Abstract
In eukaryotic cells, chromatin reorganizes within promoters of active genes to allow the transcription machinery and various transcription factors to access DNA. In this model, promoter-specific transcription factors bind DNA to initiate the production of mRNA in a tightly regulated manner. In the case of the human malaria parasite, Plasmodium falciparum, specific transcription factors are apparently underrepresented with regards to the size of the genome, and mechanisms underlying transcriptional regulation are controversial. Here, we investigate the modulation of DNA accessibility by chromatin remodeling during the parasite infection cycle. We have generated genome-wide maps of nucleosome occupancy across the parasite erythrocytic cycle using two complementary assays—the formaldehyde-assisted isolation of regulatory elements to extract protein-free DNA (FAIRE) and the MNase-mediated purification of mononucleosomes to extract histone-bound DNA (MAINE), both techniques being coupled to high-throughput sequencing. We show that chromatin architecture undergoes drastic upheavals throughout the parasite's cycle, contrasting with targeted chromatin reorganization usually observed in eukaryotes. Chromatin loosens after the invasion of the red blood cell and then repacks prior to the next cycle. Changes in nucleosome occupancy within promoter regions follow this genome-wide pattern, with a few exceptions such as the var genes involved in virulence and genes expressed at early stages of the cycle. We postulate that chromatin structure and nucleosome turnover control massive transcription during the erythrocytic cycle. Our results demonstrate that the processes driving gene expression in Plasmodium challenge the classical eukaryotic model of transcriptional regulation occurring mostly at the transcription initiation level.
With up to 1 million deaths per year (Snow et al. 2005), malaria remains one of the most deadly infectious diseases in the world. The absence of a vaccine and the spread of parasite multiple drug resistances hasten the need to better comprehend the parasite biology to identify new anti-malarial strategies. The human malaria parasite, Plasmodium falciparum, is a unicellular eukaryote with a complex life cycle, which includes an asexual erythrocytic stage associated with morbidity and mortality of the human host. Despite nearly a decade of investigation into the 23-Mb Plasmodium genome (Gardner et al. 2002), more than half of its approximately 5500 predicted genes are still annotated as “hypothetical,” and most of the promoter structures and putative regulatory regions remain indeterminate. In addition, the mechanisms driving gene expression are poorly understood. Although genome-wide gene expression analyses have shown remarkable variations of steady-state mRNA levels with a tightly regulated cascade of transcripts throughout the parasite life cycle (Bozdech et al. 2003; Le Roch et al. 2003), only a handful of transcription factors have been identified to date (Coulson et al. 2004; Balaji et al. 2005; Llinas et al. 2006). Moreover, recent studies have demonstrated that the parasite has a limited capacity to regulate the expression of its genes in response to metabolic stresses (Ganesan et al. 2008; Le Roch et al. 2008). Altogether, these findings present an enigma as to how gene expression is regulated with regards to the paucity of transcription factors and the apparent absence of gene feedback circuits. A key to this riddle could come from alterations of chromatin structure that are known to modulate DNA accessibility to DNA-binding proteins and thus regulate, to some extent, gene expression in eukaryotic cells. The presence of histones, as well as the nucleosome structure of chromatin in P. falciparum, were evidenced many years ago (Cary et al. 1994). Gene homologs coding for the core histones are present in the genome, and their transcript and protein levels vary during the parasite life cycle (Le Roch et al. 2004). A recognizable homolog of the linker histone H1, however, is absent from the parasite's genome with the expected consequence that the intensely packed chromosome as it usually appears in metaphase never occurs in Plasmodium. In Plasmodium, chromatin remodeling is believed to play a role in the mutually exclusive expression of genes responsible for antigenic variation, the var genes family, at the surface of infected red blood cells (Deitsch et al. 2001; Duraisingh et al. 2005; Chookajorn et al. 2007; Lopez-Rubio et al. 2007; Epp et al. 2009). Nonetheless, the role of chromatin structure alterations in regulating Plasmodium transcripts in a genome-wide manner remains unknown.
To discover the role of chromatin remodeling in regulating parasite transcription, we applied both formaldehyde-assisted isolation of regulatory elements (FAIRE) (Nagy et al. 2003; Hogan et al. 2006; Giresi et al. 2007), to isolate active transcriptional regulatory elements, and micrococcal nuclease digestion of mononucleosomes (here called MNase-assisted isolation of nucleosomes, or MAINE) (Bernstein et al. 2004; Gupta et al. 2008; Kaplan et al. 2009) coupled to massively parallel sequencing to dynamically and concomitantly map protein-free and nucleosome-rich regions during P. falciparum’s erythrocytic cycle. Using these two independent and complementary assays, we explored the kinetics of changes in chromatin structure with regards to transcriptional activity and progression through the malaria infection cycle. Altogether, our results picture a system where global changes of chromatin structure control the progression of the human malaria parasite through its cell cycle.
Results
Using FAIRE-seq and MAINE-seq to study an AT-rich genome
To explore chromatin architecture changes during P. falciparum’s erythrocytic cycle, we used high-throughput sequencing (Illumina Genome Analyzer) to compare genomic regions enriched by FAIRE (isolates protein-free DNA) and MAINE (isolates histone-associated DNA) at seven different time points throughout the parasite erythrocytic cycle (Supplemental Fig. S1). Genomic DNA samples, prepared by FAIRE and MAINE, were collected immediately after invasion and with a 6-h increment throughout the erythrocytic cycle. During these experiments, more than 42 million of high-quality reads were uniquely mapped, with up to two mismatches, to P. falciparum genome (sequencing metrics and a chart describing the mapping procedure are given in Supplemental Fig. S2). Depths of base coverage in FAIRE-seq and MAINE-seq were expressed per million mapped reads (pmmr) throughout the present study. Using a reference composed of a mix of each FAIRE and MAINE sample (see Methods), we found that the sequencing can retrieve more than 90% of P. falciparum’s genome, with a nucleotide covered by at least one base (Supplemental Fig. S3). These results are consistent with the coverage obtained when in silico–generated 36-bp-long reads are mapped (allowing up to two mismatches) to the genome of P. falciparum (data not shown). Nonetheless, there is still a proportion of the genome that is undetected by both FAIRE- and MAINE-seq. A first explanation could be that nonhistone proteins are bound to certain regions of the genome stably enough that cross-linking occurs. A second explanation resides in the repetitive nature of certain regions that would lead to nonunique short sequenced reads and therefore their exclusion from the data set.
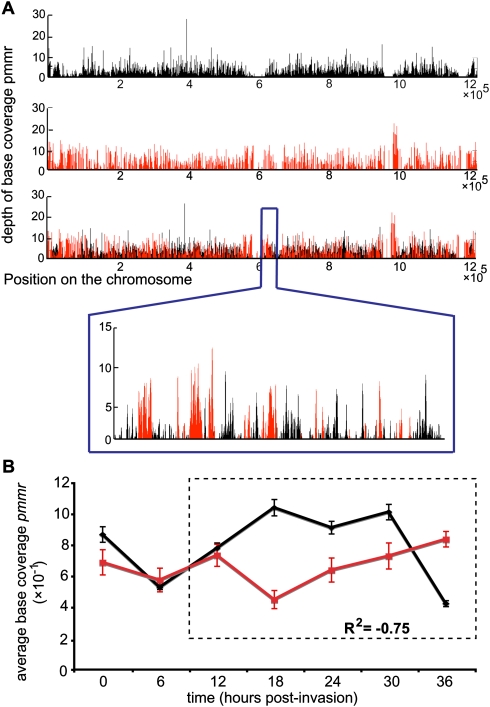
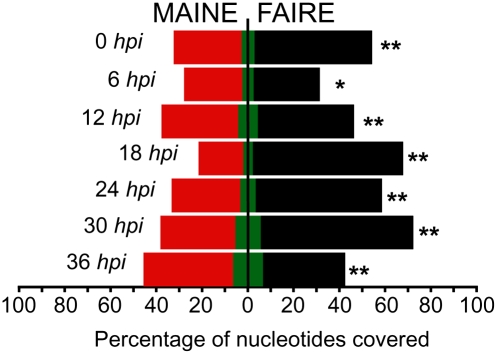
To validate the complementarity of the two methods, we compared the genome-wide distributions of mapped reads obtained with FAIRE-seq to the ones measured with MAINE-seq (Fig. 1). A visual examination of mapped reads distributions reveals that profiles of coverage for FAIRE-seq and MAINE-seq are inversely correlated: FAIRE shows high coverage where MAINE has low coverage and vice versa (Fig. 1A). To investigate objectively the extent of this inverse correlation, we compared the trends of coverage with FAIRE-seq (enrichment in protein-free DNA) to the trends of coverage with MAINE-seq (enrichment in nucleosome-bound DNA); Pearson's product-moment coefficient was computed between corresponding average base coverage pmmr across the seven investigated time points (Fig. 1B). The average correlation coefficient is strongly negative between 12 and 36 h post-invasion (hpi), indicating excellent complementation between FAIRE and MAINE (R2 = −0.75). To further investigate our data, we monitored the number of nucleotides that are actually covered by reads from FAIRE-seq, MAINE-seq, or both (hereafter “overlapping coverage”) (Fig. 2). The general genome coverage trends are consistent with our measure of the average depth of base coverage obtained previously (Fig. 1B); the least nucleotides are covered by MAINE-seq reads (red) at 18 hpi, while high nucleotide coverage is observed with FAIRE-seq (black). Overlapping coverage increases over the course of the cycle to reach a maximum (below 10% of the genome) at the latest time point. This observation is consistent with the progressive degradation of the parasites’ synchrony across time that is commonly observed in vitro. Finally, the genome is consistently more covered with FAIRE-seq than with MAINE-seq, which demonstrates a higher content in protein-free DNA. This latest observation indicates that the parasite's DNA contains more protein-free DNA than histone-bound regions and can be accessed by any type of DNA-binding proteins during most of the erythrocytic cycle, especially at the trophozoite and early schizont stages (18–30 hpi).
Figure 1.
FAIRE-seq and MAINE-seq are two complementary approaches to study nucleosome occupancy in P. falciparum. (A) Example of the complementarity between FAIRE-seq and MAINE-seq. We overlaid the coverages obtained for both FAIRE (black) and MAINE (red) at 18 hpi in chromosome 4. The chromosomal region [615,000; 635,000] is zoomed in. (B) Evaluation of the complementarity between FAIRE (black) and MAINE (red) at the genomic scale. For each time point, the average depth of base coverage pmmr in each chromosome was measured genome-wide for both FAIRE and MAINE (error bars, SD).
Figure 2.
Evolution of genome coverage across time. (Black) FAIRE; (red) MAINE. Overlapping coverage, i.e., the number of nucleotides that are covered by five reads and more in both FAIRE- and MAINE-seq, is shown in green. The genome is consistently more covered with FAIRE-seq, which demonstrates a high content in protein-free DNA, especially at 18 hpi. A t-test was performed and showed that the observed differences are significant, with P < 0.001 for all time points (**) except 6 hpi, where P < 0.01 (*). Overlapping coverage increasing at the latest time point is consistent with a progressive degradation of the parasites synchrony across time.
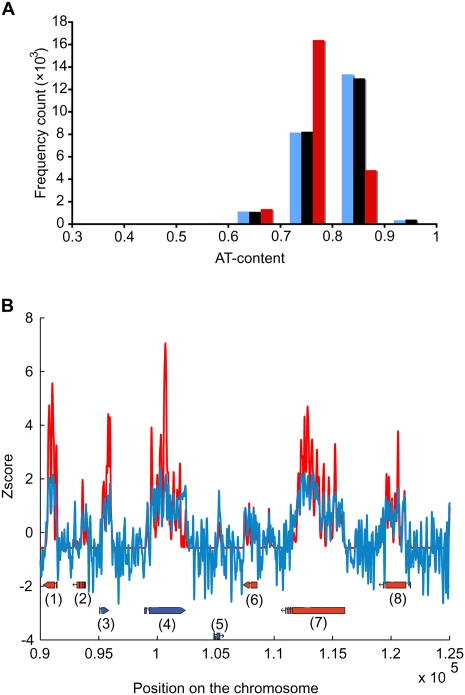
We verified that this abundance of protein-free regions was not due to an artefact caused by the extreme AT-richness of the parasite genome. First we verified the formaldehyde treatment efficiency by PCR amplification of potentially cross-linked regions (see Supplemental Methods; Supplemental Fig. S4). Then, we verified that the low complexity of AT-rich sequence has no effect on the base coverage; using a 1-kb-long non-overlapping sliding window approach (see Supplemental Methods), we calculated the AT-content in the P. falciparum genome and compared it to the AT-content within FAIRE- and MAINE-enriched regions (all time points averaged) (Fig. 3A). AT-content in FAIRE-covered regions is identical to the genome-wide one. Thus, AT-rich regions can be sequenced by the Illumina technology; the high AT-content of sequences enriched by FAIRE reflects the parasite's genome AT-content itself. On the contrary, MAINE-enriched regions have a lower AT-content, which is in agreement with lower nucleosome affinity for AT-rich regions (Field et al. 2008; Kaplan et al. 2009; Segal and Widom 2009a,b). This observation may suggest that AT-rich regions limit nucleosome binding on P. falciparum’s genome. To further explore this hypothesis, we examined the physical properties of the parasite's genomic DNA. We compared our experimental nucleosome landscape to in silico modeling of the sequence-based DNA elastic properties that determine the bendability of DNA around histones and is thought to drive nucleosome positioning in eukaryotic model organisms (Miele et al. 2008; Tolstorukov et al. 2008). An example of our results is given in Figure 3B. We found that our experimental nucleosome positioning correlates with the in silico predictions and demonstrates that the marked nucleosome preferences for regions with lower AT-content are biologically relevant. Altogether, our results demonstrate the suitability of both FAIRE-seq and MAINE-seq to study chromatin organization of the P. falciparum AT-rich genome.
Figure 3.
P. falciparum genome and its effect on nucleosome positioning. (A) AT-content distributions across the genome, FAIRE-enriched and MAINE-enriched regions were compared. AT-content was calculated in a 1000-bp-long sliding window for the P. falciparum genome (blue) and regions covered by FAIRE- and MAINE-seq (black and red, respectively). AT-content is similar genome-wide and in FAIRE-covered regions. On the contrary, regions enriched by MAINE are influenced toward less AT-rich regions, which is in agreement with the low affinity of nucleosomes for AT-rich sequences. Same distributions were obtained using sliding windows of various sizes (10, 50, 100, 1000, and 20,000 bp) (data not shown) further validating the efficiency of Illumina technology for the P. falciparum AT-rich genome. (B) Correlation between experimental and in silico nucleosome positioning was explored. MAINE profile (red) and DNA bending free energy profile (blue) were z-normalized and overlaid for the region [90,000–125,000] of chromosome 1. Oriented boxes represent gene models retrieved from http://www.plasmodb.org: (1) PFA0095c; (2) PFA0100c; (3) PFA0105w; (4) PFA0110w; (5) PFA0115w; (6) PFA0120c; (7) PFA0125c; (8) PFA0130c. Our results show that the energy profile correlates well with our experimental nucleosome landscape (average correlation coefficient = 0.65, measured using a 10-kb-long nonoverlapping sliding window approach), indicating that DNA bending properties dictates, at least partially, the parasite nucleosome positioning.
Chromatin remodeling occurs massively at the genomic scale
We observed that, in average, the parasite's DNA contains more protein-free DNA than histone-bound regions during most of the erythrocytic cycle. A similar trend is observed when chromosomes are considered individually (Supplemental Fig. S5). Several regions with relatively high coverage are observed with FAIRE-seq at ring stage, indicating insulated open chromatin. At trophozoite stage (18 hpi), the coverage with FAIRE-seq is uniformly deeper, showing completely loosened chromatin. Only a few regions, including telomeric/subtelomeric regions, have lower coverage, which could reflect the repetitive nature of these regions and therefore the limited capacity of mapping short reads to a single location in the genome. Finally, in all chromosomes, the intensity of coverage drastically drops at late schizont (36 hpi). Conversely, in MAINE-seq, the intensity of coverage reaches a minimum at trophozoite stage and a maximum at late schizont (data not shown). Altogether, these results demonstrate that the parasite's chromatin undergoes drastic changes at the early trophozoite and schizont stages of the parasite infectious erythrocytic cycle. Our observations suggest a simple model in which the chromatin has a maximal “open” state at the early trophozoite stage, and packs into a relatively “closed” state when parasites are ready for the next invasion. Interestingly, this pattern is also observed in the apicoplast genome. The apicoplast is a plastid-like organelle found in apicomplexan parasites that is homologous to the chloroplast in plants and essential for the parasite's life cycle (Goodman and McFadden 2007). While the mitochondrial DNA is entirely covered with FAIRE-seq (consistent with the fact that mitochondria lack histones) (Supplemental Fig. S6A), the apicoplast DNA show reduced FAIRE-seq coverage in ring and late schizont stages (Supplemental Fig. S6B), consistent with the presence of histone-like proteins.
Dynamic nucleosome mapping
To explore nucleosome position changes over time, the center of a nucleosome was positioned on every local maximum of MAINE-seq coverage for each time point. Since MAINE-seq enrichment at a particular locus correlates with the presence of a given nucleosome in a population of parasites, a well-positioned nucleosome will lead to a MAINE-seq peak stronger than a floating nucleosome would display. With regards to this statement, we scored the positioning of every nucleosome using the total number of reads per base involved in the nucleosome (i.e., the sum of coverage on the 147 bp around the local maximum).
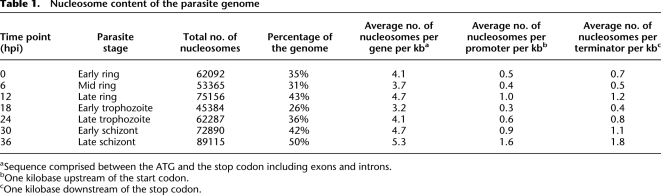
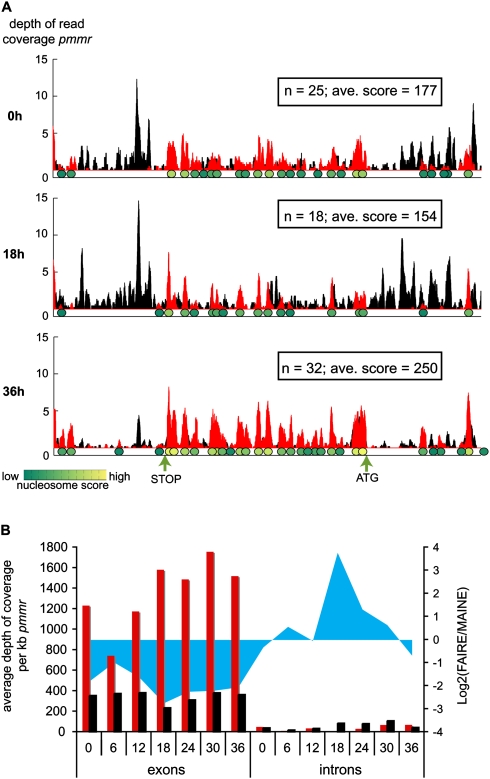
We identified about 45,000 (18 hpi) to 89,000 (36 hpi) nucleosomes genome-wide (Table 1; Supplemental Table S1), which represents 26%–50% of the genome, respectively. Figure 4A shows an example of the nucleosome content for the genomic region of the PFD0470c gene (putative replication factor A protein). Nucleosomes were detected inside the open reading frame (ORF) with a clear enrichment of nucleosomes at 36 hpi (32 nucleosomes in the considered region with an average score of 250 vs. 18 nucleosomes with an average score of 154 at 18 hpi). These observations were verified on the whole-genome scale. The average nucleosome occupancy was recorded within 1 kb upstream of the start codon of genes, within genes themselves, and 1 kb downstream of genes (Table 1). In agreement with recent results (Andersson et al. 2009; Schwartz et al. 2009; Tilgner et al. 2009), we detected an unambiguous preference of nucleosomes for coding regions. Nonetheless, variations in the number of nucleosomes over time have similar trends genome-wide. Three distinct phases can be delineated (Table 1). First, a significant density of nucleosomes is observed at ring stage (0–12 hpi). Then, a massive depletion is detected at the early trophozoite stage (18 hpi). Finally, nucleosomes are repositioned, gradually across time from 24–36 hpi (absolute maximum at 36 hpi). These observations could indicate that histones go through cycles of binding/unbinding DNA rather than sliding along the DNA strand.
Table 1.
Nucleosome content of the parasite genome
aSequence comprised between the ATG and the stop codon including exons and introns.
bOne kilobase upstream of the start codon.
cOne kilobase downstream of the stop codon.
Figure 4.
Nucleosome landscape of P. falciparum throughout the erythrocyctic cycle. (A) Example of nucleosome mapping for the region [457,031, 464,000] of chromosome 4. Coverages with FAIRE-seq (black) and MAINE-seq (red) are overlaid. The ovals under MAINE-seq peaks are nucleosomes, colored according to their score from green (low score) to yellow (high score). (B) Comparison of chromatin status within exons and introns with regards to coverage with FAIRE-seq and MAINE-seq (black and red bars, respectively, primary y-axis) and the degree of chromatin loosening [i.e., log2(FAIRE/MAINE); blue area, secondary y-axis].
To clarify the positioning of histones inside genes, the depths of read coverage per kilo-base pair pmmr were monitored in all exons and introns of the genome (Fig. 4B). As expected, exons exhibit a high MAINE-seq coverage (up to four times more coverage than FAIRE-seq), indicating high nucleosome occupancy throughout the erythrocytic cycle. This observation is confirmed by the measurement of the relative degree of chromatin opening (measured as the ratio between FAIRE, the protein-free DNA, and MAINE, the histone-bound DNA, given in log2 scale) that shows intense packaging during the entire cycle with little variations. With regards to introns, the situation is reversed and sequence enrichment is higher with FAIRE-seq. Chromatin is relatively open in introns during the entire erythrocytic cycle but the late schizont stage (36 hpi). Strikingly, chromatin seems to reach a maximum degree of opening at early trophozoite stage (18 hpi) where coverage with MAINE-seq is almost none, which is consistent with the observed genome-wide changes.
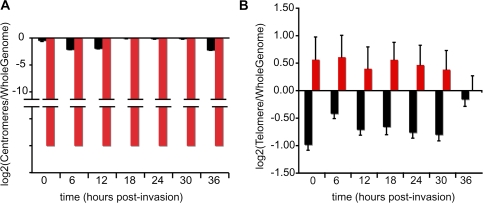
Finally, we examined the nucleosome landscape in centromeric regions. So far, identification of candidate centromeric regions has been limited to 11 of the 14 P. falciparum chromosomes (indicated by arrows in Supplemental Fig. S5) and defined as regions of ∼2 kb with extremely high (A+T) content (>97%) and imperfect short tandem repeats (Bowman et al. 1999; Kelly et al. 2006). Comparison of sequence enrichment with FAIRE and MAINE shows that centromeric regions exhibit a distinct pattern of coverage (Fig. 5A), i.e., low coverage with FAIRE-seq and no coverage at all with MAINE-seq. We verified that the low/nondetection of centromeric regions by both methods was not the consequence of the repetitive nature of these regions combined to our choice to omit nonunique reads; we aligned nonunique reads to the parasite's genome, the choice of the final location being randomly picked. We confirmed that centromeres are depleted in reads (data not shown). The fact that centromeres are not identified by both techniques is nonetheless consistent with the presence of nonhistone centromere-binding proteins. We used this pattern of low coverage to propose the identification of putative uncharacterized centromeric regions for chromosomes 10, 11, and 14. We determined scanning criteria from the 11 existing centromeres based on their AT-content and their FAIRE and MAINE coverages (see Methods). By applying blindly our method on all chromosomes, we were able to find all previously described centromeres and to discover new putative centromeric regions in chromosomes 10, 11, and 14, ranging from 2 kb to 3.5 kb in length (Supplemental Table S2).
Figure 5.
FAIRE-seq and MAINE-seq coverage in centromeric and telomeric/subtelomeric regions. Mean depths of read coverage in centromeres (A) and telomeres (B) were compared with the mean depth of read coverage genome-wide (MAINE-seq, red; FAIRE-seq, black) and displayed as a log2 scale. (A) Centromeres are depleted in nucleosomes relative to the rest of the genome. (B) Nucleosomes are consistently more abundant in telomeres than in the rest of the genome. Coding regions of the telomeres are densely packed compared with possible regulatory regions (classically located outside the genes) and other noncoding regions.
Chromatin status and parasite virulence
The subtelomeric regions are known to play a role in parasite virulence. Indeed, subtelomeric regions contain genes that code for adhesion molecules such as those of the var gene family that consists of about 60 members. var genes code for P. falciparum erythrocyte membrane protein 1 (PfEMP1) members that are expressed one at a time at the surface of infected erythrocytes, the other members being transcriptionally silent. PfEMP1 proteins are responsible for antigenic variation, allowing the parasite to escape from the host immune system and to adhere to the microvasculature, resulting in severe disease outcome (Ralph and Scherf 2005; Pasternak and Dzikowski 2009). In telomeric and subtelomeric regions, our data show an overenrichment with MAINE-seq in comparison with the rest of the genome, in particular at 18 hpi, while nucleosome content is minimal in the rest of the genome and with the exception of 36 hpi (Fig. 5B). In order to clarify the distribution of this enrichment, coverage distributions in subtelomeric regions were dissected at the gene level (Supplemental Fig. S7). For both FAIRE-seq and MAINE-seq, and consistently with our genome-wide observations described in the previous section, enrichment is linked to the presence of genes: The coverage with MAINE-seq is denser inside the ORFs, whereas the coverage with FAIRE-seq localizes within the surrounding noncoding regions.
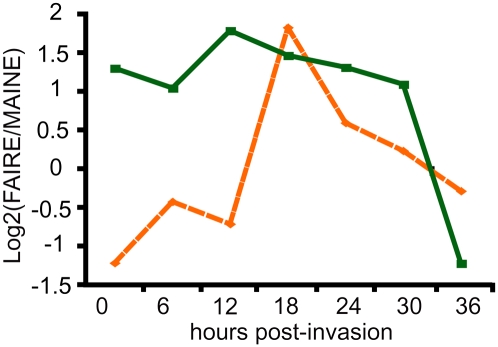
We further explored the chromatin status within putative promoter regions of var genes (up to 1.5 kb upstream of the ATG) and their intron from which a role in allelic exclusion has been proposed (Deitsch et al. 2001; Calderwood et al. 2003; Dzikowski et al. 2007); var introns are believed to pair with the promoter to promote gene silencing. We monitored the distributions of FAIRE and MAINE reads within putative promoter regions from all var genes and grouped these regions according to their profiles of nucleosome-free regions occupancy obtained with FAIRE-seq (FAIRE-seq is an indicator of the presence/absence of any tightly DNA-bound histone and nonhistone protein, but the binding of other proteins such as transcription factors is rather transient and unlikely to be detected). We delineated three groups (Supplemental Fig. S8A). The first group contains 18 genes with high density of protein-free DNA within 500 bp upstream of the ATG, the second group (three genes) shows high coverage in the more distal part of the promoter, and the third group has only low coverage (low DNA availability, which is consistent with the generally silent state of var genes in vitro). These observations suggest that regulatory sequences may be present within relatively close proximity to the ATG. With regards to introns, they are covered by FAIRE-seq on their whole length, i.e., 896 bp ± 175 bp in average, indicating no distance preference (Supplemental Fig. S8B). Then we measured the relative degree of chromatin loosening within introns and proximal promoters (ratio FAIRE over MAINE) (Fig. 6). With regards to promoter regions, the general genomic pattern of chromatin changes was observed (orange curve, dashed line), which do not follow the characteristic expression pattern for var genes. Introns (green curve), however, show a distinct trend; chromatin status is relatively loose from the ring stage through the early schizont stage and then rapidly packed at 36 hpi. Such a pattern was not found when coverage from random introns was considered (Supplemental Fig. S9), suggesting that the observed profile for the introns is specific to var genes. Finally, this pattern follows the general expression profiles for var genes. Altogether, these data support a regulatory role of var introns in the mutually exclusive transcription of var genes.
Figure 6.
Chromatin status changes in putative promoter regions and introns for var genes. Chromatin status changes in proximal promoter (orange) are compared to those of the corresponding introns (green).
We also investigated other gene families that are involved in virulence and are located in subtelomeric regions; we explored the chromatin status within putative promoter regions of rifin, stevor, and Maurer's cleft PfMC-2TM genes. Little variations were observed throughout the parasite erythrocytic stages with a general low coverage in FAIRE-seq and MAINE-seq (data not shown). These results are not surprising, considering that most of these genes seem to be heavily down regulated in vitro (Le Roch et al. 2003; Daily et al. 2005). Further investigation of the chromatin status in vivo will be required to understand the transcriptional regulation of these particular gene families.
Chromatin structure vs. gene expression
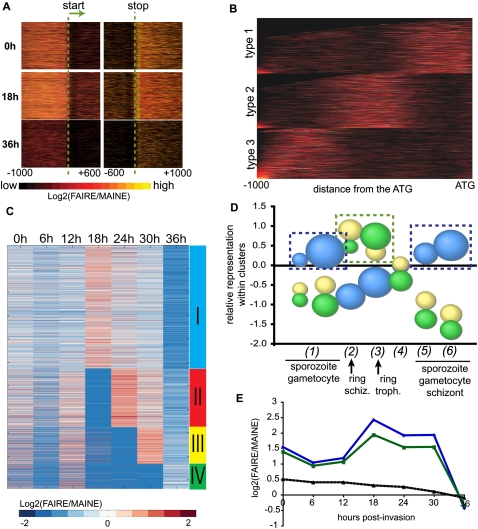
Relatively little is known about specific mechanisms that the parasite employs to control gene expression. To determine genome-wide distribution of coverage inside ORFs and their putative regulatory regions, read distributions for both FAIRE and MAINE were examined within 1600 bp with position zero centered on the start codon (range [−1000;+600]) and with position zero centered on the stop codon (range [−600;+1000]) (Fig. 7A). We monitored the relative degree of chromatin opening across time (ratio between FAIRE and MAINE coverages). For all considered time points, regions upstream and downstream the ORFs are “open,” whereas regions inside ORFs are compacted into nucleosomes (Fig. 7A), which shows that putative promoter and terminator regions are predominantly histone-free and can be accessed by a wide variety of DNA-binding proteins. The pattern of chromatin structure changes for these particular regions follows the general chromatin changes that we observe at the genome scale.
Figure 7.
Chromatin loosens in promoter regions at the trophozoite stage and massive transcription occurs. (A) Degree of chromatin opening for regions [−1000; +600] around the start codon and [−600; +1000] the stop codon [measured by log2(FAIRE/MAINE) and color coded from low (black) to high (yellow) opening]. Each line on the y-axis represents one gene. (B) K-means clustering of 1-kb-long regions upstream of the ATGs according to the localization of highly FAIRE-enriched regions. Three groups of genes are defined according to the distance of the putative promoter from the ATG. (C) K-mean clustering of chromatin status profiles within putative promoters. Four clusters are defined. (D) Enrichment of three of our four clusters with the previously published functional clusters (Le Roch et al. 2003). The size of the bubbles reflects the significance of the overrepresentation or underrepresentation (hypergeometric test). Biggest values reflect highest significance (inverse of the P-value). Our cluster I (blue bubbles) contains a high proportion of genes expressed at sporozoite and gametocyte stages and at the schizont stage. In clusters III and IV (yellow and green bubbles, respectively), genes from ring stage are overrepresented. (E) Chromatin opening profiles [log2(FAIRE/MAINE)] in the 9-bp consensus DNA motif found in the promoters of the putative targets for PFF0200c (blue) and PF14_0633 (green). The dashed black curve represents the trend obtained using random 9-bp-long stretches of nucleotides found in 200 random promoter regions genes.
We then investigated the role of chromatin structure in putative promoters in regulating parasite transcription. Typically, promoters are nucleosome-free regions located at various positions within 1 kb upstream of ATGs. Such regions should show higher enrichment with FAIRE-seq in a general manner (i.e., all time points combined). We thus optimized the locations and sizes of putative promoters relative to the ATG; we considered the 1-kb-long regions upstream of all ATGs and grouped them according to the presence of peaks and valleys of read coverage obtained with FAIRE-seq (average read coverage per base over time for each position within the 1-kb-long region). Using k-means clustering, we delineated three types of promoters (Fig. 7B): Type 1 has higher FAIRE-seq enrichment in the first 400 bp upstream of the ATG, type 2 in the 400 bp ranging from 650–250 bp upstream of the ATG, and type 3 in the last distal 400 bp. For each gene, we used the putative promoter regions defined according to its type for the rest of the analysis. We profiled the changes in chromatin status in putative promoter regions by measuring the relative degree of opening at each time point. K-means clustering was used to group genes according to the variations in their chromatin status across time. Four clusters were found, numbered I–IV (for the list of clustered genes). Hierarchical ordering of these clusters (see Supplemental Table S3; Fig. 7C) evoke the waves of expression throughout the erythrocytic cycle that were observed in previous microarray studies (Bozdech et al. 2003; Le Roch et al. 2003). The majority of the genes behave similarly: Promoters are closed at ring stage (0–12 hpi) and then open at the early trophozoite stage (18 hpi, cluster I). This enrichment in open chromatin fades with time as the parasite progresses through its erythrocytic cycle. In clusters II–IV, scattered groups of genes have promoters enriched in protein-free DNA at the ring stage. They are likely actively transcribed during the ring stage (Fig. 7C). In addition, promoters from genes in clusters II and III reopen at the late trophozoite (24 hpi) and early schizont (30 hpi) stages, respectively. In clusters I–III, all of the possible promoter regions are enriched in more compact structure at the late schizont stage (36 hpi). Genes belonging to cluster IV behave a little differently. Their promoters slightly open at ring stage and then pack at trophozoite stage to slightly reopen at 36 hpi. These observations may suggest that promoters that are open at ring stage could be regulated in a time-dependent manner. Then, the majority of the promoters could be activated at the trophozoite stage and slowly inactivated through the late trophozoite and schizont stages.
The composition of these four clusters was analyzed with regards to the 15 functional clusters (here designed at FC1–FC15) previously published based on mRNA level quantification (Le Roch et al. 2003). Le Roch et al. (2003) established that: (1) genes from FC1–FC3 are expressed in sporozoites and gametocytes; (2) genes from FC4 are corresponding to ring and schizont stages; (3) FC5–FC7 contain ring and trophozoite genes; (4) FC8–FC13 are devoted to trophozoite genes; (5) FC14 contains a mix of sporozoite, gametocyte, and schizont genes; and (6) FC15 contains schizont stage genes, in particular genes involved in the invasion of red blood cells. We performed our analysis considering these six master groups. Each of the 15 functional clusters was represented within our four groups. Nevertheless, certain functional clusters seem to be differentially represented (Fig. 7D). FC2, FC3, FC14, and FC15 are overrepresented in cluster I whereas members of FC4 abound in cluster III and FC5/FC6 are significantly overrepresented in cluster IV. Cluster II did not show any significant enrichment in any of the FCs. For clarity, the graphical display of the results for this cluster is proposed separately in Supplemental Figure S10. According to the annotation of the functional clusters (Le Roch et al. 2003), cluster I is enriched in sporozoite and gametocyte stage specific genes (respectively, the exoerythrocytic mosquito stage and sexual form of P. falciparum), whereas cluster III is enriched in genes expressed at the ring and late trophozoite/early schizont stages, and cluster IV is enriched in ring and trophozoite specific genes. In addition to the exoerythrocytic stages previously mentioned, cluster I is enriched in housekeeping genes, genes involved in DNA replication and cell cycle control, antigenic variation (rifin and stevor genes), as well as in most genes previously found in FC15 that have high steady-state mRNA levels during the schizont stage and mainly involved in invasion. This latest observation is in apparent contradiction with the classical model of chromatin remodeling upon transcriptional activation of invasion genes occurring at the schizont stage. On the contrary, in the case of ring-expressed genes, the dynamics of chromatin seems to fit with gene expression kinetics. To further validate these results, we performed a gene ontology analysis for each of our four clusters (the Gene Ontology [GO] term association was evaluated with an hypergeometric test, the threshold for significance being set to 0.0001). Cluster I is enriched with various DNA- and RNA-related processes (e.g., DNA replication and chromosome cycle, mitotic cell cycle, DNA metabolism, mRNA polyadenylation, tRNA metabolism), with genes involved in sexual development, and with sporozoite-specific genes. In addition to these classical GO terms, cluster I is significantly enriched in parasite-customized GO terms (see Zhou et al. 2008) such as “antigenic variation,” “cytoadherence,” “merozoite development,” or “protease activity” commonly associated with invasion and virulence. These results are in agreement with our previous observations.
To further investigate the possible regulatory events that occur at the late stages of the erythrocytic cycle, we analyzed the consensus DNA binding motif (9-bp-long) coverage profile of PF14_0633 and PFF0200c, two members of the Apicomplexan AP2 (ApiAP2) protein family (Balaji et al. 2005; De Silva et al. 2008). Very recently, these proteins were identified as putative P. falciparum transcription factors of late-stage genes (Balaji et al. 2005; De Silva et al. 2008). We compared the profiles of coverage within the 9 bp of their proposed targets to the profiles of 9-bp-long stretches of DNA randomly chosen within 200 random promoter regions. Both PFF0200c and PF14_0633 targets have similar coverage profiles within their consensus sequence (Fig. 7E), i.e., a peak of coverage occurs at the trophozoite stage (18–24 hpi), which are different from the profile of random sequences (dashed line) but follow the genome-wide pattern of chromatin remodeling. This observation seems to support the hypothesis of a general regulatory function for these predicted consensus DNA motifs. As a whole, these observations support a model where the vast majority of the promoters are activated at the trophozoite stage, certain sets of genes (i.e., ring or var genes) being subjected to finer regulation.
Discussion
We have analyzed the dynamic remodeling of nucleosome occupancy in P. falciparum along the erythrocytic cycle using a combination of FAIRE and MAINE coupled with high-throughput sequencing. We showed that both techniques are complementary and allow the quantification de novo of the relative enrichment in open (FAIRE-seq) or closed (MAINE-seq) chromatin for different regions of the genome. We demonstrated that the techniques are efficient for the study of the AT-rich genome of P. falciparum. We also verified that our experimental determination of nucleosome positioning is in agreement with predictions made using the DNA sequence as unique input. Given these observations, the particular AT-richness of intergenic regions in the parasite genome could serve as intrinsic signals for nucleosome positioning.
The application of these techniques on the P. falciparum genome clearly demonstrates that nucleosome distribution is restricted to coding regions. This observation is consistent with three recent analyses of nucleosome distribution in human, worms, and flies demonstrating a marked preference of nucleosomes for exons (Andersson et al. 2009; Schwartz et al. 2009; Tilgner et al. 2009). These investigators link these observations to the suspected coupling that occurs between transcription and splicing in eukaryotes nucleosome positioning being determinant for exon selection. Our results show that P. falciparum has the same features as those higher eukaryotes. However, due to the extreme AT-richness of the parasite's intergenic regions, histone depletion in noncoding regions seems to be more pronounced; this may create an extreme illustration of a eukaryotic nucleosome landscape. In addition, Plasmodium also differs from common eukaryotes in the intensity of its DNA packing. Indeed, no recognizable homolog of the linker histone H1 can be found in Plasmodium’s genome, and the intensely packed metaphase chromosome is not expected during the parasite's infection cycle. Finally, our results revealed that a maximum of 50% of the genome was found to be occupied by nucleosomes (at the late schizont stage), a value much lower than the 81% found in yeast (Lee et al. 2007). In their study, Lee et al. (2007) identified more than 1 million well-positioned nucleosomes (and as much floating nucleosomes) in the yeast genome, whereas less than 100,000 nucleosomes occupy Plasmodium’s genome at the late schizont stage (Saccharomyces cerevisiae’s and Plasmodium’s genomes are similar in size). As nucleosome occupancy is thought to be dictated by DNA sequence itself (Kaplan et al. 2009), we hypothesize that the very high (AT)-content of Plasmodium’s noncoding regions confers rigidity to the DNA and hinders the double helix from wrapping around histones. This hypothesis is consistent with the in silico prediction of P. falciparum nucleosome landscape and explains the differences with the yeast nucleosome landscape.
A drastic genome-wide nucleosome loss is observed at 18 and 24 hpi, when DNA replication and active transcriptional activity occur (Bozdech et al. 2003; Le Roch et al. 2003). This finding is consistent with the dip in histone abundance that has been previously observed at the trophozoite stage (Le Roch et al. 2004). The investigators described a loss of more than 85% of histones levels between ring and trophozoite stages. This dip is consistent with previous Plasmodium proteome analysis that demonstrated a highly connected interaction network between proteins involved in chromatin and ubiquitin metabolism (LaCount et al. 2005).
The pattern of chromatin remodeling was also observed for the apicoplast DNA. To our knowledge, our results provide the first indications of structural modifications of P. falciparum’s apicoplast DNA throughout the erythrocytic cycle. The presence of a few MAINE-enriched regions of apicoplast DNA is consistent with the recent characterization of a nuclear encoded bacterial histone-like protein PfHU, which plays a role in the compaction of the circular apicoplast DNA (Ram et al. 2008). PfHU was shown to bind preferably to supercoiled DNA in a nonsequence-specific manner. Such binding is sometimes associated with reduction of supercoils, known to affect DNA replication and transcription. In bacteria, positive supercoils could be relaxed by wrapping the DNA around histone-like octamers (Kar et al. 2006). These considerations suggest that transcription and replication in the apicoplast may be modulated by DNA architecture.
Altogether, our results are consistent with a model of global and intense changes in chromatin structure throughout the erythrocytic cycle. In this model, the chromatin opens at the early erythrocytic stage, reaching a maximum at the trophozoite stage, and an intense packaging at the late schizont stage. Such massive structural changes at the genomic scale are not found in other eukaryotes. Classically, chromatin remodeling is only observed in transcribed regions and is associated with the displacement and repositioning of one or two nucleosomes in the promoter rather than broader, region-wide, changes (Hogan et al. 2006; Lee et al. 2007). On the whole, our observations suggest that the situation in Plasmodium could represent an extremely reduced and simplified version of the eukaryotic transcriptional device, where the AT-content of the genome itself could be a major regulator. The relative degree of chromatin opening in putative promoter regions genome-wide was compared to previously published mRNA steady-state level measurements (Bozdech et al. 2003; Le Roch et al. 2003). Our results confirm that the cascade of mRNA steady-state levels observed during the parasite's erythrocytic cycle (Bozdech et al. 2003; Le Roch et al. 2003, 2004) cannot be explained by fine chromatin structure remodeling (more than half of the genes are in this situation, see cluster I), with the exceptions of ring stage-specific genes (clusters III and IV). Our data demonstrate binary changes of chromatin structure throughout the erythrocytic cycle (open/closed) and further confront the classical eukaryotic model of transcription initiation after precise removal of nucleosomes. Genes in which mRNA products were detected at the schizont stage (and involved in invasion) in previous microarray expression analyses belong to this group (Le Roch et al. 2003). The apparent paradox of a closed state of chromatin while corresponding mRNAs can be detected could be explained by the regulation of RNA stability. Indeed, previous experiments clearly demonstrate that mRNA half-life is drastically extended during the asexual erythrocytic cycle, especially at late schizont stage with a general half-life of 65 min against 9.5 min for ring stage (Shock et al. 2007). The fact that ring stage–specific genes of our data set seem to follow a more classical pattern of chromatin loosening/tightening in phase with gene expression is consistent with a shorter mRNA half-life. To our knowledge, this decay pattern seems to be unique to Plasmodium. All together these data indicate that post-transcriptional mechanisms may be major mechanisms of gene regulation in the asexual development of the parasite. The classical transcription-driven eukaryotic model of gene regulation was already challenged by the relative paucity of transcription factors (Coulson et al. 2004) and cis-acting regulatory elements detected in the parasite genome (Jason et al. 2005; De Silva et al. 2008). Moreover, even though our chromatin dynamic analyses at the binding sites of the ApiAP2 transcription factors support a role for transcriptional regulation (De Silva et al. 2008), our results indicate that these transcription factors are following the general pattern found for most genes (cluster I). In this scenario, the identified ApiAP2 could be master regulators of transcription at the trophozoite stage rather than fine regulators of targeted transcription initiation.
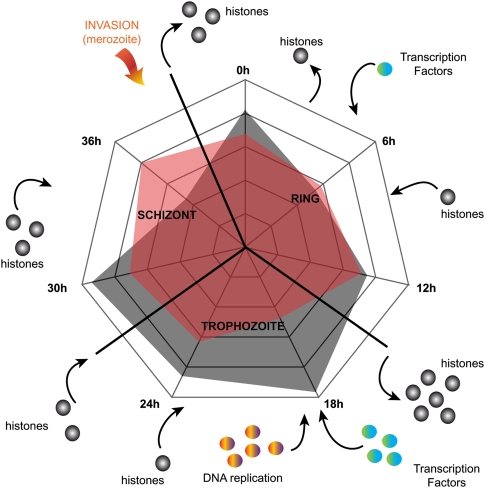
Collectively, our results are consistent with a global model of chromatin architecture changes to control the parasite development and gene expression along the erythrocytic cycle (Fig. 8). At ring stage, nucleosomes are locally removed, and transcription occurs at targeted loci in a controlled manner with the possible intervention of ring-specific transcription factors. At the trophozoite stage, histones are heavily disassembled to allow DNA replication and massive transcription to occur in most loci with the help of global master regulators such as ApiAP2s. In such a model, transcripts accumulate until the schizont and merozoite stages. At the late schizont stage (36 hpi), histones reassemble and DNA is packed at its maximal level. At the merozoite stage, the parasite can focus its efforts to invade fresh red blood cells and start a new cycle. The predominant enrichment with FAIRE-seq that we observed during the erythrocytic cycle is consistent with a relatively permissive transcriptional status of P. falciparum’s genes in general (Salcedo-Amaya et al. 2009). Such a global model to control gene expression is supported by the growing evidence that the parasite lacks transcriptional plasticity, as suggested by the absence of positive or negative feedbacks observed at the transcriptional level in response to metabolic stresses (Ganesan et al. 2008; Le Roch et al. 2008). Furthermore, it has been shown that the parasite preinitiation complex interacts with stage specific “active” and “inactive” promoters (Gopalakrishnan et al. 2009). In combination with our observation, transcription regulation could occur during the elongation level rather than the initiation one. In addition, our model is consistent with recent data demonstrating that mRNA steady-state levels and transcription rate are not correlated for about half of the parasite's genes (Sims et al. 2009). Finally, our results are consistent with the ring stage being crucial for the parasite fate. It has been long known that any kind of stress applied at the early ring stage can induce parasite sexual differentiation and gametocytogenesis.
Figure 8.
Proposed model of chromatin remodeling throughout P. falciparum’s erythrocytic cycle. Genome-wide variations of densities of coverage for MAINE-seq (red) and FAIRE-seq (black). After the invasion of a red blood cell by a parasite at its merozoite morphological stage, histones are depleted and chromatin loosens. During the ring stage, transcription occurs locally in a regulated manner. At the early trophozoite stage, histones are massively depleted. Replication and transcription factors can freely bind. DNA-related metabolic processes are permitted. After replication, DNA re-packs at the schizont stage and the parasites divide and escape the host as differentiated merozoites that are ready for a new cycle of infection.
Since Plasmodia genomes present many points of similarity, it is highly probable that the regulations that we describe here in the case of P. falciparum are conserved among other species: All Plasmodia genomes are uniform in size (23–27 Mb across 14 chromosomes) and gene number (∼5500 genes), and they reveal marked gene conservation (77% of the genes are orthologous) and gene synteny within the body of each chromosome (Carlton et al. 2008b). Among other similarities, they all appear to lack homologs for most of the specific elements of the eukaryotic transcriptional machinery and promoters but encode a very high number of proteins involved in RNA stability and chromatin structure maintenance (Coulson et al. 2004; Carlton et al. 2008b). The accumulation of such genomic specificities further indicates that gene expression regulation in Plasmodium species appears unusual compared with other eukaryotes; similar nucleosome positioning studies should be applied to other Plasmodia genomes to confirm this potentially conserved transcriptional control mechanism. Nonetheless, while it has been shown that Plasmodia genomes are well conserved, the differences in nucleotide bias can be extreme (ranging from 42.3% G+C in Plasmodium vivax and 19.4% in P. falciparum) (Carlton et al. 2008a), suggesting that the AT-richness of the P. falciparum genome might not be the only regulator of chromatin structure changes.
Taken together, our results revive the importance of post-transcriptional mechanisms in the regulation of mRNA steady-state level. The rigid and minimal transcriptional device identified in P. falciparum challenges the classical idea of eukaryotic gene regulation occurring mostly at the transcription initiation level. Our data assign malaria parasite's machinery components that control wide chromatin remodeling as putative targets for new therapeutic interventions.
Methods
For detailed methods, see Supplemental material.
P. falciparum strain and culture conditions
Sorbitol synchronized P. falciparum parasite strain 3D7 was maintained in human erythrocytes according to previously described protocols (Lambros and Vanderberg 1979; Le Roch et al. 2003). For each experiment, cultures were harvested 48 h after the first sorbitol treatment (immediately after reinvasion, time point named T0) and then every 6 h for 36 h (respectively, T6, T12, T18, T24, T30, and T36).
FAIRE procedure
The FAIRE procedure was performed as described previously (Giresi and Lieb 2009). P. falciparum cultures were harvested in phosphate buffered saline and treated with formaldehyde 1% for 20 min at room temperature. The cross-linking duration was set after optimization for the AT-rich genome of P. falciparum (Supplemental Fig. S4). Formaldehyde was quenched with glycine 125 mM for 10 min, and cultures were spun 5 min at 3600g. Pellets were washed three times with phosphate buffered saline containing 2 mM phenylmethylsulfoxide and flash frozen in liquid nitrogen. Pellets were resuspended in 1 mL of FAIRE lysis buffer (25 mM Tris-HCl at pH 7.8, 1 mM EDTA, 0.25% [v/v] Igepal CA-630, Roche Complete Mini EDTA-free protease inhibitor cocktail, 20 mM N-ethylmaleimide) per 0.4 g of cells. Samples were then sonicated for 12 sessions of 12 sec on a Sonic Dismembrator 100 (Fischer Scientific) at 20% amplitude until DNA was sheared into fragments ranging from 50 to 800 bp. Lysates were cleared by centrifugation for 20 min at 4°C and 16,000g. DNA was extracted with phenol-chloroform-isoamyl alcohol 25:24:1 and precipitated with ethanol following a previously described protocol (Giresi et al. 2007). DNA samples were then treated for 30 min at 37°C with RNase A 60 μg/mL (Promega), and finally incubated 8 h at 65°C to remove any eventual DNA–DNA cross-link.
MAINE procedure
Parasite cultures were harvested at 50% hematocrit in phosphate buffered saline containing 2 mM phenylmethylsulfoxide and three volumes of cell lysis solution (Promega) were added. After 10 min of incubation at room temperature, parasites were precipitated by 15 min of centrifugation at 2000g. DNA digestion was conducted 5 min at 37°C in 1 mL of MNase lysis buffer (10 mM Tris-HCl at pH 8.8, 2 mM CaCl2, 0.6% [v/v] Igepal CA-630, 50 mM NaCl, 2 mM phenylmethylsulfoxide, 20 mM Roche Complete Mini EDTA-free protease inhibitor cocktail, N-ethylmaleimide) using 200 units of MNase (USB Corporation). The amount of MNase that was necessary for full mononucleosome purification was optimized in our conditions (Supplemental Fig. S11). Reaction was stopped by the addition of EDTA 5 mM, and the MNase lysis buffer was discarded after 5 min of centrifugation at 16000g. Cell lysis was performed in 400 μL of parasite lysis buffer (3.75 M guanidine HCl, 0.625% [v/v] SDS, 250 μg/mL proteinase K) for 1 h at 55°C and then overnight at 4°C. DNA was extracted with phenol-chloroform-isoamyl alcohol 25:24:1 and precipitated with ethanol.
Sequencing and mapping
The manufacturer's guidelines were followed for library preparation (starting amount 3 μg of fragmented DNA), cluster generation, and single-read 36-cycle sequencing on the Illumina genome analyzer generation I (GAI) (FAIRE experiment), or generation II (GAII) (MAINE experiment). GAI sequencing was performed at the Biomedical Genomics Microarray (BIOGEM) laboratory (University of California, San Diego). GAII sequencing was performed at the Institute for Integrative Genome Biology (University of California, Riverside). Images were processed by the Illumina Pipeline version 0.3. The sequence reads generated by the Pipeline were used for P. falciparum’s genome mapping. Mapping was performed with both ELAND and RMAP software.
Nucleosome mapping
The MAINE-seq data set was used for the mapping of nucleosomes across the genome throughout the red blood cell cycle. Peaks of coverage in the coverage profiles defined the positions of the nucleosomes; the maximum of a peak gives the central position of a 147-bp-long nucleosome.
Measuring regulatory regions availability
The regions located upstream of genes, up to 1 kb long, were considered as putative promoter regions. These regions were divided into two equal parts, i.e., the first 500 bp adjacent to the ATG (proximal promoter) and the preceding 500 bp (distal promoter). Intronic regions were also considered as possible regulatory regions. The degree of chromatin opening was measured as the log2-transformed ratio between the sum of coverage with FAIRE-seq and the sum of coverage with MAINE-seq in a given region. Since all introns have different lengths, their sums of coverage were normalized per kilobase pair. Profiles of promoter or regulatory region availability across time were built using these log2 ratios. Such profiles from proximal promoter regions constituted a data set to be used for promoter availability analysis.
Analyzing changes in promoter availability
Genes with no coverage, either with FAIRE or MAINE, at any of the time points were removed from the data set (80 genes total). Most of these genes are annotated as hypothetical protein, pseudogene, or truncated. One-kilobase-long regions upstream of the ATGs were clustered according to the position of FAIRE-enriched regions using K-mean clustering. Three groups of genes were delineated, and their optimized putative regions were defined and used for the rest of the analysis. The profiles of coverage within promoters of all genes across the seven time points were used as input. Genes were grouped by K-means clustering (k = 4, number of replicates = 1000 runs, confidence measurement = silhouette value). The distribution of 15 previously published functional clusters (FC) based on mRNA levels (Le Roch et al. 2003) within our four clusters was examined and significant representation of these FCs in our clusters was tested using a hypergeometric test. For each FC in a cluster of interest, a representation score was defined as the log2 ratio between the actual gene count and the expected gene count given by the theoretical distribution.
Acknowledgments
We thank Thomas Girke, Tyler Backman, Rebecca Sun, and Barbara Walter (IIGB UC Riverside) for assistance with Illumina sequencing and Pipeline analysis. 3D7 parasites were obtained through the MR4 (MRA-102) deposited by D.J. Carucci. This work was supported in part by NIH grants DK063491, CA023100, and DK080506, and by the NSF CAREER IIS-0447773. Finally, we thank Christian Doerig for his critical review and Carol Park for her attentive proofreading.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession nos. SRA010122 and SRA010123.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.101063.109.
References
- Andersson R, Enroth S, Rada-Iglesias A, Wadelius C, Komorowski J. Nucleosomes are well positioned in exons and carry characteristic histone modifications. Genome Res. 2009;19:1732–1741. doi: 10.1101/gr.092353.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balaji S, Babu MM, Iyer LM, Aravind L. Discovery of the principal specific transcription factors of Apicomplexa and their implication for the evolution of the AP2-integrase DNA binding domains. Nucleic Acids Res. 2005;33:3994–4006. doi: 10.1093/nar/gki709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein BE, Liu CL, Humphrey EL, Perlstein EO, Schreiber SL. Global nucleosome occupancy in yeast. Genome Biol. 2004;5:R62. doi: 10.1186/gb-2004-5-9-r62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowman S, Lawson D, Basham D, Brown D, Chillingworth T, Churcher CM, Craig A, Davies RM, Devlin K, Feltwell T, et al. The complete nucleotide sequence of chromosome 3 of Plasmodium falciparum. Nature. 1999;400:532–538. doi: 10.1038/22964. [DOI] [PubMed] [Google Scholar]
- Bozdech Z, Llinas M, Pulliam BL, Wong ED, Zhu J, DeRisi JL. The transcriptome of the intraerythrocytic developmental cycle of Plasmodium falciparum. PLoS Biol. 2003;1:e5. doi: 10.1371/journal.pbio.0000005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderwood MS, Gannoun-Zaki L, Wellems TE, Deitsch KW. Plasmodium falciparum var genes are regulated by two regions with separate promoters, one upstream of the coding region and a second within the intron. J Biol Chem. 2003;278:34125–34132. doi: 10.1074/jbc.M213065200. [DOI] [PubMed] [Google Scholar]
- Carlton JM, Adams JH, Silva JC, Bidwell SL, Lorenzi H, Caler E, Crabtree J, Angiuoli SV, Merino EF, Amedeo P, et al. Comparative genomics of the neglected human malaria parasite Plasmodium vivax. Nature. 2008a;455:757–763. doi: 10.1038/nature07327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carlton JM, Escalante AA, Neafsey D, Volkman SK. Comparative evolutionary genomics of human malaria parasites. Trends Parasitol. 2008b;24:545–550. doi: 10.1016/j.pt.2008.09.003. [DOI] [PubMed] [Google Scholar]
- Cary C, Lamont D, Dalton JP, Doerig C. Plasmodium falciparum chromatin: Nucleosomal organisation and histone-like proteins. Parasitol Res. 1994;80:255–258. doi: 10.1007/BF00932684. [DOI] [PubMed] [Google Scholar]
- Chookajorn T, Dzikowski R, Frank M, Li F, Jiwani AZ, Hartl DL, Deitsch KW. Epigenetic memory at malaria virulence genes. Proc Natl Acad Sci. 2007;104:899–902. doi: 10.1073/pnas.0609084103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulson RM, Hall N, Ouzounis CA. Comparative genomics of transcriptional control in the human malaria parasite Plasmodium falciparum. Genome Res. 2004;14:1548–1554. doi: 10.1101/gr.2218604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daily JP, Le Roch KG, Sarr O, Ndiaye D, Lukens A, Zhou Y, Ndir O, Mboup S, Sultan A, Winzeler EA, et al. In vivo transcriptome of Plasmodium falciparum reveals overexpression of transcripts that encode surface proteins. J Infect Dis. 2005;191:1196–1203. doi: 10.1086/428289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deitsch KW, Calderwood MS, Wellems TE. Malaria. Cooperative silencing elements in var genes. Nature. 2001;412:875–876. doi: 10.1038/35091146. [DOI] [PubMed] [Google Scholar]
- De Silva EK, Gehrke AR, Olszewski K, Leon I, Chahal JS, Bulyk ML, Llinas M. Specific DNA-binding by apicomplexan AP2 transcription factors. Proc Natl Acad Sci. 2008;105:8393–8398. doi: 10.1073/pnas.0801993105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duraisingh MT, Voss TS, Marty AJ, Duffy MF, Good RT, Thompson JK, Freitas-Junior LH, Scherf A, Crabb BS, Cowman AF. Heterochromatin silencing and locus repositioning linked to regulation of virulence genes in Plasmodium falciparum. Cell. 2005;121:13–24. doi: 10.1016/j.cell.2005.01.036. [DOI] [PubMed] [Google Scholar]
- Dzikowski R, Li F, Amulic B, Eisberg A, Frank M, Patel S, Wellems TE, Deitsch KW. Mechanisms underlying mutually exclusive expression of virulence genes by malaria parasites. EMBO Rep. 2007;8:959–965. doi: 10.1038/sj.embor.7401063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epp C, Li F, Howitt CA, Chookajorn T, Deitsch KW. Chromatin associated sense and antisense noncoding RNAs are transcribed from the var gene family of virulence genes of the malaria parasite Plasmodium falciparum. RNA. 2009;15:116–127. doi: 10.1261/rna.1080109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field Y, Kaplan N, Fondufe-Mittendorf Y, Moore IK, Sharon E, Lubling Y, Widom J, Segal E. Distinct modes of regulation by chromatin encoded through nucleosome positioning signals. PLoS Comput Biol. 2008;4:e1000216. doi: 10.1371/journal.pcbi.1000216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganesan K, Ponmee N, Jiang L, Fowble JW, White J, Kamchonwongpaisan S, Yuthavong Y, Wilairat P, Rathod PK. A genetically hard-wired metabolic transcriptome in Plasmodium falciparum fails to mount protective responses to lethal antifolates. PLoS Pathog. 2008;4:e1000214. doi: 10.1371/journal.ppat.1000214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MJ, Hall N, Fung E, White O, Berriman M, Hyman RW, Carlton JM, Pain A, Nelson KE, Bowman S, et al. Genome sequence of the human malaria parasite Plasmodium falciparum. Nature. 2002;419:498–511. doi: 10.1038/nature01097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Lieb JD. Isolation of active regulatory elements from eukaryotic chromatin using FAIRE (formaldehyde assisted isolation of regulatory elements) Methods. 2009;48:233–239. doi: 10.1016/j.ymeth.2009.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giresi PG, Kim J, McDaniell RM, Iyer VR, Lieb JD. FAIRE (formaldehyde-assisted isolation of regulatory elements) isolates active regulatory elements from human chromatin. Genome Res. 2007;17:877–885. doi: 10.1101/gr.5533506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodman CD, McFadden GI. Fatty acid biosynthesis as a drug target in apicomplexan parasites. Curr Drug Targets. 2007;8:15–30. doi: 10.2174/138945007779315579. [DOI] [PubMed] [Google Scholar]
- Gopalakrishnan AM, Nyindodo LA, Ross Fergus M, Lopez-Estrano C. Plasmodium falciparum: Preinitiation complex occupancy of active and inactive promoters during erythrocytic stage. Exp Parasitol. 2009;121:46–54. doi: 10.1016/j.exppara.2008.09.016. [DOI] [PubMed] [Google Scholar]
- Gupta S, Dennis J, Thurman RE, Kingston R, Stamatoyannopoulos JA, Noble WS. Predicting human nucleosome occupancy from primary sequence. PLoS Comput Biol. 2008;4:e1000134. doi: 10.1371/journal.pcbi.1000134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogan GJ, Lee CK, Lieb JD. Cell cycle-specified fluctuation of nucleosome occupancy at gene promoters. PLoS Genet. 2006;2:e158. doi: 10.1371/journal.pgen.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jason LJ, Finn RM, Lindsey G, Ausio J. Histone H2A ubiquitination does not preclude histone H1 binding, but it facilitates its association with the nucleosome. J Biol Chem. 2005;280:4975–4982. doi: 10.1074/jbc.M410203200. [DOI] [PubMed] [Google Scholar]
- Kaplan N, Moore IK, Fondufe-Mittendorf Y, Gossett AJ, Tillo D, Field Y, LeProust EM, Hughes TR, Lieb JD, Widom J, et al. The DNA-encoded nucleosome organization of a eukaryotic genome. Nature. 2009;458:362–366. doi: 10.1038/nature07667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kar S, Choi EJ, Guo F, Dimitriadis EK, Kotova SL, Adhya S. Right-handed DNA supercoiling by an octameric form of histone-like protein HU: Modulation of cellular transcription. J Biol Chem. 2006;281:40144–40153. doi: 10.1074/jbc.M605576200. [DOI] [PubMed] [Google Scholar]
- Kelly JM, McRobert L, Baker DA. Evidence on the chromosomal location of centromeric DNA in Plasmodium falciparum from etoposide-mediated topoisomerase-II cleavage. Proc Natl Acad Sci. 2006;103:6706–6711. doi: 10.1073/pnas.0510363103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, Schoenfeld LW, Ota I, Sahasrabudhe S, Kurschner C, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438:103–107. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- Lambros C, Vanderberg JP. Synchronization of Plasmodium falciparum erythrocytic stages in culture. J Parasitol. 1979;65:418–420. [PubMed] [Google Scholar]
- Le Roch KG, Zhou Y, Blair PL, Grainger M, Moch JK, Haynes JD, De La Vega P, Holder AA, Batalov S, Carucci DJ, et al. Discovery of gene function by expression profiling of the malaria parasite life cycle. Science. 2003;301:1503–1508. doi: 10.1126/science.1087025. [DOI] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Florens L, Zhou Y, Santrosyan A, Grainger M, Yan SF, Williamson KC, Holder AA, Carucci DJ, et al. Global analysis of transcript and protein levels across the Plasmodium falciparum life cycle. Genome Res. 2004;14:2308–2318. doi: 10.1101/gr.2523904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Roch KG, Johnson JR, Ahiboh H, Chung DW, Prudhomme J, Plouffe D, Henson K, Zhou Y, Witola W, Yates JR, et al. A systematic approach to understand the mechanism of action of the bisthiazolium compound T4 on the human malaria parasite, Plasmodium falciparum. BMC Genomics. 2008;9:513. doi: 10.1186/1471-2164-9-513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee W, Tillo D, Bray N, Morse RH, Davis RW, Hughes TR, Nislow C. A high-resolution atlas of nucleosome occupancy in yeast. Nat Genet. 2007;39:1235–1244. doi: 10.1038/ng2117. [DOI] [PubMed] [Google Scholar]
- Llinas M, Bozdech Z, Wong ED, Adai AT, DeRisi JL. Comparative whole genome transcriptome analysis of three Plasmodium falciparum strains. Nucleic Acids Res. 2006;34:1166–1173. doi: 10.1093/nar/gkj517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez-Rubio JJ, Gontijo AM, Nunes MC, Issar N, Hernandez Rivas R, Scherf A. 5′ Flanking region of var genes nucleate histone modification patterns linked to phenotypic inheritance of virulence traits in malaria parasites. Mol Microbiol. 2007;66:1296–1305. doi: 10.1111/j.1365-2958.2007.06009.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miele V, Vaillant C, d'Aubenton-Carafa Y, Thermes C, Grange T. DNA physical properties determine nucleosome occupancy from yeast to fly. Nucleic Acids Res. 2008;36:3746–3756. doi: 10.1093/nar/gkn262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy PL, Cleary ML, Brown PO, Lieb JD. Genomewide demarcation of RNA polymerase II transcription units revealed by physical fractionation of chromatin. Proc Natl Acad Sci. 2003;100:6364–6369. doi: 10.1073/pnas.1131966100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pasternak ND, Dzikowski R. PfEMP1: An antigen that plays a key role in the pathogenicity and immune evasion of the malaria parasite Plasmodium falciparum. Int J Biochem Cell Biol. 2009;41:1463–1466. doi: 10.1016/j.biocel.2008.12.012. [DOI] [PubMed] [Google Scholar]
- Ralph SA, Scherf A. The epigenetic control of antigenic variation in Plasmodium falciparum. Curr Opin Microbiol. 2005;8:434–440. doi: 10.1016/j.mib.2005.06.007. [DOI] [PubMed] [Google Scholar]
- Ram EV, Naik R, Ganguli M, Habib S. DNA organization by the apicoplast-targeted bacterial histone-like protein of Plasmodium falciparum. Nucleic Acids Res. 2008;36:5061–5073. doi: 10.1093/nar/gkn483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salcedo-Amaya AM, van Driel MA, Alako BT, Trelle MB, van den Elzen AM, Cohen AM, Janssen-Megens EM, van de Vegte-Bolmer M, Selzer RR, Iniguez AL, et al. Dynamic histone H3 epigenome marking during the intraerythrocytic cycle of Plasmodium falciparum. Proc Natl Acad Sci. 2009;106:9655–9660. doi: 10.1073/pnas.0902515106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz S, Meshorer E, Ast G. Chromatin organization marks exon-intron structure. Nat Struct Mol Biol. 2009;16:990–995. doi: 10.1038/nsmb.1659. [DOI] [PubMed] [Google Scholar]
- Segal E, Widom J. Poly(dA:dT) tracts: Major determinants of nucleosome organization. Curr Opin Struct Biol. 2009a;19:65–71. doi: 10.1016/j.sbi.2009.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segal E, Widom J. What controls nucleosome positions? Trends Genet. 2009b;25:335–343. doi: 10.1016/j.tig.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shock JL, Fischer KF, DeRisi JL. Whole-genome analysis of mRNA decay in Plasmodium falciparum reveals a global lengthening of mRNA half-life during the intra-erythrocytic development cycle. Genome Biol. 2007;8:R134. doi: 10.1186/gb-2007-8-7-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sims JS, Militello KT, Sims PA, Patel VP, Kasper JM, Wirth DF. Patterns of gene-specific and total transcriptional activity during the Plasmodium falciparum intraerythrocytic developmental cycle. Eukaryot Cell. 2009;8:327–338. doi: 10.1128/EC.00340-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snow RW, Guerra CA, Noor AM, Myint HY, Hay SI. The global distribution of clinical episodes of Plasmodium falciparum malaria. Nature. 2005;434:214–217. doi: 10.1038/nature03342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilgner H, Nikolaou C, Althammer S, Sammeth M, Beato M, Valcarcel J, Guigo R. Nucleosome positioning as a determinant of exon recognition. Nat Struct Mol Biol. 2009;16:996–1001. doi: 10.1038/nsmb.1658. [DOI] [PubMed] [Google Scholar]
- Tolstorukov MY, Choudhary V, Olson WK, Zhurkin VB, Park PJ. nuScore: A web-interface for nucleosome positioning predictions. Bioinformatics. 2008;24:1456–1458. doi: 10.1093/bioinformatics/btn212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Ramachandran V, Kumar KA, Westenberger S, Refour P, Zhou B, Li F, Young JA, Chen K, Plouffe D, et al. Evidence-based annotation of the malaria parasite's genome using comparative expression profiling. PLoS One. 2008;3:e1570. doi: 10.1371/journal.pone.0001570. [DOI] [PMC free article] [PubMed] [Google Scholar]