sce). Jokerst et al. 9 defined conserved sequence regions (A-H) for the bacterial β′ subunit and its homologs by aligning the sequences of the Eco β′ subunit and its Sce eRNAP II and III homologs, along with the eRNAP II homologs from mouse and Drosophila melanogaster.
The X-ray crystal structure of bRNAP showed a crab claw-shaped molecule 10. One pincer of the claw comprises mostly the β′ subunit, the other mostly β. A 27 Å wide channel between the pincers accomodates downstream double-stranded DNA and the RNA/DNA hybrid at the growing end of the transcript. The enzyme active site, marked by an essential Mg2+ ion, is located on the back wall of the channel. Other features of the structure include elements positioned to maintain the upstream edge of the transcription bubble and split off the RNA transcript from the RNA/DNA hybrid, and additional channels to i) accommodate the upstream single-stranded RNA product (RNA exit channel), ii) guide the nontemplate single-stranded DNA within the transcription bubble, and iii) allow access for the nucleotide substrates into the active site (nucleotide entry channel) 11-17. The X-ray structures of eRNAP II 18 and aRNAP 19 revealed that the multi-subunit RNAPs from all three kingdoms of life share a high degree of structural similarity 1; 2; 20. In fact, there are clear homologs for all five of the core bacterial subunits in aRNAP and eRNAPs I, II, and III (Table 1). When mapped to the RNAP structure, the conserved sequence regions of β 8 and β′ 9 encompass the inner core of the two large subunits surrounding the active site, presumably in regions that govern aspects of transcription common to all classes of multi-subunit RNAPs 1; 2; 21.
Table 1.
Homologs of the bRNAP core subunits.
The multi-subunit RNAPs also contain lineage-specific domain insertions. In the case of the bRNAP β and β′ subunits, these can range in size from 50-500 amino acids. Using a small but diverse set of bRNAP sequences, Iyer et al. 22 detected and characterized bacterial lineage-specific insertions. They determined that bacterial β and β′ both contain ubiquitous as well as lineage-specific insertion domains that fall into four identifiable categories: i) Zn ribbon, ii) Sandwich Barrel Hybrid Motif (SBHM), iii) β-β′ Module 1 (BBM1), and iv) β-β′ Module 2 (BBM2). The subsequent structures of two lineage-specific domain insertions from T. aquaticus (Taq) and Eco β′ confirmed that both were SBHM domain repeats involved in important protein-protein and/or protein-nucleic acid interactions 23.
In this paper we present a large scale sequence analysis of the multi-subunit RNAP large subunits. We created comprehensive multiple sequence alignments (MSAs) for the two large subunits from the following multi-subunit RNAPs: bRNAP, pRNAP, aRNAP, eRNAPs I, II, III, as well as vRNAP. To aid in the creation of the alignments we also developed a sequence retrieval and processing system termed BlaFA (BLAST to FASTA File to Alignment). We used the alignments to better define the shared sequence regions common to all multi-subunit RNAPs. We also analyzed the intergenic gap between the bacterial rpoB and rpoC genes (encoding the β and β′ subunits, respectively), uncovering interesting examples of gene overlap and extreme spacing. In addition, we located and analyzed the bacterial lineage-specific insertions, identifying both new inserts as well as additional species and domain organizations for some of the previously identified insertions.
Results
BLAST to FASTA File to Alignment (BlaFA)
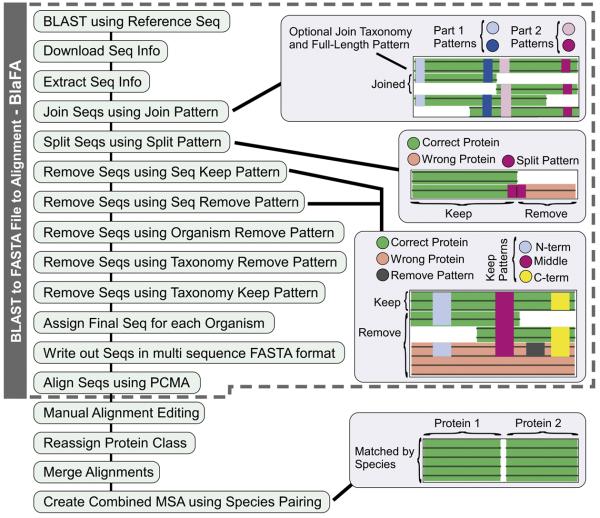
Due to the inherent complexities associated with aligning the multi-subunit RNAP large subunits, the process of sequence selection required many steps and special considerations. For example, some sequences needed to be joined since some RNAPs harbor split large subunits that are encoded by two gene products (chloroplast and cyanobacterial β′, aRNAP subunits A and B). Some sequences needed to be split since a small number of bacteria, such as Helicobacter, have β and β′ fused into a single protein product 24; 25. In addition, there are hundreds of partial large subunit sequences in the NCBI database. Simple sequence gazing was not a practical approach for identifying sequences that needed to be joined, split, or removed, due to: i) the large number of sequences (~5000-7000 BLAST hits), ii) the intrinsically large size of the large subunits (~1000-2000 amino acids each), and iii) the numerous small and large lineage-specific inserts, which caused the wholesale misalignment of large regions. Therefore, we created an automated approach, BlaFA, which allowed for custom processing using both taxonomy and sequence patterns (Fig. 1).
Fig. 1.
Sequence retrieval, processing, and alignment methodology. The creation of the bacterial β/β′ and All RNAP Large Subunit alignments required several steps. First BlaFA (gray dashed region) was used to retrieve and process the sequences, which were then aligned using PCMA, followed by manual alignment fixing. In the case of the All RNAP Large Subunit the class of the RNAPs also had to be reassigned and merged together.
BlaFA was first used to do a BLAST search to compile a list of the available NCBI sequences using Eco K12 β and β′ as representative sequences. This was followed by sequence selection, where the downloaded sequences were processed to: i) join split gene products, ii) split fused gene products, and iii) remove incorrect and partial sequences. Sequences were initially aligned using the program PCMA 26 followed by manual adjustments using PFAAT 27 to fix alignment errors and remove the lineage-specific insertions. We used BlaFA plus manual alignment adjustments to create MSAs for the bacterial β and β′ subunits, as well as for all identifiable β/β′ homologs (Table 2).
Table 2.
Number of sequences in multi-subunit RNAP MSAs
| RNAP class | number of homolog sequences |
||
|---|---|---|---|
| β | β′ | β and β′ | |
| bacterial (bRNAP) | 958 | 842 | 814 (525) |
| plastid (pRNAP) | 71 | 50 | 49 (43) |
| RNAP I | 99 | 89 | 83 (70) |
| eukaryota (eRNAP I) | 60 | 59 | 50 (45) |
| NCDLV (vRNAP I) | 39 | 30 | 33 (25) |
| RNAP II | 114 | 143 | 93 (77) |
| eukaryota (eRNAP II) | 64 | 78 | 44 (34) |
| archaea (aRNAP) | 40 | 41 | 40 (35) |
| NCDLV (vRNAP II) | 10 | 24 | 9 (8) |
| RNAP III (eRNAP III) | 57 | 63 | 50 (41) |
| total | 1299 | 1187 | 1089 (756) |
The second number in parentheses is the number of sequences nonredundant within the shared regions
Phylogenetic Analysis of the All RNAP Large Subunit MSA
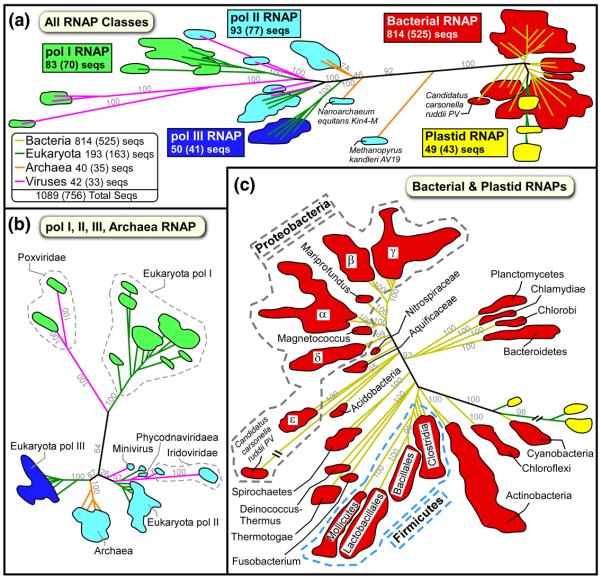
A phylogenetic tree for the all RNAP large subunit MSA (with more than 1000 large subunit sequences; Table 2) shows that each class of RNAP was clearly segregated (Fig. 2A), indicating that RNAP class assignments were accurate. As expected, the analysis showed that, although the aRNAPs clearly belong to the RNAP II class, they represent an intermediate between the eukaryotic and bacterial RNAPs. The vRNAPs from the NCLDVs are related to eukaryotic RNAPs. To our knowledge it has not been appreciated that the Iridoviridae, Phycodnaviridae, and Mimivirus families seem to have acquired an eRNAP II-like RNAP, while the Poxviridae family seems to have acquired an eRNAP I-like RNAP (Fig. 2B). It should be noted that another member of the NCLDVs, the Asfarviridae, were not included in this analysis as their RNAP sequences were relatively highly divergent. Close examination of the bRNAP branch showed that the pattern of segregation correlated with established bacterial taxonomy, demonstrating that our alignment contained sequences from a large and diverse set of bacterial species (Fig. 2C). Furthermore, it also highlighted the previously established close relationship between the cyanobacterial and pRNAPs.
Fig. 2.
Phylogenic analysis of the All RNAP Large Subunits MSA. The two All RNAP Large Subunit alignments were combined by species and the residue positions pruned to only keep the regions shared among all the sequences. The phylogenic trees were calculated using PhyML v3.0 45 and analyzed using TreeDyn 46 (see Materials and Methods). Due to the large number of sequences, only the boundaries for each group of leaves are shown colored by RNAP class: bRNAP (red), pRNAP (yellow), eRNAP I (green), eRNAP II (blue), and eRNAP III (cyan). The branches for each leaf region are colored by taxonomy: bacteria (yellow), eukaryota (green), archeaea (orange), and viruses (magenta). Due to their diversity, the proteobacteria (gray dashed region) and firmicutes (light blue dashed region) taxonomy subdivisions have been individually labeled. Selected branch support values are indicated in light grey.
A. All RNAP Classes tree. For each class, the total number of complete β/β′ homolog sequences is shown. The second number in parentheses is the number of sequences nonredundant within the shared regions.
B. eRNAP-like RNAPs (eRNAP I, II, III, aRNAP, vRNAPs).
C. Bacterial and plastid RNAPs.
Bacterial Large Subunit Fusions
The naturally occurring fusion of β and β′ in the Helicobacter species 24; 25 has been implicated in the fitness for bacterial infection as well as in the decreased sensitivity of Helicobacter RNAP to urea 28. As expected, we found fused β and β′ subunits in all of the examined Helicobacter family species, including Helicobacter pylori(Hpy) 26695 (gi:15645812), Hpy HPAG1 (gi:108563562), Hpy J99 (gi:04155718), Helicobacter hepaticus ATCC 51449 (gi:32261909), and Helicobacter acinonychis str. Sheeba (gi:109948061). We also found fused β and β′ subunits in the related Wolinella family species Wolinella succinogenes DSM 1740 (gi:34556892) 25. It is thought that all ε-proteobacteria of the Helicobacteraceae family (Helicobacter and Wolinella) harbor fused β and β′ subunits 25. We found, however, one assigned Helicobacteraceae family species, Thiomicrospira denitrificans (Tde) ATCC 33889, that seems to have separately encoded β (gi:78497094) and β′ (gi:78497095) subunits. On closer examination, we uncovered that in Tde ATCC 33889, the genes encoding β and β′ share an unusual two codon overlap also found in the closely related Campylobacteraceae species that have non-fused β and β′ subunits. In addition, based on our phylogenetic analysis Tde ATCC 33889 segregates to its own branch located directly before the branch that contains the Helicobacteraceae and Campylobacteraceae branches. Given that RNAP large subunits have been used for taxonomy classifications, we believe that our results indicate that Tde ATCC 33889 should not be considered part of the Helicobacteraceae family, but rather as its own family under the Campylobacterales (which also includes Campylobacteraceae and Helicobacteraceae), with the following proposed full taxonomy: Bacteria; Proteobacteria; Epsilonproteobacteria; Campylobacterales; Thiobacteraceae.
Surprisingly, we also discovered a previously uncharacterized β/β′ fusion in 3 of 4 sequences from the parasitic intracellular α-proteobacteria and Rickettsiaceae member Wolbachia family, including Wolbachia endosymbiont (Wen) strain TRS of Brugia malayi (gi:58419220), Wen of Drosophila melanogaster (gi:42409679), Wolbachia sp. wMel (gi:81652940), but not in Wolbachia pipientis (Wpi) (gi:15081478). However, it is important to note that on closer examination the one Wolbachia species exception, Wpi, was from a phylogenic study that only sequenced the β subunit 29. Therefore, it is reasonable to conclude that all of the Wolbachia, including Wpi, contain fused β and β′ subunits. The finding of fused β and β′ in another distant branch of the bRNAPs is intriguing and possibly represents a convergent evolutionary event. Furthermore, the sequence of the Wolbachia fusion site is not similar to the Helicobacteraceae fusion site, which contains 6 additional residues. The biological role of the Wolbachia fusion may be to increase pathogenic fitness, as in the Helicobacteraceae 28.
Bacterial rpoB/rpoC Intergenic Gap Analysis
Normally, the genes for the bacterial large subunits are transcribed as a single mRNA, with rpoB immediately preceding rpoC. The two protein-encoding genes are normally translated separately, with the rpoB translational stop codon and the rpoC translational start site separated by an untranslated 20-100 bp linker 25. As described above, the Campylobacteraceae species, which do not contain fused β and β′ subunits, have a two codon overlap between rpoB and rpoC. It has been proposed that either a 1 bp addition or a 2 bp deletion in a common ancestor could have lead to a frame shift mutation resulting in the fusion of β and β′ in the related Helicobacteraceae 25. We decided to examine the rpoB/rpoC intergenic gap in an effort to understand the Wolbachia β and β′ fusion, as well as update our understanding of this gap across the known bacterial genomes.
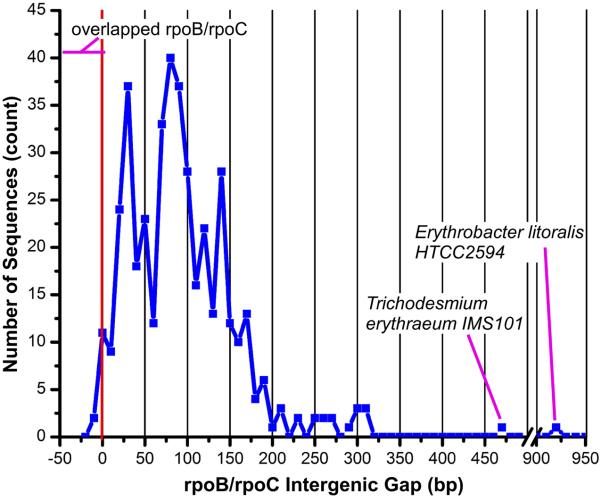
An analysis of the rpoB/rpoC intergenic gap in 426 bacterial species revealed that the intergenic gap is usually between 10-200 bp (Fig. 3), in agreement with previous analyses 25. We also found, however, a number of interesting exceptions. We found the previously known 8 bp overlap (negative intergenic gap) in Campylobacteraceae and the 4 bp overlap in Aquificae, plus additional overlaps of 14 bp in Chloroflexi, 4 bp in Candidatus Carsonella (γ-proteobacteria), 1 bp in Alcaligenaceae (β-proteobacteria), 1 bp in Clostridium novyi NT (Clostridia), and 1 bp in Thiomicrospira crunogena XCL-2 (γ-proteobacteria). We also found unusually large intergenic gaps of 462 bp in Trichodesmium erythraeum IMS101 (Cyanobacteria), and 916 bp in Erythrobacter litoralis (Elit) HTCC2594 (α-proteobacteria). In general, Cyanobacteria, which have split β′ subunits, contain a β gene followed by two sequential β′ genes. We found that in most Cyanobacteria the intergenic gap between the β gene and the first β′ gene is between 38-134 bp. The unusual Trichodesmium erythraeum IMS101 contains the same β and β′ gene organization, but for some unknown reason it has an extra long gap (462 bp) between the β gene and the first β′ gene. The extremely large gap in Elit HTCC2594 is the result of an ORF encoding an unknown, 265 amino acid gene product (gi:85375720) encoded in the same direction between rpoB and rpoC. It should be noted that the Elit HTCC2594 rpoB and rpoC genes both encode for full-length subunits. The unique rpoB/unknown ORF/rpoC gene organization in Elit HTCC2594 is extremely interesting since it seems likely that the unknown gene is co-transcribed with rpoB and rpoC and might therefore play a role in RNAP assembly or transcriptional regulation.
Fig. 3.
Bacterial rpoB and rpoC intergenic gap. The distance between the rpoB gene (encoding bRNAP β) stop codon and the rpoC gene (encoding bRNAP β′) start codon was analyzed. The number of sequences vs. intergenic gap is plotted as a blue line. The x-axis has been split between 500 and 900 bp. The red vertical line indicates an intergenic gap of zero with minus values indicating overlapping rpoB and rpoC genes. The species with fused β/β′ subunits are not shown since they do not have a true intergenic gap. Please refer to the supplemental information on our website for additional details.
We found that the species most closely related to Wolbachia contained non-overlapped but short rpoB/rpoC intergenic gaps of 11-19 bp. Therefore, we believe that although the fusion of Wolbachia did not take place exactly as in Helicobacteraceae, it is certainly possible that this small gap could have been transformed into a β/β′ fusion by the correct frame shift mutation or a small deletion. It also supports the idea that the Wolbachia and Helicobacteraceae fusions were independent evolutionary events. In addition, it would seem that persistent β/β′ fusions are rare events, since there are multiple species with small gaps and overlapping genes that to our knowledge have not resulted in closely related species with β/β′ fusions. Presumably, the persistent existence of a β/β′ fusion must confer an evolutionary advantage, as in the Helicobacteraceae 28.
Shared Sequence Regions Common to Multi-Subunit RNAPs
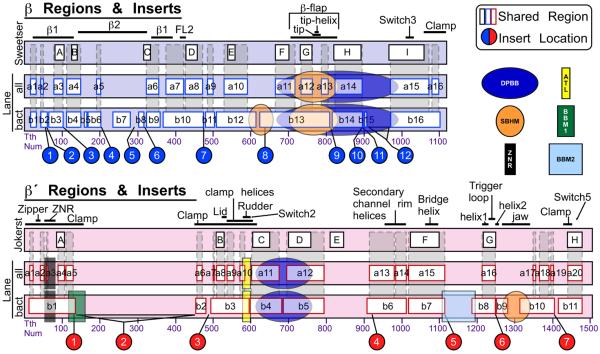
Previous analyses have established regions within the two large subunits that share significant sequence similarity across all classes of RNAP 8; 9. However, the initial β and β′ regions were established in 1987 (β) 8 and 1989 (β′) 9 using only a few sequences. It has become clear that some of the regions defined as ‘conserved’ are not conserved in many sequences (for example, the original region ‘E’ of β′), while additional regions of significant conservation have become apparent 10. There is a pressing need for a re-evaluation of these analyses, given that the numbers of sequences available has increased by several orders of magnitude. We used our comprehensive MSAs to define a new set of common sequence regions, using the positions alignable in all large subunit sequences. For β we defined 16 regions shared among all large subunit sequences (βa1 - βa16) and for β′ we defined 20 regions (β′a1 - β′a20). In general we found most of the previously established regions and for some we were able to extend the boundaries, and we have added several new regions which were previously not identified (Fig. 4). We also defined regions shared among only bRNAP sequences (βb1 - βb16, β′b1 - β′b11), which, as expected, are more extensive than the regions shared among all RNAPs. Fig. 4 shows a comparison of our shared sequence regions with the previously established regions, along with the distinct evolutionarily conserved domains identified by Iyer et al. 22; 30, locations of bacterial lineage-specific inserts (see below) as well as important structural features. Mapping of the shared sequence regions onto the bRNAP structure revealed that they comprise the center as well as parts of the outer surface of RNAP (Figs. 5 and 6). A detailed analysis of the large subunit shared regions is presented in the accompanying paper (Lane).
Fig. 4.
Shared sequence regions common to multi-subunit RNAPs. The vertical bars represent the primary sequence of the Tth (or Taq) bRNAP large subunit (β/β′) sequences. For both β (top, blue) and β′ (bottom, pink), three representations are shown. On top are the originally defined sequence regions for β 8 and β′ 9. Below are the regions common to all multi-subunit RNAPs (Lane – all), and regions common to bRNAPs (Lane – bact). Structural features are labeled above. The locations of the lineage-specific inserts (see Figs. 7, 8) are indicated below. Evolutionarily conserved domains are superimposed on the sequences according to Iyer et al. 22; 30. Domain designations are as follows: DPBB, double-psi-β-barrel; SBHM, sandwich barrel hybrid motif; ZNR, zinc ribbon; ATL, AT-hook like motif; BBM1, β-β′ specific module 1; BBM2, β-β′ specific module 2.
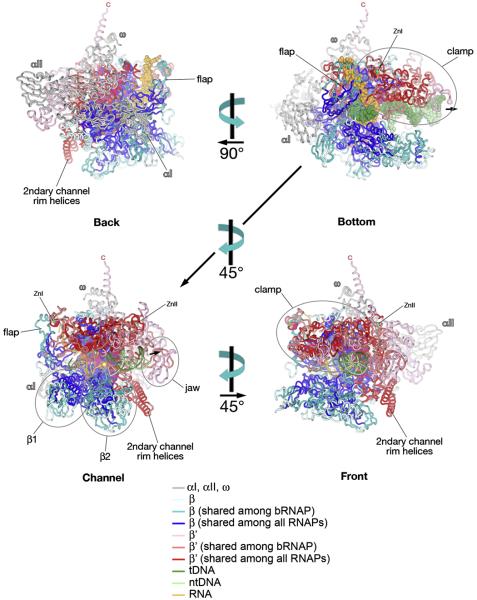
Fig. 5.
Structural Mapping of Shared Sequence Regions on the bRNAP structure; Bottom, Back, Channel, and Front views. The Tth bRNAP ternary elongation complex structure (PDB 2O5J) 15 is shown as backbone ribbons. The color-coding is shown in the key. MgI chelated at the active site is shown as a yellow sphere. The small black arrow points in the downstream direction of the template DNA. The views (Bottom, Back, Channel, Front) are as defined by Cramer et al. 18.
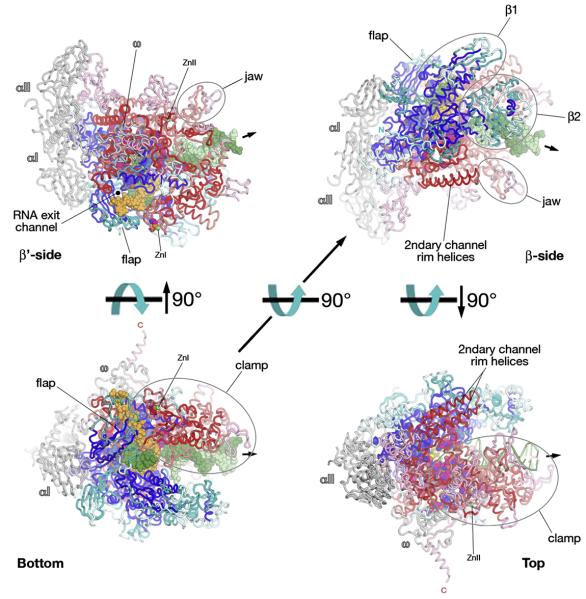
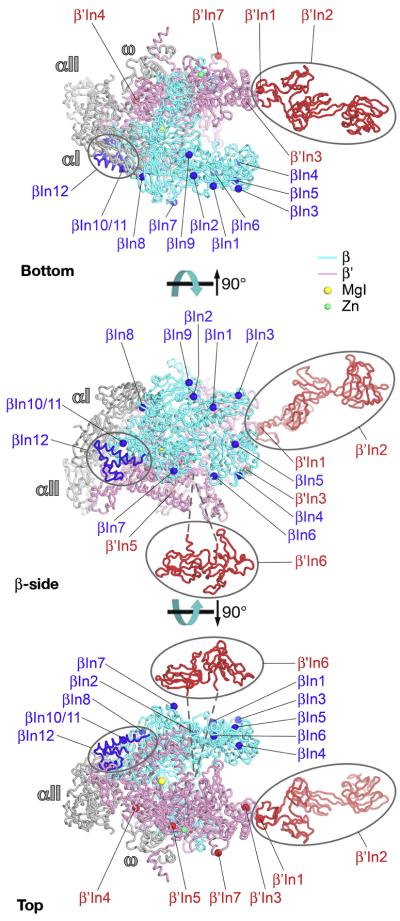
Fig. 6.
Structural mapping of shared sequence regions on the bRNAP structure; Bottom, β′-side, β-side, and Top views 18. Representation and color-coding the same as Fig. 5.
Bacterial Lineage-Specific Insertions
Iyer et al. 22 previously examined RNAP lineage-specific insertions in a small but diverse group of 42 bacterial species. Using our MSAs of the bacterial β (958 sequences) and β′ (842 sequences) subunits, we located all of the previously identified insertions along with new lineage patterns. We also identified many new insertions. We located 12 β inserts (βIn1 - βIn12; Fig. 7) and 7 β′ inserts (β′In1 - β′In7; Fig. 8). Based on the lineage-specific domain insertions (Figs. 7, 8) and the phylogenetic analysis (Fig. 2), the Acidobacteria and Nitrospirae bacterial species seem to belong to what Iyer et al. 22 defined as the Group I bacteria, which also includes Proteobacteria, Aquificae, Spirochaetes, Chlamydiae, Planctomycetes, Chlorobi, Fusobacteria, and Bacteroidetes.
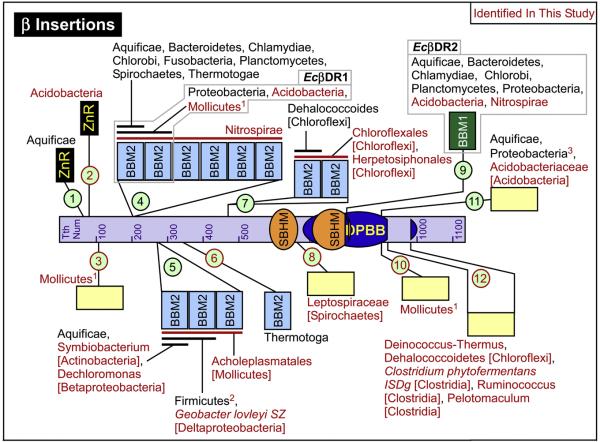
Fig. 7.
bRNAP β lineage-specific domain insertions. The locations of the β Inserts (βIn1 - βIn12) are indicated using numbered light green circles. Red text or lines indicate inserts or lineage details identified in our study. The light gray boxes indicate the identities of previously well studied inserts. The taxonomy lineage details are as inclusively broad as possible. Where subfamily taxonomy is given, the root taxonomy name to which it belongs is given in square brackets (Proteobacteria and Firmicutes are given taxonomy names one level more specific). The individual bacteria species name is given if it is the only member of a number of related bacteria to contain the insert. 1Missing in some Mollicutes. βIn4 and βIn10 contain the same Mollicutes species and are mutually exclusive with βIn3 in terms of Mollicutes species. 2Missing in some Firmicutes species. Some of the Firmicutes missing this insert represent the top 8 species with the smallest combined β/β′ sequence lengths. 3The Wolbachia species, which also have fused β/β′, have an additional 69 amino acid extension at the N-term of this insert. Please refer to the supplemental information on our website for additional details about all of the inserts.
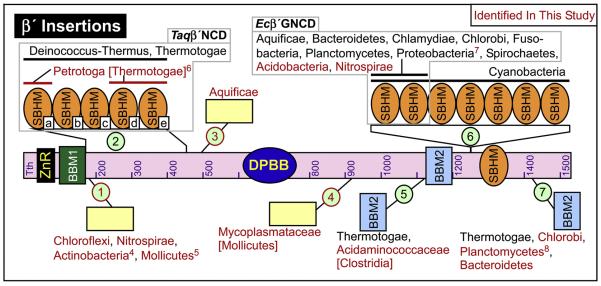
Fig. 8.
bRNAP β′ lineage-specific domain insertions. Same as Fig. 7, but with the locations of the β′ Inserts (β′In1 - β′In7). 4Missing in Symbiobacterium subfamily species. 5Missing in Acholeplasmatales subfamily species. 6Missing a region of sequence in the middle of the insert that removes domains b and c, which interestingly both extend past the σ subunit and therefore lack interactions at the interface between this insert and σ. 7The ε-Proteobacteria subfamily inserts contain ~150 additional amino acids. 8Missing in Candidatus Kuenenia stuttgartiensis. Please refer to the supplemental information on our website for additional details about all of the inserts.
We also found that the Acidobacteria β subunit contains a Zn-ribbon motif (βIn2) inserted 4 amino acids after the known Aquificae Zn-ribbon insertion (βIn1). We characterized βIn2 by using Profile Hidden Markov Model-Profile Hidden Markov Model searching using HHMpred 31; Run at http://toolkit.tuebingen.mpg.de/hhpred). HHMpred indicated that, similar to the Aquificae βIn1, the Acidobacteria βIn2 is a Zn-Ribbon (E-value=0.0062). Furthermore, manual alignment of the previously characterized Aquificae βIn1 and the newly found Acidobacteria βIn2 showed that the 4 cysteine residues (in groups of 2x CxxC), which are known to be responsible Zn binding, all aligned perfectly with each other. However, the Acidobacteria βIn2 has 15 aa insert right before the first CxxC, between amino acids 70-71 (Tth β′ numbering), while the Aquificae βIn2 is inserted between amino acids 75-76 (Tth β′ numbering). Given these differences it seems plausible that the two Zn Ribbons represent two separate and distinct horizontal gene transfer events. The additional 15 aa before the first CxxC in the Acidobacteria βIn2 might allow it to offset its different insert location to inhabit the same region as the Aquificae insert within the three-dimensional structure. Thus it is possible (or even likely) that the two Zn-ribbon insertions (βIn1 and βIn2) perform similar functions.
Interestingly, there are several instances of unusual lineage insertion patterns. We found several instances suggestive of horizontal gene transfer, where only one or at most a few species from a large group of related species contain an insertion normally only found in another lineage (e.g. βIn5, βIn12). To verify that these findings were not the result of simple clerical errors, the results of our phylogenetic analysis (Fig. 2) were used to confirm that the NCBI records for the receiving sequence contained the correct lineage. We also found several instances where a few species did not contain a lineage-specific insertion found in all other closely related species, possibly indicating incomplete penetratence or complete loss of the insertion (e.g. βIn3, βIn4, βIn5, βIn10, β′In1, β′In4, β′In5, β′In7). Furthermore, some of these exceptions are correlated across insertions, such as βIn4 and βIn10, which are both contained in the same Mollicutes species, but are mutually exclusive with the Mollicutes species that contain βIn3.
There are several examples of lineage-specific insertions either representing the addition to, or the removal from, only part of a pre-existing insertion. The Wolbachia species, which also have fused β/β′, have an additional 69 amino acid extension at the N-terminus of the shared Proteobacterial insertion βIn11. Given that a very large group of 523 related Proteobacteria contain βIn11 without the Wolbachia N-terminal extension, it is reasonable to assume that this represents an extension to a pre-existing insertion. Similarly, we found that β′In7 in the ε-Proteobacteria contains ~150 additional amino acids mostly inserted at two locations in the 1st SBHM domain. In contrast, we also found possible examples of partial loss of β′In2, with SBHM structural domains that are split and not in sequential sequence order 23. Petrotoga mobilis SJ95, which is a member of the Thermotogae family and the only examined member of the Petrotoga subfamily, is missing a stretch of amino acids in the middle of the β′In2 sequence, leading to the clean removal of β′In2 domains b and c (Fig. 7). The removal of domains b and c is particularly interesting since in the context of RNAP holoenzyme, they both extend beyond the interaction interface between the σ subunit and β′In2 SBHM domains a, d, and e 23. The discrete loss of β′In2 SBHM domains b and c is consistent with the proposal that SBHM domains b and c play a functional role independent of SBHM domains a, d, and e, which play an important role in stabilizing interactions of the σ subunit with the core RNAP 23; 32. Since only 12 of the available sequences contain β′In2, it is difficult to definitively say if the case of SBHM domains b and c represent a loss and not an extension into the middle of a pre-existing insert.
We also determined the minimal and maximal combined lengths of the bacterial β and β′ subunits. In general, the shortest β/β′ came from the Firmicutes sub-families of Bacillales and Clostridia. The 8 shortest β/β′ were Clostridia sequences missing the ~90 amino acid 2x SBHM insertion (βIn5) found in every other Firmicutes, including other Clostridia sequences. The shortest β/β′ (2248 amino acids) was from the thermophilic and halophilic bacterium Halothermothrix orenii H 168 33, which is missing an additional 22-82 amino acids when compared to the other 7 shortest Clostridia. The longest β/β′ was from the Nitrospirae and Cyanobacteria, which contain large repeated domain insertions in βIn4 and β′In6, respectively (Leptospirillum sp. Group II UBA combined β/β′, 3303 amino acids).
Several new insertions identified in this work could not be assigned to known domain motifs, despite our best attempts. Either these domains represent uncharacterized motifs, or they represent known motifs that are not identifiable due to low sequence similarity or non-sequential sequence order. Complete identification will likely require structural information in the form of either a complete RNAP or isolated insertion domain structure 23.
Mapping of the insert locations onto the bRNAP structure reveals no obvious pattern or concentration of inserts except that the inserts are located on the outer surface of RNAP (Fig. 9). With one exception (β′In6, see below), the inserts appear to be independent and structurally autonomous of the highly conserved structural core of the enzyme surrounding the active site. From a structural point of view, the inserts likely comprise independently folded, isolated domains on the RNAP surface, and this is supported by the available structural evidence 23; 34; 35; Fig. 9). From a functional point of view, the inserts are unlikely to play critical roles in RNAP assembly or basic function, but it is assumed that their presence points to roles in lineage-specific regulatory functions that, for the most part, have not been identified. Indeed, βIn4 of Eco RNAP is targeted by the bacteriophage T4-Alc protein, which selectively induces premature termination of Eco RNAP transcription on Eco DNA during phage infection 36, indicating that regulatory factors can modulate transcription via the inserts. In this view, many of the inserts may function as platforms for the interaction of regulatory factors that modulate RNAP function in a lineage-specific manner.
Fig. 9.
Structural mapping of bRNAP lineage-specific domain insertions on the bRNAP structure; Bottom, β-side, and Top views 18. The Taq core bRNAP structure, including the complete structure of Taq β′In2, is shown as backbone ribbons 23, color-coded as follows: αI, αII, ω, grey; β, cyan; β′, pink. The locations of the bRNAP lineage-specific domain insertions are labeled (according to Figs. 4, 5) and shown as spheres (β insertions, blue; β′ insertions, red), except three insertions with known structures are shown in blue (βIn12, found in the Tth and Taq bRNAP structures) and red (Taq β′In2 and Eco β′In6)23. The attachment of Eco β′In6 in the trigger loop is schematically denoted by dashed lines.
The exceptional insert, β′In6, appears to sit on the outer surface of the RNAP near the entrance of the secondary channel (Fig. 9) 23 but is inserted, via long, flexible linkers, directly in the middle of the highly conserved Trigger Loop (in β′a16, or between β′b8 and β′b9; Fig. 4), which plays a central role in the RNAP catalytic cycle 15; 37; 38; see accompanying paper, Lane), raising the possibility that β′In6 directly influences RNAP active site dynamics. Indeed, monoclonal antibodies with epitopes mapped within β′In6, as well as some deletions within β′In6, strongly inhibit the nucleotide addition reaction of RNAP 39; 40. β′In6 also appears to play a role in modulating termination 41-44. A great deal of additional detailed information derived from our analysis of the bacterial lineage-specfiic insertions can be found on our website.
Conclusions
We created comprehensive MSAs of the multi-subunit RNAP large subunits. During this process we discovered an uncharacterized fusion of the β and β′ subunits in the parasitic intracellular Wolbachia bacteria. In addition to clarifying the shared sequence regions of RNAP common to all classes of multi-subunit RNAPs, the alignment also allowed us to gain additional insights into the bacterial lineage-specific domain insertions. We identified all of the previously characterized insertions (some with expanded lineage patterns) and a number of new insertions. We also uncovered several examples of possible horizontal gene transfer of intact or partial domain insertions. By creating a comprehensive list of the intergenic gap between the bacterial β and β′ genes we revealed insights into the genesis of the β and β′ subunit fusion. We also discovered a unique β and β′ gene organization in Elit HTCC2594, which has an unknown gene product encoded in the same direction between the β and β′ genes. Our extensive alignments will provide a valuable resource for the study of multi-subunit RNAPs. In addition, we believe that our customizable sequence retrieval, processing, and alignment system, (BlaFA) along with our insertion detection methodology, will aid in the study of other large and complex protein families.
Materials and Methods
BLAST to FASTA File to Alignment (BlaFA)
Sequences were downloaded and aligned using a custom program called BlaFA which allowed for programmable automation. A representative sequence (ie from Eco K12) was used to BLAST the NCBI non-redundant (nr) dataset using NetBLAST (ftp://ftp.ncbi.nlm.nih.gov/blast/executables/LATEST/). The BLAST result list was then used to extract the NCBI Genbank ID (gi) for each potential sequence. The sequences description page (INSDSeq XML Format) was used to extract the following information: organism name, strain name, sub-strain name, taxonomy, protein product description, and protein sequence. Next each protein sequence was evaluated in order to determine if it was the correct full-length sequence. These steps were necessary since in addition to partial sequences, some species have β and β′ proteins that are naturally split in half or fused to each other.
In order to allow for a powerful and flexible system, this process was automated using custom sequence and taxonomy patterns. Possibly split sequences were first identified from the non full-length sequences lacking either a N or C-term pattern (ie N-term: G……T and C-term: KEN…G). If a sequence was identified as a potential split sequence the BLAST result list was then searched to find other sequences from the same organism. The potential halves were then identified using sequence patterns for the N-term and C-term of each half and joined by appending one to the next. In addition, since such splits usually correlate to certain taxonomies we often restricted the joins with taxonomy patterns (ie cyanobacteria). Next the system identified fused proteins, in which two proteins that are normally expressed separately in most species are instead expressed as a single large protein. Each fused sequence was evaluated using a sequence pattern unique to the fusion site (ie 2,I……..F…….|ASP..I…S.GE where 1 or 2 specifies the half to keep and “|” indicates where to split). Once a fusion site was found the correct half was used in place of the originally fused sequence. Next, incorrect and partial sequences were removed using a list of sequence keep and remove patterns. In order, to be kept a sequence had to contain all of the specified sequence keep patterns (ie an N-term and a C-term pattern to remove partial seqs) and none of the sequence remove patterns (useful for removing unwanted proteins that might pass keep patterns, like when you only want a sub-set of proteins that are part of a much larger closely related protein family). Next, sequences were removed using taxonomy keep patterns (ie general ones like Bacteria or very specific ones like Enterobacteriaceae) and remove patterns (ie Not Bacteria or not Enterobacteriaceae). The remaining sequence with the best BLAST expect score was then assigned as the final sequence for each unique species and written to a multiple sequence FASTA file, which also contained the protein sequence extracted from a known structure.
In practice identifying the various sequence patterns a priori can be very challenging. Especially since choosing the best pattern often requires optimizing between stringency and effectiveness. Therefore, it was often necessary to do a series of pattern optimizing BlaFA runs. For example to determine the join patterns, it was best to do a pattern optimizing BlaFA run for the first part as well as the second part of the protein, excluding irrelevant sequences using a taxonomy pattern. If possible we also created an additional alignment with both halves and some full-length sequences. Either way, our goal was to get an alignment where we could clearly identity a protein as either the first or the second half, allowing the generation of the sequence join patterns. For split patterns, it was best to a pattern optimizing BlaFA run to get the sequence pattern for the part of the fusion we wanted to split. We could then manually do a sub-alignment with only those sequences that look to be fused (ie those with a larger than expected sequence length). In addition, we could specify an appropriately large sequence size cutoff within BlaFA and the system will automatically prune the final sequence lists after BLAST to enrich for the potentially fused sequences. An alignment containing both fused proteins and known full-length sequences covering the first or second half of the fusion were then used to correctly determine the fusion site and generate a fusion specific sequence pattern for splitting. For generating the sequence keep and remove patterns it was often necessary to do a pattern optimizing BlaFA run from which we manually removed partial sequences. After determining the optimized patterns, we preformed a test BlaFA run, followed by an additional BlaFA run using only a species exclude pattern that excluded all of the species found in the test run. We used the second run to identify sequences that should have been included in the first test BlaFA run and adjusted our patterns accordingly. Once we were satisfied that the BlaFA patterns properly included and processed the target sequence while at the same time excluding unwanted sequences we performed the final BlaFA runs (See the information on our website for BLAST dates and final BlaFA patterns).
Although this approach required a lot of upfront manual effort, once the patterns were established they could be used in concert to quickly identify the correct sequences without having to worry about the multitude of steps where human error could have resulted in a problem. In addition, well designed patterns based on a diverse set of sequences that are not too restrictive can be used in the future to quickly identify newly available sequences.
The sequences in the multiple sequence FASTA files were then aligned using the program PCMA26 (ftp://iole.swmed.edu/pub/PCMA/). PCMA was chosen since it uses a two-stage strategy in which it first quickly pre-aligns highly identical sequences (similar to ClustalW) followed by alignment of the divergent sequences using profile-profile comparison and consistency (similar to T-Coffee). In our experience, PCMA produced the most accurate alignments in a relatively short amount of time.
Nonetheless, the alignments still needed to be manually fixed by hand, usually due to the presence of small (1-2 residue) and large (50-600 residue) lineage-specific insertions. Furthermore, the manual alignment fixing of the two large subunits of RNAP were additionally complicated due to the large number of sequences, large size of the proteins, and many different lineage-specific insertions at various locations within the sequence (Figs. 7, 8). The program PFAAT27 (http://pfaat.sourceforge.net/) was used to manually fix the alignments in order to remove lineage-specific regions and fix any misaligned positions. In general the manual alignment fixing process consisted of iterative cycles in which an alignable region was identified between two conserved boundaries created by stretches of positions that were identical or nearly identical in all of the sequences. The conserved boundaries were usually easy to spot since they were well aligned across all of the sequences by PCMA. In most cases all of the sequences contained the same number of intervening amino acids between the conserved boundaries, along with areas of high sequence similarity either between all or groups of sequences. In contrast, the positions outside of the conserved boundaries usually contained the lineage specific sequence insertions as well as stretches of low sequence similarity that PCMA tried to align by adding lots of gaps. In some cases, manual alignment fixing was successfully in properly aligning the regions outside of the conserved boundaries, since often the presence of a lineage specific insertion on the outside edge of the conserved boundary simply caused PCMA to misalign positions that were otherwise alignable in all sequences. However, there were also many sequence regions outside of the conserved boundaries that possessed low sequence similarity that could not be aligned by PCMA or manual alignment fixing. Therefore, the conserved boundaries were used to define the alignable regions which were kept, while positions outside of the conserved boundaries were either manually re-aligned or removed. In addition, we removed the lineage specific insertions due to their complicated patterns of insertion and the presence of stretches of low sequence similarly that were usually found before and after their sites of insertion. After this process we were left with our final alignments that were cleaned of non-alignable positions.
Creation of Bacterial Large Subunit Alignments
We used the Eco K12 β and β′ sequences as input reference sequences for BlaFA, along with the sequence from the Tth bRNAP structure (pdb code 2BE5). We then manually fixed the alignments and removed all regions not common to all of the bacterial sequences. In addition to using the alignment editor PFAAT, we used a custom program (msa_util.pl) that allowed us to quickly manipulate various aspects of the alignment including removing or gapping regions of the alignment by specifying the inclusion of certain sequences and/or positions.
Creation of Alignments containing All RNAP Large Subunits
The creation of alignments containing the two large subunits from all classes of multi-subunit RNAP was a multistep process. Similar to the bacterial β/β′ alignments, we first used BlaFA to determine the sequences for the two large subunits from the following classes of RNAP: eRNAP I, II, and III, and pRNAP. However, due to homology, the above sequence lists contained overlapping or incorrectly assigned RNAPs. To correct for this we used a custom program (create_reassigned_gene_and_comb_fas.pl) that read in the eRNAP I, II, and III, and pRNAP sequences and then reassigned their class according to the class of the input sequence class to which it had the best BLAST score. We then created and PCMA aligned two multiple sequence files: (1) for eRNAP I, II, and III sequences and (2) for pRNAP sequences.
To aid in the manual cleaning of the above two alignment alignments we created reference alignments with the sequence from the Tth bRNAP (pdb code 2BE5), the sequence of the Sce eRNAP II structure (pdb code 1TWF), and the sequence of Eco (gi:01790419/ 02367335) after it was manually fixed and cleaned as per the previous bRNAP alignment section. The Tth structural and Eco sequences were previously aligned in the bacterial alignment, and the yeast structural sequence was structurally aligned to the Tth structural sequence. For the plastids we added the reference sequences for the bacterial Tth structural sequence and cleaned Eco K12 sequence. For eRNAP I, II, and III we added the reference sequences for the yeast structural sequence and the Eco K12 cleaned sequence.
We used the reference sequences as guides when manually cleaning the alignment in which we removed any sequence positions that were not in the cleaned Eco sequence, since we were only interested in creating alignments with regions shared by all classes of RNAP. We also used the reference alignment to aid in manually aligning the sequences.
We next used a custom program (msa_merge.pl) that used the reference alignments to merge two alignments together. We first merged the plastid and bacterial alignments, followed by the eRNAP I, II, and III alignments. We then removed the reference sequences, leaving only the natural sequences and the Tth structural sequence, resulting in the All RNAP Large Subunit alignments.
Phylogenetic Analysis
Phylogenetic analysis was performed using only the shared sequence regions in a combined alignment created using a custom program (combined_msa_util.pl) that joined β and β′ or there homologs from the same species. We then removed sequences which were redundant in the shared regions. Table 2 lists the number of sequences in each combined alignment. We used PhyML v3.0 45 to construct the phylogenetic trees. The All RNAP Classes tree was created using LG substitution model, SPR tree improvement, and SH-Like branch support. The pol I, II, III, Archaea tree was created using LG substitution model, SPR tree improvement, and 100 replicates for boot strap branch support (SH-Like branch support was also performed with similar results). The Bacterial and Plastid RNAP tree was created using LG substitution model, SPR tree improvement, and SH-Like branch support. TreeDyn46 (http://www.treedyn.org/) was used to view and analyze the phylogenetic trees.
Intergenic Gap Analysis
We used a custom program (get_rpoB_rpoC_intergenic_gap.pl) that read in the NCBI record for each bacterial β and β′ species matched pair and extracted the gene start and stop locations for rpoB and rpoC if available. The program then verified that the two genes were going in the same direction and calculated the bp distance between the stop codon of the rpoB gene and the start codon of the rpoC gene. Unusual distances were verified by visiting the NCBI gene link for the corresponding β or β′ subunit.
Detection of Bacterial Lineage-Specific Domain Insertions
In order to detect the bacterial lineage-specific insertions, we first created individual alignments for each full-length protein sequence with its sequence from the final cleaned up alignment containing only the alignable sequence positions. In order to facilitate this we used a custom program (find_inserts.pl) to automate the creation of the individual alignments using PCMA, followed by manual correction of mismatched positions identified by the custom program. We then used a custom program (find_inserts.pl) to search through each alignment for large gaps (usually >50 residues) in the cleaned up sequence that would indicate where we removed a possible lineage-specific insertion or sequence region not contained in all of the bacterial sequences. In order to have a common frame of reference we also converted the insertion start and end positions to the Tth structure residue numbering. We then manually sorted the list of insertions to locate insertions with the same start and end points and extracted the insertion residues followed by alignment using MUSCLE47, which proved to the best alignment program for this task. We then tried to identify the sequence motifs of the insertions by comparing our results to those obtained by Iyer et al.22. We also made use of the available structural information for the Taq β′NCD (β′In2) and Eco β′GNCD (β′In6) SBHM motif lineage-specific domain insertions23.
Acknowlegements
W.J.L. was supported by National Institutes of Health MSTP grant GM07739 and The W.M. Keck Foundation Medical Scientist Fellowship. We thank Lars Westblade, Chris Lima, and Tom Muir for helpful discussions and advice. W.J.L. would also like to thank his wife for her patience and support. This work was supported by NIH GM061898 and GM053759 to S.A.D.
Abbreviations
- aRNAP
archaeal RNA polymerase
- BBM1
β-β′ Module 1
- BBM2
β-β′ Module 2
- BlaFA
BLAST to FASTA file to alignment
- bRNAP
bacterial RNA polymerase
- Eco
Escherichia coli
- Elit
Erythrobacter litoralis
- eRNAP
eukaryotic RNA polymerase
- Hpy
Helicobacter pylori
- MSA
Multiple sequence alignment
- pRNAP
plastid RNA polymerase
- RNAP
RNA polymerase
- SBHM
Sandwich barrel hybrid motif
- Sce
Saccharomyces cerevisiae
- Tde
Thiomicrospira denitrificans
- Taq
Thermus aquaticus
- Tth
Thermus thermophilus
- vRNAP
viral RNA polymerase
- Wen
Wolbachia endosymbiont
- Wpi
Wolbachia pipientis
Footnotes
Additional information
Please visit http://darstlab.org/supp/RNAP_MSA_2009 to download the BlaFA and other custom programs, RNAP BlaFA pattern files, sequence files, alignments, annotation files, phylogenetic trees, intergenic gap analysis, shared sequence region positions, and the lineage-specific insertions details.
References
- 1.Cramer P. Multisubunit RNA polymerases. Curr. Opinion Struct. Biol. 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- 2.Darst SA. Bacterial RNA polymerase. Curr. Opinion Struct. Biol. 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- 3.Initiative TAG. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature. 2000;408:796–815. doi: 10.1038/35048692. [DOI] [PubMed] [Google Scholar]
- 4.Hu J, Troxler RF, Bogorad L. Maize chloroplast RNA polymerase: the 78-kilodalton polypeptide is encoded by the plastid rpoC1 gene. Nucleic Acids Res. 1991;19:3431–3434. doi: 10.1093/nar/19.12.3431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hu J, Bogorad L. Maize chloroplast RNA polymerase: the 180-, 120-, and 38-kilodalton polypeptides are encoded in chloroplast genes. Proc. Natl. Acad. Sci. USA. 1990;87:1531–1535. doi: 10.1073/pnas.87.4.1531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Iyer LM, Balaji S, Koonin EV. Evolutionary genomics of nucleo-cytoplasmic large DNA viruses. Virus Res. 2006;117:156–184. doi: 10.1016/j.virusres.2006.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Iyer LM, Aravind L, Koonin EV. Common origin of four diverse families of large eukaryotic DNA viruses. J. Virol. 2001;75:11720–11734. doi: 10.1128/JVI.75.23.11720-11734.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sweetser D, Nonet M, Young RA. Prokaryotic and eukaryotic RNA polymerases have homologous core subunits. Proc. Natl. Acad. Sci. USA. 1987;84:1192–1196. doi: 10.1073/pnas.84.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jokerst RS, Weeks JR, Zehring WA, Greenleaf AL. Analysis of the gene encoding the largest subunit of RNA polymerase II in Drosophila. Mol. Gen. Genet. 1989;215:266–275. doi: 10.1007/BF00339727. [DOI] [PubMed] [Google Scholar]
- 10.Zhang G, Campbell EA, Minakhin L, Richter C, Severinov K, Darst SA. Crystal structure of Thermus aquaticus core RNA polymerase at 3.3 Å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 11.Gnatt AL, Cramer P, Fu J, Bushnell DA, Kornberg RD. Structural basis of transcription: An RNA polymerase II elongation complex at 3.3 Å resolution. Science. 2001;292:1876–1882. doi: 10.1126/science.1059495. [DOI] [PubMed] [Google Scholar]
- 12.Kettenberger H, Armache K-J, Cramer P. Complete RNA polymerase II elongation complex structure and its interactions with NTP and TFIIS. Mol. Cell. 2004;16:955–965. doi: 10.1016/j.molcel.2004.11.040. [DOI] [PubMed] [Google Scholar]
- 13.Korzheva N, Mustaev A, Kozlov M, Malhotra A, Nikiforov V, Goldfarb A, Darst SA. A structural model of transcription elongation. Science. 2000;289:619–625. doi: 10.1126/science.289.5479.619. [DOI] [PubMed] [Google Scholar]
- 14.Vassylyev DG, Vassylyeva MN, Perederina A, Tahirov TH, Artsimovitch I. Structural basis for transcription elongation by bacterial RNA polymerase. Nature. 2007;448:157–162. doi: 10.1038/nature05932. [DOI] [PubMed] [Google Scholar]
- 15.Vassylyev DG, Vassylyeva MN, Zhang J, Palangat M, Artsimovitch I, Landick R. Structural basis for substrate loading in bacterial RNA polymerase. Nature. 2007;448:163–168. doi: 10.1038/nature05931. [DOI] [PubMed] [Google Scholar]
- 16.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: Separation of RNA from DNA by RNA polymerase II. Science. 2004;303:1014–1016. doi: 10.1126/science.1090839. [DOI] [PubMed] [Google Scholar]
- 17.Westover KD, Bushnell DA, Kornberg RD. Structural basis of transcription: nucleotide selection by rotation in the RNA polymerase II active center. Cell. 2004;119:481–9. doi: 10.1016/j.cell.2004.10.016. [DOI] [PubMed] [Google Scholar]
- 18.Cramer P, Bushnell DA, Kornberg RD. Structural basis of transcription: RNA polymerase II at 2.8 Å resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- 19.Hirata A, Klein BJ, Murakami KS. The X-ray crystal structure of RNA polymerase from Archaea. Nature. 2008;451:851–854. doi: 10.1038/nature06530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ebright RH. RNA polymerase: Structural similarities between bacterial RNA polymerase and eukaryotic RNA polymerase II. J. Mol. Biol. 2000;293:199–213. doi: 10.1006/jmbi.2000.4309. [DOI] [PubMed] [Google Scholar]
- 21.Zhang JH, Chung TD, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J. Biomol. Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 22.Iyer LM, Koonin EV, Aravind L. Evolution of bacterial RNA polymerase: Implications for large-scale bacterial phylogeny, domain accretion, and horizontal gene transfer. Gene. 2004;335:73–88. doi: 10.1016/j.gene.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 23.Chlenov M, Masuda S, Murakami KS, Nikiforov V, Darst SA, Mustaev A. Structure and function of lineage-specific sequence insertions in the bacterial RNA polymerase β′ subunit. J. Mol. Biol. 2005;353:138–154. doi: 10.1016/j.jmb.2005.07.073. [DOI] [PubMed] [Google Scholar]
- 24.Zakharova N, Hoffman PS, Berg DE, Severinov K. The largest subunits of RNA polymerase from gastric helicobacters are tethered. J. Biol. Chem. 1998;273:19371–19374. doi: 10.1074/jbc.273.31.19371. [DOI] [PubMed] [Google Scholar]
- 25.Zakharova N, Paster BJ, Wesley I, Dewhirst FE, Berg DE, Severinov K. Fused and overlapping rpoB and rpoC genes in helicobacters, campylobacters, and related bacteria. J. Bacteriol. 1999;181:3857–3859. doi: 10.1128/jb.181.12.3857-3859.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pei J, Sadreyev R, Grishin NV. PCMA: fast and accurate ultiple sequence alignment based on profile consistency. Bioinformatics. 2003;19:427–428. doi: 10.1093/bioinformatics/btg008. [DOI] [PubMed] [Google Scholar]
- 27.Johnson JM, Mason K, Moallemi C, Xi H, Somaroo S, Huang ES. Protein family annotation in a multiple alignment viewer. Bioinformatics. 2003;19:544–545. doi: 10.1093/bioinformatics/btg021. [DOI] [PubMed] [Google Scholar]
- 28.Dallidiene D, Tan S, Ogura K, Zhang M, Lee AH, Severinov K, Berg DE. Urea sensitization caused by separation of Helicobacter pylori RNA polymerase beta and beta′ subunits. Helicobacter. 2007;12:103–111. doi: 10.1111/j.1523-5378.2007.00479.x. [DOI] [PubMed] [Google Scholar]
- 29.Taillardat-Bisch AV, Raoult D, Drancourt M. RNA polymerase beta-subunit-based phylogeny of Ehrlichia spp., Anaplasma spp., Neorickettsia spp. and Wolbachia pipientis. Int. J. Syst. Evol. Microbiol. 2003;53:455–458. doi: 10.1099/ijs.0.02411-0. [DOI] [PubMed] [Google Scholar]
- 30.Iyer LM, Koonin EV, Aravind L. Evolutionary connection between the catalytic subunits of DNA-dependent RNA polymerases and eukaryotic RNA-dependent RNA polymerases and the origin of RNA polymerases. BMC Struct. Biol. 2003;3:1–23. doi: 10.1186/1472-6807-3-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Soding J. Protein homology detection by HMM-HMM comparison. Bioinformatics. 2005;21:951–960. doi: 10.1093/bioinformatics/bti125. [DOI] [PubMed] [Google Scholar]
- 32.Kuznedelov K, Lamour V, Patikoglour G, Chlenov M, Darst SA, Severinov K. Recombinant Thermus aquaticus RNA polymerase for structural studies. J. Mol. Biol. 2006;359:110–121. doi: 10.1016/j.jmb.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 33.Mijts BN, Patel BK. Random sequence analysis of genomic DNA of an anaerobic, thermophilic, halophilic bacterium, Halothermothrix orenii. Extremophiles. 2001;5:61–69. doi: 10.1007/s007920000174. [DOI] [PubMed] [Google Scholar]
- 34.Darst SA, Opalka N, Chacon P, Polyakov A, Richter C, Zhang G, Wriggers W. Conformational flexibility of bacterial RNA polymerase. Proc. Natl. Acad. Sci. USA. 2002;99:4296–4301. doi: 10.1073/pnas.052054099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Opalka N, Mooney RA, Richter C, Severinov K, Landick R, Darst SA. Direct localization of a β subunit domain on the three-dimensional structure of Escherichia coli RNA polymerase. Proc. Natl. Acad. Sci. USA. 1999;97:617–622. doi: 10.1073/pnas.97.2.617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Severinov K, Kashlev M, Severinova E, Bass I, McWilliams K, Kutter E, Nikiforov V, Snyder L, Goldfarb A. A non-essential domain of E. coli RNA polymerase required for the action of the termination factor Alc. J. Biol. Chem. 1994;269:14254–14259. [PubMed] [Google Scholar]
- 37.Kaplan CD, Larsson K-M, Kornberg RD. The RNA polymerase II trigger loop functions in substrate selection and is directly targeted by alpha-amanitin. Mol. Cell. 2008;30:547–556. doi: 10.1016/j.molcel.2008.04.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang D, Bushnell DA, Westover KD, Kaplan CD, Kornberg RD. Structural basis of transcription: role of the trigger loop in substrate specificity and catalysis. Cell. 2006;127:941–54. doi: 10.1016/j.cell.2006.11.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zakharova N, Bass I, Arsenieva E, Nikiforov V, Severinov K. Mutations in and monoclonal antibody binding to evolutionary hypervariable region of E. coli RNA polymerase β′ subunit inhibit transcript cleavage and transcript elongation. J. Biol. Chem. 1998;273:19371–19374. doi: 10.1074/jbc.273.38.24912. [DOI] [PubMed] [Google Scholar]
- 40.Luo J, Krakow JS. Characterization and epitope mapping of monoclonal antibodies directed against the beta′ subunit of the Escherichia coli RNA polymerase. J. Biol. Chem. 1992;267:18175–18181. [PubMed] [Google Scholar]
- 41.Severinova E, Severinov K. Localization of the Escherichia coli RNA polymerase beta′ subunit residue phosphorylated by bacteriophage T7 kinase Gp0.7. J. Bacteriol. 2006;188:3470–3476. doi: 10.1128/JB.188.10.3470-3476.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zillig W, Fujiki H, Blum W, Janekovi D, Schweig M, Rahmsdorf H, Ponta H, Hirsch-Kauffmann M. In vivo and in vitro phosphorylation of DNA-dependent RNA polymerase of Escherichia coli by bacteriophage-T7-induced protein kinase. Proc. Natl. Acad. Sci. USA. 1975;72:2506–2510. doi: 10.1073/pnas.72.7.2506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rahmsdorf HJ, Pai SH, Ponta H, Herrlich P, Roskoski RJ, Schweiger M, Studier FW. Protein kinase induction in Escherichia coli by bacteriophage T7. Proc. Natl. Acad. Sci. USA. 1974;71:586–589. doi: 10.1073/pnas.71.2.586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weilbacher RG, Hebron C, Feng G, Landick R. Termination-altering amino acid substitutions in the beta′ subunit of Escherichia coli Escherichia col RNA polymerase identify regions involved in RNA chain elongation. Genes & Development. 1994;8:2913–2927. doi: 10.1101/gad.8.23.2913. [DOI] [PubMed] [Google Scholar]
- 45.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogeneis by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 46.Chevenet F, Brun C, Banuls AL, Jacq B, Christen R. TreeDyn: towards dynamic graphics and annotations for analyses of trees. BMC Bioinformat. 2006;7:439. doi: 10.1186/1471-2105-7-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Edgar RC. MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformat. 2004;5:113. doi: 10.1186/1471-2105-5-113. [DOI] [PMC free article] [PubMed] [Google Scholar]