Abstract
In eukaryotes, many chromatin proteins together regulate gene expression. Chromatin proteins often direct the genomic binding pattern of other chromatin proteins, for example, by recruitment or competition mechanisms. The network of such targeting interactions in chromatin is complex and still poorly understood. Based on genome-wide binding maps, we constructed a Bayesian network model of the targeting interactions among a broad set of 43 chromatin components in Drosophila cells. This model predicts many novel functional relationships. For example, we found that the homologous proteins HP1 and HP1C each target the heterochromatin protein HP3 to distinct sets of genes in a competitive manner. We also discovered a central role for the remodeling factor Brahma in the targeting of several DNA-binding factors, including GAGA factor, JRA, and SU(VAR)3-7. Our network model provides a global view of the targeting interplay among dozens of chromatin components.
In the eukaryotic cell nucleus, hundreds of proteins interact directly or indirectly with DNA to form the chromatin fiber, which is arguably the most intricate molecular complex in the cell. These proteins together control genome-wide transcription patterns, as well as other functions such as maintenance and replication of the genome. Proteins in the chromatin fiber can be divided into three major classes: (1) histones, which form nucleosomes that constitute the basic packaging unit of chromatin; (2) DNA binding factors (DBFs), which typically recognize specific sequence motifs, and (3) proteins that do not contact DNA directly, but interact with DNA via other proteins, which we will refer to as chromatin proteins.
Essentially all of these proteins show highly specific binding patterns along the genome. Histones are the most abundant protein component of the chromatin fiber, but nevertheless display some sequence preference (Kaplan et al. 2008) and show reduced binding at the 5′ ends of active genes (Rando and Ahmad 2007). Histones carry a multitude of post-translational modifications, many of which have specific location patterns along the genome (Berger 2007; Rando 2007). DBFs generally show focal binding patterns that are, to a large extent, dictated by the locations of their recognition motifs in the genome, but also by the accessibility of these motifs and by interactions with other proteins (Kim and Ren 2006; Morse 2007). The targeting of chromatin proteins is determined by interactions with specific histone modifications, DBFs, and other chromatin proteins. In turn, the location of histone modifications is modulated by DBFs and chromatin proteins. Thus, the genomic binding pattern of each chromatin component may be determined by a multitude of interactions with other components. How this highly complex network of interactions leads to the formation of distinct types of chromatin at different parts of the genome is still poorly understood.
In vivo genomic binding maps can provide important insights into the signals that govern the genomic targeting specificity of a chromatin component (van Steensel 2005; Kim and Ren 2006). Comparison of the binding maps of multiple proteins can be particularly informative. For example, if two proteins have highly similar distributions along the genome, this may indicate that the two proteins share a common targeting mechanism, or that one protein recruits the other. Conversely, mutually exclusive distributions suggest that the two proteins may be targeted by different, incompatible mechanisms, or that one protein prevents the other protein from binding.
Here, we describe a systematic search for targeting interactions among a broad set of chromatin components. We define a targeting interaction X→Y as an interaction between two chromatin components X and Y, such that the presence of X at a specific set of genomic loci promotes the association of Y with these loci. Note that this is a functional definition rather than a biochemical definition; i.e., a targeting interaction does not necessarily require a direct protein–protein interaction; it may also involve one or more intermediate biochemical interactions or enzymatic activities. To map targeting interactions systematically, we analyzed a broad compendium of in vivo genome-wide binding profiles of a broad set of chromatin components in Drosophila. By computational analyses and direct experimental evidence we demonstrate the high overall reliability of the predicted network of targeting interactions. We highlight several sets of interactions that illustrate how the interplay between multiple proteins determines their distribution along the genome. Specifically, we uncover distinct mechanisms that determine the genomic binding patterns of the heterochromatin components HP3 (also known as LHR) and SU(VAR)3-7, and we demonstrate that the nucleosome remodeling protein Brahma (BRM) has a central role in the targeting of various DBFs. Finally, by analysis of the genes that are bound by each chromatin protein we present evidence for compartmentalization of regulatory functions within the chromatin interaction network.
Results
A compendium of binding maps of chromatin components
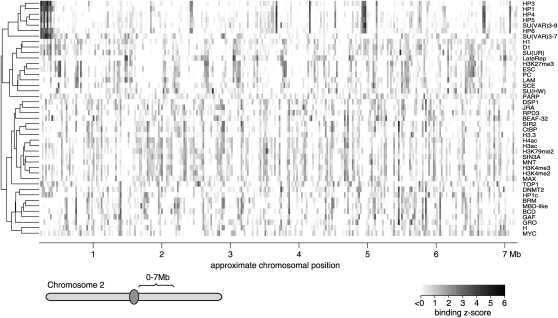
We set out to identify candidate targeting interactions among chromatin proteins by assembling a compendium of maps of 43 broadly selected chromatin components in Drosophila (Fig. 1). The maps were generated by either chromatin immunoprecipitation (ChIP) (Lieb 2003) or the DamID method (Greil et al. 2006). The set of maps was chosen to represent diverse classes of chromatin components and it is currently the broadest collection of chromatin profiles assembled in any eukaryote. The compendium includes maps of six histone modifications and one histone variant (H3.3), nine DBFs, and 26 chromatin proteins, such as classic heterochromatin proteins, Polycomb Group (PcG) proteins, nucleosome remodeling factors, high mobility group proteins, histone modifying enzymes, cofactors, and several other types (Table 1; Supplemental Data Set 1). We also included a map of late replication timing (LateRep) (Schübeler et al. 2002), which is an important attribute of chromatin. While most maps were published previously, we generated additional DamID maps for six new proteins in order to further increase the diversity of protein types (Table 1). All data were generated in the Kc167 cell line, except for histone H3.3, which was mapped in the closely related S2 cell line. All binding profiles were detected using cDNA arrays, which report protein associations at genes or in the flanking regions (∼1 kb on either side) (van Steensel et al. 2003). For 4380 genes, complete binding data were available for each of the 43 chromatin components. Hierarchical clustering of the genomic binding maps reveals similarities and dissimilarities between the protein binding patterns (Fig. 1). For example, HP1, HP3, HP4, HP5, HP6, and SU(VAR)3-9, which are all known components of classic heterochromatin (de Wit et al. 2007), show highly similar binding patterns (top six rows in Fig. 1). Likewise, the Polycomb Group components PC, ESC, SCE, and H3K27me3 cluster closely together.
Figure 1.
Chromosomal maps of 43 chromatin components. Example showing the binding levels (log2 ratios transformed to Z-scores) of 43 Drosophila chromatin components along 300 genes in the proximal part of chromosome 2R. Rows were arranged by genome-wide hierarchical clustering, as shown by the tree to the left of the binding data.
Table 1.
Overview of chromatin components analyzed in this study
aAdditional replicates done in this study.
Bayesian network inference of protein targeting interactions
Next, we employed the technique of Bayesian network inference (BNI) (Pearl 1988; Cooper and Herskovits 1992; Friedman 2004; Pe'er 2005) to build a detailed model of the targeting interactions among all 43 chromatin components. BNI has two properties that make it particularly useful for this purpose. First, it has the ability to “explain away” indirect correlations. For example, if the proteins B and C are independently recruited by protein A to a common set of target loci, then the binding maps of B and C will not only correlate with A, but also with each other. The correlation between B and C in this example does not reflect a functional interaction between these two proteins, unlike the A–B and A–C correlations. In BNI terminology, B and C are said to be conditionally independent given A. BNI models the conditional relationships among all variables, resulting in a network model of the most probable targeting interactions (i.e., the conditionally dependent relationships).
Second, under certain assumptions (Friedman et al. 2000) BNI models the most probable causality direction of interactions in the network. In the context of our chromatin data, we interpret the predicted causality direction as the most likely direction of a targeting interaction between two chromatin components (e.g., it is more likely that component X targets component Y than the inverse). Although such causality predictions should be interpreted with caution (see Methods; Friedman et al. 2000), they provide important guidance in the design of follow-up experiments (see below). BNI has previously been used in biology to predict interactions in gene and protein networks (Friedman 2004; Pe'er 2005; Missal et al. 2006; Djebbari and Quackenbush 2008; Mukherjee and Speed 2008) and interactions among histone modifications (Yu et al. 2008).
We constructed a BN using the compendium of 43 binding profiles as input data. A previously described bootstrapping approach was used to compute confidence scores for each predicted interaction (Friedman et al. 2000; Pe'er 2005) (Methods; Supplemental Data Set 2). The bootstrapping approach consists of the construction of 1000 networks that are each based on randomly picked subsets of array probes (i.e., genes). The frequency at which a given edge occurs in these 1000 networks is taken as the confidence score of this edge, which can range from 0% to 100%. Figure 2 shows the network of predicted interactions with confidence scores ≥80%. We will refer to this network as BN80. As will be discussed below, we chose 80% as a cutoff because it yields a network with high accuracy and sufficient coverage. In the graph representation of the network, each chromatin component is shown as a node, and edges between nodes indicate predicted targeting interactions. An added value of BNI with bootstrapping is that it also models the relative probability of the causality direction of the interactions. This probability is reflected in the bootstrap score for each direction and is indicated in Figure 2 by the sizes of the two arrowheads of each edge. For example, the edge between Hairy (H) and Groucho (GRO) strongly favors the H→GRO direction over the opposite direction (95.3% vs. 0.45%), predicting that H affects the targeting of GRO rather than the inverse (which is in agreement with previous data; Paroush et al. 1994). As a contrasting example, the LAM-SU(UR) edge has similar bootstrap scores for both directions (47.0% and 52.7%), meaning that BNI predicts a targeting interaction between the two proteins, but cannot resolve the causality based on the available data. In total, BN80 consists of 52 edges among 40 nodes. Of these edges, nearly half (25) show a strong directionality, with a greater than threefold higher bootstrap score in one direction compared to the opposite direction.
Figure 2.
Bayesian Network model BN80 of the targeting interactions between 43 chromatin components. Nodes represent chromatin components; edges represent predicted targeting interactions with a bootstrap score (combined for both directions) of at least 80%. The size of each arrowhead is proportional to the bootstrap score of the targeting interaction in the corresponding direction.
Importantly, BN80 recapitulates many previously known complexes and molecular associations (Fig. 2). For example, it predicts targeting interactions that are in agreement with previously reported biochemical interactions, e.g., of MNT with both SIN3A and MAX (Loo et al. 2005); SIN3A with RPD3 (Pile and Wassarman 2000); H with both GRO (Paroush et al. 1994) and CTBP (Poortinga et al. 1998); and heterochromatin protein HP1 with SU(VAR)3-9 (Schotta et al. 2002), HP3, and HP4 (Giot et al. 2003). The edge between Lamin (LAM) and LateRep is also striking, given that late-replicating DNA is typically located near the nuclear lamina (Hiratani et al. 2008). Thus, many of the predicted interactions are in agreement with previous studies, even though the BNI was solely based on genome-wide binding profiles. The proteins SU(HW), PARP, and TOP1 are the only three proteins not connected in BN80. Each of these proteins has one or more connections with boostrap scores of ∼70% (Supplemental Data Set 2), i.e., below the 80% confidence cutoff. These interactions may be of biological interest, but will be disregarded here.
Computational validation
Because very few targeting interactions have been studied so far (particularly not genome-wide), no list of confirmed targeting interactions is available that could be used as a “gold standard” to estimate the performance of our BNI approach. We therefore used two public databases as surrogate reference lists. First, we took the BioGRID database (Breitkreutz et al. 2008), which is a compilation of known physical and genetic interactions. Although the Drosophila section of this database is sparse—it only lists 17 interactions among the 43 chromatin components—we found that BN80 recovers seven of these 17 interactions (41%). This is substantially more than expected by random chance (random permutation of nodes typically yields zero or one interaction). In addition, BN80 predicts 46 interactions that are not in BioGRID. We note that targeting interactions between two proteins may also occur in the absence of known physical or genetic interactions, or vice versa, and therefore one may not expect a perfect correspondence between BioGRID and BNI.
Second, we queried the PubMed database for publications that mention combinations of chromatin components in their abstract or title (“co-citations”). We found that 48% (25/52) of the BN80-predicted interactions overlap with PubMed co-citations (Supplemental Fig. 1). In contrast, among all pairs of the 43 chromatin components not linked by an edge in BN80, only 13% have PubMed co-citations, which demonstrates the highly nonrandom nature of BN80. Obviously, co-citation of chromatin components in PubMed is only a crude approximation of the existence of targeting interactions; however, given the large number of possible pairs of components (903), manual curation of these PubMed co-citations is not feasible. Lowering of the BNI bootstrap confidence threshold increases the total number of predicted interactions, but with a larger percentage of interactions not listed in the public databases (Supplemental Figs. 1, 2), and therefore may result in a lower overall accuracy. Taken together, these analyses indicate that BN80 predicts a large number of biologically relevant interactions, about half of which involve pairs of proteins that have been previously linked according to a variety of biochemical or functional assays, and the other half being completely novel.
Experimental validation strategy
An X→Y targeting interaction can be experimentally validated by monitoring the binding pattern of component Y after knockdown or deletion of component X: If the loss of X causes a specific reduction of the binding of Y at the original target genes of X, then the targeting interaction is (by definition) confirmed. According to these criteria we confirmed several of the novel interactions experimentally, as described below.
Competitive targeting of HP3 by HP1 and HP1C
First, we considered the predicted targeting interactions in the dense cluster of classic heterochromatin proteins centered around HP1 in BN80 (Fig. 2). One such protein is HP3, a recently discovered heterochromatin component that is thought to play a role in Drosophila speciation (Brideau et al. 2006; de Wit et al. 2007). The HP1→HP3 edge predicted that HP1 mediates targeting of HP3, which is consistent with the reported physical interaction between the two proteins (Giot et al. 2003; Brideau et al. 2006). Reanalysis of previously published data (Greil et al. 2007) shows that HP3 is indeed selectively lost from the original HP1 target genes after HP1 depletion (Fig. 3A), but not from HP1 nontarget genes. This confirms that the HP1→HP3 edge represents a bona fide targeting interaction.
Figure 3.
Experimental validation of predicted HP3 and HP6 targeting interactions. Density plots (“smoothed histograms”) showing the changes in binding levels of HP3 (A) and HP6 (B) after RNAi knockdown of HP1, compared to a control knockdown. Changes in binding are shown for subsets of genes as indicated. The y-axes show relative frequency (plots in each panel are normalized to have the same surface area). Binding data after knockdown are from (Greil et al. 2007). Nontargets are bound by neither HP1 nor HP1C. P-values are according to two-sided Wilcoxon tests. (C) Cartoon depicting the competitive targeting of HP3 by HP1 and HP1C, as well as the targeting of HP6 by HP1.
Interestingly, BN80 predicts that HP3 is also targeted by HP1C, which is a homolog of HP1. The genomic binding patterns of HP1C and HP1 are essentially nonoverlapping (only ∼5% of their target genes are shared), suggesting that the two proteins may compete for HP3 interaction. Indeed, depletion of HP1 causes HP3 to shift toward HP1C target genes (Fig. 3A), strongly supporting this competition model.
We also analyzed the predicted HP1→HP6 interaction. Again, this targeting interaction was confirmed by a specific loss of HP6 from HP1 target genes upon depletion of HP1 (Fig. 3B). Unlike HP3 however, HP6 does not relocate significantly to HP1C target genes, in agreement with the lack of an edge between the HP6 and HP1C nodes. Taken together, BN80 correctly predicts two targeting interactions involving HP1, and identifies a competition mechanism involving HP1→HP3 and HP1C→HP3 interactions, the balance of which determines the distribution of HP3 along the genome (Fig. 3C).
BRM targets several DBFs
With six edges, the nucleosome remodeling protein Brahma (BRM) is the most highly connected node in BN80, suggesting a central role in the network. Interestingly, five of the six BRM-linked proteins are known or predicted to contact DNA directly [JRA, GAF, MBD-like, SU(VAR)3-7, histone H3.3] (Jaquet et al. 2002; Marhold et al. 2004). For each of these five proteins the edge is strongly oriented away from BRM, predicting that BRM is necessary for the targeting of these DNA-binding proteins. We experimentally tested this prediction for JRA, GAF, and SU(VAR)3-7. As a negative control we also tested HP1, which is not linked to BRM in BN80. The results of these experiments show that depletion of BRM causes a reduction in the binding levels of all three proteins at BRM target genes, but not at other genes (Fig. 4A–D; Supplemental Data Set 3), confirming that BRM is needed for their correct targeting. This effect of BRM is specific, because HP1 shows no detectable change in its binding levels at BRM target genes after knockdown of BRM (Fig. 4E).
Figure 4.
BRM mediates targeting of GAF, JRA, and SU(VAR)3-7. (A) Western blot of BRM protein after RNAi knockdown of brm. RNAi of white (w), a gene that is not expressed in Kc cells, served as a control. Asterisk marks a nonspecific band. Lamin was probed as a loading control. (B–D) Changes in binding levels of GAF, JRA, and SU(VAR)3-7 at BRM target genes (black) and nontarget genes (gray) upon knockdown of brm, relative to control knockdown. (E) No significant changes in HP1 relocation after brm knockdown. (F) brm knockdown does not affect SU(VAR)3-7 binding at heterochromatic genes, defined as HP1 target genes. P-values are according to Wilcoxon tests.
SU(VAR)3-7 is located in both euchromatin and pericentric heterochromatin of polytene chromosomes. The targeting to these different regions requires different protein domains in SU(VAR)3-7, strongly suggesting two distinct molecular mechanisms (Jaquet et al. 2002). Consistent with the exclusively euchromatic localization of BRM (Armstrong et al. 2002) we find that BRM only contributes to the targeting of SU(VAR)3-7 to euchromatic genes and not to heterochromatic genes, as after knockdown of BRM SU(VAR)3-7 association does not change at targets it shares with HP1 (Fig. 4F). The targeting mechanism of SU(VAR)3-7 to pericentric heterochromatin is not immediately clear from BN80 because this network lacks any edges between SU(VAR)3-7 and the heterochromatin protein cluster centered around HP1. Underrepresentation of pericentric DNA sequences on our microarrays could in part account for this. The pericentric targeting of SU(VAR)3-7 may be mediated by a physical interaction with HP1 (Delattre et al. 2000), and possibly also by HP3, which according to BNI has a predicted weak targeting interaction with SU(VAR)3-7 (bootstrap score 54%; Supplemental Data Set 2). The latter scenario is supported by the fact that HP3 and SU(VAR)3-7 both contain a BESS domain, which is a rare dimerization domain (Jaquet et al. 2002).
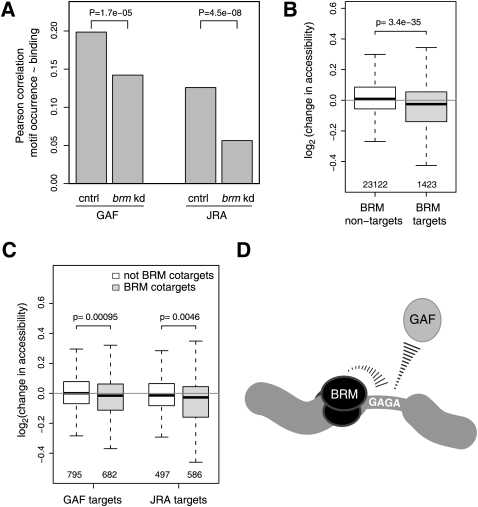
How might BRM contribute to the targeting of several DBFs? First, we investigated whether BRM targets these DBFs by enhancing their interactions with their binding motifs in DNA. If so, then depletion of BRM should lead to a loss of the DBFs from motif-containing binding sites rather than from sites lacking these motifs. Indeed, for both GAF and JRA, we observed that knockdown of brm caused a significant loss of the correlation between binding of these factors and the occurrence of the respective motifs (Fig. 5A). Thus, both proteins require BRM for efficient interaction with their binding motifs. We could not do the same analysis for SU(VAR)3-7, because no binding motif is known for this protein.
Figure 5.
BRM promotes the interaction of JRA and GAF with their binding motifs through enhancement of chromatin accessibility. (A) Depletion of BRM causes a reduction in the correlation between binding and recognition motif occurrence for GAF and JRA, indicating that BRM enhances binding of these DBFs to their recognition motifs. cntrl, control knockdown; brm kd, brm knockdown. (B) Depletion of BRM causes a slight decrease in chromatin accessibility at BRM target genes. Depicted are changes in accessibility per DpnI-fragment that overlaps with indicated target or nontarget reporters of microarrays used for DamID profiles. (C) Same as B, but for GAF and JRA target genes that are either BRM targets or nontargets. (D) Cartoon model depicting the targeting of GAF through the combined action of BRM and GAGA motifs. A similar model may apply to JRA and SU(VAR)3-7. P-values are according to a test for difference between two dependent correlations (Chen and Popovich 2002) (A) or two-sided Wilcoxon tests (B,C).
Next, we considered that BRM may promote these direct DBF-DNA contacts by virtue of its ability to open up chromatin (Cairns 2007). To test this hypothesis, we directly measured the effects of BRM on chromatin accessibility in vivo using freely diffusing DNA adenine methyltransferase (Dam) as a probe (Gottschling 1992; Wines et al. 1996; Vogel et al. 2009). We constructed high-resolution genome-wide maps of methylation by Dam, comparing cells in which BRM was depleted to control-treated cells (Fig. 5B). Indeed, we found that loss of BRM typically caused a mild decrease of local chromatin accessibility. Specifically, targets of GAF and JRA that were also targets of BRM became significantly less accessible than non-BRM targets (Fig. 5C). This result suggests that BRM mediates binding of GAF and JRA by virtue of its chromatin remodeling activity, making the cognate binding sites of these DBFs more accessible (Fig. 5D).
Taken together, BN80 identifies BRM as an important player in the targeting of several DBFs. A recent genome-wide study in yeast also pointed to a central role for chromatin remodeling complexes in the control of transcription factor activity (Steinfeld et al. 2007), suggesting that this may be an evolutionarily conserved mechanism.
Network compartmentalization of regulatory function
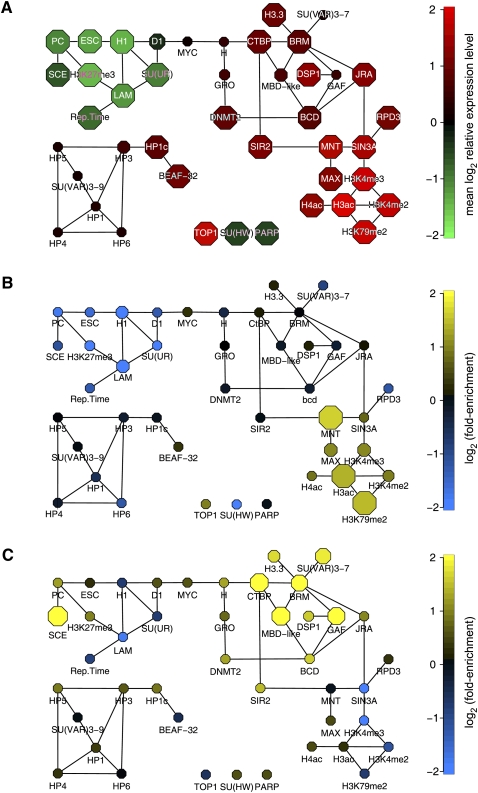
Besides the prediction of many specific targeting interactions, BNI also offers insights into the global organization of the targeting interaction network. Visualization of the average expression level of the target genes of each protein in Kc167 cells (the same cell type in which the chromatin components were mapped) reveals that the overall network structure is closely linked to regulatory function (Fig. 6A). Strong repression of gene activity is found in the subnetwork that includes the PcG proteins, Lamin and histone H1, in agreement with previous reports (Pickersgill et al. 2006; Sparmann and van Lohuizen 2006). Highest transcriptional activity occurs at genes targeted by acetylated histones and by H3K79 and H3K4 methylation, which is also consistent with other data (Rando 2007). Genes bound by the cluster of heterochromatin proteins including HP1 and SU(VAR)3-9 show overall intermediate expression levels, underscoring the emerging notion that these proteins are not strong repressors at their natural target genes (de Wit and van Steensel 2009).
Figure 6.
Examples of compartmentalization of regulatory function in the chromatin network. (A) BN80 with the same layout as in Figure 2, with nodes colored according to the mean expression level (in Kc cells) of the target genes of each chromatin component. (B,C) Same as in A, but node colors depicting enrichment (yellow) or depletion (blue) of genes that are expressed in embryonic trunk mesoderm (B) or yolk cells (C). Node sizes in A–C depict the statistical significance of the observed expression level (A, two-sided Wilcoxon test) or the observed enrichment or depletion (B,C, two-sided binomial test), ranging from P > 10−3 (smallest nodes) to P ≤ 10−8 (largest nodes). Tissue expression data in B and C are from (Tomancak et al. 2007).
We further investigated the nature of the target genes of each chromatin component by studying their developmental expression patterns (Tomancak et al. 2007). Interestingly, the genes bound by proteins in the different subnetworks show a striking degree of coregulation during development. For example, genes expressed in embryonic trunk mesoderm are specifically enriched among target genes of the protein subnetwork containing MNT, MAX, and the active histone marks (Fig. 6B), while genes expressed in yolk cells are preferential targets of the subnetwork centered around BRM, CTBP, and MBD-like (Fig. 6C). Various other sets of tissue-specific genes show a nonhomogeneous distribution across the network (Supplemental Fig. 3). These results provide evidence for a substantial degree of compartmentalization of regulatory function within the chromatin protein network, with different subnetworks being dedicated to groups of genes with distinct tissue-specific functions in the developing embryo.
Discussion
Here, we report the first network model of protein targeting interactions in chromatin. Not only does this model predict many novel targeting interactions, it also provides insights into the overall architecture of the chromatin protein network. As such, it offers a powerful framework for tackling the complexity of targeting interactions in chromatin. The merits of BNI are particularly well illustrated by the examples that we studied in more depth: It provides insight into the previously poorly understood targeting of proteins with complex genomic distributions [e.g., SU(VAR)3-7], uncovers novel competitive targeting mechanisms (e.g., HP1 and HP1C compete for HP3), and identifies a chromatin remodeling enzyme (BRM) that is a key player in the targeting of a broad set of DBFs.
The experimental validation of all five BNI-predicted targeting interactions that we tested, together with the strong overall enrichment of BN80 for known interactions detected by other assays, indicates that our network model is highly robust. This robustness is in part due to the favorable balance between the number of observations (4380 genes) and the relatively small number of nodes in the network (43), unlike some previous BNI studies of gene networks in which the number of nodes (i.e., genes) far outweighs the number of available observations (e.g., experimental conditions) (Friedman et al. 2000; Pe'er et al. 2001). Despite this robustness, we note that BNI is probabilistic by nature and has some intrinsic restrictions (see Methods).
For lack of a gold standard, it is difficult to assess the number of real targeting interactions that are missed by our network prediction. Not all known biochemical interactions are recoved in BN80. For example, GRO and RPD3 have been reported to form a complex (Chen et al. 1999), yet they are not linked by BNI. This may be because other interactions are quantitatively dominant, consistent with genetic evidence that GRO requires interactions with other partners for its repressive activity (Mannervik and Levine 1999). SU(VAR)3-7 is known to be partially located in pericentric heterochromatin (Jaquet et al. 2002), but no high-confidence targeting interaction with any other heterochromatin protein is predicted by BN80. As mentioned, this may be because heterochromatic sequences are underrepresented, and other interactions (particularly with BRM) dominate the connectivity of SU(VAR)3-7. Finally, we did not detect a targeting interaction between MYC (the product of dm) and its heterodimerization partner MAX. This is explained by previous evidence that the endogenous expression level of MAX in Kc cells may be too low to drive joint binding of MYC and MAX to their E-box binding motif (Orian et al. 2003). Instead, BN80 predicts that the high mobility group protein D1 may play an unexpected role in the targeting of MYC.
Besides the targeting interactions that we confirmed experimentally, BN80 predicts several other interactions with strong directionality that will be worthy of further experimental analysis. For example, histone H1, a still poorly understood component of repressive chromatin, is predicted to regulate the targeting of several other chromatin components [ESC, LAM, SU(UR), and D1]. Another interesting prediction is that the histone modification H3K79me2 is regulated by the other “active” marks H3ac and H3K4me2. Finally, the LAM→LateRep is of particular interest because late-replicating DNA is typically found near the nuclear lamina (Hiratani et al. 2008). The strongly directed edge suggests that LAM (or another closely associated component of the nuclear lamina) may help to position late-replicating DNA at the nuclear periphery, or to regulate replication timing. These and other edges in BN80 represent testable hypotheses for future research.
We note that the limited resolution afforded by cDNA microarrays does not permit resolution of differential binding across the length of a gene. This is not a severe limitation for our analysis because targeting interactions between, for instance, one protein binding to transcription start sites and another protein binding to downstream regions of genes may be functionally relevant. Currently, sufficiently broad sets of high-resolution maps of chromatin components are unavailable. Once such high-resolution data sets become available they will enable the construction of interaction networks that focus on distinct genomic features, such as promoters, enhancers, and coding regions. Bayesian logic can be extended to the integration of other types of data (Barash and Friedman 2002; Jansen et al. 2003; Djebbari and Quackenbush 2008), such as known physical or genetic interactions. We expect that expansion and increased resolution of the compendium of binding profiles, together with the inclusion of additional information, will progressively lead to an accurate computational model of the complex network of molecular interactions in chromatin.
Methods
DamID
Generation of new protein location maps as marked in Table 1 was done by DamID in Kc167 cells as previously described (de Wit et al. 2008). Dam-su(Hw), Dam-Mt2 (DNMT2), Dam-Parp, Dam-Top1, and BEAF-32-Dam constructs were made for this study, RPD3 profiles were newly generated with a published construct (Moorman et al. 2006), and the DSP1 profile is the average of two previously published and four new replicates (de Wit et al. 2008). Experiments were done in duplicate or triplicate and normalized data were averaged. RNA interference was performed as described (Greil et al. 2007) with modifications as follows. On day 0, 5 × 106 logarithmically growing cells were seeded in serum-free BPYE with 150 μg of dsRNA. After 1 h, 5 mL of BPYE with 10% fetal calf serum was added. On days 2 and 4, cells were washed with serum-free BPYE and again treated with dsRNA. On day 5, cells were transfected with DamID constructs or mock transfected for protein isolation and incubated for 24 h with fresh dsRNA. brm dsRNA was produced with primers for amplicon DRSC26226 of the Harvard Drosophila RNAi Screening Center. New DamID data sets are available from the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo/) accession number GSE15807.
Accessibility assay
Probing of accessibility by diffusible Dam methylase was performed as for DamID, except that material from white and brm RNAi-treated cells transfected with unfused Dam was co-hybridized to genomic tiling microarrays (Choksi et al. 2006). Log ratios of duplicate experiments were loess normalized, median centered, and averaged. Values from probes fully overlapping the same DpnI fragments in FlyBase release 5 were averaged. Data are available from the Gene Expression Omnibus, http://www.ncbi.nlm.nih.gov/geo/, accession number GSE15807.
BNI
All data preprocessing and statistical analysis was done using the R package (http://www.r-project.org). The binding profiles (average log2 ratios) of all chromatin components used in this study were combined into a single data matrix (Supplemental Data Set 1). Because the BNI algorithm cannot handle missing values, array probes with missing data were removed. The resulting matrix has 4380 rows (genes) and 43 columns (chromatin components).
Because BNI on continuous data is computationally expensive, we first discretized the data by setting the top 5% most strongly bound genes for each chromatin component to 1 (“target genes”) and the remainder to 0 (“nontarget genes”). The binarized data were used as input for the BNI algorithm. See below for a justification of this discretization scheme. Next, static BNI was performed using Banjo 2.0 (http://www.cs.duke.edu/∼amink/software/banjo/). A file listing the parameter settings is provided as Supplemental Data Set 4. We empirically found that more than ∼200,000 search iterations typically did not yield any new networks with substantially better overall network scores; for this reason we limited each search (i.e., each single bootstrap, see below) to 300,000 cycles. Banjo can accommodate substantially larger input data sets than the one we used; for example, dummy tests indicate that an input matrix of 300 proteins × 50,000 loci can be processed in ∼15 min on a single CPU with 10-Gbyte memory.
Bootstrapping
As reviewed elsewhere (Pe'er 2005), exact calculation of the posterior probability of a targeting interaction between two chromatin components is not feasible. We therefore used a previously described nonparametric bootstrapping strategy as an approximation (Friedman et al. 2000). We ran 1000 Banjo searches (of 300,000 cycles each), each time using as input a data matrix consisting of all 43 columns (chromatin components), but with 4380 rows selected by random sampling of the original 4380 rows (genes) with replacement. Thus, we computed 1000 networks, each based on slightly perturbed input data. These 1000 bootstrap runs required in total ∼10 h on a 1.83 GHz dual processor Apple Mac Mini. Next, for each possible pair of chromatin components, the occurrence of the corresponding directed edge in the 1000 Banjo networks was counted, yielding the bootstrap confidence score for the directed edge linking this pair of components. The 43 × 43 matrix of confidence scores is available as Supplemental Data Set 2.
Comparison to previously known interactions
In the absence of a gold standard to estimate the performance of our BNI approach, we used two different public databases as surrogate reference lists. First, we used the BioGRID database (Breitkreutz et al. 2008) release 2.0.45, which is a compilation of known physical and genetic interactions. BioGRID interactions among the 43 chromatin components are listed in Supplemental Data Set 5. Second, we systematically searched PubMed (version 28 October 2008) for the co-occurrence of pairs of chromatin components mentioned in either the abstract or the title of previous publications. Supplemental Data Set 5 lists the search terms used for each chromatin component, and the resulting co-citation frequencies for each pair. Supplemental Figure S1 shows the degrees of overlap between BN and either BioGRID interactions or PubMed co-citations, as a function of the bootstrap score cutoff applied.
Optimization of input profile discretization
The discretization of the binding profiles prior to BNI can be done in various ways, and this may affect the BNI outcome (Smith et al. 2002; Yu et al. 2004). In the absence of a gold standard, we reasoned that the most optimal discretization scheme should yield a BN with the highest possible overlap with BioGRID interactions and PubMed co-citations. We compared seven different discretization schemes. Four of these schemes used a quantile threshold to binarize the data into “target gene” (value 1) or “nontarget gene” (value 0) categories. We compared “target” definitions corresponding to the 90th–100th, 95th–100th, 98th–100th, and 99th–100th percentile of the binding log ratios for each chromatin component. In addition, three schemes employed discretization into three categories: “low” (value 0), “medium” (value 1), and “high” (value 2). Here, the ranges were either the 0th–33rd, 34th–65th, or 66th–100th percentile, the 0th–fifth, sixth–94th, and 95th–100th percentile, or the 0th–10th, 11th–89th, and 90th–100th percentile for each chromatin component. For all of these seven schemes we conducted BNI with 1000 bootstrapping cycles.
To compare these discretization schemes, we calculated “performance curves” that visualize the overlap with BioGRID or PubMed interactions for every possible bootstrap score cutoff (Supplemental Fig. S2). An example of such curves for binarized data based on the 95th percentile threshold (Supplemental Fig. S2A,B) shows that with an increasing bootstrap cutoff, fewer of the previously known interactions (listed in the public databases) are recovered, but at the same time a larger fraction of BNI-predicted interactions overlaps with previously known interactions, suggesting a higher specificity. Such a trade-off between sensitivity and specificity is typical of many prediction algorithms. Comparison of performance curves for the seven discretization schemes (Supplemental Figure S2C,D) indicates that binarization based on the 95th percentile threshold yields a BN with the most optimal correspondence to previously known interactions. We therefore concluded that this particular discretization scheme yields the most reliable prediction of targeting interactions. This binarization was also used for the definition of target and nontarget genes in the analyses shown in Figures 3–6.
Robustness of BNI
The topology of BN80 is meant to evolve as binding data will accumulate. In the ideal case, all the key components are already in the network, and the addition of new components will merely turn predicted direct interactions into indirect ones. However, it is also conceivable that the addition of a key targeting component will have dramatic effects on the whole topology of BN80. To assess the impact of nonavailable data on the topology of BN80, we took the following approach: Assuming that we started with less than 43 components, we studied the changes caused by progressively adding new information. Combinations of 1–5 chromatin components (nodes) were removed from the original data set (each time 50 random combinations, except for 1 node, where all 43 nodes were deleted individually), and new networks were constructed by BNI, again using a bootstrap confidence threshold of 80%. The change in overlap of these trimmed networks relative to the original 43-node network (Fig. 2) was calculated and plotted. To compute the scores, we defined the “target network” as the original BN80 where the given subset of nodes has been deleted, and the “common subnetwork” as the undirected intersection of the target network and the newly inferred network. The change in connection (Supplemental Fig. S4A) was defined as the ratio of the number of differing edges between the inferred network and the target network (i.e., present in one and absent from the other) to the number of edges of the target network. The change in orientation (Supplemental Fig. S4B) was defined as the ratio of the number of edges present in the inferred network and the target network, but with a different orientation (i.e., oriented in one and not in the other, or oriented in an opposite way) to the number of edges of the common subnetwork. In this analysis only 100 bootstraps were done for each BNI because 1000 bootstraps would have required > 3000 CPU hours in total. These results show that on average, for every added/deleted node, a change of connectivity occurs on ∼2% of the edges, and a change of orientation on ∼1%. This high robustness suggests that our current predictions on targeting interactions will likely remain valid when the BN80 is updated.
Gene expression analysis
Gene expression levels in Kc cells were taken from Pickersgill et al. (2006). Embryonic tissue gene expression data were taken from Tomancak et al. (2007). Enrichment of expressed genes among target gene sets (again defined as the top 5% genes ranked by binding ratio for each chromatin protein) was calculated using a two-sided binomial test.
Sequence motif analysis
We used GAGA as the consensus binding motif for GAF (van Steensel et al. 2003). The consensus motif for JRA was inferred from in vitro binding data (Perkins et al. 1988) as TG[A/C][C/G][A/T][C/A]A.
Some cautionary notes regarding BNI
The use of BNI to model biological regulatory systems is subject to inherent limitations. Concerning the data presented in this study, the following points deserve mention. First, many chromatin components are not yet represented in our compendium of binding profiles. Such “hidden variables” may affect both the presence and directionality of some predicted edges in the network. Only the generation of a complete set of binding profiles for all chromatin components will ultimately solve this issue. Second, the current implementation of our BNI approach does not detect inhibitory interactions (i.e., protein A locally prevents the binding of protein B). Third, the theoretical framework underlying BNI only allows for the modeling of acyclic directed graphs. As a result, loops of interactions (e.g., A→B→C→A) are always resolved as a different topology in the BN. While these artifacts raise the false positive and false negative rates of BNI, our computational and experimental evidence suggest that BN80 is strongly enriched in bona fide targeting interactions. As such it can be used as a guide to formulate useful and testable hypotheses.
Acknowledgments
We thank Elzo de Wit for preliminary analyses that inspired this work; Romeo Lascaris for initial studies of chromatin accessibility using Dam; the NKI microarray facility for microarray hybridizations; Jeff Delrow for providing cDNA arrays; Frauke Greil, Joachim Marhold, and Frank Lyko for the Dnmt2 profile; Peter Verrijzer for Brahma antibody; members of our labs for helpful suggestions. This work was supported by the Netherlands Genomics Initiative and an EURYI Award to B.v.S., and grant R01 ES014811 to T.I. from the NIH National Institute of Environmental Health Sciences.
Author contributions: B.v.S. and T.I. conceived and designed this study; B.v.S. conducted BNI and other computational analyses; U.B. performed all follow-up experiments, generated new DamID maps, and analyzed data; G.J.F. and M.C. did computational analyses that helped us with the interpretation of BNI results; and J.G.v.B. contributed DamID maps.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE15807.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.098822.109.
References
- Armstrong JA, Papoulas O, Daubresse G, Sperling AS, Lis JT, Scott MP, Tamkun JW. The Drosophila BRM complex facilitates global transcription by RNA polymerase II. EMBO J. 2002;21:5245–5254. doi: 10.1093/emboj/cdf517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barash Y, Friedman N. Context-specific Bayesian clustering for gene expression data. J Comput Biol. 2002;9:169–191. doi: 10.1089/10665270252935403. [DOI] [PubMed] [Google Scholar]
- Berger SL. The complex language of chromatin regulation during transcription. Nature. 2007;447:407–412. doi: 10.1038/nature05915. [DOI] [PubMed] [Google Scholar]
- Bianchi-Frias D, Orian A, Delrow JJ, Vazquez J, Rosales-Nieves AE, Parkhurst SM. Hairy transcriptional repression targets and cofactor recruitment in Drosophila. PLoS Biol. 2004;2:E178. doi: 10.1371/journal.pbio.0020178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brideau NJ, Flores HA, Wang J, Maheshwari S, Wang X, Barbash DA. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science. 2006;314:1292–1295. doi: 10.1126/science.1133953. [DOI] [PubMed] [Google Scholar]
- Cairns BR. Chromatin remodeling: Insights and intrigue from single-molecule studies. Nat Struct Mol Biol. 2007;14:989–996. doi: 10.1038/nsmb1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen PY, Popovich PM. Correlation: Parametric and nonparametric measures. Sage; Thousand Oaks, CA: 2002. [Google Scholar]
- Chen G, Fernandez J, Mische S, Courey AJ. A functional interaction between the histone deacetylase Rpd3 and the corepressor groucho in Drosophila development. Genes & Dev. 1999;13:2218–2230. doi: 10.1101/gad.13.17.2218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choksi SP, Southall TD, Bossing T, Edoff K, de Wit E, Fischer BE, van Steensel B, Micklem G, Brand AH. Prospero acts as a binary switch between self-renewal and differentiation in Drosophila neural stem cells. Dev Cell. 2006;11:775–789. doi: 10.1016/j.devcel.2006.09.015. [DOI] [PubMed] [Google Scholar]
- Cooper GF, Herskovits E. A Bayesian method for the induction of probabilistic networks from data. Mach Learn. 1992;9:309–347. [Google Scholar]
- Delattre M, Spierer A, Tonka CH, Spierer P. The genomic silencing of position-effect variegation in Drosophila melanogaster: Interaction between the heterochromatin-associated proteins Su(var)3-7 and HP1. J Cell Sci. 2000;113:4253–4261. doi: 10.1242/jcs.113.23.4253. [DOI] [PubMed] [Google Scholar]
- de Wit E, van Steensel B. Chromatin domains in higher eukaryotes: Insights from genome-wide mapping studies. Chromosoma. 2009;118:25–36. doi: 10.1007/s00412-008-0186-0. [DOI] [PubMed] [Google Scholar]
- de Wit E, Greil F, van Steensel B. High-resolution mapping reveals links of HP1 with active and inactive chromatin components. PLoS Genet. 2007;3:e38. doi: 10.1371/journal.pgen.0030038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit E, Braunschweig U, Greil F, Bussemaker HJ, van Steensel B. Global chromatin domain organization of the Drosophila genome. PLoS Genet. 2008;4:e1000045. doi: 10.1371/journal.pgen.1000045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djebbari A, Quackenbush J. Seeded Bayesian networks: Constructing genetic networks from microarray data. BMC Syst Biol. 2008;2:57. doi: 10.1186/1752-0509-2-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman N. Inferring cellular networks using probabilistic graphical models. Science. 2004;303:799–805. doi: 10.1126/science.1094068. [DOI] [PubMed] [Google Scholar]
- Friedman N, Linial M, Nachman I, Pe'er D. Using Bayesian networks to analyze expression data. J Comput Biol. 2000;7:601–620. doi: 10.1089/106652700750050961. [DOI] [PubMed] [Google Scholar]
- Giot L, Bader JS, Brouwer C, Chaudhuri A, Kuang B, Li Y, Hao YL, Ooi CE, Godwin B, Vitols E, et al. A protein interaction map of Drosophila melanogaster. Science. 2003;302:1727–1736. doi: 10.1126/science.1090289. [DOI] [PubMed] [Google Scholar]
- Gottschling DE. Telomere-proximal DNA in Saccharomyces cerevisiae is refractory to methyltransferase activity in vivo. Proc Natl Acad Sci. 1992;89:4062–4065. doi: 10.1073/pnas.89.9.4062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, van der Kraan I, Delrow J, Smothers JF, de Wit E, Bussemaker HJ, van Driel R, Henikoff S, van Steensel B. Distinct HP1 and Su(var)3-9 complexes bind to sets of developmentally coexpressed genes depending on chromosomal location. Genes & Dev. 2003;17:2825–2838. doi: 10.1101/gad.281503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greil F, Moorman C, van Steensel B. DamID: Mapping of in vivo protein-genome interactions using tethered DNA adenine methyltransferase. Methods Enzymol. 2006;410:342–359. doi: 10.1016/S0076-6879(06)10016-6. [DOI] [PubMed] [Google Scholar]
- Greil F, de Wit E, Bussemaker HJ, van Steensel B. HP1 controls genomic targeting of four novel heterochromatin proteins in Drosophila. EMBO J. 2007;26:741–751. doi: 10.1038/sj.emboj.7601527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiratani I, Ryba T, Itoh M, Yokochi T, Schwaiger M, Chang CW, Lyou Y, Townes TM, Schubeler D, Gilbert DM. Global reorganization of replication domains during embryonic stem cell differentiation. PLoS Biol. 2008;6:e245. doi: 10.1371/journal.pbio.0060245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan NJ, Chung S, Emili A, Snyder M, Greenblatt JF, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- Jaquet Y, Delattre M, Spierer A, Spierer P. Functional dissection of the Drosophila modifier of variegation Su(var)3-7. Development. 2002;129:3975–3982. doi: 10.1242/dev.129.17.3975. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, Leproust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2008;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim TH, Ren B. Genome-wide analysis of protein–DNA interactions. Annu Rev Genomics Hum Genet. 2006;7:81–102. doi: 10.1146/annurev.genom.7.080505.115634. [DOI] [PubMed] [Google Scholar]
- Lieb JD. Genome-wide mapping of protein–DNA interactions by chromatin immunoprecipitation and DNA microarray hybridization. Methods Mol Biol. 2003;224:99–109. doi: 10.1385/1-59259-364-X:99. [DOI] [PubMed] [Google Scholar]
- Loo LW, Secombe J, Little JT, Carlos LS, Yost C, Cheng PF, Flynn EM, Edgar BA, Eisenman RN. The transcriptional repressor dMnt is a regulator of growth in Drosophila melanogaster. Mol Cell Biol. 2005;25:7078–7091. doi: 10.1128/MCB.25.16.7078-7091.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mannervik M, Levine M. The Rpd3 histone deacetylase is required for segmentation of the Drosophila embryo. Proc Natl Acad Sci. 1999;96:6797–6801. doi: 10.1073/pnas.96.12.6797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marhold J, Kramer K, Kremmer E, Lyko F. The Drosophila MBD2/3 protein mediates interactions between the MI-2 chromatin complex and CpT/A-methylated DNA. Development. 2004;131:6033–6039. doi: 10.1242/dev.01531. [DOI] [PubMed] [Google Scholar]
- Missal K, Cross MA, Drasdo D. Gene network inference from incomplete expression data: Transcriptional control of hematopoietic commitment. Bioinformatics. 2006;22:731–738. doi: 10.1093/bioinformatics/bti820. [DOI] [PubMed] [Google Scholar]
- Mito Y, Henikoff JG, Henikoff S. Genome-scale profiling of histone H3.3 replacement patterns. Nat Genet. 2005;37:1090–1097. doi: 10.1038/ng1637. [DOI] [PubMed] [Google Scholar]
- Moorman C, Sun LV, Wang J, de Wit E, Talhout W, Ward LD, Greil F, Lu XJ, White KP, Bussemaker HJ, et al. Hotspots of transcription factor colocalization in the genome of Drosophila melanogaster. Proc Natl Acad Sci. 2006;103:12027–12032. doi: 10.1073/pnas.0605003103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morse RH. Transcription factor access to promoter elements. J Cell Biochem. 2007;102:560–570. doi: 10.1002/jcb.21493. [DOI] [PubMed] [Google Scholar]
- Mukherjee S, Speed TP. Network inference using informative priors. Proc Natl Acad Sci. 2008;105:14313–14318. doi: 10.1073/pnas.0802272105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Orian A, van Steensel B, Delrow J, Bussemaker HJ, Li L, Sawado T, Williams E, Loo LW, Cowley SM, Yost C, et al. Genomic binding by the Drosophila Myc, Max, Mad/Mnt transcription factor network. Genes & Dev. 2003;17:1101–1114. doi: 10.1101/gad.1066903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paroush Z, Finley RL, Jr, Kidd T, Wainwright SM, Ingham PW, Brent R, Ish-Horowicz D. Groucho is required for Drosophila neurogenesis, segmentation, and sex determination and interacts directly with hairy-related bHLH proteins. Cell. 1994;79:805–815. doi: 10.1016/0092-8674(94)90070-1. [DOI] [PubMed] [Google Scholar]
- Pearl J. Probabilistic reasoning in intelligent systems: Networks of plausible inference. Morgan Kaufmann; San Francisco, CA: 1988. [Google Scholar]
- Pe'er D. Bayesian network analysis of signaling networks: A primer. Sci STKE. 2005;2005:pl4. doi: 10.1126/stke.2812005pl4. [DOI] [PubMed] [Google Scholar]
- Pe'er D, Regev A, Elidan G, Friedman N. Inferring subnetworks from perturbed expression profiles. Bioinformatics. 2001;17:S215–S224. doi: 10.1093/bioinformatics/17.suppl_1.s215. [DOI] [PubMed] [Google Scholar]
- Perkins KK, Dailey GM, Tjian R. Novel Jun- and Fos-related proteins in Drosophila are functionally homologous to enhancer factor AP-1. EMBO J. 1988;7:4265–4273. doi: 10.1002/j.1460-2075.1988.tb03324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickersgill H, Kalverda B, de Wit E, Talhout W, Fornerod M, van Steensel B. Characterization of the Drosophila melanogaster genome at the nuclear lamina. Nat Genet. 2006;38:1005–1014. doi: 10.1038/ng1852. [DOI] [PubMed] [Google Scholar]
- Pile LA, Wassarman DA. Chromosomal localization links the SIN3–RPD3 complex to the regulation of chromatin condensation, histone acetylation and gene expression. EMBO J. 2000;19:6131–6140. doi: 10.1093/emboj/19.22.6131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pindyurin AV, Moorman C, de Wit E, Belyakin SN, Belyaeva ES, Christophides GK, Kafatos FC, van Steensel B, Zhimulev IF. SUUR joins separate subsets of PcG, HP1 and B-type lamin targets in Drosophila. J Cell Sci. 2007;120:2344–2351. doi: 10.1242/jcs.006007. [DOI] [PubMed] [Google Scholar]
- Poortinga G, Watanabe M, Parkhurst SM. Drosophila CtBP: A hairy-interacting protein required for embryonic segmentation and hairy-mediated transcriptional repression. EMBO J. 1998;17:2067–2078. doi: 10.1093/emboj/17.7.2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rando OJ. Global patterns of histone modifications. Curr Opin Genet Dev. 2007;17:94–99. doi: 10.1016/j.gde.2007.02.006. [DOI] [PubMed] [Google Scholar]
- Rando OJ, Ahmad K. Rules and regulation in the primary structure of chromatin. Curr Opin Cell Biol. 2007;19:250–256. doi: 10.1016/j.ceb.2007.04.006. [DOI] [PubMed] [Google Scholar]
- Schotta G, Ebert A, Krauss V, Fischer A, Hoffmann J, Rea S, Jenuwein T, Dorn R, Reuter G. Central role of Drosophila SU(VAR)3-9 in histone H3-K9 methylation and heterochromatic gene silencing. EMBO J. 2002;21:1121–1131. doi: 10.1093/emboj/21.5.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schübeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M. Genome-wide DNA replication profile for Drosophila melanogaster: A link between transcription and replication timing. Nat Genet. 2002;32:438–442. doi: 10.1038/ng1005. [DOI] [PubMed] [Google Scholar]
- Schübeler D, MacAlpine DM, Scalzo D, Wirbelauer C, Kooperberg C, van Leeuwen F, Gottschling DE, O'Neill LP, Turner BM, Delrow J, et al. The histone modification pattern of active genes revealed through genome-wide chromatin analysis of a higher eukaryote. Genes & Dev. 2004;18:1263–1271. doi: 10.1101/gad.1198204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith VA, Jarvis ED, Hartemink AJ. Evaluating functional network inference using simulations of complex biological systems. Bioinformatics. 2002;18:S216–S224. doi: 10.1093/bioinformatics/18.suppl_1.s216. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Steinfeld I, Shamir R, Kupiec M. A genome-wide analysis in Saccharomyces cerevisiae demonstrates the influence of chromatin modifiers on transcription. Nat Genet. 2007;39:303–309. doi: 10.1038/ng1965. [DOI] [PubMed] [Google Scholar]
- Tolhuis B, de Wit E, Muijrers I, Teunissen H, Talhout W, van Steensel B, van Lohuizen M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat Genet. 2006;38:694–699. doi: 10.1038/ng1792. [DOI] [PubMed] [Google Scholar]
- Tomancak P, Berman BP, Beaton A, Weiszmann R, Kwan E, Hartenstein V, Celniker SE, Rubin GM. Global analysis of patterns of gene expression during Drosophila embryogenesis. Genome Biol. 2007;8:R145. doi: 10.1186/gb-2007-8-7-r145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Steensel B. Mapping of genetic and epigenetic regulatory networks using microarrays. Nat Genet. 2005;37:S18–S24. doi: 10.1038/ng1559. [DOI] [PubMed] [Google Scholar]
- van Steensel B, Delrow J, Bussemaker HJ. Genomewide analysis of Drosophila GAGA factor target genes reveals context-dependent DNA binding. Proc Natl Acad Sci. 2003;100:2580–2585. doi: 10.1073/pnas.0438000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel MJ, Pagie L, Talhout W, Nieuwland M, Kerkhoven RM, van Steensel B. High-resolution mapping of heterochromatin redistribution in a Drosophila position-effect variegation model. Epigenetics Chromatin. 2009;2:1. doi: 10.1186/1756-8935-2-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wines DR, Talbert PB, Clark DV, Henikoff S. Introduction of a DNA methyltransferase into Drosophila to probe chromatin structure in vivo. Chromosoma. 1996;104:332–340. doi: 10.1007/BF00337221. [DOI] [PubMed] [Google Scholar]
- Yu J, Smith VA, Wang PP, Hartemink AJ, Jarvis ED. Advances to Bayesian network inference for generating causal networks from observational biological data. Bioinformatics. 2004;20:3594–3603. doi: 10.1093/bioinformatics/bth448. [DOI] [PubMed] [Google Scholar]
- Yu H, Zhu S, Zhou B, Xue H, Han JD. Inferring causal relationships among different histone modifications and gene expression. Genome Res. 2008;18:1314–1324. doi: 10.1101/gr.073080.107. [DOI] [PMC free article] [PubMed] [Google Scholar]