Abstract
Objective
Approximately 10% of patients undergoing cardiac surgery suffer perioperative myocardial injury (PMI). A recent genome-wide association study identified an association between myocardial infarction in non-surgical populations and common genetic variants on chromosome 9p21. We hypothesized that these variants are also associated with PMI after isolated primary coronary artery bypass graft (CABG) surgery.
Methods
In a prospective observational study of 846 Caucasian primary CABG surgery patients at 2 US centers, we genotyped 61 linkage disequilibrium tagging single nucleotide polymorphisms (SNPs), encompassing 436 kbp of the 9p21 region. A multivariable logistic model was used to adjust for previously identified clinical covariates of PMI. PMI was defined as a postoperative day 1 cardiac Troponin I (cTnI) in the top 10th percentile (> 9.13 μg/L) of the cohort. Multiple testing of SNPs was corrected for with family-wise (FW) errors.
Results
Prior myocardial infarction and longer cardiopulmonary bypass time were significant independent predictors of PMI. Levels of postoperative cTnI were incrementally increased for each additional copy of the risk alleles of three SNPs: rs10116277, rs6475606 and rs2383207. Adjusted additive odds ratios ranged between 1.64 and 1.79 (asymptotic P value between 3.7 × 10-3 - 6×10-4) and remained significantly associated with PMI even after accounting for clinical covariates including severity of coronary disease, and multiple comparisons.
Conclusions
We have now demonstrated that common genetic variants in 9p21 previously known to be associated with myocardial infarction in non-surgical populations, are also associated with PMI after CABG. Further investigation is warranted to elucidate functional mechanisms.
Ultramini-Abstract
We identified an association between common genetic variants in the 9p21 region in primary CABG surgery patients and levels of postoperative cTnI, which were incrementally increased for each additional copy of the risk alleles. The association remained highly significant even after accounting for clinical covariates including severity of coronary disease.
Introduction
Multiple epidemiologic studies have documented a significant heritability for coronary artery disease (CAD) and myocardial infarction (1). Using an unbiased genome-wide approach, recent studies identified an association between cardiovascular disease, CAD or myocardial infarction, with common genetic variants on chromosome 9p21 in non-surgical populations (2-4). Though the responsible molecular genetic determinants remain largely unidentified, these variants are adjacent to genes for the cyclin-dependent kinases CDKN2A/B, which play a critical role in regulating cell aging, cell proliferation, and apoptosis (5-8), as well as ANRIL, a large anti-sense non-coding RNA gene, shown to be expressed in cell types integral to atherosclerosis (9-11).
Perioperative myocardial injury (PMI), which afflicts nearly one million people annually during or after non cardiac surgery worldwide(12), has conventionally been associated with transient myocardial ischemia and reperfusion due to aortic occlusion, cardiotomy, and an obligatory acute inflammatory response associated with cardiopulmonary bypass (CPB). While a few limited studies have implicated a heritable risk of PMI (13, 14), variants in the 9p21 region have not yet been examined for an association with this adverse perioperative outcome. Therefore, we hypothesized that variants in the 9p21 region are also independently associated with PMI after isolated primary coronary artery bypass graft (CABG) surgery with CPB.
Methods
Two institutions (Brigham and Women’s Hospital [BWH] and Texas Heart Institute [THI]), recruited patients within a single study structure known as the CABG Genomics Program (http://clinicaltrials.gov/show/NCT00281164). After August 2001, we prospectively enrolled patients aged 20-90 years undergoing non-emergent primary CABG surgery utilizing CPB, without other concurrent surgery. Patients with a preoperative hematocrit < 25% or transfusion of leukocyte-rich blood products within 30 days before surgery were not enrolled. In order to avoid potential population stratification, analysis was restricted to subjects who self-reported four Caucasian grand-parental ancestry. Study protocols were approved by respective Institutional Review Boards, and participants were enrolled following informed written consent. The funding agencies had no involvement in study design, data interpretation or data analysis.
Data Collection
At each site, we recorded patient demographics, perioperative risk factors, medications, and postoperative outcomes using study-specific case report forms. Blood samples were drawn prior to induction of general anesthesia, after administration of post-CPB protamine, and on the mornings of postoperative days (POD) 1-5. Serum and plasma were stored in vapor-phase liquid nitrogen until analysis for cardiac Troponin I (cTnI), B-type natriuretic peptide (BNP), and creatine kinase MB fraction on a sandwich immunoassay on a Triage® platform using monoclonal and polyclonal antibodies (Biosite Inc., San Diego, CA) at a single core facility. Myocardial injury was defined as postoperative day (POD) 1 cTnI in the top 10th percentile (> 9.13 μg/L).
Genotyping
DNA was extracted from white blood cells using standard procedures. Linkage-disequilibrium (LD) tagging SNPs between chromosome 9 positions 21,930,588-22,366,970 (NCBI Genome Build 36 assembly) encompassing the originally-identified 9p21 region with minor allele frequencies ≥5% in the HapMap Caucasian cohort were identified using Tagger (15). Overall sixty-one tagging SNPs that described 70.8% of the variation with a mean r2 of 0.8 in the 394 HapMap-identified SNPs were genotyped (Supplemental Table 1) using the Golden Gate assay with an Illumina Bead Station 500G system (Illumina, San Diego, CA), in accordance with the manufacturer’s standard recommendations.. Analysis of the raw data was performed with the clustering algorithm of the Illumina BeadStudio software and individual examination of all intensity plots, with manual curation of genotype calls. SNPs with genotyping call rate <95%, with significant deviation from Hardy-Weinberg equilibrium (P < 0.001 in controls) and non-random missingness (P<0.05) between cases and controls were excluded from subsequent analysis.
Statistical Analysis
Categorical and continuous demographic characteristics were compared between groups with likelihood ratio chi-squared and Wilcoxon rank sum tests, respectively. A multivariable logistic model was used to derive perioperative and demographic variables associated with PMI, including severity of CAD. Factors previously associated with PMI after cardiac surgery in prior studies, including acute coronary syndrome, were forced into the logistic regression model along with clinically relevant variables using stepwise selection. PLINK (version 1.04) and SAS (version 9.1.3, SAS Institute, Cary, NC), were used for genetic association analysis. Hardy-Weinberg equilibrium was evaluated using an exact test. After application of genotype quality control criteria, univariate analyses were carried out for each SNP to test the null hypothesis of no association between marker polymorphism and PMI, based on log-additive genetic models. Adjusted odds ratios (OR) of every SNP for PMI were estimated using the above-mentioned multivariable logistic regression model. To correct for multiple testing, tests of significance are reported as family-wise empirical P values based on permuting case/control status.
Results
Between 8/2001 and 8/2006, 877 Caucasian patients were genotyped as described in the methods. 31 patients had genotyping rates <90% or were missing biomarkers and were excluded.
Patient characteristics stratified by PMI status after primary CABG surgery are shown in Table 1. Eighty-five patients (top 10th percentile) had a POD1 cTnI level >9.13mcg/L, and therefore were classified as having PMI. Patients who had a past history of myocardial infarction, lower preoperative left ventricular ejection fraction, longer CPB duration or aortic cross clamp time, or were not receiving preoperative ACE inhibitors, were more likely to develop PMI. Similarly, elevated preoperative cTnI and CKMB, but not preoperative BNP, was associated with a higher risk of PMI. The degree of coronary artery stenosis by angiography or the number of vessels grafted intraoperatively was not significantly different between patients who developed PMI and those who did not. Patients with PMI were hospitalized for one additional day, but did not have significant differences in all-cause mortality. Multivariable logistic model identified recent myocardial infarction and longer duration of CPB time as significant independent predictors of PMI (Table 2).
Table 1. Demographics and clinical characteristics.
| Demographics | cTnI < 9.13mcg/L | cTnI ≥9.13mcg/L | |
|---|---|---|---|
| (N=761) | (N=85) | P-Value | |
| Gender (N, % male) | 628 (83) | 67 (79) | 0.374 |
| Institution (N, %) | 615 (81) | 70 (82) | 0.884 |
| Age (years) | 64 (57-72) | 65 (59-74) | 0.291 |
| BMI (kg/m2) | 28.5 (25.8-31.9) | 28.3 (25.0-32.7) | 0.972 |
| Past Medical History | |||
| LVEF preop (%) | 55 (45-60) | 50 (45-60) | 0.014 |
| Diabetes (Insulin or non-insulin dependent; %) | 203 (27) | 16 (19) | 0.150 |
| Pulmonary disease (COPD, Asthma; %) | 32 (4) | 3( 4) | 0.767 |
| Creatinine (mg/dL) | 1 (0.9-1.2) | 1.1 (0.9-1.3) | 0.401 |
| Hematocrit (%) | 40 (34-44) | 40 (37-43) | 0.520 |
| Hypertension (%) | 570 (75) | 60 (71) | 0.360 |
| Hypercholesterolemia ( %) | 581 (76) | 62 (74) | 0.591 |
| Coronary Stenosis (>50%) regions | |||
| ≤2 | 165 (22) | 14 (16) | |
| 3 | 388 (51) | 49 (58) | |
| ≥ 4 | 208 (27) | 22 (26) | 0.417 |
| Previous myocardial infarction | 309 (41) | 46 (54) | 0.021 |
| Myocardial infarction within prior 2 weeks (%) | 119 (16) | 26 (31) | 0.001 |
| Medications - preoperative (N, %) | |||
| ACE inhibitor | 30 (35) | 362 (48) | 0.039 |
| β-blocker | 587 (77) | 66 (78) | 0.915 |
| Ca++ antagonist | 103 (14) | 8 (9) | 0.396 |
| Aspirin | 568 (75) | 60 (71) | 0.434 |
| HMG CoA reductase inhibitor | 583 (77) | 60 (71) | 0.229 |
| Biomarkers - preoperative | |||
| BNP (μg/L) | 18 (5-56) | 26 (6-72) | 0.112 |
| CKMB (μg/L) | 0.5 (0.2-1.2) | 0.9 (0.3-3.0) | <0.001 |
| cTnI (μg/L) | 0.01 (0-0.03) | 0.03 (0-0.9) | <0.001 |
| Surgery | |||
| Number of grafts (N, %) | |||
| ≤2 | 119 (16) | 13 (15) | |
| 3 | 356 (47) | 41 (48) | |
| ≥4 | 286 (38) | 31 (36) | 0.968 |
| CPB duration (min) | 94 (66-117) | 108 (85-138) | <0.001 |
| Aortic cross clamp duration (min) | 71 (41-90) | 82 (61-107) | <0.001 |
| Postoperative Data | |||
| HLOS (days) | 7 (6-9) | 8 (6-10) | 0.001 |
| Mortality % (N) up to 5 yrs | 52 (7) | 10 (12) | 0.120 |
BMI, Body mass index; LVEF, left ventricular ejection fraction; ACE, angiotensin converting enzyme; HMG, 3-hydroxy-3-methyl-glutaryl-CoA reductase; BNP, B-type natriuretic peptide; CKMB creatinine kinase MB fraction; cTnI, cardiac troponin I; CPB, cardiopulmonary bypass; HLOS, hospital length of stay.
Table 2. Multivariable clinical predictor model of perioperative myocardial injury.
| Predictor | Odds Ratio (95% CI) | P-Value |
|---|---|---|
| Age (Decile) | 1.11 (0.88-1.39) | 0.378 |
| Gender (Male) | 1.25 (0.69-2.28) | 0.457 |
| Institution | 0.66 (0.86-3.2) | 0.131 |
| Preoperative HMG Co-A reductase inhibitor use | 0.73 (0.44-1.22) | 0.232 |
| Myocardial infarction within previous 2 weeks | 2.29 (1.37-3.84) | 0.002 |
| Coronary Stenosis (>50%) regions | ||
| ≤2 | 1 | |
| 3 | 1.14 (0.60-2.18) | |
| ≥4 | 0.97 (0.47-2.01) | 0.80 |
| CPB time | 1.14 (1.08-1.21) | <0.0001 |
| Preoperative Creatinine | 1.56 (0.16-15.5) | 0.70 |
Important clinical and demographic variables from table 1 selected by stepwise regression and entered into multivariable model.
CPB, cardiopulmonary bypass, in increments of 10-mimutes
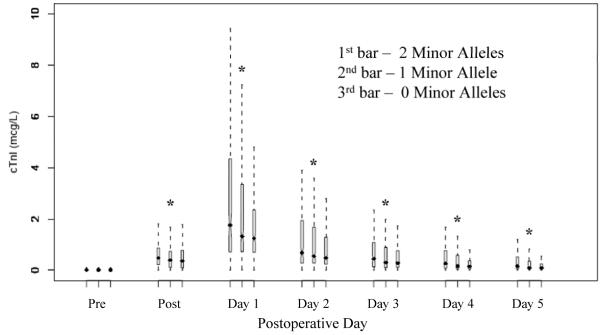
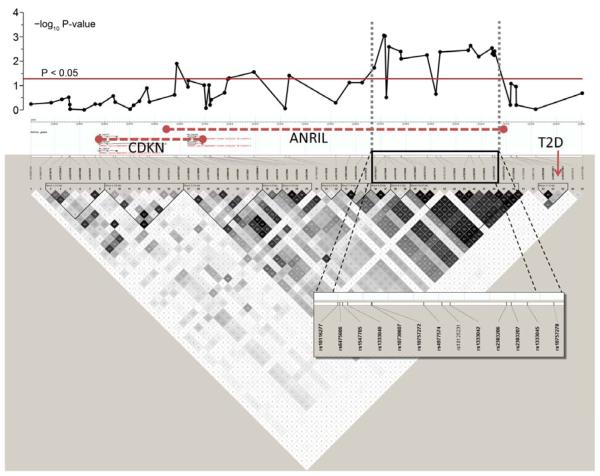
All 61 SNPs passed quality control by genotyping call rate, Hardy-Weinberg equilibrium and non-random missingness. One SNP had an observed minor allele frequency of 1% and was excluded from further analysis leaving a total of 60 remaining SNPs that were included in the analysis (Supplementary Table 1). Levels of postoperative cTnI were incrementally increased for each additional copy of the risk alleles of rs10116277 (Figure 1), rs6475606 and rs2383207. These three SNPs within the coding region of ANRIL were also significantly associated with PMI, even after accounting for clinical covariates and multiple comparisons using an additive genetic model (Table 3). Careful inspection of model fit statistics, based on commonly used statistical criteria (Likelihood Ratio, Nagelkerke R2, Akaike information criterion, and Schwarz’s Bayesian information criterion), revealed very substantial incremental improvement of adding these 9p21 variants into the clinical predictive multivariable model of PMI. Significant pair-wise linkage disequilibrium was present in the region that precludes identification of a single associated variant (Figure 2). The SNP rs10811661, previously associated with type 2 diabetes (16), had no association with PMI in our cohort and is also not in LD with SNPs associated with PMI (Figure 2).
Figure 1. Cardiac Troponin I level for 9p21 risk allele.
The SNP rs10116277 plotted against cardiac troponin I values on the seven perioperative time points. The bars on an individual day represent the copies of the risk allele; the first bar represents 2 copies, the second bar one copy, and the third bar no copy of the risk allele. Each bar represents median with inter-quartile range and outliers shown by the dotted lines.
“*”= P<0.01
cTnI, cardiac troponin I; Pre, preoperatively; Post, immediately post protamine
Table 3. Selected 9p21 SNPs associated with PMI based logistic regression model.
| Minor Allele Frequency |
Covariate-adjusted additive genetic model* |
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Chr 9 position | Allele (Minor/Major) | Cases | Controls | Odds Ratio (95% CI) | Asymptotic P-value † | FWER permuted P-value ‡ | Prior non-surgical MI studies |
| rs10116277 | 22,071,397 | G/T | 0.53 | 0.40 | 1.79 (1.29 - 2.51) | 0.001 | 0.019 | (7) |
| rs6475606 | 22,071,850 | C/T | 0.53 | 0.40 | 1.79 (1.28 - 2.50) | 0.001 | 0.020 | |
| rs1333040 | 22,073,404 | C/T | 0.46 | 0.33 | 1.66 (1.20, 2.28) | 0.002 | 0.054 | (7) |
| rs2383206 | 22,105,026 | A/G | 0.50 | 0.38 | 1.67 (1.20, 2.32) | 0.002 | 0.061 | (2) |
| rs2383207 | 22,105,959 | A/G | 0.50 | 0.37 | 1.71 (1.23 - 2.38) | 0.001 | 0.040 | (7) |
| rs10757278 | 22,114,477 | A/G | 0.54 | 0.42 | 1.70 (1.22 - 2.38) | 0.001 | 0.052 | (7) |
Six most significant SNPs by asymptotic P-value selected. For entire SNP list, see supplement table 1.
SNP, single nucleotide polymorphism; Chr, chromosome; FWER, family-wise error rate; CI, confidence interval; MI, myocardial infarction
The cohort-specific clinical model consists of predictors from the multivariable model in Table 2.
Asymptotic P-value covariate adjusted
Family-wise empirical P value is used to adjust for multiple comparisons.
Figure 2. Linkage disequilibrium (LD) plot of 9p21 region.
The upper panel shows negative log10 P-values for the association of the individual SNPs at their physical location with perioperative myocardial injury (PMI), after adjustment in multivariable regression model. The lower panel summarizes the LD structure in the HapMap database (CEU European ancestry). Regions of LD are shaded in grey (moderate LD) or black (strong LD). Predicted haplotype blocks are framed by triangles. The physical location of the genes CDKN2A and 2B and ANRIL are shown in the HapMap info tack above the SNPs. The type 2 diabetes (T2D) locus is seen on the far right side of the LD plot, with a non-significant association with PMI.
CDKN= cyclin dependent kinase
ANRIL= anti-sense non-coding RNA gene
T2D= type 2 diabetes
Discussion
We have shown that variants on 9p21 previously associated with cardiovascular disease, CAD or myocardial infarction in ambulatory non-surgical populations, are also associated with PMI after CAGB surgery with CPB. Furthermore, we confirmed that this association is independent of type 2 diabetes, and with the SNP rs10811661, previously associated with type 2 diabetes (16). Additionally, though the 9p21 variants have been associated with CAD in non-surgical patients, the association we observed between 9p21 SNPs and PMI in our cohort was independent of CAD severity assessed by coronary angiogram. To our knowledge, this is the first report demonstrating a unique and novel association between 9p21 variants and any adverse perioperative outcome.
Despite considerable efforts by geneticists, the underlying mechanisms of the association of 9p21 with cardiovascular disease, CAD and myocardial infarction have not been elucidated. The 9p21 region encompassing the identified SNPs is flanked by two recombination hotspots and is adjacent to the cyclin-dependent kinases CDKN2A and CDKN2B, which have important roles in cell cycle regulation (11). Through their role in TGF-β- induced growth inhibition, the CDKNs have been implicated in the pathogenesis of atherosclerosis (17), although their role in acute events such as myocardial infarction has yet to be elucidated. A recent study found that probands with the 9p21 risk allele had decreased expression of the CDKN2A/2B locus in peripheral blood leucocytes (18) and that inactivation of CDKN2B and/or CDKN2A allows cells to escape cell cycle arrest, or become senescent (11). Senescent cells are resistant to programmed cell death, leading to accumulation of these cells in injured tissue with substantial effects on healthy neighboring tissue (19). Hypothetically, ischemic injury may unmask and accelerate the effects of the 9p21 variants with at risk patients more susceptible to senescent cell accumulation. Also of interest, the associated SNPs lie within ANRIL, a large antisense non-coding RNA gene, considered to be operating in the transcriptional control repertoire of the cell and expressed in tissues and cell types affected by atherosclerosis (10). Experimental evidence suggests possible coordinated transcriptional regulation of ANRIL mediated by CDKN (10).
In addition to being a risk factor for CAD and having an association with myocardial infarction, variation in 9p21 has also been associated with carotid atherosclerosis, progression of atherosclerosis (20) and abdominal aortic aneurysm (AAA) (21, 22). Surprisingly, 9p21 variants are also associated with intracranial aneurysms (22), a disease not mediated by atherosclerosis. However, the role of variants in 9p21 does not appear to be mediated through vascular reactivity.(23) This lack of association with vascular reactivity, along with the relation of the 9p21 locus to aneurysms, suggests a mechanism more complex than simply promoting atherosclerotic plaque development.
The etiology of PMI occurring during cardiac surgery is markedly different from myocardial infarction occurring in ambulatory populations. Non-surgical myocardial infarction is predominantly associated with prolonged stress-induced myocardial ischemia (i.e. non-ST-segment elevation myocardial infarction) or coronary plaque disruption (i.e. ST-segment elevation myocardial infarction) (24-26). By contrast, CABG surgery with CPB requires aortic cross-clamping and the use of cardioplegic solutions with obligate ischemia and profound coagulation and inflammatory responses. Thus it is perhaps surprising that the genetic evidence of common 9p21 association with both myocardial infarction in non-surgical populations and PMI, supports common molecular mechanisms in both phenotypes.
Similar to the previously published genome-wide association studies in the ambulatory population, the most likely mode of inheritance in our population is an additive model. In contrast, the odds ratios per copy of the risk allele between 1.66 and 1.79 that we observed were significantly higher than the average of 1.3 in associations with cardiovascular disease in the GWAS in Caucasians(6). The variability in risk may be attributed to interaction with environmental factors and the examination of a provoked phenotype; all patients, cases and controls, had CAD severe enough to warrant CABG surgery. Compared to the non-surgical setting, the incidence of myocardial injury in our population is extremely high.
While genetic variation in the 9p21 locus is associated with incident CVD across many different populations and has a high frequency of the risk allele with a substantial increase in the probability of developing disease with each allele, the utility of screening for this polymorphism to improve risk prediction remains unclear. We observed very substantial incremental improvement of adding 9p21 variants into clinical predictive models of PMI. Similarly, in the Caucasian population of the Atherosclerosis Risk in Communities (ARIC) study (n=10,004), adding 9p21 variation to traditional risk factors improved CAD risk prediction and reclassification, particularly in higher risk categories. Conversely, in the Women’s Genome Health Study (n=22,129), genetic variation in 9p21 did not improve discrimination or classification of predicted risk (27). Therefore, further investigation is warranted before the genetic variation in 9p21 can be routinely incorporated into perioperative risk prediction models.
Limitations
We have not yet replicated our findings in a separate cardiac surgical cohort. However, the 9p21 region is the most replicated locus for CAD and myocardial infarction to date across multiple populations. Furthermore, despite examining the entire region, we have observed consistent and strong effect sizes at the same SNPs previously described.
Population stratification, a limitation to all genetic association studies, was addressed by including patients in the analysis who self reported four generations of Caucasian grand-parental ancestry. In addition, we tested genomic controls and tested for differences between Northern and Southern Europeans. Accordingly, we believe that this association is not driven by cryptic heterogeneity.
In a study of this magnitude across two centers and many practitioners, differences in treatment will exist. Institutional differences are unlikely to confound the observed association between 9p21 variants and PMI as (i) personnel and patient were blinded to genotype and perioperative cTnI measurements performed as part of the study; (ii) within this cohort, we did not observe any association between genotype and risk factors for PMI; (iii) within this cohort, it is highly unlikely that concomitant therapies were associated with genotype with the possible exception of gender which was included in all models.
Our analysis was limited to Caucasians and therefore these results are not generalizable to other ethnic groups with variable allele frequencies.
Conclusion
We have identified common genetic variants in 9p21 within or adjacent to genes CDKN2A/B and ANRIL, involved in cell-cycle regulation and atherosclerosis, which are significantly and independently associated with PMI in patients undergoing CABG with CPB. Further investigation is warranted to elucidate the mechanisms by which the variants exert their effects on the development of PMI.
Supplementary Material
Acknowledgements
We acknowledge the outstanding contributory efforts of the CABG Genomics research staff: James Gosnell, RN; Kujtim Bodinaku, MD; Jai Madan, MD, MPH; Svetlana Gorbatov, MPH, James Chen, RN and Isabella Candelaria. We thank all study subjects who participated in the CABG Genomics Program and surgeons who identified their patients.
Sources of funding
These studies were supported in part by the Bayer® Fellowship in Blood Conservation, Biosite Inc. (San Diego, CA); NIH (HL-068774 and NCRR M01 02558) and a Society of Cardiovascular Anesthesiologists Research Starter Grant.
Footnotes
Disclosures
These studies were supported in part by the Bayer® Fellowship in Blood Conservation, Biosite Inc., San Diego, CA; NIH (HL-068774 and NCRR M01 02558) and a Society of Cardiovascular Anesthesiologists Research Starter Grant
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
CABG Genomics Research Study; http://clinicaltrials.gov/show/NCT00281164
References
- 1.Yusuf S, Hawken S, Ounpuu S, Dans T, Avezum A, Lanas F, et al. Effect of potentially modifiable risk factors associated with myocardial infarction in 52 countries (the INTERHEART study): case-control study. Lancet. 2004 Sep 11-17;364(9438):937–52. doi: 10.1016/S0140-6736(04)17018-9. [DOI] [PubMed] [Google Scholar]
- 2.McPherson R, Pertsemlidis A, Kavaslar N, Stewart A, Roberts R, Cox DR, et al. A common allele on chromosome 9 associated with coronary heart disease. Science. 2007 Jun 8;316(5830):1488–91. doi: 10.1126/science.1142447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Consortium WTCC Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007 Jun 7;447(7145):661–78. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, Mayer B, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007 Aug 2;357(5):443–53. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Larson MG, Atwood LD, Benjamin EJ, Cupples LA, D’Agostino RB, Sr., Fox CS, et al. Framingham Heart Study 100K project: genome-wide associations for cardiovascular disease outcomes. BMC Med Genet. 2007;8(Suppl 1):S5. doi: 10.1186/1471-2350-8-S1-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schunkert H, Gotz A, Braund P, McGinnis R, Tregouet DA, Mangino M, et al. Repeated replication and a prospective meta-analysis of the association between chromosome 9p21.3 and coronary artery disease. Circulation. 2008 Apr 1;117(13):1675–84. doi: 10.1161/CIRCULATIONAHA.107.730614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Helgadottir A, Thorleifsson G, Manolescu A, Gretarsdottir S, Blondal T. A common variant on chromosome 9p21 affects the risk of myocardial infarction. Science. 2007 Jun 8;316(5830):1491–3. doi: 10.1126/science.1142842. [DOI] [PubMed] [Google Scholar]
- 8.Abdullah KG, Li L, Shen GQ, Hu Y, Yang Y, MacKinlay KG, et al. Four SNPS on chromosome 9p21 confer risk to premature, familial CAD and MI in an American Caucasian population (GeneQuest) Ann Hum Genet. 2008 Sep;72(Pt 5):654–7. doi: 10.1111/j.1469-1809.2008.00454.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Broadbent HM, Peden JF, Lorkowski S, Goel A, Ongen H, Green F, et al. Susceptibility to coronary artery disease and diabetes is encoded by distinct, tightly linked SNPs in the ANRIL locus on chromosome 9p. Hum Mol Genet. 2008 Mar 15;17(6):806–14. doi: 10.1093/hmg/ddm352. [DOI] [PubMed] [Google Scholar]
- 10.Pasmant E, Laurendeau I, Heron D, Vidaud M, Vidaud D, Bieche I. Characterization of a germ-line deletion, including the entire INK4/ARF locus, in a melanoma-neural system tumor family: identification of ANRIL, an antisense noncoding RNA whose expression coclusters with ARF. Cancer Res. 2007 Apr 15;67(8):3963–9. doi: 10.1158/0008-5472.CAN-06-2004. [DOI] [PubMed] [Google Scholar]
- 11.Kim WY, Sharpless NE. The regulation of INK4/ARF in cancer and aging. Cell. 2006 Oct 20;127(2):265–75. doi: 10.1016/j.cell.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 12.Devereaux PJ, Goldman L, Cook DJ, Gilbert K, Leslie K, Guyatt GH. Perioperative cardiac events in patients undergoing noncardiac surgery: a review of the magnitude of the problem, the pathophysiology of the events and methods to estimate and communicate risk. CMAJ. 2005 Sep 13;173(6):627–34. doi: 10.1503/cmaj.050011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Podgoreanu MV, White WD, Morris RW, Mathew JP, Stafford-Smith M, Welsby IJ, et al. Inflammatory gene polymorphisms and risk of postoperative myocardial infarction after cardiac surgery. Circulation. 2006 Jul 4;114(1 Suppl):I275–81. doi: 10.1161/CIRCULATIONAHA.105.001032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Collard CD, Shernan SK, Fox AA, Bernig T, Chanock SJ, Vaughn WK, et al. The MBL2 ‘LYQA secretor’ haplotype is an independent predictor of postoperative myocardial infarction in whites undergoing coronary artery bypass graft surgery. Circulation. 2007 Sep 11;116(11 Suppl):I106–12. doi: 10.1161/CIRCULATIONAHA.106.679530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005 Jan 15;21(2):263–5. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 16.Saxena R, Voight BF, Lyssenko V, Burtt NP, de Bakker PI, Chen H, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007 Jun 1;316(5829):1331–6. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 17.Kalinina N, Agrotis A, Antropova Y, Ilyinskaya O, Smirnov V, Tararak E, et al. Smad expression in human atherosclerotic lesions: evidence for impaired TGF-beta/Smad signaling in smooth muscle cells of fibrofatty lesions. Arterioscler Thromb Vasc Biol. 2004 Aug;24(8):1391–6. doi: 10.1161/01.ATV.0000133605.89421.79. [DOI] [PubMed] [Google Scholar]
- 18.Liu Y, Sanoff HK, Cho H, Burd CE, Torrice C, Mohlke KL, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS ONE. 2009;4(4):e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Raffetto JD, Leverkus M, Park HY, Menzoian JO. Synopsis on cellular senescence and apoptosis. J Vasc Surg. 2001 Jul;34(1):173–7. doi: 10.1067/mva.2001.115964. [DOI] [PubMed] [Google Scholar]
- 20.Ye S, Willeit J, Kronenberg F, Xu Q, Kiechl S. Association of genetic variation on chromosome 9p21 with susceptibility and progression of atherosclerosis: a population-based, prospective study. J Am Coll Cardiol. 2008 Jul 29;52(5):378–84. doi: 10.1016/j.jacc.2007.11.087. [DOI] [PubMed] [Google Scholar]
- 21.Thompson AR, Golledge J, Cooper JA, Hafez H, Norman PE, Humphries SE. Sequence variant on 9p21 is associated with the presence of abdominal aortic aneurysm disease but does not have an impact on aneurysmal expansion. Eur J Hum Genet. 2008 Oct 15; doi: 10.1038/ejhg.2008.196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Helgadottir A, Thorleifsson G, Magnusson KP, Gretarsdottir S, Steinthorsdottir V, Manolescu A, et al. The same sequence variant on 9p21 associates with myocardial infarction, abdominal aortic aneurysm and intracranial aneurysm. Nat Genet. 2008 Feb;40(2):217–24. doi: 10.1038/ng.72. [DOI] [PubMed] [Google Scholar]
- 23.Samani NJ, Raitakari OT, Sipila K, Tobin MD, Schunkert H, Juonala M, et al. Coronary artery disease-associated locus on chromosome 9p21 and early markers of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008 Sep;28(9):1679–83. doi: 10.1161/ATVBAHA.108.170332. [DOI] [PubMed] [Google Scholar]
- 24.Priebe HJ. Perioperative myocardial infarction--aetiology and prevention. Br J Anaesth. 2005 Jul;95(1):3–19. doi: 10.1093/bja/aei063. [DOI] [PubMed] [Google Scholar]
- 25.Landesberg G, Luria MH, Cotev S, Eidelman LA, Anner H, Mosseri M, et al. Importance of long-duration postoperative ST-segment depression in cardiac morbidity after vascular surgery. Lancet. 1993 Mar 20;341(8847):715–9. doi: 10.1016/0140-6736(93)90486-z. [DOI] [PubMed] [Google Scholar]
- 26.Braunwald E, Antman EM, Beasley JW, Califf RM, Cheitlin MD, Hochman JS, et al. ACC/AHA guidelines for the management of patients with unstable angina and non-ST-segment elevation myocardial infarction. A report of the American College of Cardiology/American Heart Association Task Force on Practice Guidelines (Committee on the Management of Patients With Unstable Angina) J Am Coll Cardiol. 2000 Sep;36(3):970–1062. doi: 10.1016/s0735-1097(00)00889-5. [DOI] [PubMed] [Google Scholar]
- 27.Paynter NP, Chasman DI, Buring JE, Shiffman D, Cook NR, Ridker PM. Cardiovascular disease risk prediction with and without knowledge of genetic variation at chromosome 9p21.3. Ann Intern Med. 2009 Jan 20;150(2):65–72. doi: 10.7326/0003-4819-150-2-200901200-00003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.