Abstract
Comparative studies of gene regulation suggest an important role for natural selection in shaping gene expression patterns within and between species. Most of these studies, however, estimated gene expression levels using microarray probes designed to hybridize to only a small proportion of each gene. Here, we used recently developed RNA sequencing protocols, which sidestep this limitation, to assess intra- and interspecies variation in gene regulatory processes in considerably more detail than was previously possible. Specifically, we used RNA-seq to study transcript levels in humans, chimpanzees, and rhesus macaques, using liver RNA samples from three males and three females from each species. Our approach allowed us to identify a large number of genes whose expression levels likely evolve under natural selection in primates. These include a subset of genes with conserved sexually dimorphic expression patterns across the three species, which we found to be enriched for genes involved in lipid metabolism. Our data also suggest that while alternative splicing is tightly regulated within and between species, sex-specific and lineage-specific changes in the expression of different splice forms are also frequent. Intriguingly, among genes in which a change in exon usage occurred exclusively in the human lineage, we found an enrichment of genes involved in anatomical structure and morphogenesis, raising the possibility that differences in the regulation of alternative splicing have been an important force in human evolution.
Changes in gene regulation are thought to play an important role in adaptive evolution and speciation (Britten and Davidson 1971; King and Wilson 1975; Jin et al. 2001; Carroll 2003, 2008; Abzhanov et al. 2004; Iftikhar et al. 2004; Shapiro et al. 2004; Taron et al. 2004; Wray 2007). In support of this notion, comparative genome-wide studies of gene regulation within and between populations and species have revealed evidence consistent with the action of both stabilizing as well as directional selection on gene expression levels (Oleksiak et al. 2002; Lemos et al. 2005; Rifkin et al. 2005; Gilad et al. 2006; Whitehead and Crawford 2006). Most of these studies, however, focused on estimates of overall gene expression levels, probably because prior to the development of next-generation sequencing, it was very challenging to characterize expression level variation of individual exons on a genome-wide scale.
Indeed, previous studies of alternative splicing patterns in mammalian species focused on relatively small numbers of exons and genes. For example, Su et al. (2008) studied variation in exon usage and alternative splicing in liver samples from a number of mouse strains from both sexes, by using a custom microarray designed to probe the expression levels of 25,760 exons and exon–exon junctions from 1312 genes. By analyzing the exon-level data (without correcting for overall gene expression level), Su et al. (2008) found that 14% of exons are differentially expressed between sexes. Similarly, using computational searches for alternative splicing events, Pan et al. (2005) estimated that more than 11% of human and mouse cassette alternative exons are skipped in one species but used constitutively in the other. The species-specific alternative splicing events were predicted to modify conserved domains in proteins more often than alternative splicing events that were shared across species. In turn, Calarco et al. (2007) studied alternative splicing differences between humans and chimpanzees using both computational analysis and primary data generated using a custom microarray platform, which included probes designed to detect 3126 alternative splicing events in 2647 genes. Using this combination of approaches, Calarco et al. (2007) found that at least 6% of the exons they tested displayed significant differences in splicing levels between humans and chimpanzees. Moreover, they found that the genes containing these exons were typically not differentially expressed between the two species.
These observations suggest that interspecies and sexually dimorphic variation in the regulation of alternative splicing may be common. However, the studies mentioned above notwithstanding, computational analyses of alternative splicing are typically limited to highly sequenced genomes with an abundance of publicly available expressed sequence tag (EST) data. In turn, microarrays are not an optimal platform for studying variation in alternative splicing because detection is limited to predesigned probes, which requires prior knowledge of all possible exon boundaries as well as exon–exon junctions. In addition, differences in microarray probe composition result in large effects due to variability in hybridization kinetics (Oshlack et al. 2007), and cross-hybridization makes it difficult to distinguish closely related transcripts (Draghici et al. 2006). Perhaps because of these limitations, the studies discussed above focused on only a small number of transcripts, and as a result, we still know relatively little about variation in exon usage and alternative splicing within or between species.
Recent developments in sequencing technology have made it possible to use sequence-based approaches for gene expression profiling (an approach recently termed RNA-seq; Marioni et al. 2008; Mortazavi et al. 2008; Fu et al. 2009). In contrast to microarrays, these new approaches do not rely on specific predesigned probes and can thus provide a more detailed picture of gene regulatory variation. In particular, RNA-seq data can be used to study differences in exon usage, alternative splicing, and allele-specific expression levels among samples (Wang et al. 2009). Thus, sequencing approaches have the potential to provide insight into the mechanisms of regulatory change across species at unprecedented resolution.
Results
We used RNA-seq to study transcript regulation in humans, chimpanzees, and rhesus macaques, using liver RNA samples from three males and three females from each species, sequencing each sample independently in two lanes of Illumina's Genome Analyzer II (for more details on samples, data collection, and associated protocols, see Methods; Supplemental Tables S1, S2; Supplemental Fig. S1). Using this study design, we obtained on average 5.9 million, 6.6 million, and 7.2 million short (35-bp) sequence reads per lane of human, chimpanzee, and rhesus macaque samples, respectively (Supplemental Table S3).
To compare exon and gene expression levels across species, we used BLAT (Kent 2002) to identify human exons for which clear orthologs exist in the other two species. To avoid biases due to mapping problems, we removed exons for which multiple plausible orthologs or highly similar paralogs exist (Supplemental Methods). This resulted in the identification of 150,107 orthologous exons in 20,689 genes. We then used MAQ (Li et al. 2008) to align reads to their corresponding genome sequences and counted the number of reads that mapped to orthologous exons in each sample. We performed extensive quality control analyses, including an assessment of the number of genes detected as expressed in each species (13,267, 13,275, and 13,105 genes in humans, chimpanzees, and rhesus macaques, respectively), as well as a comparison of the RNA-seq data to previously collected analogous microarray data (Supplemental Methods; Supplemental Figs. S2–S12; Supplemental Tables S3, S4).
Lineage-specific and sex-specific patterns of gene regulation
To estimate the expression level of a gene, we summed the number of reads mapping to its exons. We analyzed these gene expression levels using a Poisson mixed-effects model, controlling for the total number of reads in each lane, and including fixed effects for species, sex, and sex-by-species interactions, as well as an individual-specific random effect to account for interindividual variability. To identify genes that are differentially expressed between pairs of species, we used a likelihood-ratio test statistic (Table 1; Supplemental Figs. S13, S14). As expected, in both sexes, the number of differentially expressed genes between humans and chimpanzees is much lower than between either humans and rhesus macaques or between chimpanzees and rhesus macaques (Tables 1; Supplemental Table S1). That said, we note that (as seen in other studies; Lemos et al. 2005; Gilad et al. 2006), overall, our data are consistent with the action of stabilizing selection on gene regulation. Indeed, most of the variation in gene expression can be seen between individuals within species (e.g., the variation in gene expression among humans and chimpanzees is just 20% higher then the variation in gene expression among individuals from the same species; Supplemental Fig. S15).
Table 1.
Numbers of differentially expressed genes between species at FDR < 0.05
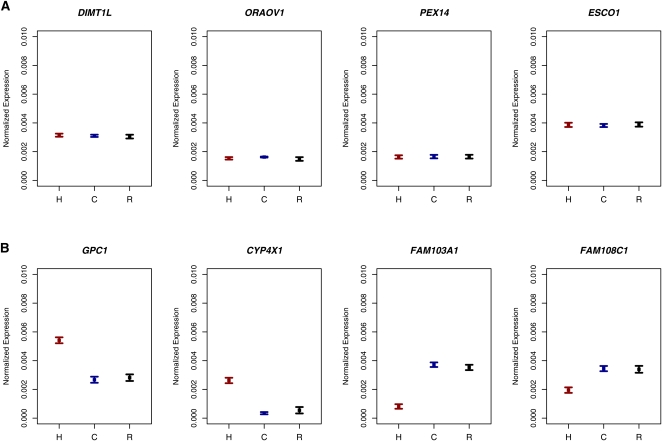
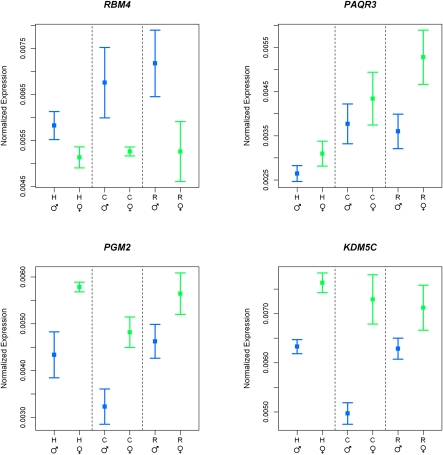
Our next analysis therefore aimed to identify individual genes whose regulation likely evolved under natural selection in primates. To do so, we used Poisson mixed-effects models corresponding to expectations under three different evolutionary scenarios (Gilad et al. 2006; Whitehead and Crawford 2006; see Supplemental Methods). Specifically, we looked for: (1) Genes whose expression levels likely evolved under stabilizing selection, regardless of the sex; we expect such genes to have little variation in gene expression levels among individuals and species (Fig. 1A). (2) Genes whose expression levels evolved under directional selection in the human lineage, regardless of the sex; we expect such genes to have little variation in expression levels within and between chimpanzee and rhesus macaque individuals and a significantly different expression level in humans (Fig. 1B). (3) Genes with conserved sexually dimorphic expression patterns; we expect that the expression levels of such genes will differ significantly between sexes in a consistent direction in all three species (Fig. 2).
Figure 1.
Examples of gene expression patterns that are consistent with the action of natural selection on gene regulation. Gene expression profiles from the three species are plotted for genes whose regulation has likely evolved under stabilizing (A) or directional selection (B) in the human lineage. In all panels, mean (±SEM) normalized expression levels (y-axis) of each species (x-axis) are plotted.
Figure 2.
Examples of conserved sexually dimorphic gene expression patterns. In all panels, mean (±SEM) normalized expression levels (y-axis) of each species (x-axis) are plotted separately for males (blue) and females (green).
We used a combination of statistical analyses to produce ranked lists of genes whose expression patterns best fit these three different scenarios (Supplemental Methods). At the top of the lists, we expect an enrichment of genes whose regulation evolved under natural selection. To simplify subsequent analyses, we defined statistical cutoffs for each list, thus identifying 1391 and 887 genes whose regulation we classified as likely evolving under stabilizing selection, or directional selection in the human lineage, respectively, and 627 genes classified as having conserved sexually dimorphic expression patterns. Importantly, since any particular cutoff is somewhat arbitrary, we confirmed that all qualitative properties of the data reported below are robust to the choice of cutoff (Supplemental Table S12).
To examine the biological functions of genes whose regulation likely evolves under natural selection, we used Gene Ontology (GO) annotations (The Gene Ontology Consortium 2000). Using this approach, among genes whose regulation likely evolved under natural selection in humans, we observed an enrichment of genes involved in transcriptional regulation and genes involved in metabolic pathways (P < 0.001 by Fisher's exact test) (Supplemental Tables S5, S6; see Supplemental Table S7 for results of a similar analysis for genes whose regulation evolves under directional selection in the chimpanzee lineage). These results are consistent with previous observations (Blekhman et al. 2008).
In turn, among genes that show a conserved sexually dimorphic expression pattern, we found, as expected (Rinn and Snyder 2005), an enrichment of genes located on the X chromosome (P = 0.022). In addition, when we focused on autosomal genes, we found conserved regulatory differences between the sexes, which may contribute to phenotypic differences between males and females (Supplemental Tables S8, S9). For example, among the subset of genes that are highly expressed in females compared to males in all three species, we found an enrichment of genes involved in metabolism and catabolism of lipids (including steroid metabolism and biosynthesis; P < 0.005), as well as enrichment of genes with ATPase activity (P < 0.01). Both female and male sex hormones have been found to control ATPase activity (Shima 1992; Dzurba et al. 1997), and previous observations suggest that ATPase expression levels are sexually dimorphic (Quintas et al. 1997; Fekete et al. 2004). Among genes that are highly expressed in males compared to females in all three species, we found a significant over-representation of genes involved in RNA splicing, RNA binding, and RNA processing (P < 0.01) (see Supplemental Table S9). This latter observation is consistent with the notion that sexually dimorphic alternative splicing is an important biological mechanism (Stolc et al. 2004; McIntyre et al. 2006).
Analysis of exon usage and alternative splicing
We next examined patterns of exon usage within and between sexes and species. Specifically, we used likelihood ratio tests within the framework of the Poisson mixed-effects model to test, for each individual exon, whether it is differentially expressed between sexes or species after controlling for overall expression levels of the gene.
In a comparison of males and females, 144 exons in 140 genes were differentially expressed between the sexes, regardless of species (at P < 0.001; false discovery rate [FDR] = 0.35) (for examples, see Supplemental Fig. S20). Among genes with sexually dimorphic exon usage, we observed a depletion of genes that regulate transcription (P < 0.003) and an enrichment of genes involved in immune system processes and inflammatory response (P < 10−4) (Supplemental Table S10). These results, together with the observation of an enrichment of genes involved in RNA splicing among sexually dimorphic genes, may point to functionally conserved sexually dimorphic alternative splicing in primates. Indeed, such a mechanism has been previously reported in flies (McIntyre et al. 2006). That said, due to the large FDR in our analysis and the potential for unobserved confounding factors that might affect FDR calculations (Leek and Storey 2007), these results should be treated with caution.
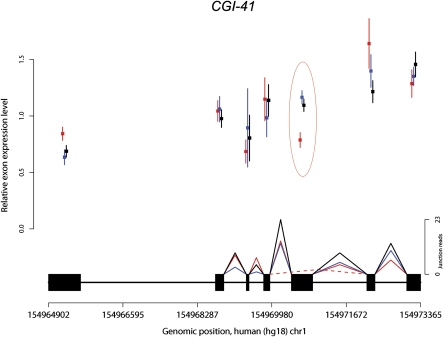
When we compared exon usage across species, we identified 256, 565, and 837 genes with evidence of alternative transcripts that are significantly more abundant in humans, chimpanzees, or rhesus macaques, respectively (at P < 0.001; exon-level FDR < 0.08 in all species) (for an example, see Fig. 3; for more information, see Supplemental Table S13). In this case, the FDR associated with our analysis is reasonably small. In addition, quantitative PCR data for eight of 10 tested differences in exon usage between human and chimpanzee were consistent with the RNA-seq data (see Supplemental Table S14; Methods).
Figure 3.
An example of human-specific change in exon usage. Mean (±SEM) relative exon expression levels (y-axis) are plotted separately for each species; (red) human; (blue) chimpanzee; (black) rhesus macaque. The gene structure appears above the x-axis, which denotes the genomic coordinates. Splice junctions identified for each species are shown as triangles connecting pairs of exons, solid lines between consecutive exons, and dotted lines between alternatively spliced exons. A typical difference in exon usage between humans and the non-human primates, which is also supported by junction reads, is circled.
Based on the position of the exons that are differentially used across species, we further classified such genes as: (1) those that are consistent with alternative use of transcription start or end sites (76, 226, and 225 genes in humans, chimpanzees, or rhesus macaques, respectively), and (2) those that are consistent with alternative splicing (180, 339, and 612 genes in humans, chimpanzees, or rhesus macaques, respectively) (Fig. 3).
Interestingly, among the latter class of genes in humans, there is a strong enrichment for genes involved in metabolic processes (P < 10−4), as well as an over-representation of genes that play a role in morphological development (P < 0.002) (Supplemental Table S11). These observations are intriguing from an evolutionary standpoint as they indicate that differences in the regulation of alternative splicing may have played an important role in human adaptations.
Conserved alternative splicing in primates
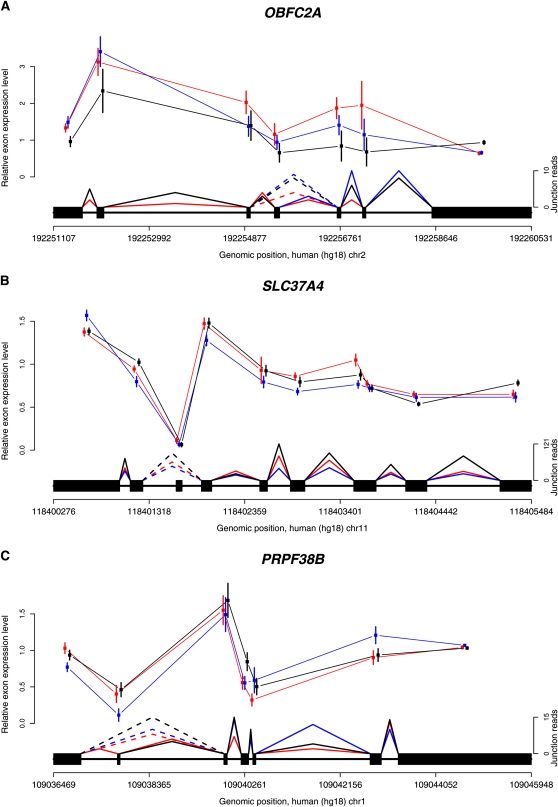
Finally, we also looked for evidence of conserved alternative splice forms in the three species. This analysis is more delicate because the inability to reject the null model of no difference in exon usage between species does not provide strong support for conservation. Thus, instead of analyzing exon expression levels, we mapped exon–exon junction reads to look for cases of conserved alternative splicing. The identification of junction reads in species for which exons are not well annotated (including chimpanzee and rhesus masque) is difficult, since the exact exon boundaries are not well defined. Nonetheless, using TopHat (Trapnell et al. 2009) with our orthologous exon definitions, we identified 79,391 pairs of adjacent exons spanned by at least one read, as well as 3478 pairs of non-adjacent exons spanned by at least one read (i.e., exon-skipping events) across the three species (Supplemental Table S15). Of the identified junctions, a larger number of exon skipping events are shared between human and chimpanzee (950) than between either human (808) or chimpanzee (783) and rhesus macaque, as might be expected given the known phylogeny of these species. Moreover, 42,610 (54%) and 631 (18%) adjacent and skipping events, respectively, were observed in all three species (for examples, see Fig. 4).
Figure 4.
Examples of conserved exon usage and alternative splicing across the three species. Mean (±SEM) relative exon expression levels (y-axis) are plotted separately for each species; (red) human; (blue) chimpanzee; (black) rhesus macaque. The gene structure appears above the x-axis, which denotes the genomic coordinates. Splice junctions identified for each species are shown as triangles connecting pairs of exons, solid lines between consecutive exons, and dotted lines between alternatively spliced exons.
To provide further support that alternative splicing occurs in the 631 genes in which we infer conserved exon skipping events, we examined known human transcripts in the Ensembl database. Of 515 genes with at least one known transcript that includes the exon we infer to be skipped in all three species, 298 (58%) also have at least one known transcript in Ensembl that includes the flanking but not the skipped exon (4% are expected by chance alone; P < 2.2 × 10−16). The evidence for splicing events that are also supported by previously annotated transcripts is slightly stronger than the evidence for splicing events not supported by previously annotated transcripts (a median of 18 compared with a median of 14 exon junction reads, across all lanes), suggesting that the false discovery rate for splice forms not supported by previous observations may be higher.
Discussion
We used RNA sequencing to compare patterns of gene expression and exon usage, in both sexes across three primate species. One potential pitfall of gene expression studies in primates is the reliance on small sample sizes (as primate tissues are rare). However, a comparison of results from the present study to those obtained by Blekhman et al. (2008) suggest that data from small samples are often quite informative. Indeed, of the 18 liver samples we used in this study, only seven (two from human, three from chimpanzee, and two from rhesus macaque) were also used in a previous microarray study (Blekhman et al. 2008), and none of the total RNA preparations were shared across studies. Reassuringly, estimates of gene expression levels across the two studies are highly consistent. (The Spearman correlations between estimates of gene expression levels from the array and RNA-seq studies are 0.75, 0.74, and 0.75, for data from humans, chimpanzees, and rhesus macaques, respectively [Supplemental Figs. S7–S9].) As typical correlations between different array platforms in controlled experiments, using the same RNA samples, are ∼0.70 (Shi et al. 2006), the level of consistency across studies in this case is satisfying (for examples of concordance of estimates of interspecies relative gene expression levels across studies, see Supplemental Fig. S10). Indeed, the consistency across the two studies validates not only the RNA-seq approach, but also the assumption that gene expression estimates based on six individuals inform us about species-wide gene expression patterns.
Exon-level analysis
Beyond estimates of overall gene expression levels, the RNA-seq data also allowed us to study conservation and differences in exon usage and alternative splicing between sexes and across species. The inference of conserved alternative splicing patterns relies on sequence reads that span exon junctions, which indicate a consistent exon-skipping event in all three species. Since the number of junction reads identified in our analysis is ultimately limited by sequence coverage, we likely underestimated the number of conserved alternative splice forms. Of the inferred conserved exon skipping events, 58% were also supported by previously annotated human transcripts—a substantial fraction, especially given that most alternative splice forms are likely not currently known (Wang et al. 2008).
In addition to inferring conservation of alternative splicing patterns, we found that 7% of genes expressed in the liver undergo differential alternative splicing between humans and chimpanzees or may have different transcription start or end sites in the two species. This estimate is consistent with the observations of Calarco et al. (2007), who estimated that the alternative splicing patterns of 6%–8% of genes that are expressed in frontal cortex and/or heart are different between humans and chimpanzees. That said, it is likely that both Calarco et al. (2007) and the present study underestimate the proportion of differential alternative splicing between humans and chimpanzees due to lack of statistical power.
Our analysis also suggests, somewhat unexpectedly, that the number of human-specific changes in exon usage is smaller than that seen in chimpanzee. This pattern is only partly explained by the interspecies differences in the number of mapped reads, which affect the power to detect lineage-specific changes in exon expression levels (Oshlack and Wakefield 2009). In our data there are, on average, fewer reads mapping to orthologous exons in human samples (median = 1.64 million) than chimpanzees (median = 1.77 million) and macaques (median = 2.30 million). Indeed, when we subsampled 1 million reads from each lane of sequenced data, the differences in the numbers of genes with evidence of alternative transcripts that are significantly more abundant in each species are somewhat less pronounced (173, 329, and 384 such genes in human, chimpanzee, and rhesus macaque, respectively).
A second property of the data that may contribute to this pattern is the lack of independent exon annotation in the genomes of chimpanzee and rhesus macaques. Indeed, all orthologous exon definitions in our data originate with exons annotated in the human genome. Thus, while our data set is expected to include exons that are used in humans but not in chimpanzees or rhesus macaques, we are unlikely to include exons that are used in either chimpanzees or rhesus macaque, but not in humans. In many of these cases, only one non-human primate may have lost the exon. Using our approach, a lineage-specific lack of exon expression will be interrupted as a linage-specific change in exon usage. Thus, larger numbers of apparent linage-specific changes in exon usage are expected to be observed in the non-human primates.
Sexually dimorphic transcript expression patterns
Our data allowed us to identify genes with expression patterns that are consistent with conserved sexually dimorphic gene regulation. Among these, we found an enrichment of genes involved in lipid metabolism (using the gene-level analyses) and immune response (using the exon-level analyses). Both these observations are consistent with gene expression data from mouse livers in Yang et al. (2006), who found an enrichment of genes involved in carboxylic acid metabolism, immune response, lipid metabolism, steroid biosynthesis, and steroid metabolism among genes with sexually dimorphic expression patterns (Table 2 in Yang et al. 2006). Put together, these observations suggest that sexual dimorphism in the regulation of genes involved in lipid metabolism may have been maintained in mammals over a long evolutionary period.
We also compared our results with those of Su et al. (2008), who analyzed a total of 1020 genes in mice, and tested individual exons for expression differences between the sexes, without controlling for overall gene expression levels (so identified differences may reflect differences in overall gene expression, rather than in exon usage). To make the comparison we analyzed our data in the same way. Among the 746 human–mouse orthologous genes common to the two studies, there are 232 genes in Su et al. (2008) data, and 97 genes in our data, with evidence for a difference in expression level of at least one exon between sexes (Bonferroni-corrected P < 0.01), with an overlap of 37 genes (a moderate enrichment over the overlap expected by chance; P ≈ 0.07, Fisher's exact test). The smaller number of differences identified in our data may partly reflect the pooling of three different species.
Summary
Put together, our results are consistent with previous observations (Xing and Lee 2005; Calarco et al. 2007) and indicate that alternative splicing is tightly regulated. We found large numbers of exons that are consistently skipped in livers from humans, chimpanzees, and rhesus macaques, as well as evidence for lineage-specific shifts in the composition of alternative transcripts. Indeed, the evolution of gene function through the regulation of alternative splicing is an intuitive mechanism and is consistent with a growing number of cases where mutations that affect splicing were found to be associated with human diseases (for review, see Tazi et al. 2009). We expect that further RNA-seq data will allow more detailed assessment of the relative contribution of such mechanisms to adaptation through changes in gene regulation.
Methods
Sample collection, study design, and RNA sequencing
In this study, we used RNA sequencing (RNA-seq) to compare gene expression levels and exon usage across sexes within and between species. We chose to work with liver tissue samples because livers are among the most homogeneous tissue with respect to cellular composition (more than 70% of the cells are hepatocytes) (Balashova and Abdulkadyrov 1984). As a result, it is unlikely that observed differences in gene expression levels and/or exon usage between sexes or species can be explained by corresponding systematic differences in cell composition between samples.
We obtained samples from several sources (for details on all samples, see Supplemental Table S2). Non-human liver tissues were collected for us by the Yerkes primate center and the Southwest Foundation for Biomedical Research (SFBR). Additional primate tissues were provided by Anne Stone (Arizona State University, Tempe). The human adult tissue samples were collected for us by the National Disease Research Interchange (NDRI). All samples were collected from healthy adult individuals for all three species (for the non-human primates, tissues were collected when chimpanzees or rhesus macaques died of natural causes such as accidents or fights, or were euthanized due to an illness unrelated to the liver).
We extracted RNA from each tissue sample using TRIzol (Invitrogen) and confirmed that the RNA was of high quality both by visualizing the RNA on a gel, and by analyzing it using Agillent's Bioanalyzer 2100. We then prepared samples for RNA sequencing using Illumina's technology (Solexa) by using our previously published RNA-seq protocol (Marioni et al. 2008; the detailed protocol is available at http://giladlab.uchicago.edu). Based on results from our previous study (Marioni et al. 2008), we decided to sequence each sample using two lanes of the Genome-Analyzer II (GA2), yielding a total of 36 lanes, distributed over multiple flow cells. The sequencing study design and the distribution of samples over the flow-cells are illustrated and detailed in Supplemental Figure S1 and Supplemental Table S3.
Read mapping, normalization, and estimates of gene expression levels
The Illumina GA2 output files were mapped to the human, chimpanzee, and rhesus macaque genomes, as appropriate, using MAQ (Li et al. 2008) version 0.6.8. Specifically, the files were transformed into the fastq format using MAQ's sol2sanger script, and then to the bfq format using MAQ's fastq2bfq script. The sequence reads were mapped to the appropriate reference genome using the MAQ match script with default parameters. Output files were transformed to the mapview format using the MAQ mapview script.
To obtain a measure of exon and gene expression levels, we counted the number of reads that mapped within each exon from each lane of sequence. We excluded reads that (1) did not overlap any of the three-species orthologous exons; (2) had a MAQ mapping quality lower than 20, which might indicate errors or ambiguous mapping; or (3) mapped to more than a single exon in our list. A summary of the data from each lane is available in Supplemental Table S3.
Once we obtained the final list of reads that mapped unambiguously to orthologous exons in each lane, we estimated relative gene expression levels for each lane of data by summing the number of reads mapped to all the exons within the gene and dividing by the total number of reads mapped to genes in that lane. Furthermore, in all plots, we consider the square root of the proportions to aid both visualization and to stabilize the variance of the observations.
Using this approach, we found 13,267, 13,275, and 13,105 expressed genes in the livers of humans, chimpanzees, and rhesus macaques, respectively (using an arbitrary classification of genes as “expressed” when the median number of reads mapped to the gene across all the lanes in which the same species is sequenced is higher than 0; see Supplemental Fig. S2).
A statistical framework for identifying differentially expressed genes
In an earlier study (Marioni et al. 2008), we showed that when the same cDNA library is sequenced in multiple lanes (either on the same, or in different flow cells) the number of reads mapping to a gene in each lane can be modeled as independent Poisson random variables. Consequently, we extended this framework to model the number of reads mapping to each gene (i.e., the sum of reads mapped to all exons within a gene) in our study by using a Poisson mixed-effects model, where a random effect is incorporated to estimate variability between individuals.
Specifically, if ygs,i,l denotes the number of reads mapped to gene g for individual i of species s in replicate lane l, Cs,i,l denotes the total number of reads mapping to genes in the lth replicate lane in which individual i of species s is sequenced, and l, Cs,i,l denotes the total length of gene g in species s, we assume that:
where
and μg is an intercept term (representing the average “overall” expression of a gene across all individuals), θgs is a species-specific fixed effect, δgsex(i) is a sex-specific fixed effect, δgsex(i) is a sex-by-species interaction term, and γgi is a per-individual random effect that follows an N(0, σg2) distribution, where the variance, σg2, has to be estimated. When making inferences about differences in gene expression levels between either species or sexes, we use the above model by restricting the values that the fixed-effects parameters can take, and assessing how well various models describing different evolutionary scenarios fit the data. To fit the model (under all parameterizations of μgs,i), we maximized the likelihood (estimated using a Laplace approximation) as implemented in the R library, lme4.
Interspecies differences in gene expression
To test whether a gene is differentially expressed between pairs of species, we considered data from the three possible pairwise combinations separately. We assessed how well the model defined in Equation 1 fitted the data under the two parameterizations of μgs,i (see Equation 2) defined below:
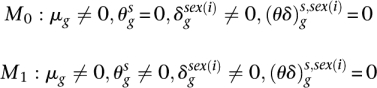 |
Here, M0 is the null model where we assume no difference in gene expression across species (when accounting for gene length, number of reads in each lane, and after controlling for a sex effect that is common across species). In turn, M1 describes an alternative model where the expression of gene g differs between the pair of species. We determine whether there is significant evidence of a difference in expression between the two species by comparing the difference in the likelihood (calculated at the maximum likelihood estimates of the parameters) between M0 and M1 and calculating a likelihood ratio statistic. Subsequently, to obtain P-values, we compared the test statistic with a chi-square distribution with one degree of freedom. Finally, to correct for multiple testing, we calculated an FDR using the approach of Storey and Tibshirani (2003). Genes with a q-value < 0.05 were considered differentially expressed between species. See Table 1 for the numbers of differentially expressed genes, Supplemental Figure S13 for expression patterns of genes DE between human and chimpanzee, and Supplemental Figure S14 for a Venn diagram depicting the overlap in DE genes between the three species.
We used a similar approach (with the same FDR) to identify genes that are differentially expressed between species within each sex by considering only lanes from one sex (in this analysis we did not include the sex effect δgsex(i)). For details of the parameterizations of the models that were used to identify genes under various selective pressures, see the Supplemental Methods.
Splice junction identification
To identify reads that overlap exon–exon junctions, we used TopHat (Trapnell et al. 2009) version 1.0.8 with Bowtie (Langmead et al. 2009) version 0.9.9.3. We first created Bowtie index files for the three reference genomes (hg18, panTro2, or rheMac2) using the Bowtie-build program with default parameters. We then created three GFF files (one for each species) containing all possible exon junctions (within each gene) using our orthologous exon definitions. Next, we ran TopHat separately for all the lanes from each species and sex (six runs, six lanes for each run). We used the GFF and no-novel-juncs options and inputted the appropriate GFF file and reference genome Bowtie index. Finally, we summarized the junction reads in the output into three categories: (1) reads supporting splicing of consecutive exons; (2) reads supporting skipping events, where one exon is skipped; and (3) reads supporting other alternative splice forms (skipping of more than one exon). The numbers of junctions, reads, and genes in each category for each species are given in Supplemental Table S15.
Identifying exon-level expression differences between species and sexes
To identify differences in expression at the exon level, we first considered a modification of the model defined in Equations 1 and 2. Let 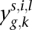 denote the number of reads mapped to exon k of gene g in replicate lane l for individual i of species s, and Cs,i,l denote the length of exon k of gene g in species s. Then, with Cs,i,l defined as before, we assume that
denote the number of reads mapped to exon k of gene g in replicate lane l for individual i of species s, and Cs,i,l denote the length of exon k of gene g in species s. Then, with Cs,i,l defined as before, we assume that
Here
This model is very similar to the one described in Equations 1 and 2. In essence, this formulation assumes that the number of reads mapping to each exon (conditional on its length) can be modeled by conditioning on the overall expression of the gene. To this end, the following gene-wide parameters (common across all exons) are included: an intercept term, a different effect for each species, a sex effect, and a sex-by-species interaction. Furthermore, we incorporate an individual random effect (as defined previously), and one exon-specific term, ϕg,k, which allows for differences in the mean expression of each exon (common across each sex and species) to be incorporated into the model.
We fitted this model (using the lme4 R package) to all genes with more than one exon and to all genes where the median number of reads across all 36 lanes is greater than 1. Subsequently, to determine whether specific exons showed different expression levels between either sexes or species, we considered the standardized residuals [defined as 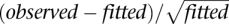 ] where the fitted values were extracted from the mixed-effects model defined above. Let
] where the fitted values were extracted from the mixed-effects model defined above. Let 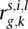 denote the (standardized) residual for the k-th exon of gene g in replicate lane l for individual i of species s. Since we have controlled for gene-wide sex and species effects (as well as differences in the mean expression level of each exon across sexes and species), correlation of the residuals for a particular exon with either sex or species will suggest differences in exon usage between sexes or across species (for more details, see Supplemental Methods).
denote the (standardized) residual for the k-th exon of gene g in replicate lane l for individual i of species s. Since we have controlled for gene-wide sex and species effects (as well as differences in the mean expression level of each exon across sexes and species), correlation of the residuals for a particular exon with either sex or species will suggest differences in exon usage between sexes or across species (for more details, see Supplemental Methods).
Analysis of enrichment in functional categories
Throughout this study, we have used Gene Ontology (GO) annotations (The Gene Ontology Consortium 2000) to examine the biological functions of genes whose regulation likely evolves under natural selection. We recognize, however, that a global analysis of all GO terms is somewhat difficult to interpret since many functional annotations are not mutually exclusive at any level of the GO hierarchy and are often not very informative. Consequently, we focused on enriched categories at the top of ranked lists for each analysis (Supplemental Tables S5–S11) and only reported qualitative results that are either supported by several observations or are consistent with data from other studies. Moreover, to confirm that our results are robust with respect to the statistical cutoffs used to identify genes whose regulation evolves under selection, we repeated the GO analyses using two additional statistical cutoffs for each class and confirmed that the qualitative results are unchanged (Supplemental Methods; Supplemental Table S12). Finally, since the power to detect differences in exon usage using RNA-seq data is related to the length of the exon and the number of exons in a gene (Oshlack and Wakefield 2009), we also confirmed that the results of the GO analysis for the categories of genes with either species or sexually dimorphic exon usage are not biased toward enrichment of long genes (as there may be an association between functional categories and gene length). To do so, we repeated the enrichment analysis using a background set containing only genes whose length is within the 20th and 80th percentiles of the distribution of gene lengths in the test set. The qualitative results have not changed (Supplemental Methods).
Quantitative RT-PCR analysis of interspecies differences in exon usage
To provide further support for the inference of interspecies differences in the expression level of alternative splice forms, we performed quantitative PCR. To do so, we selected 10 genes for which we inferred a human-specific (either reduced or elevated) level of exon usage and tested the expression level of this exon in two humans and two chimpanzees (one male and one female from each species). To account for overall gene expression difference between the species, we also assayed the expression of a control exon, chosen such that the mean levels of expression in the two species, although not significantly differential expressed, show the opposite trend compared with the test exon.
PCR primers for all exons were designed in genomic regions that are identical between human and chimpanzee. As templates, we used a new RNA extraction from the human and chimpanzee livers different from the one that was used for the RNA-seq experiments. Quantitative RT-PCR was performed in a 25-μL reaction containing 2× SYBR master mix (Sigma), 0.2 pM each primer, and 1 μL of cDNA template. PCR was performed in a 7900HT Fast Real-Time PCR System (Applied Biosystem, Inc.), in three technical replicates for each sample. The detection threshold cycle for each reaction was determined using a standard curve. For a summary of the results, see Supplemental Table S14.
Acknowledgments
We thank the Yerkes primate center, the Southwest Foundation for Biomedical Research, MD Anderson Cancer Center, and Anne Stone for providing primate tissue samples. We thank Z. Gauhar, N. Zeuss, G. Coop, K. Thornton, J. Pritchard, K. Bullaughey, A. Oshlack, three anonymous reviewers, and all members of the Gilad lab for discussions and/or for comments on the manuscript. This work was supported by a Sloan fellowship to Y.G.; by NIH grants GM077959 to Y.G.; GM08153 to Charles Lee, Anne Stone, and Y.G.; and HG002585 to M.S.
Footnotes
[Supplemental material is available online at http://www.genome.org. The RNA-seq data have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) under series accession no. GSE17274.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.099226.109.
References
- Abzhanov A, Protas M, Grant BR, Grant PR, Tabin CJ. Bmp4 and morphological variation of beaks in Darwin's finches. Science. 2004;305:1462–1465. doi: 10.1126/science.1098095. [DOI] [PubMed] [Google Scholar]
- Balashova VA, Abdulkadyrov KM. Cellular composition of hemopoietic tissue of the liver and spleen in the human fetus. Arkh Anat Gistol Embriol. 1984;86:80–83. [PubMed] [Google Scholar]
- Blekhman R, Oshlack A, Chabot AE, Smyth GK, Gilad Y. Gene regulation in primates evolves under tissue-specific selection pressures. PLoS Genet. 2008;4:e1000271. doi: 10.1371/journal.pgen.1000271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Britten RJ, Davidson EH. Repetitive and non-repetitive DNA sequences and a speculation on the origins of evolutionary novelty. Q Rev Biol. 1971;46:111–138. doi: 10.1086/406830. [DOI] [PubMed] [Google Scholar]
- Calarco JA, Xing Y, Caceres M, Calarco JP, Xiao X, Pan Q, Lee C, Preuss TM, Blencowe BJ. Global analysis of alternative splicing differences between humans and chimpanzees. Genes & Dev. 2007;21:2963–2975. doi: 10.1101/gad.1606907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll SB. Genetics and the making of Homo sapiens. Nature. 2003;422:849–857. doi: 10.1038/nature01495. [DOI] [PubMed] [Google Scholar]
- Carroll SB. Evo-devo and an expanding evolutionary synthesis: A genetic theory of morphological evolution. Cell. 2008;134:25–36. doi: 10.1016/j.cell.2008.06.030. [DOI] [PubMed] [Google Scholar]
- Draghici S, Khatri P, Eklund AC, Szallasi Z. Reliability and reproducibility issues in DNA microarray measurements. Trends Genet. 2006;22:101–109. doi: 10.1016/j.tig.2005.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzurba A, Ziegelhoffer A, Vrbjar N, Styk J, Slezak J. Estradiol modulates the sodium pump in the heart sarcolemma. Mol Cell Biochem. 1997;176:113–118. [PubMed] [Google Scholar]
- Fekete A, Vannay A, Ver A, Vasarhelyi B, Muller V, Ouyang N, Reusz G, Tulassay T, Szabo AJ. Sex differences in the alterations of Na+, K+-ATPase following ischaemia-reperfusion injury in the rat kidney. J Physiol. 2004;555:471–480. doi: 10.1113/jphysiol.2003.054825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu X, Fu N, Guo S, Yan Z, Xu Y, Hu H, Menzel C, Chen W, Li Y, Zeng R, et al. Estimating accuracy of RNA-Seq and microarrays with proteomics. BMC Genomics. 2009;10:161. doi: 10.1186/1471-2164-10-161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The Gene Ontology Consortium. Gene Ontology: Tool for the unification of biology. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilad Y, Oshlack A, Rifkin SA. Natural selection on gene expression. Trends Genet. 2006;22:456–461. doi: 10.1016/j.tig.2006.06.002. [DOI] [PubMed] [Google Scholar]
- Iftikhar R, Kladney RD, Havlioglu N, Schmitt-Graff A, Gusmirovic I, Solomon H, Luxon BA, Bacon BR, Fimmel CJ. Disease- and cell-specific expression of GP73 in human liver disease. Am J Gastroenterol. 2004;99:1087–1095. doi: 10.1111/j.1572-0241.2004.30572.x. [DOI] [PubMed] [Google Scholar]
- Jin W, Riley RM, Wolfinger RD, White KP, Passador-Gurgel G, Gibson G. The contributions of sex, genotype and age to transcriptional variance in Drosophila melanogaster. Nat Genet. 2001;29:389–395. doi: 10.1038/ng766. [DOI] [PubMed] [Google Scholar]
- Kent WJ. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- King MC, Wilson AC. Evolution at two levels in humans and chimpanzees. Science. 1975;188:107–116. doi: 10.1126/science.1090005. [DOI] [PubMed] [Google Scholar]
- Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leek JT, Storey JD. Capturing heterogeneity in gene expression studies by surrogate variable analysis. PLoS Genet. 2007;3:1724–1735. doi: 10.1371/journal.pgen.0030161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lemos B, Meiklejohn CD, Caceres M, Hartl DL. Rates of divergence in gene expression profiles of primates, mice, and flies: Stablizing selection and variability among functional categories. Evolution. 2005;59:126–137. [PubMed] [Google Scholar]
- Li H, Ruan J, Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–1858. doi: 10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marioni JC, Mason CE, Mane SM, Stephens M, Gilad Y. RNA-seq: An assessment of technical reproducibility and comparison with gene expression arrays. Genome Res. 2008;18:1509–1517. doi: 10.1101/gr.079558.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntyre LM, Bono LM, Genissel A, Westerman R, Junk D, Telonis-Scott M, Harshman L, Wayne ML, Kopp A, Nuzhdin SV. Sex-specific expression of alternative transcripts in Drosophila. Genome Biol. 2006;7:R79. doi: 10.1186/gb-2006-7-8-r79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- Oleksiak MF, Churchill GA, Crawford DL. Variation in gene expression within and among natural populations. Nat Genet. 2002;32:261–266. doi: 10.1038/ng983. [DOI] [PubMed] [Google Scholar]
- Oshlack A, Wakefield MJ. Transcript length bias in RNA-seq data confounds systems biology. Biol Direct. 2009;4:14. doi: 10.1186/1745-6150-4-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oshlack A, Chabot AE, Smyth GK, Gilad Y. Using DNA microarrays to study gene expression in closely related species. Bioinformatics. 2007;23:1235–1242. doi: 10.1093/bioinformatics/btm111. [DOI] [PubMed] [Google Scholar]
- Pan Q, Bakowski MA, Morris Q, Zhang W, Frey BJ, Hughes TR, Blencowe BJ. Alternative splicing of conserved exons is frequently species-specific in human and mouse. Trends Genet. 2005;21:73–77. doi: 10.1016/j.tig.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Quintas LE, Lopez LB, Souccar C, Noel F. Na+/K+-ATPase density is sexually dimorphic in the adult rat kidney. Ann N Y Acad Sci. 1997;834:552–554. doi: 10.1111/j.1749-6632.1997.tb52316.x. [DOI] [PubMed] [Google Scholar]
- Rifkin SA, Houle D, Kim J, White KP. A mutation accumulation assay reveals a broad capacity for rapid evolution of gene expression. Nature. 2005;438:220–223. doi: 10.1038/nature04114. [DOI] [PubMed] [Google Scholar]
- Rinn JL, Snyder M. Sexual dimorphism in mammalian gene expression. Trends Genet. 2005;21:298–305. doi: 10.1016/j.tig.2005.03.005. [DOI] [PubMed] [Google Scholar]
- Shapiro MD, Marks ME, Peichel CL, Blackman BK, Nereng KS, Jonsson B, Schluter D, Kingsley DM. Genetic and developmental basis of evolutionary pelvic reduction in threespine sticklebacks. Nature. 2004;428:717–723. doi: 10.1038/nature02415. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de Longueville F, Kawasaki ES, Lee KY, et al. The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shima S. Effects of androgen treatment on adenylate cyclase system in rat hepatic membranes. Pharmacol Toxicol. 1992;70:429–433. doi: 10.1111/j.1600-0773.1992.tb00502.x. [DOI] [PubMed] [Google Scholar]
- Stolc V, Gauhar Z, Mason C, Halasz G, van Batenburg MF, Rifkin SA, Hua S, Herreman T, Tongprasit W, Barbano PE, et al. A gene expression map for the euchromatic genome of Drosophila melanogaster. Science. 2004;306:655–660. doi: 10.1126/science.1101312. [DOI] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. Statistical significance for genomewide studies. Proc Natl Acad Sci. 2003;5:9440–9445. doi: 10.1073/pnas.1530509100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su WL, Modrek B, GuhaThakurta D, Edwards S, Shah JK, Kulkarni AV, Russell A, Schadt EE, Johnson JM, Castle JC. Exon and junction microarrays detect widespread mouse strain- and sex-bias expression differences. BMC Genomics. 2008;9:273. doi: 10.1186/1471-2164-9-273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taron M, Rosell R, Felip E, Mendez P, Souglakos J, Ronco MS, Queralt C, Majo J, Sanchez JM, Sanchez JJ, et al. BRCA1 mRNA expression levels as an indicator of chemoresistance in lung cancer. Hum Mol Genet. 2004;13:2443–2449. doi: 10.1093/hmg/ddh260. [DOI] [PubMed] [Google Scholar]
- Tazi J, Bakkour N, Stamm S. Alternative splicing and disease. Biochim Biophys Acta. 2009;1792:14–26. doi: 10.1016/j.bbadis.2008.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Pachter L, Salzberg SL. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Gerstein M, Snyder M. RNA-Seq: A revolutionary tool for transcriptomics. Nat Rev Genet. 2009;10:57–63. doi: 10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead A, Crawford DL. Neutral and adaptive variation in gene expression. Proc Natl Acad Sci. 2006;103:5425–5430. doi: 10.1073/pnas.0507648103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray GA. The evolutionary significance of cis-regulatory mutations. Nat Rev Genet. 2007;8:206–216. doi: 10.1038/nrg2063. [DOI] [PubMed] [Google Scholar]
- Xing Y, Lee C. Evidence of functional selection pressure for alternative splicing events that accelerate evolution of protein subsequences. Proc Natl Acad Sci. 2005;102:13526–13531. doi: 10.1073/pnas.0501213102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]