Abstract
Post-translational modification of proteins by covalent attachment of a small protein ubiquitin or a polymeric chain of ubiquitin molecules (called polyubiquitin) is involved in controlling a vast variety of processes in eukaryotic cells. The question of how different polyubiquitin signals are recognized is central to understanding the specificity of various types of polyubiquitination. In polyubiquitin, the monomers are linked to each other via an isopeptide bond between the C-terminal glycine of one ubiquitin and a lysine of the other. The functional outcome of polyubiquitination depends on the particular lysine involved in the chain formation and appears to rely on linkage-dependent conformation of polyubiquitin. Thus, K48-linked chains, a universal signal for proteasomal degradation, under physiological conditions adopt a closed conformation where functionally important residues L8, I44, and V70 are sequestered at the interface between the two adjacent ubiquitin monomers. By contrast, K63-linked chains, which act as a non-proteolytic, regulatory signal, adopt an extended conformation that lacks the hydrophobic inter-ubiquitin contact. Little is known about functional roles of the so-called “non-canonical” chains, linked via K6, K11, K27, K29, K33, or head-to-tail; and no structural information on these chains is available, except for the crystal structure of the head-to-tail linked diubiquitin. In this study, we use molecular modeling to examine whether any of the non-canonical chains can adopt a closed conformation similar to that in K48-linked polyubiquitin. Our results show that the eight possible di-ubiquitin chains can be divided into two groups: K6-, K11-, K27-, and K48-linked chains are predicted to form a closed conformation, whereas chains linked via K29, K33, K63, or head-to-tail are unable to form such a contact due to steric occlusion. These predictions are validated by the known structures of K48-, K63-, and head-to-tail linked chains. Our study also predicts structural models for di-ubiquitins linked via K6, K11, 3 and K27. Implications of these findings for linkage-selective recognition of the non-canonical polyubiquitin signals by various receptors are discussed.
Keywords: polyubiquitin, non-canonical linkage, isopeptide linkage, head-to-tail, modeling
Introduction
Ubiquitin (Ub) is a highly conserved 76 amino-acid protein found throughout the eukaryotic cells. A vast number of cellular processes, including targeted protein degradation, cell cycle progression, DNA repair, protein trafficking, inflammatory response, virus budding, and receptor endocytosis1,2, are regulated by Ub-mediated signaling, where the target protein is tagged by a monomeric Ub or a polymeric chain of Ubs. This post-translational modification is tightly controlled by several enzymes (E1, E2, and E3) and occurs through an isopeptide bond between the C-terminal G76 of Ub and a specific lysine residue in the ubiquitinated protein. Ubiquitin polymers (polyUb) are formed in a similar manner, in which consecutive Ub monomers are linked through an isopeptide bond via one of the seven Ub’s lysines. Remarkably, the outcome of polyubiquitination depends on the particular lysine involved in the chain formation and appears to rely on linkage-dependent differences in the conformation/topology of polyUb chains3. For instance, Lys48-linked polyUb, a universal signal for proteasomal degradation, adopts a closed conformation wherein functionally important residues L8, I44, and V70, forming the so-called hydrophobic patch on Ub surface4, are sequestered at the interface between the two adjacent ubiquitin moieties5–8. By contrast, Lys63-linked chains, which act as a regulatory rather than degradative signal in different signaling pathways, adopt an extended conformation in solution, with no direct contact between the hydrophobic patches9,10. Similar conformations of K63-linked chains were recently observed in crystals11,12. As a consequence of these conformational differences, the recognition signals presented by the two types of chains are quite distinct, as illustrated by the different modes in which K48- or K63-linked di-ubiquitin (Ub2) chains bind the UBA-2 domain of hHR23a9,13.
One of the fundamental questions in Ub biology is how the amazing diversity in Ub signaling is achieved. The uniqueness of polyUb as a molecular signal is due to the fact that it is not just a simple chemical group (like e.g. the methylation of phosphorylation signal) or a single protein (e.g. SH2 or SH3 domain, or Ub itself) but a polymeric chain. This not only enhances (via the local concentration effect) the signal carried by each individual Ub monomer but – due to the chain’s conformational flexibility – provides an additional level of structural adaptability which could lead to a greater variety of specific recognition events involving polyUb. Importantly, many known Ub-receptor proteins contain multiple Ub-binding domains, and their avid binding to different Ub units could provide a mechanism for chain-linkage or length selectivity14,15. Thus, knowledge of the conformational properties of polyUb chains holds the key to our understanding of their ability to act as diverse molecular signals. The importance to understand the structural and signaling properties of polyUb has been strengthened further recently by the finding of Ub chains linked via the other five lysines16,17 or even branched (forked) chains18,19. Yet very little is known about the functional roles of these ‘non-canonical’ chains and their structural and recognition properties20. None of these chains has been isolated so far for in vitro studies. Moreover, their structural analysis is hindered by the current unavailability of the E2/E3 enzymes that would allow precise assembly of such chains in vitro. Thus, although structures of K48- and K63-linked as well as linear chains are already available,5,7,8,11,12,21–24 the lack of functional and structural information on the non-canonical linkages limits our understanding of the full scope of Ub signaling and leaves many important questions unanswered. For instance, what are the molecular requirements for specific recognition of the various polyubiquitin signals? What are the structural and recognition properties of the non-canonical chains? Is the linkage via K48 unique or do other linkages also allow close non-covalent contacts between the adjacent Ub units in the chain? Is the extended conformation of K63-linked Ub2 due, as hypothesized in9, to the steric occlusion that prevents direct contact between the hydrophobic patches on the two Ub units?
In this study, we address some of these questions by means of molecular modeling/docking. Specifically, we investigate which of the seven possible isopeptide-linked di-ubiquitin chains can form a closed conformation via direct contact between the hydrophobic patches on the adjacent Ub monomers. Additionally, we perform similar analysis of linear chains where Ub monomers are connected head-to-tail (via their C and N-termini). It was suggested3 that the conformational properties of head-to-tail chains are similar to those of K63-linked chains. Our approach makes use of the HADDOCK software25 and its documented ability8 to account for both ambiguous and unambiguous distance constraints to model the structure of Ub2 chains. Our results show that the eight possible Ub2 chains can be divided into two groups: K6-, K11-, K48-linked chains, and to a somewhat lesser extent the K27-linked one, are predicted to form a closed conformation, whereas the chains with K33-, K63-, K29-linkages as well as head-to-tail are unable to form such a close contact. Our study also provided, for the first time, structural models of di-ubiquitin chains linked via K6, K11, and K27.
Results
The concept
Our approach to modeling polyUb chains is based on the following concept. The two main features of Ub as a monomer (forming a polyUb chain) are the hydrophobic patch (L8-I44-V70) on the β-sheet face of Ub surface and the flexible C terminus (residues R72-G76). All the experimental data available to date indicate that the hydrophobic patch is the “binding hot spot” in Ub: it is directly involved in Ub’s interactions with most of Ub-binding domains26,27. Moreover, this hydrophobic patch is directly involved in Ub/Ub interaction and forms the Ub/Ub interface in K48-linked Ub2 and Ub45–7. Importantly, the recent structure of K48-Ub4, based both on X-ray and NMR data7, shows that the Ub4’s structure is a dimer of Ub2s formed by pair-wise contacts between the neighboring Ub monomers. Therefore we hypothesize that a similar mechanism is applicable to other polyUb chains, in that the hydrophobic patch acts as a molecular “velcro” that forms Ub/Ub contacts. Therefore, if a polyUb chain can have a definitive structure, it has to be formed by the hydrophobic contacts between Ub monomers mediated by their hydrophobic patches. In this study we focus only on di-ubiquitins, as the shortest possible polyUb chain. The question we set to address here is whether any of the possible chain linkages could allow the hydrophobic patch-to-patch contact between the two adjacent Ub monomers.
Chain modeling
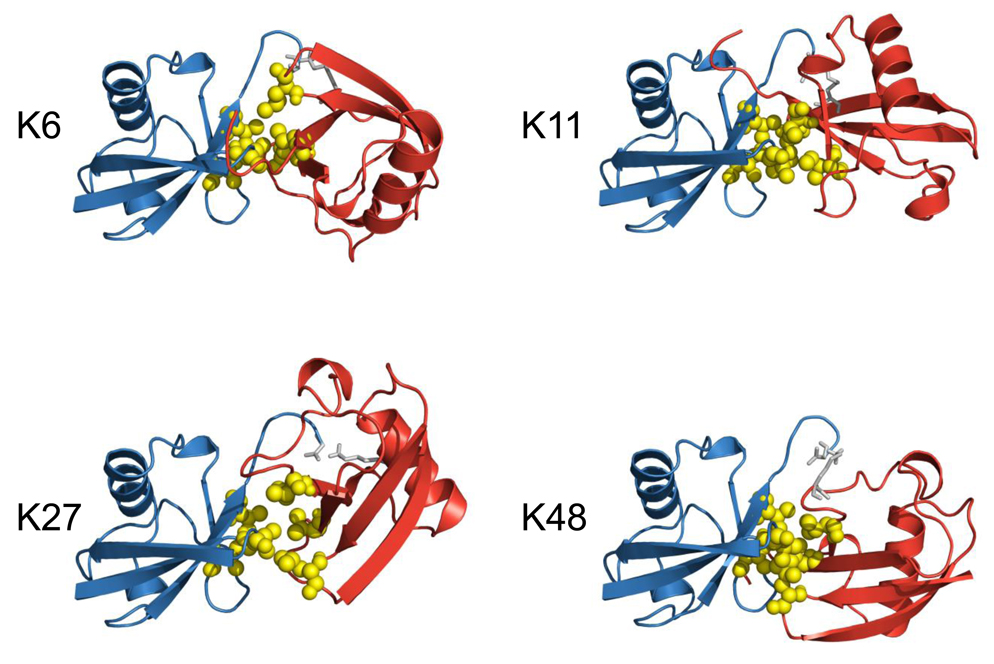
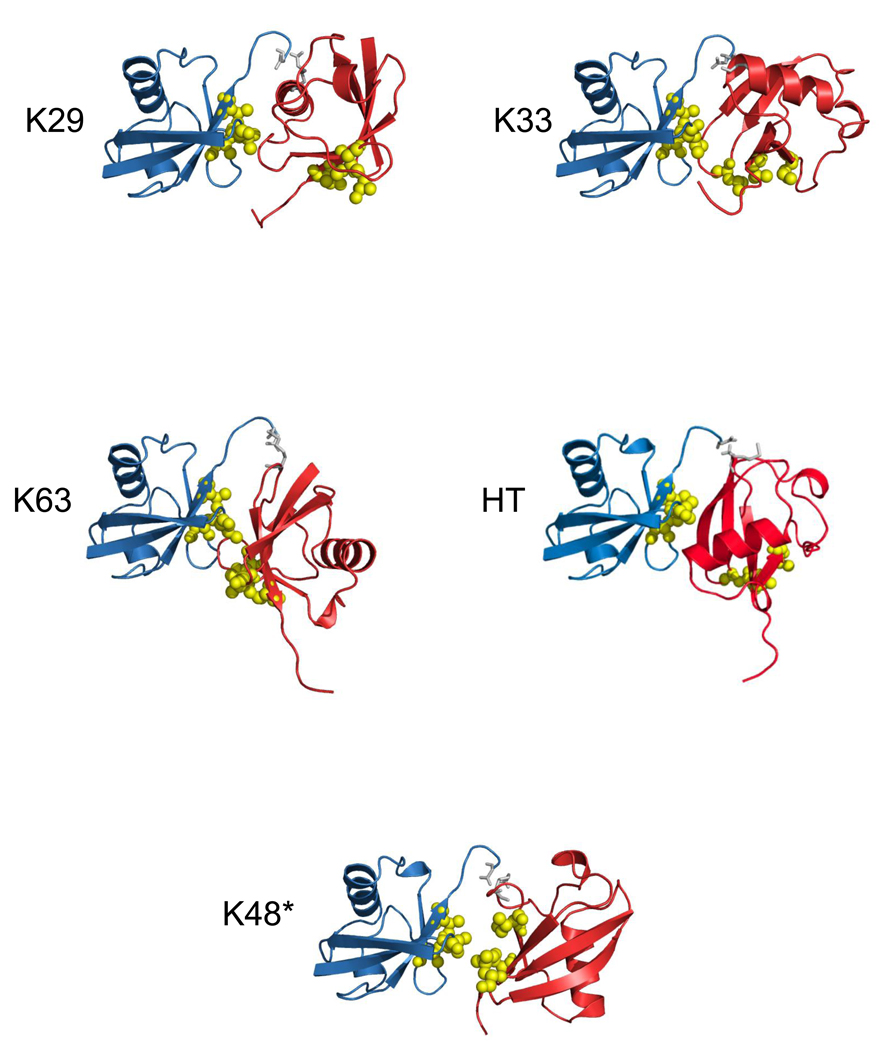
Ub2 chains of all seven possible isopeptide linkages as well as head-to-tail linked chains were generated as described in Materials and Methods. Briefly, to account for the possible close contact between the hydrophobic patches (residues L8, I44, V70) on both Ubs in the chain, we incorporated ambiguous restraints where active and passive residues on each Ub were defined using the procedure described previously25. The covalent Ub-Ub linkage was introduced using unambiguous distance restraints based on typical interatomic distances for a peptide bond in crystal structures, as described8. For each linkage, the resulting 200 structures were subjected to clustering. The clusters were ranked according to their HADDOCK score (Hscore) and the ten best structures in each cluster were retained and analyzed in terms of Hscore (Table 1). In all cases, cluster 1 has the lowest average value of Hscore and also contains the lowest intermolecular energy structure. For all the clusters, the electrostatic energy Eelec was the major energy contribution to the intermolecular energy Einter, whereas the energy associated with unambiguous restraints made the major contribution to Erest. Here Erest quantifies the ability of each chain to satisfy both the unambiguous distance restraints associated with the isopeptide bond and the ambiguous restraints representing the hydrophobic interdomain contacts. Average values of the buried surface area (BSA) for the various clusters range from 736 Å2 to 1458 Å2. For chains linked via K48, K6, K29, K27, and K11, the BSA values are consistent with the typical buried surface area found in protein complexes28. For chains linked via K6 and K33, only one cluster was found, whereas for K48- and K27-linked chains, the average RMSD values to the lowest energy structure are relatively close for both clusters, indicating that the conformations of the two clusters are not dramatically different. For the sake of comparison, the ten best structures of the best cluster in terms of the HADDOCK score were extracted for each chain and will be used in the following discussion. Their respective characteristics are presented in Table 2 whereas the best structure for the best cluster for each generated chain is shown in Figure 1 and Figure 2.
Table 1.
Statistical analysis of HADDOCK results for the generated Ub2 chains of all eight linkages after clustering. Clusters are sorted according to their HADDOCK scores (Hscore). Standard deviations are shown in parentheses.
| Chain linkage |
Clustera | RMSD-Eminb (Å) |
Einterc (kcal.mol−1) |
Evdwd (kcal.mol−1) |
Eelecd (kcal.mol−1) |
BSAe (Å2) | Erestf (kcal.mol−1) |
Eunambg (kcal.mol−1) |
Eambh (kcal.mol−1) |
Hscore (a.u.) |
|---|---|---|---|---|---|---|---|---|---|---|
| K48 | Cluster 1 [117] | 0.5 (0.2) | −280.3 (56.1) | −10.8 (6.8) | −269.6 (57.6) | 1458 (74) | 3.7 (0.8) | 2.9 (0.7) | 0.8 (0.1) | −119.0 (7.7) |
| Cluster 2 [83] | 1.9 (0.1) | −264.3 (53.0) | −3.5 (6.2) | −260.8 (55.1) | 1247 (50) | 6.1 (1.0) | 3.5 (0.9) | 2.7 (0.6) | −108.0 (6.8) | |
| K63 | Cluster 1 [124] | 0.9 (0.4) | −115.6 (17.1) | −6.2 (4.9) | −109.4 (20.3) | 912 (30) | 29.3 (3.9) | 3.4 (0.8) | 25.9 (3.9) | −60.4 (7.2) |
| Cluster 2 [76] | 3.8 (0.1) | −76.2 (17.4) | −7.0 (6.4) | −69.2 (18.4) | 979 (42) | 32.8 (1.4) | 2.7 (0.5) | 30.0 (1.2) | −51.3 (14.9) | |
| K6 | Cluster 1 [200] | 0.7 (0.3) | −136.0 (33.2) | −7.6 (4.9) | −128.4 (32.2) | 1064 (32) | 7.1 (0.8) | 3.3 (0.7) | 3.8 (0.4) | −72.0 (8.5) |
| K33 | Cluster 1 [200] | 0.6 (0.3) | −77.0 (12.9) | 8.6 (4.8) | −85.6 (10.4) | 780 (81) | 20.7 (0.9) | 3.1 (0.5) | 17.6 (0.7) | −59.0 (5.2) |
| K29 | Cluster 1 [112] | 0.6 (0.2) | −256.4 (29.1) | −2.7 (6.5) | −253.7 (33.3) | 1095 (62) | 55.2 (1.3) | 2.6 (0.4) | 52.6 (1.0) | −64.3 (10.4) |
| Cluster 2 [36] | 11.5 (0.1) | −211.2 (45.4) | 1.0 (3.8) | −212.1 (45.9) | 890 (91) | 58.1 (0.9) | 2.8 (0.5) | 55.3 (0.7) | −63.6 (5.4) | |
| Cluster 3 [29] | 9.9 (0.1) | −155.4 (25.3) | −1.0 (4.6) | −154.4 (28.8) | 815 (49) | 52.6 (1.6) | 2.9 (0.6) | 49.7 (1.3) | −51.0 (8.8) | |
| Cluster 4 [16] | 12.1 (0.1) | −131.9 (53.5) | −9.1 (7.1) | −122.8 (54.5) | 736 (77) | 58.4 (1.5) | 3.2 (0.6) | 55.1 (1.7) | −49.0 (4.3) | |
| K27 | Cluster 1 [122] | 0.9 (0.5) | −152.4 (40.2) | −9.4 (3.0) | −143.0 (41.2) | 1058 (85) | 12.5 (0.9) | 4.6 (0.4) | 7.8 (0.8) | −94.2 (7.7) |
| Cluster 2 [78] | 1.1 (0.2) | −127.6 (22.0) | −11.2 (2.8) | −116.4 (21.8) | 1002 (40) | 12.7 (0.5) | 4.7 (0.3) | 8.0 (0.5) | −70.9 (10.3) | |
| K11 | Cluster 1 [166] | 0.8 (0.4) | −117.1 (35.5) | 1.3 (7.3) | −118.4 (35.5) | 1279 (101) | 3.6 (0.4) | 2.4 (0.3) | 1.2 (0.3) | −58.9 (10.4) |
| Cluster 2 [34] | 2.7 (0.1) | −103.8 (34.0) | 5.7 (4.8) | −109.4 (34.0) | 809 (85) | 14.4 (1.2) | 3.1 (0.6) | 11.3 (0.8) | −53.3 (5.8) | |
| HTi | Cluster 1 [70] | 1.7 (0.3) | −256.3 (29) | −24.6 (5.0) | −231.7 (31.2) | 1184 (57) | 65.1 (1.8) | 3.5 (0.7) | 61.6 (1.2) | −32.4 (2.8) |
| Cluster 2 [41] | 0.9 (0.4) | −221.2 (25.5) | −19.7 (5.7) | −201.4 (27.5) | 1169 (53) | 66.4 (1.2) | 2.9 (0.8) | 63.4 (1.3) | −25.8 (7.2) | |
| Cluster 3 [35] | 1.7 (0.3) | −214.1 (34.7) | −21.4 (4.5) | −192.7 (36.3) | 1150 (53) | 66.6 (2.2) | 3.16 (0.9) | 63.5 (1.8) | −19.5 (5.8) | |
| Cluster 4 [30] | 2.33 (0.1) | −196.9 (35.5) | −20.2 (5.0) | −176.7 (37.0) | 1136 (77) | 65.6 (1.8) | 3.5 (1.2) | 62.0 (0.9) | −18.2 (2.7) | |
| Cluster 5 [24] | 1.9 (0.3) | −157.6 (23.5) | −11.2 (6.0) | −146.4 (23.4) | 971 (50) | 68.5 (1.6) | 3.2 (1.00) | 65.3 (1.1) | −2.6 (7.0) | |
| K48*j | Cluster 1 [152] | 7.5 (0.2) | −114.9 (27.2) | 10.2 (2.9) | −125.0 (25.9) | 284 (37) | 3.7 (0.7) | N/A | 3.7(0.7) | −47.9 (2.6) |
| Cluster 2 [20] | 11.4 (0.2) | −57.1 (18.3) | 9.8 (3.7) | −66.9 (15.8) | 440 (37) | 1.9 (0.5) | N/A | 1.9 (0.5) | −36.4 (8.0) | |
| Cluster 3 [13] | 12.1 (0.2) | −43.0 (27.1) | 18.1 (4.2) | −61.1 (23.7) | 249 (50) | 2.3 (0.6) | N/A | 2.3 (0.6) | −30.0 (9.1) | |
| Cluster 4 [9] | 12.5 (0.2) | −66.2 (16.9) | 13.8 (4.7) | −80.0 (16.3) | 228 (37) | 2.5 (0.7) | N/A | 2.5 (0.7) | −25.3 (10.4) | |
| Cluster 5 [6] | 13 (0.2) | −46.4 (20.4) | 7.2 (5.1) | −53.6 (17.2) | 533 (60) | 2.3 (0.6) | N/A | 2.3 (0.6) | −22.4 (13.8) | |
Sorting of the generated structures for each linkage into clusters. The corresponding cluster size is indicated in square brackets.
Average RMSD and standard deviation from the lowest-Hscore structure, for the 10 best structures for each cluster.
Intermolecular energy: sum of the van der Waals and electrostatic energies.
The nonbonded energies were calculated with the OPLS parameters using a 8.5 Å cut-off.
Total buried surface area: sum of the BSA for each ubiquitin subunits.
Restraints energy: sum of the unambiguous and ambiguous energies.
Unambiguous energy, accounts for the isopeptide-bond-related restraints.
Ambiguous energy, accounts for restraints associated with the interdomain contacts between the hydrophobic patches.
Statistical analysis for the head-to-tail linked Ub2 chain.
Statistical analysis for the K48-linked Ub2 generated without the restraints that account for the interdomain hydrophobic contacts.
Table 2.
Summary of HADDOCK parameters averaged over the 10 best structures of the best cluster for each of the eight linkages. Standard deviations are shown in parentheses.
| Chain linkage |
Einter (kcal/mol) |
Evdwa (kcal/mol) |
Eeleca (kcal/mol) |
BSA (Å2) |
Erest (kcal/mol) |
Eunambb (kcal/mol) |
Eambc (kcal/mol) |
Hbonds | Hydrod | Ebinde | Hscore (a.u.) |
ASAhpf (Å2) |
Hbonds per 100 Å2 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| K48 | −280.3 (56.1) |
−10.8 (6.8) |
−269.6 (57.6) |
1458 (74) |
3.7 (0.8) |
2.9 (0.7) |
0.8 (0.1) |
7.4 (2.4) |
14.4 (1.7) |
−11.2 (0.5) |
−119.0 (7.7) |
59.8 (6.4) |
0.51 (0.18) |
| K63 | −115.6 (17.1) |
−6.2 (4.9) |
−109.4 (20.3) |
912 (30) |
29.3 (3.9) |
3.4 (0.8) |
25.9 (3.9) |
3.0 (1.2) |
0.4 (0.5) |
−6.9 (0.2) |
−60.4 (7.2) |
268.9 (9.5) |
0.33 (0.13) |
| K6 | −136.0 (33.2) |
−7.6 (4.9) |
−128.4 (32.2) |
1064 (32) |
7.1 (0.8) |
3.3 (0.7) |
3.8 (0.4) |
3.9 (1.3) |
10.9 (1.1) |
−8.9 (0.2) |
−72.0 (8.5) |
53.9 (8.2) |
0.37 (0.12) |
| K33 | −77.0 (12.9) |
8.6 (4.8) |
−85.6 (10.4) |
780 (81) |
20.7 (0.9) |
3.1 (0.5) |
17.6 (0.7) |
3.0 (1.2) |
0.4 (0.5) |
−5.2 (0.2) |
−59.0 (5.2) |
259.2 (14.2) |
0.40 (0.20) |
| K29 | −256.4 (29.1) |
−2.7 (6.5) |
−253.7 (33.3) |
1095 (62) |
55.2 (1.3) |
2.6 (0.4) |
52.6 (1.0) |
4.8 (1.1) |
0.0 (0.0) |
−7.2 (0.3) |
−64.3 (10.4) |
250.9 (14.3) |
0.44 (0.10) |
| K27 | −152.4 (40.2) |
−9.4 (3.0) |
−143.0 (41.2) |
1058 (85) |
12.5 (0.9) |
4.6 (0.4) |
7.8 (0.8) |
6.0 (1.2) |
9.1 (1.8) |
−8.7 (0.4) |
−94.2 (7.7) |
192.2 (17.8) |
0.57 (0.12) |
| K11 | −117.1 (35.5) |
1.3 (7.3) |
−118.4 (35.5) |
1279 (101) |
3.6 (0.4) |
2.4 (0.3) |
1.2 (0.3) |
4.0 (1.2) |
13.6 (1.7) |
−10.7 (0.7) |
−58.9 (10.4) |
151.0 (9.0) |
0.31 (0.08) |
| HTg | −256.3 (29.0) |
−24.6 (5.0) |
−231.7 (31.2) |
1184 (57) |
65.1 (1.8) |
3.5 (0.7) |
61.6 (1.2) |
6.0 (2.0) |
0.0 (0.0) |
−8.9 0.7) | −32.4 (2.8) |
272.2 (10.7) |
0.50 (0.16) |
| K48*h | −114.9 (27.2) |
10.2 (2.9) |
−125.0 (25.9) |
284 (37) |
3.7 (0.7) |
N/A | 3.7 (0.7) |
1.0 (0.0) |
0.0 (0.0) |
−4.9 (0.2) |
−47.9 (2.6) |
269.8 (10.0) |
0.36 (0.05) |
The nonbonded energies were calculated with the OPLS parameters using a 8.5 Å cut-off.
Unambiguous energy, accounts for the isopeptide bond-related restraints.
Ambiguous energy, accounts for restraints associated with the intermolecular contacts between the hydrophobic patches.
Number of hydrophobic contacts between all atoms belonging to residues V8, L44, and I70 only.
Binding free energy calculated with the DCOMPLEX software46.
Sum of the accessible surface areas for residues V8, L44, and I70 for both Ub units in the chain
Head-to-tail linked Ub2 chain.
K48-linked Ub2 generated without the restraints that account for the interdomain hydrophobic contacts.
Figure 1.
Cartoon representations of the best structures of the best cluster for di-ubiquitin chains which are able to form close contacts between the neighboring Ub units. In all these structures, the distal domain is colored blue, the proximal domain is red, the G76-KX linkage between the two Ubs is colored gray. The hydrophobic patch residues L8, I44, and V70 are shown in ball-and-stick, colored yellow.
Figure 2.
Cartoon representations of the best structures of the best cluster for di-ubiquitin chains which are not able to form close contacts between the neighboring Ub units. The K48* structure was generated without any interdomain constraints between the hydrophobic patches. In all these structures, the distal domain is colored blue, the proximal domain is red, the G76-KX or G76-M1 linkage between the two Ubs is colored gray. The hydrophobic patch residues L8, I44, and V70 are shown in ball-and-stick, colored yellow. HT indicates the head-to-tail linked chain. Note that these are merely computer-generated models under specific set of restraints, not bona fide structures.
Validation of the approach
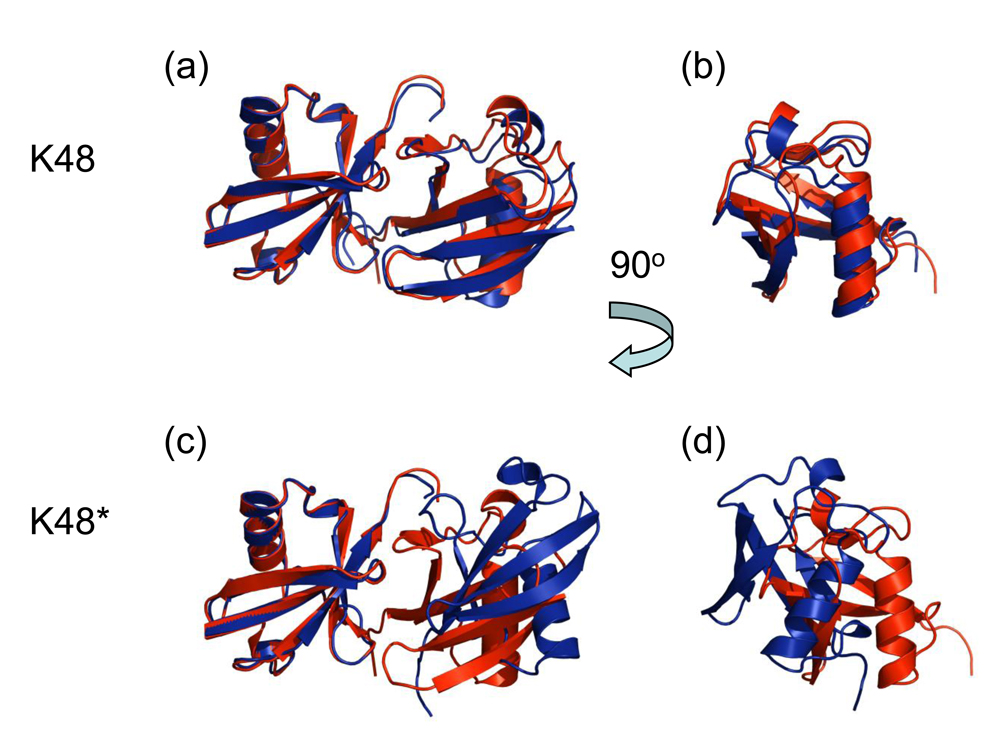
To validate our approach, we performed two different calculations for the K48-linked Ub2. The first one included both the distance restraints associated with the linker (isopeptide bond) and the restraints responsible for the intermolecular contact between the hydrophobic patches on the two Ub moieties. In the second calculation (here referred to as K48*-Ub2, see Fig. 2), the hydrophobic contact restraints were not included, while the linker-related restraints were still present. Differences between the two sets of generated clusters are remarkable, as evident from Table 1, and clearly reflect the conformational difference between the two generated structures (see Fig. 3). The predicted K48-linked Ub2 chain superimposes well with the corresponding crystal structure (Fig. 3a,b): the backbone RMSD is 0.93Å (unstructured C-terminal residues 72–76 were excluded), as well as with the two Ub2 modules from the Ub4 structure7 (RMSDs are 1.18 Å and 1.25Å). By contrast, the predicted K48*-Ub2 structure, where no patch-to-patch interdomain constraints have been included, differs from the crystal structure by a 45° twist of one of the Ub units, resulting in the total backbone RMSD of 5.0 Å to the crystal structure (Fig. 3c,d). It is worth mentioning here that including only a small number of ambiguous interdomain restraints (three active and two passive residues on each Ub unit, Table 3) resulted in the K48-linked Ub2 structure that is similar to the HADDOCK-generated structure based on a complete set of experimentally-derived ambiguous constraints from chemical shift perturbation mapping8. Thus, the inclusion in HADDOCK2.0 of both linker-related restraints and ambiguous restraints representing hydrophobic patch-to-patch interdomain contacts was both necessary and sufficient for predicting the closed conformation of K48-linked Ub2 with good accuracy.
Figure 3.
Experimental validation of the HADDOCK-based approach to generate Ub2 structures used in this study. Shown is the overlay of the crystal structure of K48-linked Ub2 (red, PDB code 1AAR) with the structures (blue) of this chain generated with (a, b) and without (c, d) constraints that mimic the hydrophobic contact between the L8-I44-V70 patches of the two Ub units. All these structures were aligned by the distal Ub only. Panels b and d show the structures from panels a and c, respectively, rotated by 90° about the vertical axis, to facilitate their comparison. Both the relative position and orientation of the two Ub units in the generated Ub2 chain in panels a, b are in good agreement with the crystal structure. By contrast, in the K48*-Ub2 chain (c, d) the interdomain orientation is twisted by 45° compared to the experimental structure.
Table 3.
Details of the restraints included in the HADDOCK simulations
| Distal Uba | |
| Active residuesb | L8, I44, V70 |
| Passive residuesb | G10, L71 |
| Flexible segmentsc | K6-T12 ; R42-A46 ; H66-A72 |
| Fully flexible segmentc | R72-G76 |
| Proximal Uba | |
| Active residuesb | L8, I44, V70 |
| Passive residuesb | G10, L71 |
| Flexible segmentsc | K6-T12 ; R42-A46 ; H66-A72 |
| Fully flexible segmentc | R72-G76 ; K6 or K11 or K27 or K29 or K33 or K48 or K63 |
| Isopeptide Bond (G76 of the distal Ub to the lysine X of the proximal Ub)d |
Unambiguous Restraint Distance (Å) |
| O-NZ | 2.25 ± 0.05 |
| C-NZ | 1.35 ± 0.05 |
| C-CE | 2.45 ± 0.05 |
| CA-NZ | 2.45 ± 0.05 |
The Ub monomers, which are linked to one another via a G76-KX (X=6,11,27,29,33,48,63) isopeptide bond or G76-M1 (head-to-tail), are designated as the distal (containing G76) and proximal (containing KX or M1 and the free C-terminus), respectively.
The docking protocol uses active residues (V8, L44, and I70, forming the hydrophobic patch on each Ub unit) and passive residues (see ref.25). Ambiguous interaction restraints are defined from each active residue of one Ub unit to all active and passive residues of the other Ub, using a 2 Å upper distance bound.
Fully flexible segments are the parts of the molecules that are free to move during all stages of simulated annealing.
The distal and proximal Ubs are connected via an isopeptide bond between the carbonyl C of G76 and the NZ atom of KX (see Materials and Methods).
As a separate validation of the method used here, the generated structure of K63-linked Ub2 (Fig. 2) shows no close contacts between the hydrophobic patches L8-I44-V70, in complete agreement with experimental NMR and SAXS data in solution9,10. The absence of a patch-to-patch contact between adjacent Ub monomers predicted here also agrees well with the recent crystal structures of K63-linked Ub211 and Ub412. It is worth emphasizing in this regard that in the absence of a direct Ub:Ub contact, the flexibility of the Ub-Ub linker29 would allow Ub monomers to undergo rotations that could change their relative orientation. This makes the conformations of such chains somewhat random and affected by external forces originating from crystal packing or ligand interactions. In fact, the presence of such motions is clearly demonstrated by the recent SAXS studies of K63-linked Ub4 in solution12 which showed that additional conformations, distinct from that observed in the crystals, are necessary in order to fit the scattering data measured in solution. These results indicate that K63-linked chains do not form a single well-defined conformation in solution, whereas in the crystals the chain is locked in one particular conformation due to crystal packing (see ref.12). Importantly, in agreement with our predictions, none of these additional conformations presented in12 shows a hydrophobic patch-to-patch contact between Ub monomers.
Thus the generated structures of both K48- and K63-linked chains serve as an indication that our HADDOCK-based approach is suitable for addressing the question raised in this study, i.e. whether a particular Ub-Ub linkage can result in a chain conformation that allows close hydrophobic-patch contacts between adjacent Ub units.
Classification of the resulting structures
Rigorous comparison and classification of the generated Ub2 structures of different Ub-Ub linkages is not trivial and requires taking into account multiple parameters reflecting various aspects of interdomain contacts. For this purpose we chose twelve different descriptors, summarized in Table 2 and briefly detailed below. Note that we make no a priori assumption which of these variables are more and which are less important.
One of the descriptors, the intermolecular energy (Einter), was in the range from −115 to −280 kcal/mol except for K33-linked Ub2 which showed a higher Einter value of −77 kcal/mol. As evident from Table 2, the main contribution to the intermolecular energy arises from electrostatic interactions. This is not unexpected given that the hydrophobic patch on Ub surface is surrounded by polar and charged (mostly basic) side chains, which would contribute to interactions at the Ub/Ub interface.
Direct interactions at the interface, like hydrogen bonds and hydrophobic contacts, are also important factors for the stability of intermolecular complexes. Due to their predominantly polar character and energies of 3–7 kcal/mol, hydrogen bonds are often held to confer specificity on protein-protein interactions. Several studies attempted to rely the number of hydrogen bonds to the buried surface area. The number of hydrogen bonds varies widely from one interface to another depending on its size and its hydrophobicity. The average number of hydrogen bonds per 100 Å2 spans values between 0.35 and 0.59 for homo- as well as heterodimers30–32. Our analysis shows that K48- and K27-linked Ub2s exhibit the highest number of average hydrogen bonds, ranging from 0.5 to 0.57 per 100 Å2. Moreover, K48-, K6-, and K11-linked chains have the highest number of average hydrophobic contacts (from 14.4 to 10.9) between residues L8, I44, and V70 of the two Ub units (a detailed account for each best structure can be found in Supplementary Material). Conversely, head-to-tail, K29-, K33-, and K63-linked chains do not show any hydrophobic contacts between Ub monomers. Note that, in addition, K63- and K33-linked Ub2 have the lowest average number of hydrogen bonds. This combination of unfavorable criteria decreases the stability of the interface and hence, makes a closed conformation of these chains unfavorable33,34. As mentioned above, the result for K63-Ub2 is in good agreement with solution NMR data for this chain9 that detected no close contacts between the two Ub units and, a fortiori, between the hydrophobic patches. Note that an extended conformation of K63-linked chains (Ub2, Ub4) compared to the K48-linked chains is also supported by small angle X-ray scattering data10. As for K29-linked Ub2, it has 0.44 hydrogen bonds per 100 Å2 of buried area and no intermolecular hydrophobic contacts between the L8-I44-V70 patches.
As mentioned above, the restraint energy (Erest) accounts for the (unambiguous) distance restraints for the linker as well as for the (ambiguous) distance restraints between the residues that are expected to form the interdomain interface. For all the generated structures, the unambiguous constraints tend to be well satisfied, whereas violations of the ambiguous constraints show substantial dispersion (Table 2). While K48-, K11-, and even K6-linked Ub2s display low violations of ambiguous constraints, other chains: K63-, K33-, K29-, and particularly head-to-tail-linked Ub2, have the highest values of the ambiguous energy. This further supports the conclusion that the latter four Ub2 chains cannot allow neighboring Ub units to contact each other via their hydrophobic patches.
Another factor controlling intermolecular contacts is their shape complementarity, usually accounted for by means of BSA. On average, the formation of homo- or heterodimers buries 12% of the accessible surface area, although variations are large and could span a range from 6% to 29% of their accessible surface area35. For the Ub2 chains generated in this study the average BSA varied from 780 to 1458 Å2, with K33-, K63-, and K29-linked Ub2s burying only 9, 10, and 11%, respectively, of their accessible surface area, compared to 15% in the case of K48-linked Ub2 (Supplementary Material, Table S1).
To complement the ambiguous energy, which accounts for the proximity of the hydrophobic patches of the two Ubs, we also calculated the all-atoms accessible surface area for residues L8, I44, and V70 (further referred to as ASAhp) of the two Ub units in each chain (Table 2, see also Supplementary Material). This information provides an additional means to assess the degree of (desirable) intermolecular contact, as the latter residues would become buried (hence lower ASAhp) when the two Ub moieties enter a close contact. Consistent with the abovementioned results, K48- and K6-linked Ub2s have the lowest ASAhp, whereas K29-, K63-, K33-, and head-to-tail-linked chains exhibit the highest ASAhp values. Moreover, it is worth mentioning that ASAhp of 269.8 Å2 (273.8 Å2 for the best K48* structure) obtained for the unconstrained, K48*-Ub2 chain is one of the highest among all predicted structures. This indicates that constraints between the hydrophobic patches have to be included in order to form a close contact between the two Ub units.
Principal Component Analysis and clustering
Because the descriptors chosen in this study represent various (sometimes partially overlapping) aspects of the multifaceted nature of protein-protein interactions, it is difficult (if possible at all) to choose a single one of them to draw rigorous conclusions regarding the ability of some chains to form close contacts between the hydrophobic patches. Therefore we turned to Principal Component Analysis (PCA)36–38, which is a classical technique to reduce the dimensionality of a data set by transforming it to a new set of variables (the principal components, PCs) that summarize the essential features of the data. Briefly, the PCA works by decomposing the matrix of data into a product of two matrices called the loading and score matrices. The loading matrix contains the principal component coefficients whereas the score matrix contains the original data in a rotated coordinate system. The traditional PCA approach is to use the first few (i.e. the largest) PCs for the analysis because they usually account for most of the variation in the original data set. In contrast, the last few (the smallest) PCs are often assumed to represent only the residual “noise” in the data. A common rule of thumb here is to choose the smallest number of PCs such that they account for a given percentage of the total variance.
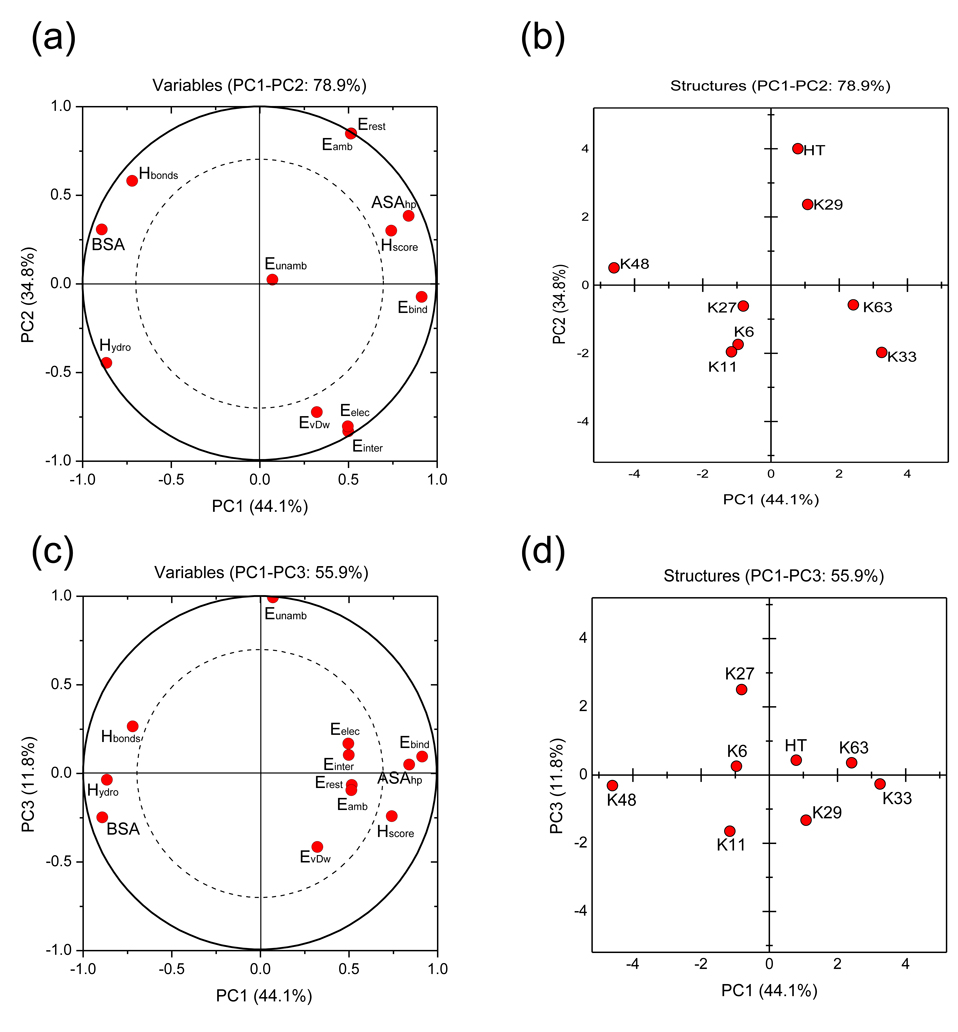
The PCA analysis was performed by using the average data over the 10 best structures of the best cluster for each chain. In our case, the first two PCs (PC1 and PC2) explain 44.1% and 34.8%, respectively, of the variance in the data set. Thus, the principal plane built by PC1 and PC2 contains 78.9% of the total variance. Adding a third PC (PC3) results in 90.6% coverage of the total variance (Supplementary Material, Table S3). Hence, the structure of the original data is well preserved in the first three PCs. Except for the unambiguous energy, all variables are well represented already in the PC1-PC2 principal plane, as can be seen from the loading plots (Fig. 4a and 4c),. The buried surface area (BSA), the accessible surface area for hydrophobic residues (ASAhp), the number of hydrophobic contacts (Hydro), and the free energy of binding (Ebind) are the variables that contribute the most to PC1, whereas the intermolecular (Einter), restraint (Erest), and ambiguous (Eamb) energies contribute mainly to PC2 (see Supplementary Material). In addition, unambiguous energy (Eunamb) is the main contributor of PC3. The binding free energy (Ebind) and ASAhp are close to each other in the loading plots (Fig. 4a,c), indicating that they have a positive correlation. On the other hand, these variables (ASAhp and Ebind) are negatively correlated with the number of hydrophobic contacts (Hydro) and the buried surface area (BSA). Thus, the separation of the generated structures into two different groups is a consequence of an opposite variation of these variables. Indeed, high solvent accessibility of hydrophobic residues is concomitant with a relatively small number of hydrophobic contacts between these residues. In the same way, a small buried surface area is concomitant with a higher energy of binding. The representation of the generated structures in the PC1-PC2-PC3 principal axes system (Fig. 4b,d) shows a clear separation of these structures into two groups along the PC1 axis. One of them comprises K48-, K6-, K11-, and K27-linked chains, which are able to form close contacts between the neighboring Ub units. The other group includes head-to-tail, K29-, K33-, and K63-linked chains, which do not allow such a contact. It should be mentioned here that score plots of the rest of the PCs (not shown) did not reveal any additional separation. To verify the robustness of our method, the principal component analysis has been applied to two other data sets. The first data set takes into account an ensemble including the ten best structures of the best cluster for each generated chain whereas the second data set uses the best structure of the best cluster for each generated chain. Both cases yield the same conclusion i.e the separation of the structures into two sub-groups including K48-, K6-, K11- and K27-linked chains from one side and head-to-tail, K29-, K33- and K63-linked chains from the other side. The complete analysis can be found in Supplementary Material.
Figure 4.
Principal component analysis of the generated structures. Shown are the loading plots (a, c) and the score plots (b, d). Analysis has been applied to the 12 descriptors (Table 2) averaged over the 10 best structures of the best cluster for each generated chains. The loading plots represent coordinates of each descriptor in the principal axes system of the selected PC components, whereas the score plots represent coordinates of the original data in the PC principal axes system (see Supplementary Material). The score plots clearly show a separation of the data along the PC1 axis, allowing the clustering of Ub2 chains into two groups. The meaning of the different labels is as follows: Einter: intermolecular energy; EvDw: van der Waals energy; Eelec: electrostatic energy; BSA: buried surface area; Erest: total restraints energy; Eunamb: unambiguous-restraints energy; Eamb: ambiguous-restraints energy; Hbonds: number of interdomain hydrogen bonds at the interface; Hydro: number of hydrophobic contacts involving residues V8, I44, V70; Ebind: binding free energy; Hscore: HADDOCK score; ASAhp: accessible surface area of residues L8, I44, and V70. HT indicates the head-to-tail linked chain. The solid circle (radius = 1) in panels a and c encompasses the area that includes all the data, while the dashed circle includes 70% of the data. In the geometric sense, the circumferences of these two circles represent projections of a vector that is in-plane or tilted 30° away from the projection plane, respectively. It is usually assumed that variables lying between the two circles are well represented in the corresponding plane.
Discussion
All polyUb chains are composed of the same monomer, and yet they are recognized as specific signals in a vast variety of distinct pathways. This ought to do with the structure and conformational adaptability of the chains as well as of the receptor molecules that recognize them. Although experimental studies of K48- and K63-linked polyUb chains have provided important insights into linkage-dependence of the conformational and ligand-binding properties of polyUb, our understanding of the molecular/structural basis of the amazing ability of polyUb to act as a versatile signal in a variety of cellular events is still very limited. The issue of linkage specificity of polyUb signal recognition has become particularly important with the discovery of the non-canonical chains, linked via other lysines (K6, K11, K27, K29, and K33) or head-to-tail. Understanding the structural properties of these chains is the necessary first step in addressing this issue. Since the experimental studies of such chains are currently hindered by the unavailability of unanchored chains containing non-canonical linkages, here we used computer-based protein docking to model the conformations of such chains, as well as of K48- and K63-linked chains, as controls. Specifically, we asked the question, whether polyUb chains of various linkages could allow a close contact between the hydrophobic patch surfaces on the neighboring Ub units: these surfaces mediate Ub’s interactions with most of Ub-binding proteins, including Ub/Ub interactions.
The results of our modeling show a clear separation of all possible Ub2 chains into two groups: the chains that can form such contacts (K6, K11, K27, and K48) and those where steric occlusion prevents the adjacent Ub units from forming a hydrophobic patch-to-patch contact (K29, K33, K63 and head-to-tail). Our approach is validated by the good agreement with the known structures of K48-linked chains (Ub2 and Ub4)7,21 and with experimental data for K63-linked chains. In the latter case, both solution NMR data10,24 and the recent crystal structures of K63-linked Ub211 and Ub412, as well as SAXS data12 all show no hydrophobic-patch contacts between Ub monomers in K63-linked chains. Also our prediction that in linear (head-to-tail) chains the adjacent Ubs are not able to make a patch-to-patch contact agrees with the recent crystal structure of such chains11. Furthermore, our study provides, for the first time, structural models for K6-, K11-, and K27-linked Ub2 chains, and thus sets the stage for experimental verification of these predictions.
Extrapolated to longer chains, our results suggest that in solution polyUb chains linked via K6, K11, and K27 could exist in a closed conformation in which adjacent Ub units contact each other via their hydrophobic patches. Because the L8-I44-V70 hydrophobic patch is involved in Ub’s interactions with most of the known Ub-binding molecules, burying these patches at the Ub/Ub interface has several functionally important implications. First, ligand binding to these chains would require domain motions that open the interdomain interface, like in the case of K48-linked polyUb13,39. Second, the analogy with K48-linked Ub2 suggests that these chains might be able to bind ligands in a sandwich-like fashion, like in the case of hHR23a UBA-2/Ub2 interaction13. Third, we expect the conformation of these chains to be pH dependent, like in the case of K48-linked polyUb6,40,41. While the closed conformation (predicted in this study) is expected to dominate at neutral and basic pH, lowering the pH should cause these chains to open, as the protonation of H68 (located near the center of the hydrophobic patch) on each Ub will cause strong electrostatic repulsion of the two Ub monomers that could overcome their hydrophobic attraction.
By contrast, Ub2 chains linked via K29, K33, and K63, as well as head-to-tail, are predicted to always be in an open conformation, and thus have their hydrophobic patches readily available to interact with Ub-binding molecules. These conformational properties combined with the linker flexibility could be the determining factors for selective (possibly avid) binding of such chains to various receptor molecules containing multiple Ub-binding units, like e.g. Rap8042. It should be pointed out that in this study we examined the possibility of direct hydrophobic patch-to-patch contacts between the adjacent Ubs in polyUb. Conformational flexibility of the Ub-Ub linker could promote contacts between non-adjacent Ub units (e.g. i and i+2, i+3 etc.) in longer chains, which could further enhance linkage-specific structural and receptor recognition properties of the polyUb signal.
Materials and Methods
The docking calculations were performed using the standard protocols implemented in HADDOCK2.043. For each run, 2000 rigid-body docking solutions were first generated by energy minimization. The 200 best solutions according to the AIR restraint energy (defined in25) were subjected to semi-flexible simulated annealing with flexibility introduced first on side chains and then on the backbone, followed by a final refinement in explicit water. Non-bonded energies were calculated with an 8.5 Å distance cut-off using the OPLS non-bonded parameters44. The overall score was calculated as a weighted sum of different terms, using the default HADDOCK2.0 values for the weights. After the final step, the resulting 200 structures were subjected to clustering with a 2.5 Å cutoff using the backbone RMSD of both Ub units calculated after positional least-square fitting on the first Ub only.
Starting structures
As the starting structure of Ub monomer we used the atom coordinates from the crystal structure of K48-linked Ub2 (Protein Data Bank entry 1AAR)5. This is justified by (i) the close structural similarity among both Ub units in this structure and the crystal structure of monomeric Ub (PDB entry 1UBQ) (the backbone RMSD is 0.65–0.66 Å for residues 2–70) and (ii) the fact that solution NMR spectra and residual dipolar couplings indicate no substantial structural difference between Ub in the isolated form and in the context of Ub2 (K48- or K63-linked) or K48-linked tetra-ubiquitin (Ub4)6,9. To avoid any bias that could result from the actual positioning of Ub monomers in the crystal structure of Ub2, the two Ubs were translated 150 Å away from each other and randomly rotated.
Definition of Restraints
The restraints that were used in this study are listed in Table 3. The Ub monomers, which are linked to one another via a G76-KX (X=6, 11, 27, 29, 33, 48, or 63) isopeptide bond, are designated ‘distal’ (containing G76) and ‘proximal’ (containing the bonded KX as well as the free C-terminus), respectively. The same restraints were used to build the G76(distal)-M1(proximal) linkage in the case of a linear, head-to-tail chain. To account for the possible close contact between the hydrophobic patches (residues L8, I44, V70) on both Ubs in the chain, we incorporated ambiguous restraints where active and passive residues on each Ub were defined using the procedure described previously25. The G76-KX isopeptide bond was modeled by including a set of distance restraints based on typical interatomic distances for a peptide bond in crystal structures, as described8. Backbone and side chain flexibility was also included to account for possible conformational rearrangement that could occur at the interface between the two Ub moieties (see Table 3). The interface amino acids that constitute the flexible segments were defined as the hydrophobic residues L8, I44, V70 plus two sequential neighbors on each side.
Analysis of Intermolecular Contacts
Intermolecular contacts (hydrogen bonds and non-bonded interactions) were analyzed with DIMPLOT, part of the LIGPLOT software45, using the default settings: 3.9 Å heavy-atom distance cutoff for non-bonded contacts; 2.7 and 3.35 Å proton-acceptor and donor-acceptor distance cutoffs, respectively, with minimum 90° angles (D-H-A, H-A-AA, D-A-AA) for hydrogen bonds. A contact is defined to be present in the solution structure if it is found in at least four out of the ten best structures.
The binding free energy was computed using the DCOMPLEX web server46. The principal component analysis (PCA) was performed using an in-house program implemented in Matlab and available from the authors upon request. Complete details of the analysis can be found in Supplementary Material.
Supplementary Material
Acknowledgements
Supported by NIH grant GM065334 to D.F. We thank Ranjani Varadan for useful discussions regarding di-ubiquitin chains. We also thank Pierre Lanteri and Jean-Yves Gauvrit for usefull discussion regarding PCA analysis.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wilkinson K. Ubiquitin: a Nobel protein. Cell. 2004;119:741–745. doi: 10.1016/j.cell.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A, Ciechanover A. The ubiquitin system. Annu Rev Biochem. 1998;67:425–479. doi: 10.1146/annurev.biochem.67.1.425. [DOI] [PubMed] [Google Scholar]
- 3.Pickart CM, Fushman D. Polyubiquitin chains: polymeric protein signals. Curr Opin Chem Biol. 2004;8:610–616. doi: 10.1016/j.cbpa.2004.09.009. [DOI] [PubMed] [Google Scholar]
- 4.Beal R, Deveraux Q, Xia G, Rechsteiner M, Pickart C. Surface hydrophobic residues of multiubiquitin chains essential for proteolytic targeting. Proc Natl Acad Sci U S A. 1996;93:861–866. doi: 10.1073/pnas.93.2.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cook WJ, Jeffrey LC, Carson M, Chen Z, Pickart CM. Structure of a diubiquitin conjugate and a model for interaction with ubiquitin conjugating enzyme (E2) J Biol Chem. 1992;267:16467–16471. doi: 10.2210/pdb1aar/pdb. [DOI] [PubMed] [Google Scholar]
- 6.Varadan R, Walker O, Pickart C, Fushman D. Structural properties of polyubiquitin chains in solution. J Mol Biol. 2002;324:637–647. doi: 10.1016/s0022-2836(02)01198-1. [DOI] [PubMed] [Google Scholar]
- 7.Eddins MJ, Varadan R, Fushman D, Pickart CM, Wolberger C. Crystal structure and solution NMR studies of Lys48-linked tetraubiquitin at neutral pH. J Mol Biol. 2007;367:204–211. doi: 10.1016/j.jmb.2006.12.065. [DOI] [PubMed] [Google Scholar]
- 8.Dijk A, Fushman D, Bonvin A. Various strategies of using residual dipolar couplings in NMR-driven protein docking: application to Lys48-linked di-ubiquitin and validation against 15N-relaxation data. Proteins. 2005;60:367–381. doi: 10.1002/prot.20476. [DOI] [PubMed] [Google Scholar]
- 9.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart C, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 10.Tenno T, Fujiwara K, Tochio H, Iwai K, Morita EH, Hayashi H, Murata S, Hiroaki H, Sato M, Tanaka K, Shirakawa M. Structural basis for distinct roles of Lys63- and Lys48-linked polyubiquitin chains. Genes Cells. 2004;9:865–875. doi: 10.1111/j.1365-2443.2004.00780.x. [DOI] [PubMed] [Google Scholar]
- 11.Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D. Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep. 2009;10:466–473. doi: 10.1038/embor.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Datta AB, Hura GL, Wolberger C. The structure and conformation of Lys63-linked tetraubiquitin. J Mol Biol. 2009;392:1117–1124. doi: 10.1016/j.jmb.2009.07.090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Varadan R, Assfalg M, Raasi S, Pickart CM, Fushman D. Structural determinants for selective recognition of a Lys48-linked polyubiquitin chain by a UBA domain. Mol Cell. 2005;18:687–698. doi: 10.1016/j.molcel.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 14.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sims JJ, Haririnia A, Dickinson BC, Fushman D, Cohen RE. Avid interactions underlie the Lys63-linked polyubiquitin binding specificities observed for UBA domains. Nat Struct Mol Biol. 2009;16:883–889. doi: 10.1038/nsmb.1637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng J, Schwartz D, Elias JE, Thoreen CC, Cheng D, Marsischky G, Roelofs J, Finley D, Gygi SP. A proteomics approach to understanding protein ubiquitination. Nat Biotechnol. 2003;21:921–926. doi: 10.1038/nbt849. [DOI] [PubMed] [Google Scholar]
- 17.Xu P, Peng J. Dissecting the ubiquitin pathway by mass spectrometry. Biochim Biophys Acta. 2006;1764:1940–1947. doi: 10.1016/j.bbapap.2006.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim HT, Kim KP, Lledias F, Kisselev AF, Scaglione KM, Skowyra D, Gygi SP, Goldberg AL. Certain pairs of ubiquitin-conjugating enzymes (E2s) and ubiquitin-protein ligases (E3s) synthesize nondegradable forked ubiquitin chains containing all possible isopeptide linkages. J Biol Chem. 2007;282:17375–17386. doi: 10.1074/jbc.M609659200. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Ye Y. Polyubiquitin chains: functions, structures, and mechanisms. Cell Mol Life Sci. 2008;65:2397–2406. doi: 10.1007/s00018-008-8090-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ikeda F, Dikic I. Atypical ubiquitin chains: new molecular signals. 'Protein Modifications: Beyond the Usual Suspects' review series. EMBO Rep. 2008;9:536–542. doi: 10.1038/embor.2008.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cook WJ, Jeffrey LC, Kasperek E, Pickart CM. Structure of tetraubiquitin shows how multiubiquitin chains can be formed. J Mol Biol. 1994;236:601–609. doi: 10.1006/jmbi.1994.1169. [DOI] [PubMed] [Google Scholar]
- 22.Phillips CL, Thrower J, Pickart CM, Hill CP. Structure of a new crystal form of tetraubiquitin. Acta Crystallogr D Biol Crystallogr. 2001;57:341–344. doi: 10.1107/s090744490001800x. [DOI] [PubMed] [Google Scholar]
- 24.Varadan R, Assfalg M, Haririnia A, Raasi S, Pickart CM, Fushman D. Solution conformation of Lys63-linked di-ubiquitin chain provides clues to functional diversity of polyubiquitin signaling. J Biol Chem. 2004;279:7055–7063. doi: 10.1074/jbc.M309184200. [DOI] [PubMed] [Google Scholar]
- 25.Dominguez C, Boelens R, Bonvin A. HADDOCK: a protein-protein docking approach based on biochemical or biophysical information. J Am Chem Soc. 2003;125:1731–1737. doi: 10.1021/ja026939x. [DOI] [PubMed] [Google Scholar]
- 26.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 27.Hurley JH, Lee S, Prag G. Ubiquitin-binding domains. Biochem J. 2006;399:361–372. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ponstingl H, Kabir T, Gorse D, Thornton JM. Morphological aspects of oligomeric protein structures. Prog Biophys Mol Biol. 2005;89:9–35. doi: 10.1016/j.pbiomolbio.2004.07.010. [DOI] [PubMed] [Google Scholar]
- 29.Fushman D, Varadan R, Assfalg M, Walker O. Determining domain orientation in macromolecules by using spin-relaxation and residual dipolar coupling measurements. Progress NMR Spectroscopy. 2004;44:189–214. [Google Scholar]
- 30.Lo Conte L, Chothia C, Janin J. The atomic structure of protein-protein recognition sites. J Mol Biol. 1999;285:2177–2198. doi: 10.1006/jmbi.1998.2439. [DOI] [PubMed] [Google Scholar]
- 31.Bahadur RP, Chakrabarti P, Rodier F, Janin J. A dissection of specific and non-specific protein-protein interfaces. J Mol Biol. 2004;336:943–955. doi: 10.1016/j.jmb.2003.12.073. [DOI] [PubMed] [Google Scholar]
- 32.Bahadur RP, Chakrabarti P, Rodier F, Janin J. Dissecting subunit interfaces in homodimeric proteins. Proteins. 2003;53:708–719. doi: 10.1002/prot.10461. [DOI] [PubMed] [Google Scholar]
- 33.Sheinerman FB, Norel R, Honig B. Electrostatic aspects of protein-protein interactions. Curr Opin Struct Biol. 2000;10:153–159. doi: 10.1016/s0959-440x(00)00065-8. [DOI] [PubMed] [Google Scholar]
- 34.Norel R, Sheinerman F, Petrey D, Honig B. Electrostatic contributions to protein-protein interactions: fast energetic filters for docking and their physical basis. Protein Sci. 2001;10:2147–2161. doi: 10.1110/ps.12901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kleanthous C. Protein-protein Recognition. In: Glover BDHaDM., editor. Frontiers in Molecular Biology. Oxford University Press; 2000. [Google Scholar]
- 36.Ringnér M. What is principal component analysis? Nat Biotech. 2008;26:303–304. doi: 10.1038/nbt0308-303. [DOI] [PubMed] [Google Scholar]
- 37.Jackson JD. A user's guide to principal components. Wiley; 1991. [Google Scholar]
- 38.Brereton RG. Chemometrics: Data Analysis for the Laboratory and Chemical Plant. Wiley; 2003. [Google Scholar]
- 39.Dickinson BC, Varadan R, Fushman D. Effects of cyclization on conformational dynamics and binding properties of Lys48-linked di-ubiquitin. Protein Sci. 2007;16:369–378. doi: 10.1110/ps.062508007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ryabov Y, Fushman D. Interdomain mobility in di-ubiquitin revealed by NMR. Proteins. 2006;63:787–796. doi: 10.1002/prot.20917. [DOI] [PubMed] [Google Scholar]
- 41.Ryabov Y, Fushman D. A model of interdomain mobility in a multidomain protein. J Am Chem Soc. 2007;129:3315–3327. doi: 10.1021/ja067667r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sims JJ, Cohen RE. Linkage-specific avidity defines the lysine 63-linked polyubiquitin-binding preference of rap80. Mol Cell. 2009;33:775–783. doi: 10.1016/j.molcel.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vries S, Dijk A, Krzeminski M, Dijk M, Thureau A, Hsu V, Wassenaar T, Bonvin AM. HADDOCK versus HADDOCK: new features and performance of HADDOCK2.0 on the CAPRI targets. Proteins. 2007;69:726–733. doi: 10.1002/prot.21723. [DOI] [PubMed] [Google Scholar]
- 44.Jorgensen WL, Tirado-Rives J. The OPLS [optimized potentials for liquid simulations] potential functions for proteins, energy minimizations for crystals of cyclic peptides and crambin. J Am Chem Soc. 1988;110:1657–1666. doi: 10.1021/ja00214a001. [DOI] [PubMed] [Google Scholar]
- 45.Wallace AC, Laskowski RA, Thornton JM. LIGPLOT: a program to generate schematic diagrams of protein-ligand interactions. Protein Eng. 1995;8:127–134. doi: 10.1093/protein/8.2.127. [DOI] [PubMed] [Google Scholar]
- 46.Liu S, Zhang C, Zhou H, Zhou Y. A physical reference state unifies the structure-derived potential of mean force for protein folding and binding. Proteins. 2004;56:93–101. doi: 10.1002/prot.20019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.