Abstract
Linear DNAs of any sequence can be packaged with high efficiency in vitro into empty viral procapsids by the phage T4 terminase. Packaging substrates of 5 and 50 kbps, terminated by energy transfer dye pairs, were constructed from plasmid and lambda phage DNAs. Nuclease and fluorescence correlation spectroscopy (FCS) assays showed that ~20% of the substrate DNA was packaged and the DNA dye ends of the packaged DNA were protected from nuclease digestion. Both 5 and 50 kbp DNAs upon packaging produced comparable FRET (fluorescence resonance energy transfer) between the Cy5 and Cy5.5 two-dye terminated DNAs. Single molecule FRET (smFRET) and photobleaching analysis shows that the FRET is intramolecular rather than intermolecular upon packaging of most procapsids and demonstrates that single molecule detection (SMD) allows mechanistic analysis of packaging in vitro. FCS- and sm-FRET measurements are comparable, and show that both the 5 and the 50 kbp packaged DNA ends are held within 8–9 nm of each other, within the dimensions of the long axis of the procapsid portal. The calculated distribution of FRET distances is relatively narrow for both FRET-FCS and sm-FRET, suggesting that the two packaged DNA ends are held at the same fixed distance relative to each other in most capsids. Because one DNA end is known to be positioned for ejection through the portal, it can be inferred that both DNAs ends are held in proximity to the portal entrance and ejection channel. The analysis suggests that a DNA loop rather than a DNA end is translocated by the packaging motor to fill the procapsid.
Keywords: Bacteriophage Portal, Terminase, DNA packaging, Capsid DNA condensate, Fluorescence Correlation Spectroscopy, single molecule FRET
Introduction
Viral genomes are enclosed within a capsid for protection, and for transfer and infection of host cells. Nucleic acid translocation into an empty procapsid is a conserved capsid assembly mechanism found among diverse DNA and RNA viruses 1. Many large dsDNA viruses employ high force-generating molecular motors to package their genomes into procapsids by a broadly conserved molecular mechanism 2. Packaging of DNA into most dsDNA bacteriophage capsids requires a two component motor consisting of a portal-containing procapsid docked with a terminase 3; 4; 5. The viral DNA packaging machine is thought to be an ancient invention that is found in all kingdoms, and its mechanism appears to be highly conserved among most tailed dsDNA bacteriophages as well as among many DNA viruses including Herpes and related viruses 6; 7. DNA is often packaged in conjunction with formation of the mature genome ends from a replicative concatemer within bacteriophage or Herpes infected cells by the packaging terminase.
The exact three-dimensional structure and properties of the packaged double-stranded DNA in bacteriophage capsids are fascinating unsolved problems. The final structure found within many bacteriophage capsids results from the high force ATP-energized packaging motor-driven translocation of DNA into a procapsid to yield a highly condensed structure (~500 mg/ml DNA) comparable to that of liquid crystalline DNA. The packaged condensate structure found among numerous phage capsids displays an ordered 2.5 nm duplex to duplex DNA spacing that arises from the energetic packaging mechanism 8. A number of models have been proposed to account for the broadly conserved structural features of such capsid condensates in which DNA is generally organized in concentric shells [summarized in 9; 10]. However, definitive evidence to choose among these structural models appears to be lacking. In fact, despite some generally similar features such as the ordered 2.5 nm duplex spacings found among DNAs packaged into various phage capsids, other structural, chemical, and biological features of packaged DNA condensates found among a number of phages argue against a single unitary packaged capsid DNA structure. Among significant differences found among a number of packaged phage capsids are the following. Depending upon the phage capsid, DNA is oriented transverse (T7)11 or parallel (T4)12; 13 to the head-tail axis of the phage particle; the first DNA end packaged is the last end out (λ, T7)14; 15 or the first end packaged is the first end out (T4)16; and although packaged to comparable 500 mg/ml density, DNA is found in the capsid as a protein-free condensate (λ), found together with a discrete internal protein core structure (T7)11, or is packed together with several thousand molecules of mobile internal proteins9 (T4). These and other significant differences, such as functional DNA-containing “giant” capsids13, argue against a single deterministic packaged phage capsid DNA structure.
In addition both to significant similarities and differences, a number of fundamental properties of packaged DNA remain to be determined for any bacteriophage capsid. Thus, where determined, it is known that one unique end of the packaged phage DNA descends through the portal and into the tail to be injected into the infected host. Such a DNA duplex descending through the portal has been imaged by cryoelectron microscopy17; 18 and a single end is known to be attached to the phage lambda tail following disruption by classical electron microscopy19. However the positions of both mature DNA ends in the capsid have not been determined for any phage. Evidently knowledge of the locations of both packaged DNA ends has an important bearing on the DNA packaging mechanism as well as on the packaged DNA structure itself.
In this work we have specifically labeled one end of a substrate DNA with a single fluorescent donor dye and the other end with a single acceptor dye employing optimal Förster energy transfer distances for the dye pair. We demonstrate that such dye pair DNAs can be efficiently packaged in vitro by the phage T4 large terminase subunit to yield encapsidated DNA where both the dyes and the DNA are protected from nuclease release or digestion. Following efficient in vitro packaging of 5 kbp and 50 kbp DNAs doubly dye-labeled in this way, we have determined by FCS-FRET and smFRET analysis that both ends of the substrate DNAs are held in close proximity to each other within the voluminous T4 phage capsid. From the energy transfer between the pair of dyes we are able to infer that both ends of the DNA are held in proximity to the portal entry and exit channel of the capsid. In addition, our analysis of the in vitro packaged procapsid demonstrates the power of a fluorescence SMD approach to analyzing the DNA packaging mechanism.
Results
Construction and packaging of DNA substrates with a specific fluorescent dye at each end
A 5 kbp linear DNA packaging substrate was constructed with an energy transfer pair of dyes at the two ends by PCR using primers labeled with Cy5 and Cy5.5 (Fig. 1a). We used plasmid pL16 20 as the PCR template with 26 residue primers complementing gene 16 sequences separated by 4,764 bps in this plasmid to yield a substrate predicted to be 4,816 bps (See Materials and methods). PCR amplification led to ~5 kbp DNA products either with two different dyes (Cy5 and Cy5.5) at the ends, or, where one unlabeled and one labeled primer was employed, to ~5 kbp DNA substrates with only a single dye (either Cy5 or Cy5.5) at one end.
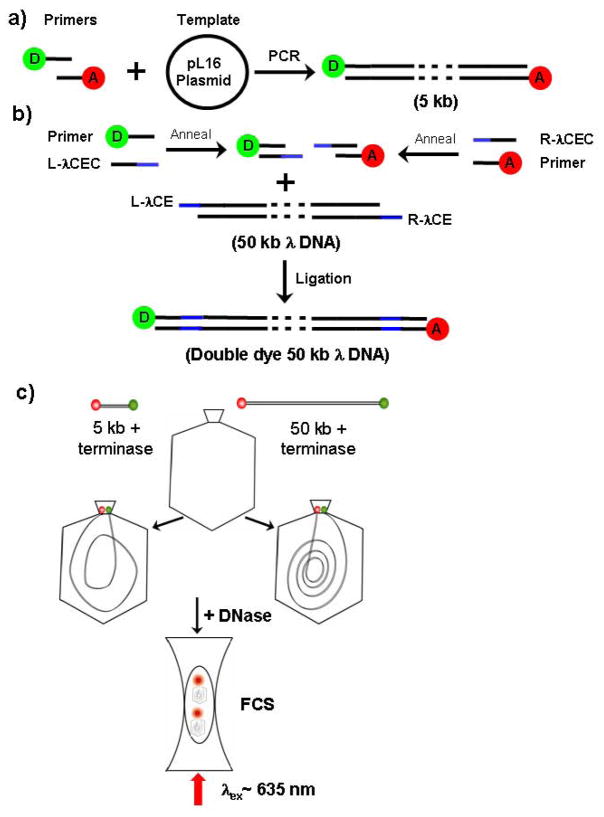
Figure 1. Preparation of double dye-labeled packaging substrate DNAs.
a) The 5 kbp DNA product was obtained by PCR using plasmid pL16 (4851 bps) with primers labeled with Cy5 and Cy5.5.
b) By annealing two 35 residue oligonucleotides (L-λCEC) and (R-λCEC) complementary to the sequences of the left and right single stranded cohesive ends of λ DNA and to the Cy5 and Cy5.5 dye-labeled pL16 PCR primers used in a, we obtained two 35 bp duplex DNAs. Then these products were ligated to linear λ DNA to obtain specific Cy5 and Cy5.5 end-labeled 50 kbp λ DNA. green ball, Cy5 pL16 primer: red ball, Cy5.5 pL16 primer; Blue bars: λ DNA cohesive ends, L-λCE: λ left cohesive end; R-λCE: λ right cohesive end; L-λCEC: left cohesive end complement; R-λCEC: right cohesive end complement.
c) FCS observation of packaged 5 kbp and 50 kbp DNA substrates: Following limit DNA packaging of single or double dye labeled DNA substrates, the unpackaged DNA was digested to short, fast diffusing dye labeled oligonucleotides, whereas the packaged dye labeled substrate DNAs are protected by the procapsid. The packaging reaction solution was then observed in a femtoliter chamber by laser light excitation for measurement of diffusion and FRET energy transfer of the Cy5 to Cy5.5 dyes by FCS.
To construct a dye-labeled λ DNA-size packaging substrate, we synthesized two 35 residue oligonucleotides (L-λCEC and R-λCEC, see Fig. 1b) that were complementary to the λ left and right cohesive ends at their 5′ ends and that were complementary to the dye-labeled pL16 primers at their 3′ ends. Following annealing of the left and right end specific oligonucleotides and primers, we added in excess the resulting two dye-labeled duplex DNAs to linear lambda DNA with free cohesive ends. Following annealing and ligation to the λ DNA left and right cohesive ends, a λ DNA substrate with 35 bp duplex extensions with 3 base 5′ single strand ends specifically labeled with Cy5 and Cy5.5 dyes at the left and right λ ends respectively, were synthesized and purified.
Nuclease and FCS assay of in vitro packaging of dye-labeled DNA fragments
Gel purified dye-labeled 5 and 50 kbp DNA substrates were employed for packaging in vitro employing highly purified empty large procapsids and terminase large subunit gp17 as previously described21. The phage components (buffer, procapsids and terminase) and DNA were mixed together and generally incubated overnight at room temperature to maximize packaging. Procapsids were added in molar excess to DNA molecules to favor single packaging events per procapsid. After incubation, DNAase was added to the mixture to selectively digest free DNA in solution, whereas DNA packaged into the procapsid is protected from digestion21. The packaged material is released by subsequent SDS-Proteinase K digestion of the procapsid and gel analysis is then used to determine the amount of translocated DNA and to measure packaging efficiency by comparison to the amount of DNA originally added to the packaging mixture. The DNAase digested samples were also employed for FCS measurements of the diffusibility of the packaged and unpackaged dyes, the latter expected to be increased to the diffusibility of dye-bound short oligonucleotides (See Fig. 1c).
Employing two dye-labeled 5 kb DNA or one dye labeled 5 kb DNA, we found by nuclease assay that both DNAs could be packaged. If there were no procapsids, we could not see any protected DNA from the gel analysis (data not shown). Comparable results were obtained using two dye-labeled lambda DNA. The packaging efficiencies of the two dye-labeled 5 kbp DNA and 50 kbp λ DNA are 17% and 20% respectively (Fig. 2e and Fig. 3e. The smears below the substrates in lanes 4 are due to SDS-denatured procapsid proteins). The results therefore showed that DNAs labeled with dyes at both ends could be packaged efficiently in vitro. Previously we had characterized efficient DNA packaging of only DNAs labeled at a single end with dye 22; 23
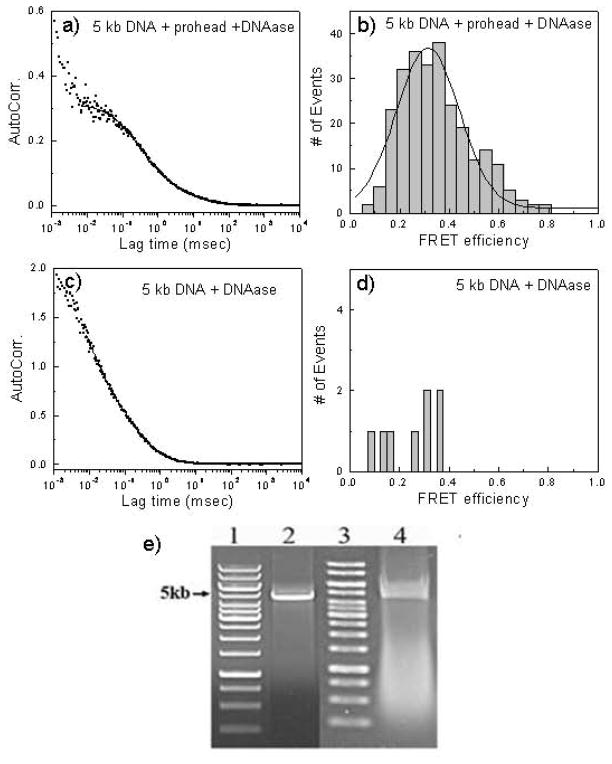
Figure 2. FCS and gel analysis of packaging of double dye-labeled 5kbp DNA.
a) FCS analysis is of an overnight packaging reaction with two dye-labeled 5kbp DNA following DNAase treatment by the decay of the autocorrelation function over time.
b) FRET analysis of the packaged two dye-labeled 5kbp DNA.
c) FCS analysis of the two dye-labeled 5kbp DNA in the packaging reaction buffer without packaging (minus procapsids) after DNAase digestion.
d) FRET analysis of the two dye-labeled 5kbp DNA in the packaging reaction buffer without packaging (minus procapsids) after DNAase digestion.
e) Gel analysis of the two dye-labeled 5kbp input and packaged DNA. Lane 1: 1 kbp DNA ladder; lane 2: two dye-labeled 5kbp input DNA; lane 3: 1 kbp DNA ladder; lane 4: the packaged two dye-labeled 5kbp DNA remaining after DNAase digestion.
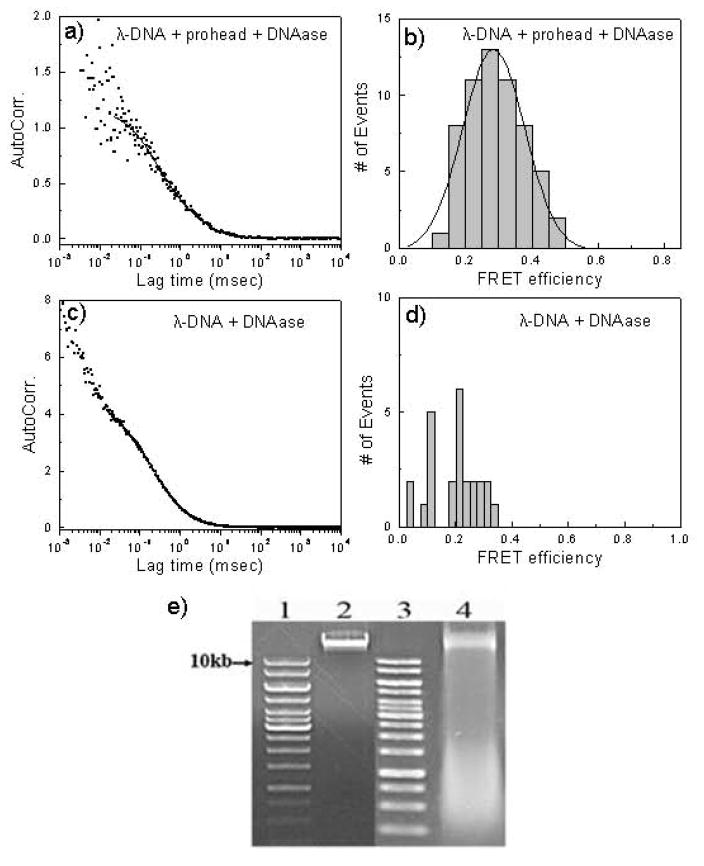
Figure 3. FCS and gel analysis of packaging of double dye-labeled λ DNA.
a) FCS analysis of an overnight packaging reaction with Cy5 and Cy5.5 dye-labeled λ DNA monomer and the decay of the autocorrelation function over time.
b) FRET analysis of the packaged Cy5 and Cy5.5 dye-labeled λDNA.
c) FCS analysis of Cy5 and Cy5.5 dye-labeled λ DNA in the packaging reaction buffer without packaging (minus procapsids) after DNAase digestion.
d) FRET analysis of Cy5 and Cy5.5 dye-labeled λ DNA in the packaging reaction buffer without packaging (minus procapsids) after DNAase digestion.
e) Gel analysis of Cy5 and Cy5.5 dye-labeled λ DNA input and packaged DNA. Lane 1: 1 kbp DNA ladder; lane 2: Cy5 and Cy5.5 dye-labeled λ input DNA; lane 3: 1 kbp DNA ladder; lane 4: the packaged double dye labeled λ DNA remaining after DNAase digestion.
Packaging in vitro of 5 kb and lambda DNA monomers characterized by fluorescence correlation spectroscopy
We used one and two dye-labeled 5kbp DNA packaging substrates for FCS and FRET-FCS. The trace presented in Fig. 2a shows the fluorescence autocorrelation curve of the packaged substrate. The autocorrelation curve was fitted with the following three-dimensional diffusion model. The autocorrelation function for n fluorescent species traversing a 3D Gaussian volume with radius ω0 and half axial z0 is given by:
| (1) |
where τ is the lag time, N is the number of molecules in the volume and ρi are the fractions of the corresponding diffusion coefficients Di. The autocorrelation of the 5kbp DNA substrate in a negative reaction control, one lacking procapsids and treated with DNAase, could be fitted by a single species diffusion model, consistent with a single fluorescent species with a diffusion coefficient of around 115 μm2/sec (Fig. 2c). In contrast, the correlation obtained after the packaging reaction containing procapsids in addition to the other packaging components required two component fitting, a faster species with diffusion coefficient of 108 μm2/sec with a relative abundance of 78%, and a slower species with a diffusion coefficient of 7 μm2/sec and a relative abundance of 22% (Fig. 2a). Correlation curves for the single dye-labeled 5 kbp DNA substrate were similar. The diffusion coefficient of the fast species thus was close to that of free dye, while the slower species had a diffusion coefficient approximating that previously calculated for the T4 procapsid22. This analysis showed that about 22% of the input 5 kbp DNA is sequestered in the procapsid after an overnight packaging reaction, close to the gel-determined packaging efficiency value.
Packaging of two-dye labeled 50 kbp λ DNA yielded comparable FCS results. With all packaging components present FCS curves yielded calculated diffusion coefficients of 10 and 118 μm2/sec with a relative abundance of 21% and 79 % respectively (Fig. 3a); without procapsids only the fast diffusing species was observed (Fig. 3c). These results suggest that the two DNA substrates (5 kbp and 50 kbp) had comparable efficiencies of packaging as judged by FCS and nuclease protection.
FRET-FCS of packaged double dye labeled 5 and 50 kbp DNAs
The collected single photon data was binned by 1 msec bin in each channel (donor or acceptor) which resulted in intensity-time traces and count-rate histograms. Threshold values in each channel were used to identify the single molecule bursts from the corresponding background signal level. Significant signals from adjacent intervals were binned together to get a sum photon number for each burst. Thus, fluorescence bursts were recorded simultaneously in donor and acceptor channels and FRET efficiencies were calculated using
| (2) |
where nD and nA are the sums of donor counts and acceptor counts for each burst, taking into account the possible difference in the detection efficiency (γ) in two separate channels. Energy transfer efficiency (E), donor-to-acceptor distance (r) and Förster Distance (R0) are related by
| (3) |
For the Cy5 (donor) and Cy5.5 (acceptor) pair the value of R0 is ~ 8 nm. We have estimated the donor-to-acceptor distance using Eq. 3. For fluorophores bound to macromolecules, segmental motions of the donor and acceptor tend to randomize the orientations. We assume that a range of static donor-acceptor orientations are present and these orientations do not change during the lifetime of the excited state. Accordingly orientation factor changes in the donor-acceptor fluorophores bound to a rigid procapsid can account for at most a minor portion of the observed Förster Distance changes.
Single dye labeled 5 kbp DNAs produced little or no FRET-FCS following packaging. In addition, packaging a mixture of Cy5 and Cy5.5 single dye terminated 5 kbp DNA substrates led to little FRET. In contrast, packaging of the double dye-labeled DNAs (5 kbp and 50 kbp λ DNA) produced substantial FRET when packaging was observed (Figs. 2b and 3b). However, in the absence of packaging the same double dye substrates showed much less and scattered FRET (Figs. 2d and 3d), suggesting that packaging is required to yield the correlated FRET events. In the presence of packaging the energy transfer efficiencies of the 5 kbp and 50 kbp λ DNAs were comparable. The average value of FRET efficiencies for both double-dye labeled 5 kbp and 50 kbp λ-DNAs is ~0.3. The distance r between the Cy5 and Cy5.5 dye can be calculated using equation 3 and the value in this case is ~ 9.2 nm.
Single molecule observations of immobilized packaged double dye labeled 5 and 50 kbp DNAs
Multiple copies of short dye-labeled DNAs or longer plasmid DNAs can be packaged in vitro into each procapsid 22; 24. Although we carried out packaging reactions with procapsid/DNA ratios that should minimize multiple packaging into one procapsid, it is difficult to exclude the possibility that our FRET-FCS measurements might reflect inter- rather than intramolecular dye interactions within a packaged procapsid. Attempts to resolve this issue by purifying two dye-labeled λ trimers (near to a 170 kbp T4 headful) were inconclusive. Therefore we employed a SMD fluorescence strategy to analyze individual packaged 5 and 50 kbps DNA containing procapsids (Fig. 4). This approach permits a determination of the number of packaged dye-labeled DNAs in the procapsid by the number of photobleaching steps required to eliminate fluorescence.
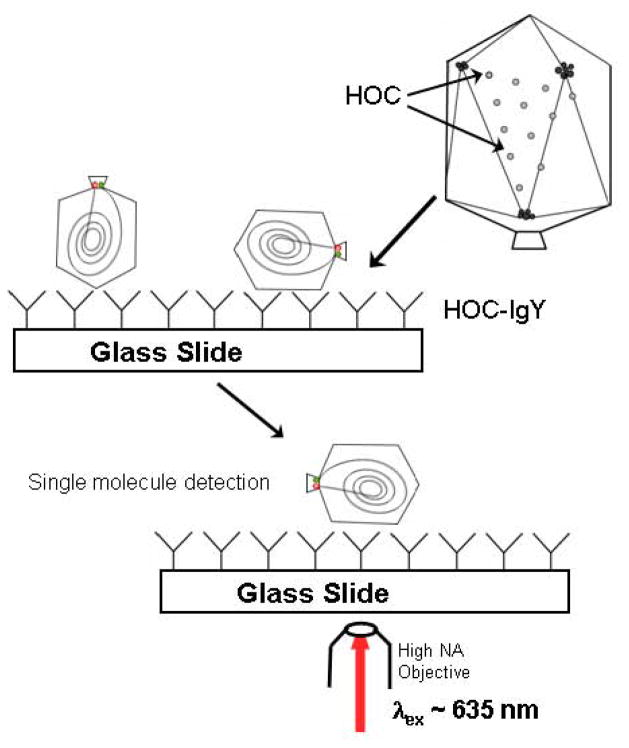
Figure 4.
Surface immobilization strategies for single molecule FRET experiments of packaged Cy5-Cy5.5 labeled 5 kbp DNA or 50 kbp λ-DNA. HOC-IgY polyclonal antibodies bind procapsids to the support glass slide at the 155 HOC molecules decorating the capsid situated at the center of each capsomer. Individual immobilized double dye-labeled particles are then imaged following laser excitation as described.
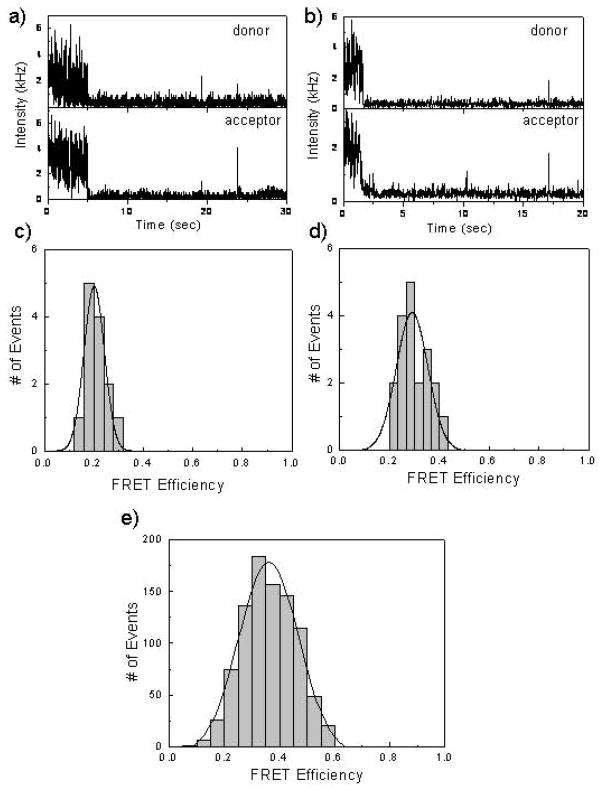
The intensity-time traces shown in Fig. 5 are representative of the donor-acceptor labeled 5 kbp DNA packaged in the procapsids that we examined. The acceptor emission decreased simultaneously with the photobleaching of the donor emission, indicating that most of the acceptor emission is the result of FRET and not direct excitation. The sudden drop in the emission intensity (e.g. Fig. 5a and b) is due to the photobleaching of fluorophores. The results observed for an individual molecule might not be representative of the entire sample. Hence, we examined a large number of individual molecules and used the resulting data to understand the overall properties of the ensemble. We selected individual donor-acceptor pairs and recorded the intensity traces for each molecule. The efficiency of energy transfer was estimated from the total photons in the donor (Cy5) and acceptor (Cy5.5) channels for each individual fluorescence spot. A correction factor was used for the detection efficiencies of the detectors and for cross-talk between the detection channels. A histogram of the FRET efficiencies (Fig. 5e) was constructed using data from about 150 single-molecule time traces. Only spots displaying single-step photobleaching which represented ~ 90% of the total observations corresponding to single DNA molecules were included in the analysis. The average value of FRET efficiencies for double-dye labeled 5kbp DNA is ~0.4. The distance r between the Cy5 and Cy5.5 dye can be calculated using equation 3 and the value in this case is ~8.6 nm.
Figure 5.
(Panels a, b) Single molecule fluorescent intensity time-traces of immobilized packaged Cy5-Cy5.5 labeled 5kbp DNA as seen in the donor and acceptor channels recorded simultaneously by two SPAD detectors. (c,d) Corresponding FRET efficiencies from the double dye labeled packaged 5kbp DNAs. e) Single molecule FRET efficiency of packaged Cy5-Cy5.5 labeled 5 kbp DNA. Data in this bottom panel was compiled from about 150 individual time-traces.
Single molecule fluorescence analysis of the packaged 50 kbp λ DNA containing procapsids was comparable to that of packaged 5 kbp double dye-labeled DNA. Fig. 6a and 6b show two representative intensity-time traces of the Cy5-Cy5.5 labeled λ DNA packaged into the procapsid and the corresponding FRET efficiencies are given in Fig. 6c and 6d. A histogram of the FRET efficiencies constructed using data from about 120 single-molecule time traces is shown in Fig. 6e. Again only molecules displaying single-step photobleaching representing more than 90% of the total population were included in the analysis. The average value of FRET efficiencies for double-dye labeled 50 kbp DNA is ~0.4. From this value the distance r between the Cy5 and Cy5.5 dyes is again calculated to be ~8.6 nm.
Figure 6.
(Panels a, b) Single molecule fluorescent intensity time-traces of immobilized packaged Cy5-Cy5.5 labeled λ-DNA as seen in the donor and acceptor channels recorded simultaneously by two SPAD detectors. (c,d) Corresponding FRET efficiencies from the double dye labeled packaged λ DNAs. e) Single molecule FRET efficiency of packaged Cy5-Cy5.5 labeled 50 kbp λ DNA. Data in this bottom panel was compiled from about 120 individual time-traces.
Discussion
In this work we demonstrate by FRET-FCS and by single molecule FRET that the two ends of the DNA packaged by the phage T4 motor are held in proximity in the procapsid. Similar results for 5 and 50 kbp DNAs suggest that full length 170 kbp T4 genomic DNA has a comparable packaged structure; indeed, the underpackaged 5 and 50 kbp DNA ends presumably would have greater mobility within the capsid. The Cy5 to Cy5.5 FRET distances calculated for both the packaged 5 kbp and 50 kbp DNAs as observed by FCS and SMD measurements are 9.3 and 8.6 nm respectively, thus a distance of 8–9 nm separates the two ends of both packaged DNAs. The calculated distribution of FRET distances is relatively narrow for both FRET-FCS and sm-FRET, suggesting that the two packaged DNA ends are held at the same fixed distance relative to each other in most capsids.
It should be emphasized that our analysis should apply to most of the packaged DNAs in the population. Our FRET-FCS measurements can be derived quantitatively from the acceptor dye molecules channel that show the diffusibility of packaged proheads. The nuclease results also support the FCS quantitation, both at ~20%. Moreover the bulk of over 100 packaged immobilized single proheads in smFRET measurements yield comparable FRET values buttressing the FCS quantitation and showing that the results apply to most packaged molecules. We note that it is highly unlikely that the two dyes at the ends of the packaged DNA are in proximity due to hydrophobic interactions which distort the normal packaged DNA structure; were this the case the spacing should be far closer than 8–9 nm.
The phage T4 expanded procapsid is 100 nm long and 75 nm across, whereas the phage T4 portal is a turbine-shaped dodecamer of 740 kDa with a hollow cylinder 14 nm long and 7 nm in diameter with a central channel of about 3 nm 25. There is strong and direct evidence in the case of phage T426, and as previously discussed in other phages as well, that an end of the DNA is held in the narrow channel of the external end of the procapsid portal ready for injection. If the end to be ejected is held by the portal in this location, our FRET distance measurements strongly suggest that the other end of the packaged DNA is also held close to or within it. The ε15 and P22 portals apparently show a single duplex descending through the portal17; 18. Could the other end also fit into this structure? It is established for λ phage that a DNA duplex containing a 19 base heteroduplex loop can pass through the portal27, thus the DNA fit may not be as tight as it appears in these images, or the portal may hold the second end less tightly and in a more disordered conformation so that it is invisible in the cryo-EM reconstruction. The portion of the portal found inside the capsid also contains a more sizable chamber at its crown where one of the two ends might fit28; 29. Thus the X-ray and cryo-EM structures are consistent with our co-localization of both DNA ends within the portal at a separation of 8–9 nm, within the top to bottom dimension of the phage T4 portal, with a channel measured at 14nm from exit to crown. We can probably not rigorously exclude the possibility that one of the two ends of the DNA lies within the capsid but fixed outside and near to the portal, although we consider this unlikely, since there is no structure within the capsid that is known to be available to fix a DNA end in such a position.
Our determination that both ends of the packaged DNA are in proximity in the capsid is in agreement with a number of other less direct lines of evidence. DNA packaging proceeds, following an initiating cut in the concatemer by the terminase, until the head is full, and after packaging one headful length of DNA, a second endonucleolytic cleavage disconnects the terminase–DNA complex from the packed capsid, allowing addition of DNA end protection proteins (gp2), and neck and tail components to complete infectious virion assembly 30. Because of headful cutting, the second DNA end should lie near to the terminase docked to its portal binding site exposed on the procapsid. The first end packaged into the phage T4 procapsid was determined to be the first end ejected by 32P decay disrupted transfer into the host16, opposite to most sequence specific packaging phages4. The T4 result requires that both ends in the full capsid should be close to each other and to the portal and is consistent with our direct determination in this work. Research on other phages is in agreement with this conclusion. The DNA ends of P2 and P4 heads can be ligated together within packaged capsids or after extraction from the capsid to form complex knotted DNAs, suggesting the DNA ends are in proximity 31; 32; 33; 34. Ion etching results suggest that both ends of the packaged lambda DNA genome are found at the periphery of the DNA mass 35. Overall, because both DNA ends are apparently held in the portal, and because both the portal and the packaged DNA undergo considerable structural rearrangement following packaging, it appears that either end could be selected for ejection, depending upon the phage.
In relation to the packaging mechanism, our work strongly suggests that a DNA loop rather than an end is translocated into the procapsid by the packaging machinery, since if an end were translocated into and moved freely within the capsid, the probability of co-localizing both ends in the packaged structure would be low. However our results do not require that two duplexes lie side-by-side in the narrow portal egress channel, likely one DNA end is situated in this channel whereas the other is anchored in the wider portal crown channel. Our smFRET analysis of packaged dye labeled DNA yields results comparable to the FCS analysis, and suggests that the packaging mechanism can be further analyzed by applying SMD technologies to immobilized procapsids initiating DNA translocation, undergoing packaging, or stalled in DNA packaging.
Materials and methods
DNA packaging substrate primers
First, a 5 kbp pL16-derived PCR substrate pair of primers was synthesized: 5kbp-F: AGAGGTTCATACACTTTGATTTCCTC, and 5kbp-R: ACCGTACTCCAGACTTAGAAGATGAT. Then the 5kbp-F primer was labeled with Cy5 at the 5′end, and the 5kbp-R was also labeled with Cy5.5 at the 5′end. i.e., Cy5-5kbp-F: Cy5-AGAGGTTCATACACTTTGATTTCCTC, Cy5.5-5kbp-R: Cy5.5-ACCGTACTCCAGACTTAGAAGATGAT (synthesized by Operon).
5 kbp DNA products with a single specific dye molecule at each end
Using primers given above, the 5 kbp DNA substrate was obtained by PCR using as template plasmid pL16 (4851 bps). Using primers Cy5-5kbp-F and Cy5.5-5kbp-R, we got 5kbp DNA products with dyes at both ends; using primers Cy5-5kbp-F and 5kbp-R, we got one dye (Cy5) labeled 5kbp DNA fragments; and using primers of 5kbp-F and Cy5.5-5kbp-R, we got single Cy5.5 labeled 5kbp DNA substrates with the same sequences. All these fragments were purified from amplification reactions using a PCR purification kit (Qiagen).
Lambda DNA with a single specific dye molecule at each end
λ-DNA monomers were purified from CsCl purified bacteriophage λ particles by standard methods. We synthesized two 35 residue oligos to include sequences complementary to each of the cohesive ends of λ DNA, while the remainder of each oligonucleotide could pair to the pL16 primers Cy5-5kbp-F or Cy5.5-5kbp-R. 5kbp-F+COS: GGGCGGCGACCTGAGGAAATCAAAGTGTATGAACC; 5kbp-R+COS: AGGTCGCCGCCCATCATCTTCTAAGTCTGGAGTAC. Then equimolar Cy5-5kbp-F and 5kbp-F+COS were mixed in one tube, and Cy5.5-5kbp-R and 5kbp-R+COS in another tube. Following heating of the mixture at 92°C for 5 minutes, slow cooling to room temperature led to two short duplex DNAs with Cy5 or Cy5.5 dyes, each complementary to one of the two lambda cohesive ends. Two dye-labeled λ DNA was prepared as follows: first, a dye-containing DNA duplex and λ DNA were heated to 65°C for 10 minutes in a single tube to melt any cohesive termini that had reannealed during DNA purification. The tube was placed in ice-water immediately to chill the DNA solution to 0°C before the remainder of the ligation reagents were added. Reaction tubes were incubated for several hours at room temperature after all ligation reagents were added. Then the other dye-labeled DNA duplex was added to the reaction solution and the ligation was repeated. Finally double dye-labeled λ DNA was purified from the ligation components and unreacted short DNA fragments by gel extraction and beta-Agarase I (New England Biolabs) digestion.
Packaging assay
The packaging reaction buffer comprised 50 mM Tris-HCl pH 7.5, 6 mM MgCl2, 100 mM NaCl, 1.25 mM spermidine, 0.1 mg/ml BSA, 1.5 mm DTT, 1.5 mm ATP, 2% PEG (Fluka, 20kDa), 216 nM gp17 (monomer), 9.6 × 109 molecules 5 kbp DNA or 8.2 × 108 molecules of λ DNA substrate and 2 × 1010 purified procapsids in 16 μL volumes. We carried out two packaging reactions for each sample. Reaction tubes were incubated overnight at room temperature. Then 1 μL of 5 mg/ml Pancreatic DNAase I was added and tubes were incubated at 37°C for 30 minutes. One reaction of each sample was employed for FCS analysis, to the other was added a 1:1:1 mix of 0.5 M EDTA, 5 mg/ml proteinase K and 10% SDS (3 μL), and followed by incubation at 65°C for 30 minutes. Protected DNA was resolved on 0.7% agarose gels, bands were detected by EthBr staining, and images were obtained and quantitated on a UVP Epichem3 darkroom apparatus.
FRET-FCS measurements and data analyses
A Picoquant MicroTime 200 confocal microscope was used for FCS measurements. Fluorescence signal was collected through the main optical unit of Picoquant microtime system coupled to an Olympus IX71 microscope. As excitation source, a 635 nm pulsed laser diode (LDH-P-635) with a repetition rate of 40 MHz was used. The excitation laser was reflected by a dichroic mirror (Q655LP, Chroma Technology) to an Olympus UPlanAPO 100x (NA ~ 1.3) oil immersion objective and focused onto the solution sample. The fluorescence that passed a dichroic mirror was focused onto a 75 μm pinhole for spatial filtering to reject out-of-focus signals. The donor and acceptor emissions were separated using a 50/50 nonpolarizing beam-splitter plate and were then focused onto two single-photon avalanche diodes (SPADs) (SPCM-AQR-14, Perkin-Elmer). High quality bandpass and longpass emission filters were used to eliminate the residual excitation and to minimize spectral cross-talk while collecting the donor and acceptor fluorescence in two separate detection channels. The donor emission was collected in the spectral region of 655–685 nm and the acceptor emission was recorded above 710 nm. These set of filters reduced the cross-talk between the Cy5 and Cy5.5 emission significantly. All measurements and data analysis was performed using the PicoQuant SymPhoTime Software.
Single molecule fluorescence assay
Preparation of chicken IgY anti-HOC polyclonal antiserum and its reaction with HOC on phage T4 particles was characterized previously by immuno-gold labeling36. For immobilization of procapsids, cleaned corning glass cover slips were noncovalently coated with Hoc IgY antibodies (10 mg/ml) for two hours at room temperature. The chambers were then washed with PBST (137 mM NaCl; 2.7 mM KCl; 10 mM Na2HPO4; 2 mM KH2PO4, pH 7.4, 0.1% Tween 20 detergent) 3 times. The packaging samples treated with DNAase were applied to the separate chambers and incubated at room temperature for one hour. Next the chambers were washed several times with PBST buffer again as previously. The packaging solution was adjusted to nanomolar concentration to give an appropriate surface density for single-molecule studies. Finally, to reduce photobleaching, 10 μl of imaging buffer (an Oxygen Scavenger System consisting of 0.1 mg/ml glucose oxidase, 0.02 mg/ml catalase, 1% 2-mercaptoethanol, and 3% glucose) were loaded onto the chamber. Without this imaging buffer addition, photobleaching was too rapid to perform smFRET measurements. The samples were then sealed immediately with plastic covers for laser excitation and observation. In the absence of anti-HOC IgY at least 100 fold less fluorescent procapsids was observed to be immobilized on the support slide.
All single-molecule measurements were performed using a time-resolved confocal microscope (Micro-Time 200, PicoQuant). The excitation laser (635 nm) was reflected by a dichroic mirror to a high numerical aperture (NA) oil objective (100x, NA 1.3) and focused to a diffraction limited spot (~300 nm) on the sample surface. The laser power used for single-molecule experiments was maintained at 1 μW. The fluorescence that passed a dichroic mirror (HQ655LP, Chroma) was focused onto a 75-μm pinhole for spatial filtering to reject out-of-focus signals. The donor and acceptor emissions were separated using a 50/50 nonpolarizing beam-splitter plate and were then focused onto two SPADs. Emission filters were used to eliminate the residual excitation and to minimize spectral cross-talk. Intensity-time trajectories were obtained by positioning the excitation beam above the double-dye labeled 5 kbp DNA or 50 kbp λ-DNA packaged individual procapsid molecules immobilized on the cover glass slide. The smFRET data analyses were performed using PicoQuant symphotime software.
Acknowledgments
This work was supported by NIH grants AI011676 and NHGRI HG002655.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Kainov DE, Tuma R, Mancini EJ. Hexameric molecular motors: P4 packaging ATPase unravels the mechanism. Cell Mol Life Sci. 2006;63:1095–105. doi: 10.1007/s00018-005-5450-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Smith DE, Tans SJ, Smith SB, Grimes S, Anderson DL, Bustamante C. The bacteriophage straight phi29 portal motor can package DNA against a large internal force. Nature. 2001;413:748–52. doi: 10.1038/35099581. [DOI] [PubMed] [Google Scholar]
- 3.Johnson JE, Chiu W. DNA packaging and delivery machines in tailed bacteriophages. Current Opinion in Structural Biology. 2007;17:237–243. doi: 10.1016/j.sbi.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 4.Black LW. DNA packaging in dsDNA bacteriophages. Annu Rev Microbiol. 1989;43:267–92. doi: 10.1146/annurev.mi.43.100189.001411. [DOI] [PubMed] [Google Scholar]
- 5.Rao VB, Feiss M. The Bacteriophage DNA Packaging Motor. Annu Rev Genet. 2008 doi: 10.1146/annurev.genet.42.110807.091545. [DOI] [PubMed] [Google Scholar]
- 6.Salmon B, Baines JD. Herpes simplex virus DNA cleavage and packaging: association of multiple forms of U (L) 15-encoded proteins with B capsids requires at least the U(L)6, U(L)17, and U(L)28 genes. J Virol. 1998;72:3045–3050. doi: 10.1128/jvi.72.4.3045-3050.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sheaffer AK, Newcomb WW, Gao M, Yu D, Weller SK, Brown JC, Tenney DJ. Herpes simplex virus DNA cleavage and packaging proteins associate with the procapsid prior to its maturation. J Virol. 2001;75:687–98. doi: 10.1128/JVI.75.2.687-698.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Earnshaw WC, Casjens SR. DNA packaging by the double-stranded DNA bacteriophages. Cell. 1980;21:319–31. doi: 10.1016/0092-8674(80)90468-7. [DOI] [PubMed] [Google Scholar]
- 9.Mullaney JM, Black LW. Activity of foreign proteins targeted within the bacteriophage T4 head and prohead: implications for packaged DNA structure. J Mol Biol. 1998;283:913–29. doi: 10.1006/jmbi.1998.2126. [DOI] [PubMed] [Google Scholar]
- 10.Comolli LR, Spakowitz AJ, Siegerist CE, Jardine PJ, Grimes S, Anderson DL, Bustamante C, Downing KH. Three-dimensional architecture of the bacteriophage phi29 packaged genome and elucidation of its packaging process. Virology. 2008;371:267–77. doi: 10.1016/j.virol.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 11.Cerritelli ME, Cheng N, Rosenberg AH, McPherson CE, Booy FP, Steven AC. Encapsidated conformation of bacteriophage T7 DNA. Cell. 1997;91:271–80. doi: 10.1016/s0092-8674(00)80409-2. [DOI] [PubMed] [Google Scholar]
- 12.Lepault J, Dubochet J, Baschong W, Kellenberger E. Organization of double-stranded DNA in bacteriophages: a study by cryo-electron microscopy of vitrified samples. Embo J. 1987;6:1507–12. doi: 10.1002/j.1460-2075.1987.tb02393.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Earnshaw WC, King J, Harrison SC, Eiserling FA. The structural organization of DNA packaged within the heads of T4 wild-type, isometric and giant bacteriophages. Cell. 1978;14:559–68. doi: 10.1016/0092-8674(78)90242-8. [DOI] [PubMed] [Google Scholar]
- 14.Hartman PS, Eisenstark A, Pauw PG. Inactivation of phage T7 by near-ultraviolet radiation plus hydrogen peroxide: DNA-protein crosslinks prevent DNA injection. Proc Natl Acad Sci U S A. 1979;76:3228–32. doi: 10.1073/pnas.76.7.3228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sternberg N, Weisberg R. Packaging of prophage and host DNA by coliphage lambda. Nature. 1975;256:97–103. doi: 10.1038/256097a0. [DOI] [PubMed] [Google Scholar]
- 16.Black LW, Silverman DJ. Model for DNA packaging into bacteriophage T4 heads. J Virol. 1978;28:643–55. doi: 10.1128/jvi.28.2.643-655.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lander GC, Tang L, Casjens SR, Gilcrease EB, Prevelige P, Poliakov A, Potter CS, Carragher B, Johnson JE. The structure of an infectious P22 virion shows the signal for headful DNA packaging. Science. 2006;312:1791–5. doi: 10.1126/science.1127981. [DOI] [PubMed] [Google Scholar]
- 18.Jiang W, Chang J, Jakana J, Weigele P, King J, Chiu W. Structure of epsilon15 bacteriophage reveals genome organization and DNA packaging/injection apparatus. Nature. 2006;439:612–6. doi: 10.1038/nature04487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Thomas JO. Chemical linkage of the tail to the right-hand end of bacteriophage lambda DNA. J Mol Biol. 1974;87:1–9. doi: 10.1016/0022-2836(74)90555-5. [DOI] [PubMed] [Google Scholar]
- 20.Lin H, Simon MN, Black LW. Purification and characterization of the small subunit of phage T4 terminase, gp16, required for DNA packaging. J Biol Chem. 1997;272:3495–501. doi: 10.1074/jbc.272.6.3495. [DOI] [PubMed] [Google Scholar]
- 21.Black LW, Peng G. Mechanistic coupling of bacteriophage T4 DNA packaging to components of the replication-dependent late transcription machinery. J Biol Chem. 2006;281:25635–43. doi: 10.1074/jbc.M602093200. [DOI] [PubMed] [Google Scholar]
- 22.Sabanayagam CR, Oram M, Lakowicz JR, Black LW. Viral DNA packaging studied by fluorescence correlation spectroscopy. Biophys J. 2007;93:L17–9. doi: 10.1529/biophysj.107.111526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Oram M, Sabanayagam C, Black LW. Modulation of the packaging reaction of bacteriophage t4 terminase by DNA structure. J Mol Biol. 2008;381:61–72. doi: 10.1016/j.jmb.2008.05.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Leffers G, Rao VB. A discontinuous headful packaging model for packaging less than headful length DNA molecules by bacteriophage T4. J Mol Biol. 1996;258:839–50. doi: 10.1006/jmbi.1996.0291. [DOI] [PubMed] [Google Scholar]
- 25.Driedonks RA, Engel A, tenHeggeler B, van D. Gene 20 product of bacteriophage T4 its purification and structure. J Mol Biol. 1981;152:641–62. doi: 10.1016/0022-2836(81)90121-2. [DOI] [PubMed] [Google Scholar]
- 26.Zachary A, Black LW. Isolation and characterization of a portal protein-DNA complex from dsDNA bacteriophage. Intervirology. 1992;33:6–16. doi: 10.1159/000150225. [DOI] [PubMed] [Google Scholar]
- 27.Pearson RK, Fox MS. Effects of DNA heterologies on bacteriophage lambda packaging. Genetics. 1988;118:5–12. doi: 10.1093/genetics/118.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Simpson AA, Tao Y, Leiman PG, Badasso MO, He Y, Jardine PJ, Olson NH, Morais MC, Grimes S, Anderson DL, Baker TS, Rossmann MG. Structure of the bacteriophage phi29 DNA packaging motor. Nature. 2000;408:745–50. doi: 10.1038/35047129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lebedev AA, Krause MH, Isidro AL, Vagin AA, Orlova EV, Turner J, Dodson EJ, Tavares P, Antson AA. Structural framework for DNA translocation via the viral portal protein. Embo J. 2007;26:1984–94. doi: 10.1038/sj.emboj.7601643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Black LW, Showe MK, Steven AC. Morphogenesis of the T4 head. In: Karam JD, editor. Molecular Biology of Bacteriophage T4. ASM Press; Washington, DC: 1994. pp. 218–258. [Google Scholar]
- 31.Liu LF, Calendar LPR, Wang JC. Knotted DNA from bacteriophage capsids. Proc Natl Acad Sci U S A. 1981;78:5498–5502. doi: 10.1073/pnas.78.9.5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wolfson JS, GLM, Hooper DC, Swartz MN. Knotting of DNA molecules isolated from deletion mutants of intact bacteriophage P4. Nucleic Acids Research. 1985;13:6695–6702. doi: 10.1093/nar/13.18.6695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu LF, JLD, Calendar R. Novel topologically knotted DNA from bacteriophage P4 capsids: studies with DNA topoisomerases. Nucleic Acids Research. 1981;9:3979–3989. doi: 10.1093/nar/9.16.3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Arsuaga J, Vazquez M, McGuirk P, Trigueros S, Sumners D, Roca J. DNA knots reveal a chiral organization of DNA in phage capsids. Proc Natl Acad Sci U S A. 2005;102:9165–9. doi: 10.1073/pnas.0409323102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mendelson EC, Newcombe WN, Brown JC. Ar+ Plasma-Induced Damage to DNA in Bacteriophage A: Implications for the Arrangement of DNA in the Phage Head. JOURNAL OF VIROLOGY. 1992;66:2226–2231. doi: 10.1128/jvi.66.4.2226-2231.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Baumann RG, Mullaney J, Black LW. Portal fusion protein constraints on function in DNA packaging of bacteriophage T4. Mol Microbiol. 2006;61:16–32. doi: 10.1111/j.1365-2958.2006.05203.x. [DOI] [PubMed] [Google Scholar]