Abstract
The biological impact of transposons on the physiology of the host depends greatly on the frequency and position of integration. Previous studies of Tf1, a long terminal repeat retrotransposon in Schizosaccharomyces pombe, showed that integration occurs at the promoters of RNA polymerase II (Pol II) transcribed genes. To determine whether specific promoters are preferred targets of integration, we sequenced large numbers of insertions using high-throughput pyrosequencing. In four independent experiments we identified a total of 73,125 independent integration events. These data provided strong support for the conclusion that Pol II promoters are the targets of Tf1 integration. The size and number of the integration experiments resulted in reproducible measures of integration for each intergenic region and ORF in the S. pombe genome. The reproducibility of the integration activity from experiment to experiment demonstrates that we have saturated the full set of insertion sites that are actively targeted by Tf1. We found Tf1 integration was highly biased in favor of a specific set of Pol II promoters. The overwhelming majority (76%) of the insertions were distributed in intergenic sequences that contained 31% of the promoters of S. pombe. Interestingly, there was no correlation between the amount of integration at these promoters and their level of transcription. Instead, we found Tf1 had a strong preference for promoters that are induced by conditions of stress. This targeting of stress response genes coupled with the ability of Tf1 to regulate the expression of adjacent genes suggests Tf1 may improve the survival of S. pombe when cells are exposed to environmental stress.
Retrotransposons are elements of mobile DNA that constitute a substantial portion of eukaryotic genomes. In the yeasts Schizosaccharomyces pombe and Saccharomyces cerevisiae, long terminal repeat (LTR) transposons make up 1% and 3% of the genome, respectively (Kim et al. 1998; Bowen et al. 2003), while in mammals, retrotransposons comprise nearly half of the genome (Lander et al. 2001; Mouse Genome Sequencing Consortium 2002). The continued ability of these elements to proliferate depends on integration strategies that do not compromise the survival of the host. In S. cerevisiae, retrotransposons avoid inflicting damage to the host by directing integration to regions of the genome that lack coding potential. Ty1 and Ty3 integrate just upstream of RNA polymerase III (Pol III) transcribed genes, and Ty5 inserts into regions of heterochromatin (Lesage and Todeschini 2005). The retrotransposon Tf1 of S. pombe has a mechanism that is distinct from the other transposons of yeast in that its integration clusters in a window 500 nt upstream of ORFs (Behrens et al. 2000; Singleton and Levin 2002; Bowen et al. 2003).
A study of Tf1 integration in plasmids that contained individual genes showed that the insertion sites corresponded to positions where transcription factors bind (Leem et al. 2008). The dominant positions of integration in fbp1 occurred 30 and 40 nt downstream of upstream activating sequence 1 (UAS1), the position where the activator Atf1p binds. This directed integration adjacent to UAS1 is disrupted when mutations are placed in the binding site of Atf1p or when the gene encoding Atf1p is deleted (Leem et al. 2008). These data indicate that it is the promoters of Pol II transcribed genes that are the targets of Tf1 integration.
The result that integration in the genome of S. pombe is directed to the promoters of genes raises several key questions about the biological impact of Tf1 integration. Are all promoters recognized equally or is integration directed to specific sets of promoters? If specific sets of promoters are preferred targets, what distinguishes the preferred promoters from those not recognized by Tf1? In addition, it is also important to test whether Tf1 integrates into sites other than Pol II promoters. To address these questions, large numbers of integrations throughout the genome of S. pombe must be sequenced. The revolutionary new methods for high-throughput pyrosequencing make it possible to characterize extraordinarily large numbers of integration events (Wang et al. 2007).
We report here the use of ligation-mediated PCR and 454 sequencing to determine the position of Tf1 insertions throughout the genome of S. pombe. Four independent collections of transposition were sequenced and from these we identified the positions of 21,848, 14,242, 16,188, and 20,847 insertions. This expansive set of data revealed that greater than 95% of integration occurred in intergenic sequences. The position of the insertions heavily clustered upstream of ORFs. The overwhelming majority (76%) of the insertions were distributed in ∼1000 of the intergenic sequences, which in turn contained about 31% of the promoters of S. pombe. Importantly, we found 80%–88% of the intergenic regions with insertions identified in one experiment also had integration in an independent library of transposition. This high level of overlap from independent experiments demonstrates that we have obtained a genome-wide profile of integration activity for each intergenic sequence of S. pombe. Analysis of the integration levels in each of the intergenic regions revealed that Tf1 had a strong preference for promoters that are induced by conditions of stress.
Results
High-throughput pyrosequencing positioned 73,125 independent insertion events
To create a genome-wide profile of integration sites we sequenced large numbers of Tf1 inserts using the pyrosequencing technology of 454 Life Sciences (Roche). Cells were induced for the expression of Tf1 containing neo (Tf1-neo), and media containing G418 was used to select for the cells with integration events. As developed for sequencing insertion sites of human immunodeficiency virus 1, (HIV-1) and murine leukemia virus (MLV), we applied ligation-mediated PCR to generate libraries of Tf1-neo associated with the downstream flanking DNA (Schroder et al. 2002; Wu et al. 2003). Unlike the cases of HIV-1 and MLV, there are hundreds of preexisting transposon LTRs in S. pombe with sequences identical to that of Tf1-neo (Bowen et al. 2003). To distinguish new integration sites from the preexisting elements, a unique tag of substituted nucleotides was introduced in the U5 sequence of Tf1-neo at a position previously shown to be unimportant for transposition (Lin and Levin 1998). In addition, a SpeI restriction site was introduced just after the upstream LTR to allow us to block the PCR amplification of the internal sequences of Tf1-neo (Supplemental Fig. S1).
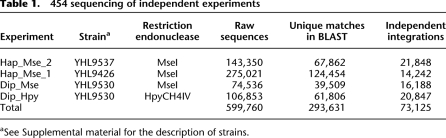
In this study, we performed four independent transposition experiments (Hap_Mse_1, Hap_Mse_2, Dip_Mse, and Dip_Hpy), which were named according to the strains (haploid or diploid) and restriction enzymes (MseI or HpyCH4IV) used to digest the genomic DNA from the cells with integration events. The cut libraries of DNA were ligated to linkers, digested with SpeI, and subjected to barcoded PCR. The amplified products, consisting of the downstream LTRs and their flanking DNA, were size selected and submitted to 454 Life Sciences for sequencing.
All together we obtained 599,760 high quality sequence reads that were then analyzed with BLAST to determine the chromosomal location of the insertions. Many sequence reads mapped to identical positions and corresponded to insertions in the same orientation. These duplicate events were not included in our analyses because they could have been the result of sibling amplification in the yeast cells or in PCR. Other sequence reads were disregarded because they matched sequences that were duplicated in the genome. In all, we identified 73,125 independent Tf1 integration events in unique positions of the S. pombe genome (Table 1). The integrations were distributed in 34,511 sites on the three chromosomes. Since there could be integrations in either orientation at a specific site, and we completed four independent experiments, one site could have up to eight independent insertions. We found 18,216 sites containing integrations in both orientations and 874 sites containing eight independent integrations. These numbers suggest our collection of integration sites approached the level of saturation.
Table 1.
454 sequencing of independent experiments
aSee Supplemental material for the description of strains.
The chromosomal distribution of Tf1 integration was broadly distributed but nonrandom
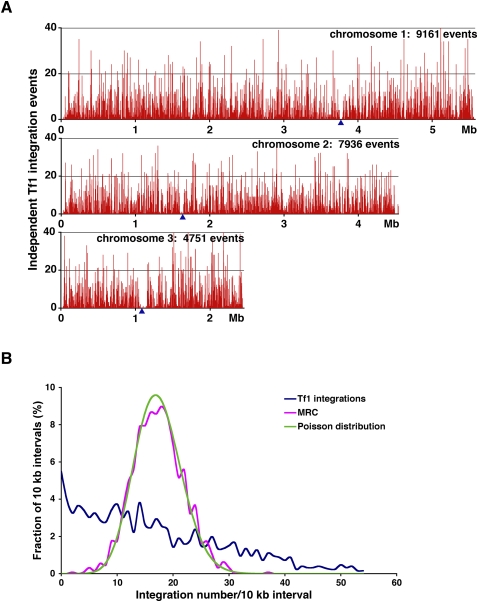
To obtain our first library of integration events we induced transposition of Tf1-neo in haploid cells arrayed into patches. We selected for cells with integration using G418, isolated genomic DNA, and digested the DNA to completion with MseI. This material was then processed as described in Methods for pyrosequencing. The BLAST results of this sequence identified 21,848 independent insertions in this experiment termed Hap_Mse_2. Figure 1A is a histogram of the integration events divided into 1 kb intervals of the three chromosomes. The density of integration events was broadly distributed across each chromosome. However, there were many intervals with high levels of integration. To test the distribution of the events for bias, we divided the genome into 10-kb intervals and plotted the fraction of the intervals that had various numbers of insertions (Fig. 1B, blue line). The resulting population of intervals had a mean of 17.4 inserts/10-kb interval. If these insertions were randomly distributed throughout the genome they would have a Poisson distribution with a mean of 17.4 inserts/10 kb shown in Figure 1B (green line). Clearly, there were many more intervals with greater numbers of inserts than predicted by a Poisson distribution. This divergence from a Poisson distribution resulted in a variance of 204, which was substantially higher than what is expected for a Poisson distribution where the variance equals the mean. This result indicates the integration had a high level of aggregation.
Figure 1.
The distribution of Tf1 integration in the genome of S. pombe. (A) The numbers of independent insertion events from the Hap_Mse_2 experiment are shown within 1 kb intervals of the three chromosomes of S. pombe. The positions of centromeres are indicated by blue triangles. The total number of independent integration events in each chromosome are labeled. (B) The distribution of Tf1 integrations within10-kb intervals of the S. pombe genome is shown for the Hap_Mse_2 experiment. Also shown are the distribution of the random control (MRC of Hap_Mse_2, magenta) and the Poisson distribution (green) based on the mean of the integration data.
One explanation for the strong bias we observed was that the MseI sites themselves had an aggregated distribution that biased the insertions detected by linker ligation. To test the integration data for this bias, as well as others that could occur during ligation and PCR, we generated a matched random control (MRC) data set. Each insertion was matched with a randomly chosen site in the genome that was constrained to have the same distance to an MseI site as the authentic insertion (Wang et al. 2007). As seen in Figure 1B (magenta line) the distribution of the MRC sites matched the Poisson distribution, indicating that the strong clustering of Tf1 integration we detected was not due to the position of the MseI sites.
Tf1 integration clustered upstream of ORFs
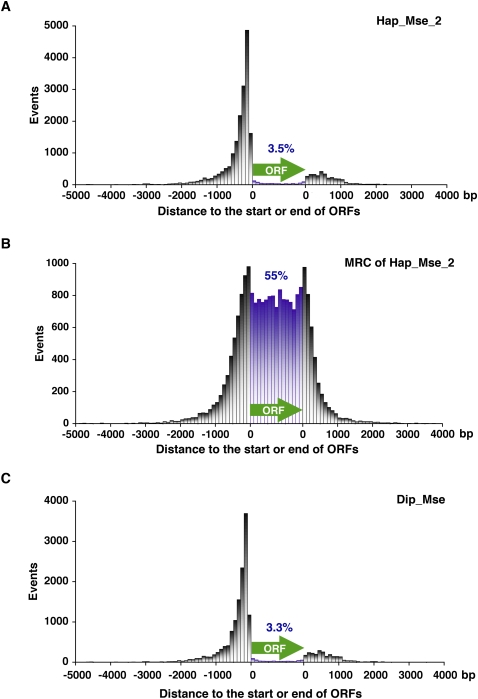
Two studies reported the position of Tf1 integration throughout the genome of S. pombe (Behrens et al. 2000; Singleton and Levin 2002). A total of 78 insertions were sequenced and their positions showed a strong preference for regions upstream of ORFs. To test whether a substantially larger set of insertion data would reveal different preferences for integration, all 21,848 insertion sites from the Hap_Mse_2 experiment were mapped relative to ORFs (Fig. 2A). The distance from the insertions to the closest ORF was determined. The inserts closer to the 5′ end of an ORF were mapped upstream of the ORF in Figure 2A and inserts closer to the 3′ ends of an ORF were positioned downstream in the diagram. Insertions within ORFs were placed into 15 bins based on their relative position. The integration from Hap_Mse_2 showed a clear preference for the first 500 nucleotides (nt) upstream of ORFs. No other significant bias was evident. To test whether this bias resulted from a disproportionate number of MseI sites upstream of ORFs, we mapped the position of the MRC sites relative to ORFs (Fig. 2B). The even distribution of MRC sites throughout the ORF is clearly different from the integration data and demonstrates that the clustering of inserts upstream of ORFs is not due to a bias in the position of MseI sites. The high number of MRC events on either side of ORFs is due to the higher AT nucleotide content and resulting number of MseI sites in the intergenic sequences compared to the ORFs.
Figure 2.
The distance from Tf1 integration sites to the nearest ORF. The x coordinate is the distance from the 5′ and 3′ ends of ORFs. The y coordinate shows the number of integration events within bins of 100 bp. Insertions closer to the 5′ end of an ORF were plotted upstream of the ORF (green arrow), while insertions closer to the 3′ end of an ORF were plotted downstream of ORF. Insertions within ORFs were tabulated within 15 bins of equal proportion. The percentage of the independent integrations in ORF was labeled. (A) Hap_Mse_2; (B) MRC for Hap_Mse_2; (C) Dip_Mse.
The previous studies of Tf1 integration did not detect any integration within ORFs. However, the magnitude of the Hap_Mse_2 data set revealed a full 3.5% of the inserts occurred within ORFs. This finding reveals that Tf1 does have a mechanism of integration that can disrupt coding sequences.
Although integration was detected within ORFs, it occurred at levels significantly lower than in the intergenic sequences. One potential contribution to the low integration in ORFs could be that, in a haploid strain, cells with insertions in key coding sequences would not grow on the medium used to select integration events. To test this possibility we conducted a separate integration experiment in a strain that was isogenic except that it was diploid. The integration data for the diploid experiment, Dip_Mse, resulted in 16,188 independent insertions (Table 1). Despite the diploid nature of the strain, the insertions exhibited the same low level of events in the ORF (3.3% vs. 3.5%) and the same strong preference for the upstream region (Fig. 2C). These results indicate that the pattern of integration detected in our high-throughput experiments resulted from the mechanism of integration, not a selection for survival.
Insertion libraries provided a saturated profile of integration activity for each intergenic sequence
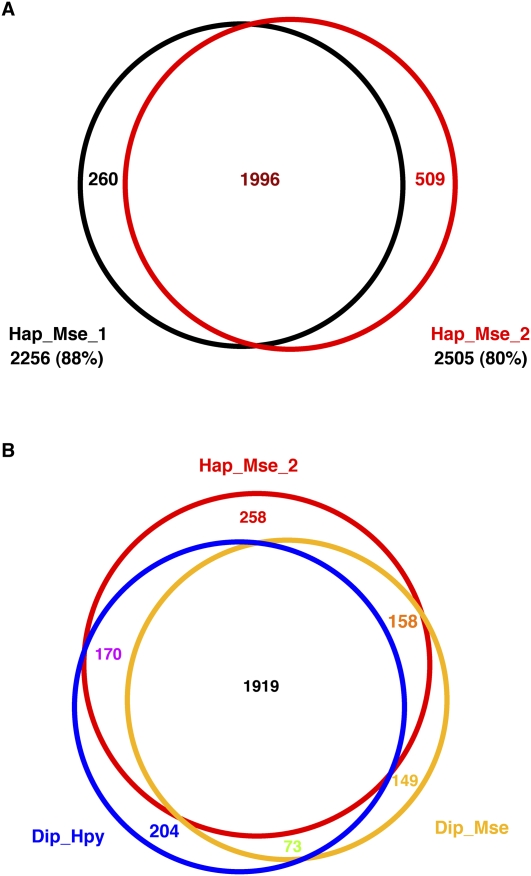
The profile of integration across the genome revealed substantial variation with some intervals containing 35 to 40 insertions per kb, while many others had zero to five insertions per kb (Fig. 1A). The key question about this variation in integration is whether it was due to intrinsic differences in integration efficiency between different sequences in the genome or whether the size of our cultures and the PCR amplification limited our ability to sample the integration potential of each sequence. To distinguish between these two possibilities we tested whether the levels of integration in individual intergenic sequences were reproducible between two independent experiments. The Hap_Mse_2 experiment identified 21,848 insertions and these fell into 2505 intergenic regions. In an independent experiment also using a haploid strain (Hap_Mse_1), 14,242 insertions were isolated and these were distributed within 2256 intergenic regions (Table 1). In each of these experiments, insertions occurred in approximately half of the 5045 intergenic regions of S. pombe (Fig. 3A). More importantly, there was a high level of agreement between the two experiments. Of the 2256 intergenic sequences of Hap_Mse_1 that had one or more insertions, 88% had at least one integration in the Hap_Mse_2 collection. Conversely, of the 2505 intergenic regions that had one or more insertion in the Hap_Mse_2 experiment, 78% were also integration targets in the Hap_Mse_1 experiment. This strong level of congruence between the two independent experiments argues that the size of the yeast cultures and the volumes of the PCRs were sufficient to sample the integration potential of each intergenic region.
Figure 3.
Venn diagrams showing the number of intergenic regions that had integration events in both the Hap_Mse_1 and Hap_Mse_2 experiments. Under the name of each experiment is listed the total number of intergenic regions that had at least one insertion. (A) Comparison of Hap_Mse_1 and Hap_Mse_2; (B) Comparison of Hap_Mse_2, Dip_Mse and Dip_Hpy.
The two integration experiments Hap_Mse_2 and Hap_Mse_1 were both conducted using DNA digested with MseI, an enzyme that cuts TTAA. As a result, the strong correspondence between these two experiments might have resulted from a bias in favor of a subset of insertions in A/T rich DNA. To test the influence of a specific restriction enzyme on the pattern of integration detected, we generated an additional set of integration events from DNA digested with Hpy CH4 IV, an enzyme that cuts a G/C containing sequence (ACGT). This experiment, Dip_Hpy, was generated with a diploid, and resulted in 20,847 independent insertions (Table 1). The data of Dip_Hpy were compared directly to that of an experiment with independently generated inserts (Dip_Mse), for which 16,188 were isolated in diploid DNA cut with MseI. In the Dip_Hpy experiment, 2,366 intergenic regions had one or more insertions and 84% of these regions were targets of integration in the Dip_Mse experiment (Fig. 3B, blue and yellow). Similarly, with Dip_Mse, 87% of the intergenic regions with inserts were also targets of integration in the collection of Dip_Hpy. This high level of overlap between independently generated integration sets in DNA cut with different restriction enzymes demonstrates there was little bias introduced by the recognition sequence of the enzyme. In addition, comparison of the intergenic sequences with integration between experiments with a haploid versus a diploid (Hap_Mse_2 vs. Dip_Mse) also showed high levels of correspondence (Fig. 3B, red and yellow). This result indicated that the ploidy of the strain did not influence which intergenic regions had insertions.
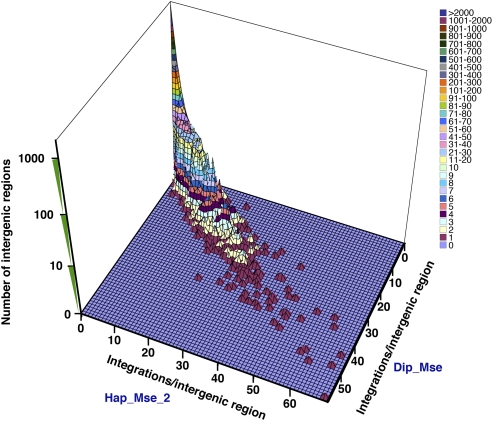
The concordance between the different experiments presented in Figure 3 indicated that the same intergenic regions were active for integration in independent experiments. However, that analysis did not address whether the amount of integration in intergenic regions was consistent between experiments. Figure 4 compares the numbers of integration events in the intergenic regions of the Hap_Mse_2 experiment to the numbers of integration events from the Dip_Mse experiment. Each intergenic region was plotted using the number of integration events identified in the Hap_Mse_2 experiment as the x coordinate and the number of inserts recorded in Dip_Mse experiment as the y coordinate. Because each of the 5,045 intergenic regions was plotted, and many intergenic regions had the same x,y coordinates, we used the z coordinate to indicate the number of the intergenic regions that had the same x,y coordinates. The planar distribution of the data points shows that the amount of integration in each intergenic region is similar between the two independent experiments. The R-value for the data in Figure 4 is 0.95 (R2 = 0.91), indicating there is strong correlation of the integration levels between the two experiments. This comparison was performed between all pairs of the four experiments and the plots showed similar correlations (data not shown).
Figure 4.
Comparison between two experiments (Hap_Mse_2 and Dip_Mse) of the number of insertions within each intergenic region. Each unit of the surface represents a group of intergenic regions. The x coordinate shows the number of integrations/intergenic region in the Hap_Mse_2 data. The y coordinate shows the number of integrations/intergenic region in the Dip_Mse data. The z coordinate has a log scale and shows the number of intergenic regions with the same x and y coordinates. The colors of the surface and the associated key represent values of the z coordinate.
Integration strongly favors intergenic regions that contain Pol II promoters
The genome of S. pombe contains intergenic sequences that can be classified as divergent, tandem, or convergent, depending on the direction of transcription of the two ORFs flanking the intergenic sequence. The previous studies of Tf1 integration in the genome found a strong bias in favor of the divergent and tandem intergenic regions (Behrens et al. 2000; Singleton and Levin 2002). Although this preference indicated Tf1 was directed to the intergenic regions with Pol II promoters, these studies were based on fewer than 100 insertions. For a comprehensive analysis of integration in intergenic regions, we examined the insertions from the 454 data sets. Of the 21,848 independent insertions from the Hap_Mse_2 data, 11,224 (51%) occurred within divergent intergenic sequences, 9135 (42%) were in tandem sequences, and 703 (3.2%) were in convergent sequences. The level of integration in the convergent sequences was substantially less than 27%, the proportion of the intergenic regions that are convergent. This demonstrates that Tf1 strongly favors integration into the divergent and tandem intergenic sequences. This is consistent with the earlier reports, and the model that integration is directed to the Pol II promoters within the intergenic sequences (Behrens et al. 2000; Singleton and Levin 2002). Further support that it is the promoters that are recognized by Tf1 was that the average number of inserts per divergent region is 8.2, which is almost exactly twice the average number of inserts per tandem region (4.0).
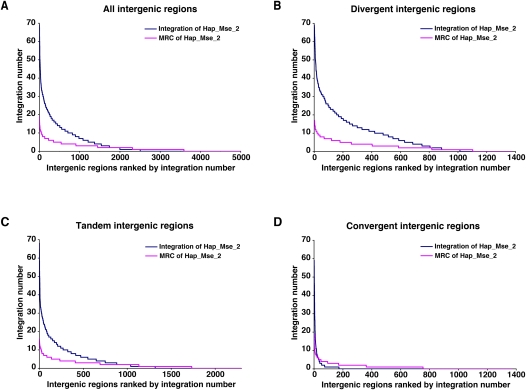
As an independent method of testing Tf1 for targeting bias we analyzed the distribution of inserts in the intergenic regions. The frequency of integration in individual intergenic regions varied greatly. To visualize the distribution of integration we sorted all the intergenic regions by the number of insertions they contained in the Hap_Mse_2 experiment. The other data sets gave similar results (data not shown). Figure 5A shows the number of insertions in each intergenic region on the y-axis when, on the x-axis the individual intergenic regions were ranked and sorted by the numbers of insertions. The intergenic region with the most integration contained 69 insertions, and 2538 intergenic sequences had no integration at all (Supplemental Table S1). The 1000 intergenic regions with the highest levels of integration were 20% of all the intergenic regions, and they contained 76% of all the insertion events. To test whether this distribution was biased we compared it to the distribution of the MRC_Hap_Mse_2 data, the random insertions matched for the distances to MseI sites. When we sorted the intergenic regions from highest number of inserts to lowest, there were many fewer inserts in the top 1000 intergenic regions in the random control data than the experimental data (Fig. 5A). This difference represented the extent to which Tf1 integration was biased.
Figure 5.
The ranking of intergenic regions based on their number of insertions detected in the Hap_Mse_2 experiment. (A) All intergenic regions of S. pombe were plotted on the x-axis in order of their number of insertions (blue). (Magenta) A distribution based on the random control data of MRC Hap_Mse_2. (B) The divergent intergenic regions were plotted on the x-axis in order of their number of insertions (blue). (Magenta) The corresponding random control MRC Hap_Mse_2. (C) The tandem intergenic regions were plotted on the x-axis in order of their number of insertions (blue). (Magenta) The corresponding distribution of random control MRC Hap_Mse_2. (D) The convergent intergenic regions were plotted on the x-axis in order of their number of insertions (blue). (Magenta) The random control MRC Hap_Mse_2.
To determine which types of sequences were preferred for integration we plotted the distribution of integration in the divergent, tandem, and convergent intergenic regions, again sorted from highest to lowest number of inserts (Fig. 5B–D). The level of integration in the divergent and tandem intergenic sequences was clearly greater than the random control (MRC_Hap_Mse_2). In comparison, the level of integration in the convergent intergenic regions was substantially less in the experimental data than the random control (Fig. 5D).
The integration preference for Pol II promoters revealed the position of nonannotated genes
If Pol II promoters are the targets of Tf1 integration then convergent intergenic sequences should not have inserts. Yet, 51 convergent regions of S. pombe were targets of integration in all four independent experiments (Supplemental Table S2). Eight of the convergent sequences had greater numbers of inserts (>19) in the Hap_Mse_2 data than predicted for the random control, MRC_Hap_Mse_2 (Fig. 5D; Supplemental Table S2). One explanation for this is that these convergent regions actually had Pol II promoters that were not annotated. A recent study of the S. pombe transcriptome identified 29 transcripts in convergent intergenic regions (Wilhelm et al. 2008). Indeed, all eight convergent intergenic regions with greater numbers of inserts than in the random control corresponded to nonannotated transcripts detected by Wilhelm and colleagues (Supplemental Table S2). These results indicate that many of the 51 convergent regions targeted in all four experiments contain nonannotated promoters.
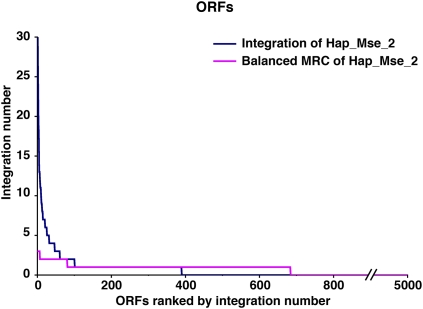
If Tf1 integration were specific for Pol II promoters no integration would occur in ORFs. However, the frequency of integration in ORFs was between 3.3% and 4.4%, depending on the particular experiment. These integration events may be in nonannotated genes with promoters overlapping an annotated ORF or alternatively, there may be a second integration mechanism with a different class of targets. To address this question we analyzed the distribution of insertions within ORFs when ranked by their number of integrations. To compare the distribution of inserts within ORFs to what would be expected from random integration, we increased the total number of events from the random control MRC_Hap_Mse_2 to match the number of insertions in ORFs from the Hap_Mse_2 data (Fig. 6). Interestingly, the 100 ORFs with the highest level of integration had more inserts than the top 100 ORFs from the matched random control. As an indication of reproducible integration activity, there were 39 ORFs that had the exact same insertion sites disrupted in all four independent experiments (Supplemental Table S3). These highly targeted sites suggest that promoters are present within regions annotated as coding sequences. Many of these sequences may actually not encode proteins as 16 of the 39 ORFs are annotated as dubious or sequence orphans. Also, seven more of these positions were in the extreme N termini of coding sequences that were not conserved. These N-terminal insertions suggest that the true start codons may actually be further downstream. Indeed, the annotation for one of these ORFs was recently changed so that the position of the start codon is now believed to be downstream of the insertions (Supplemental Table S3, see rpa2). Although 23 of the 39 highly active targets annotated as ORFs may actually not be in coding sequence, the remaining 16 are in conserved genes and may be unusual cases of internal promoters. Supporting this possibility is that three of the active targets are in intron sequences.
Figure 6.
All ORFs of S. pombe ranked by the number of insertions in the Hap_Mse_2 experiment. The ORFs were plotted on the x-axis in order of their number of insertions (blue). (Magenta) The distribution of the corresponding random control MRC Hap_Mse_2 increased in number to have the same number of insertions in ORFs as in the Hap_Mse_2 data.
Excluding the 100 ORFs with the highest number of inserts there were ∼300 ORFs with a single insertion and the remaining ORFs had no integration (Fig. 6). This distribution was significantly lower than that of the original “unbalanced” set of random controls (MRC_Hap_Mse_2; data not shown). But when the number of inserts from the random controls was matched to the number of true inserts in ORFs, and when the top 100 ORFs were excluded, the two data sets had very similar distributions (Fig. 6). This similarity indicates that for the integration that occurs in most ORFs, there appears to be no bias.
Promoters of stress-induced genes are preferred targets of Tf1 integration
As described above for the Hap_Mse_2 experiment, 76% of all the insertion events occurred in just 20% of the intergenic sequences (see Fig. 5A). This strong bias is a consequence of the integration preference for a specific set of promoters. One possibility was that Tf1 integrated into the promoters with the highest transcription activity. We tested this hypothesis by plotting the number of insertions in each tandem intergenic sequence against its level of transcription, based on Affymetrix expression-chip hybridization signals (Wilhelm et al. 2008). No correlation was observed between transcription and integration (the correlation coefficient R = 0.08; Supplemental Fig. S2).
In another effort to determine what distinguishes promoters that had high levels of insertions from the promoters that did not, we asked whether the genes associated with the targeted promoters contributed to specific classes of biological function. For this, we identified the 219 tandem intergenic regions with the highest numbers of inserts from the Hap_Mse_2 data and tested their downstream genes for patterns of gene ontology. The Gene Ontology (GO) term enrichment tool of the AmiGO consortium found 47 of the 219 genes were classified with the GO term “response to stimulus” as their biological process. This clustering was highly significant with a P-value of 5.1 × 10−4. Among the 47 genes identified, 30 belonged to the subclassification “cellular response to stress.” These 30 genes of the 219 in the query constituted 13.7% compared to 7.4%, the percent of all genes of S. pombe that have the GO term “response to stress.”
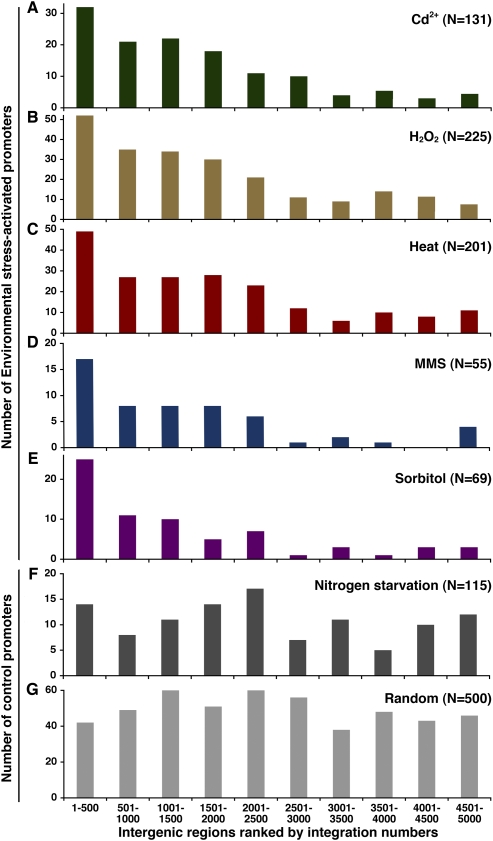
The results of the gene ontology analysis suggested that genes regulated by environmental stress were among the strongest targets of integration. To examine this further we sorted all the intergenic sequences from highest number of insertions to the lowest using the Hap_Mse_2 data. Using this order, the intergenic regions were placed into bins of 500 each. We then used published microarray data to tabulate how many of the intergenic regions in each bin contained promoters that are induced at least threefold by conditions of stress (Chen et al. 2003). As seen in Figure 7A, the bin containing the 500 intergenic regions with the most integration contained the highest number of genes induced by cadmium. The bins with successively lower amounts of integration contained fewer promoters that are induced by cadmium. This relationship indicates that integration has a preference for promoters that are induced by cadmium. Similar preferences were observed for genes induced when cells are treated with hydrogen peroxide or by heat (Fig. 7B,C). Particularly strong preferences for integration into promoters induced by MMS or sorbitol were observed for the first bin of 500 intergenic regions (Fig. 7D,E). When the same bins of intergenic sequences were examined for promoters induced by nitrogen starvation (Mata et al. 2002), no correlation with integration numbers was observed, indicating that Tf1 exhibited integration preferences for specific sets of promoters (Fig. 7F). In comparison, when 500 promoters were chosen at random and their distribution with the bins was tabulated, they showed no correlation with the amount of integration (Fig. 7G).
Figure 7.
Promoters of stress-induced genes are preferred targets of integration. The intergenic regions were ranked by numbers of insertion and then placed into bins of 500. (A–E) The numbers of promoters induced by various environmental stresses in each bin were tabulated. The total number of promoters induced by each stress (N) is listed. (F) The number of promoters induced when cells are starved for nitrogen. (G) As a control, the bins were tabulated for the numbers of promoters they contained from a set of 500 promoters that were selected at random.
Integration activity in repeated sequences was low
When we assembled the data sets of insertion sites, sequence reads that matched repeat regions in the genome were excluded. As a result, the four data sets in Table 1 lacked insertions in any sites with repeats such as the centromere, telomere, or rDNA sequences. To explore the level of integration within these repeat regions we reexamined the sequence reads from the Hap_Mse_2 that matched repeated sequences. A graph of the centromere in chromosome 1 (cen1) with the total number of independent insertion events that matched the repeat sequences shows that these repeats are active targets (Supplemental Fig. S3A). For this and other graphs of repeated sequences, the inserts were tabulated in 1-kb intervals. Although it is not possible to map the insertions to specific copies of repeated sequences, we graphed the average level of insertion within the repeat sequences. This was done by dividing the total number of insertions that matched a specific repeat by the total copy number of that repeat in the genome. This graph for cen1, as well as similar graphs for cen2 and cen3, showed that there was very little integration in the centromeric repeats compared to the integration in the flanking genes (Supplemental Fig. S3A–C). Even when considering the total number of inserts that matched the centromeric repeats, the level of integration was lower than that in the unique sites flanking the centromeres.
The telomeres are another set of repeat-rich regions of the genome. Although the highly repeated telomere sequences are dynamic and not included in the reference sequence of S. pombe, we did tabulate the inserts within the subtelomeric repeats on the left end of chromosome 1 (Supplemental Fig. S3D). In this region there are unique sequences interspersed with several segments that are duplicated elsewhere in the genome. Many regions in the subtelomeric repeats were active for integration and the insertion sites corresponded to promoter regions. The left telomere of chromosome 3 contains ∼100 copies of the rDNA repeats (Schaak et al. 1982). Using the genome sequence and the three copies of the rDNA repeats that are included (Wood et al. 2002), we found that rDNA repeats were targets of integration (Supplemental Fig. S3E). However, the average integration activity in the rDNA repeats is very low since the overall integration numbers in the rDNA repeats must be divided by 100, the approximate number of cDNA repeats in the genome.
The genome of 972, the reference strain of S. pombe contains 13 full-length copies of Tf2, a retrotransposon that differs from Tf1 primarily within Gag (Levin et al. 1990; Weaver et al. 1993; Wood et al. 2002). By tabulating the total number of insertions within Tf2 sequence we found that the transposon sequence was a strong target of integration. Even after dividing the total number of insertions by 13 to get the average level of integration for Tf2 elements, the center region of the elements averaged as many as 18 inserts/kb (Supplemental Fig. S3F). This pattern of integration suggests that there are promoters internal to Tf2. Although it is also possible that integration within Tf2 elements was due to a different mechanism.
Discussion
The application of high-throughput pyrosequencing in this article revealed in S. pombe the position of 73,125 independent insertion events from a total of four independent experiments. These data demonstrated that sequences upstream of ORFs were the dominant sites of integration, as 93% of all insertions occurred in tandem and divergent intergenic regions. This overwhelming preference for tandem and divergent regions provides strong support for the conclusion that Pol II promoters are the targets of Tf1 integration (Leem et al. 2008).
The specificity of Tf1 integration for Pol II promoters raised questions about the 51 convergent regions that were strong targets of integration. That the eight convergent sequences with the highest level of integration were recently found to actually contain Pol II promoters indicates that integration activity can be used to discover new Pol II promoters. The discovery of highly active integration sites within ORFs not only identified new potential promoters, but also raised questions about whether these sequences are ORFs. Most of the 39 ORFs with reoccurring insertion sites were annotated as dubious and many of the other ORFs had strong insertion sites at the extreme N-termini of the ORFs, suggesting the true starts of translation were downstream. These integration data provide strong motivation to reevaluate the annotations of both the convergent regions and the ORFs that had high numbers of insertions.
The biological impact of transposons on the physiology of the host depends greatly on the frequency and position of integration. Previous studies of transposon integration in eukaryotes documented relatively small numbers of insertions. It has therefore been difficult to identify the scope of insertion sites throughout a genome or quantify the integration activity of specific sites. The size and number of the integration experiments reported here resulted in reproducible measures of integration for each intergenic region and ORF in the S. pombe genome. Despite the use of different restriction enzymes in cutting the genomic DNA and the different ploidies of S. pombe used, very similar integration levels were detected across the four experiments for each intergenic and ORF sequence. The reproducibility of the integration activity of each intergenic and ORF sequence from experiment to experiment demonstrates that we have saturated the full set of insertion sites that are actively targeted by Tf1. To our knowledge, this is the first time such a profile of integration data has been assembled.
The highly active insertion sites, representing 76% of all integration, were positioned in just 20% of the intergenic regions of the genome. The analysis of these data demonstrates that Tf1 integration is highly biased in favor of a specific set of Pol II promoters. However, there was no correlation between the amount of integration at the promoters and their level of transcription (Supplemental Fig. S2). This indicates that the proteins responsible for directing integration are not likely to be general factors of transcription, but are specific for the targeted promoters. Such a model is consistent with our previous findings that when promoters are included in target plasmids, transcription is not required for integration and the stress response transcription factor Atf1p mediates integration at the promoter of fbp1 (Leem et al. 2008). Importantly, the finding that Tf1 integrates preferentially into stress response genes supports the model that integration is directed to target sites by transcription factors that induce the stress response genes. It will be important in future studies to test known stress response transcription factors for a role in mediating integration. We will also test the formal possibility that Tf1 transposition induces a stress response and that is why the stress response genes are targets of integration.
The association of Tf1 integration with the response to environmental stress is just one of a vast number of examples of how transposons evolved to specifically react to stress (Wessler 1996; Weiner 2002; Lesage and Todeschini 2005; Haniford 2006; Ebina and Levin 2007; Beauregard et al. 2008). It was her pioneering study of transposons in corn that led Barbara McClintock to propose that cells under stress use transposons to reorganize their genomes in a way that alters gene expression and allows them to overcome a threat to their survival (McClintock 1984). Recent examples of stress-induced activation of transposons have provided molecular understanding of such mechanisms. The transcription of Tf2 is greatly stimulated by the activator Sre1p when cells are deprived of oxygen (Sehgal et al. 2007). Ty5, an LTR retrotransposon that integrates specifically into heterochromatin in S. cerevisiae, targets its integration into genes when cells are starved for nutrients. This integrase possesses a specific phosphorylation that is required for it to bind the heterochromatin factor Sir4p and direct integration to the sites of insertion (Dai et al. 2007). It is the stress of nutrient depravation that diminishes the phosphorylation and disrupts the interaction between integrase and Sir4p. The targeting of Tf1 to stress induced promoters represents a unique response that may function to specifically alter expression levels of stress response genes. Although there is no systematic data, integration of Tf1 into the promoter of ade6 and bub1 does stimulate transcription (Leem et al. 2008).
Of all the integration we detected, 3.9% occurred within ORFs. Although this is a small fraction of the total, it represents 2827 inserts. The position of these events suggests they occur by a mechanism that differs significantly from the process that mediates integration into promoters. Although there were about 100 sites of insertion in ORFs that had higher frequencies than predicted by the random control (Fig. 6), the remaining ORFs in the genome had frequency distributions that were close to what would be expected if integration were random. This indicates that Tf1 possesses a second integration mechanism that appears to be responsible for a low frequency of integration that is distributed randomly throughout the genome.
Methods
Media
Cells were grown on agar plates containing EMM (Forsburg and Rhind 2006) supplemented with 2 gm/L dropout mix (an equal weight mix of all amino acids plus 2.5 times more adenine than the amino acids; no uracil was present) and a final concentration of 10 μM thiamine to repress the transcription of Tf1 driven by the nmt1 promoter. To eliminate the plasmid with Tf1-neo, EMM agar contained 1 g/L 5-fluoroorotic acid (5-FOA), the dropout mix, and 50 mg/mL of uracil. Transposition was measured by a final replica print to YES (YE plus dropout mix) plates supplemented with 1 g/L 5-FOA and 0.5 g/L G418.
Sample preparation and 454 sequencing
The plasmid containing Tf1-neo, pHL2673, was generated by placing a unique sequence tag in the U5 region of pHL891 (Lin and Levin 1997). For the sequence see Supplemental Fig. S1B. The yeast strains are described in Supplemental Table S4. The transposition experiments were performed essentially as described previously (Lin and Levin 1998). For each experiment, 12 independent patches of cells were induced for transposition on each plate and a minimum of 20 plates were processed (see Supplemental Methods for detailed description). The patches of cells on the plates containing YES 5-FOA/G418 were harvested and genomic DNA was isolated (Supplemental Methods; Supplemental Fig. S1). The samples of genomic DNA were digested with MseI or HpyCH4IV and ligated to linkers. Next, the ligation products were digested with SpeI and amplified by PCR with primers containing the A and B tag sequences required for the bridged PCR of 454 sequencing. The PCR products were gel-purified, pooled, and sent to 454 Life Sciences (Roche) for sequencing.
Accession numbers for sequence data
The 454 DNA sequence from the Hap_Mse_1 experiment was submitted to the Short Read Archive (SRA) at NCBI under the accession number SRA009282. However, this data will initially be available from a provisional FTP (ftp://ftp-trace.ncbi.nlm.nih.gov/sra/Submissions/SRA009/SRA009282/provisional/C489SID3069_HL070510_read.tar). The sequence from the remaining experiments (Hap_Mse_2, Dip_Mse, and Dip_Hpy) resulted from a single 454 sequence run and was submitted to the SRA with the accession number SRA009354.
Mapping Tf1 integration sites on the genome of S. pombe
Sequence reads were screened for those containing the end of the LTR. Then the LTR and any linker sequences were trimmed. The trimmed sequences were positioned in the genome using the NCBI BLAST software on a local computer. The S. pombe genome database used in BLAST was the Feb. 2007 version of the chromosome contigs from the Wellcome Trust Sanger Institute. (ftp://ftp.sanger.ac.uk/pub/yeast/pombe/Chromosome_contigs/OLD/20070206/). The BLAST results were filtered to collect matches with genomic sequence that started from the first nucleotide after the LTR and with identities greater than or equal to 95% and expect (E) values less than or equal to 0.05. Then, of the matches that met these criteria, the one with the highest bit score was used to obtain the coordinates for the unique insertion sites. Sequences that were from the same experiment and were found to have the same insertion coordinate and the same orientation, were considered to be duplicate reads, and were considered as only one independent integration event. The program scripts for screening raw sequences or filtering the BLAST results were written in Perl or Visual BASIC (VB).
Other bioinformatic analysis
The CDS coordinates for the S. pombe genome were from Wellcome Trust Sanger Institute, the Feb. 2007 version. The coordinates within intergenic regions or ORFs and the distance to the start or end of the nearest ORF of each integration site were calculated with scripts written with Perl or VB. The Poisson distribution was generated with a function in Excel (Microsoft Office).
Gene Ontology analyses were conducted using AmiGO, the GO Consortium's annotation and ontology toolkit, database release 2008-08-12 (Carbon et al. 2009).
Acknowledgments
This research was supported by the Intramural Research Program of the NIH from the Eunice Kennedy Shriver National Institute of Child Health and Human Development. Additional support was provided by the Intramural AIDS Targeted Antiviral Program. We thank Xiaolin Wu for advice with bioinformatic analyses and Dan Voytas for helpful discussions about the manuscript. We also thank Dr. Jurg Bahler for the discussions we had about genome-wide transcription levels.
Footnotes
[Supplemental material is available online at http://www.genome.org. The sequence data from this study have been submitted to the NCBI Short Read Archive (http://www.ncbi.nlm.nih.gov/Traces/sra/sra.cgi) under accession nos. SRA009282 and SRA009354. See note in Methods about provisional FTP.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.099648.109.
References
- Beauregard A, Curcio MJ, Belfort M. The take and give between retrotransposable elements and their hosts. Annu Rev Genet. 2008;42:587–617. doi: 10.1146/annurev.genet.42.110807.091549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behrens R, Hayles J, Nurse P. Fission yeast retrotransposon Tf1 integration is targeted to 5′ ends of open reading frames. Nucleic Acids Res. 2000;28:4709–4716. doi: 10.1093/nar/28.23.4709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowen NJ, Jordan I, Epstein J, Wood V, Levin HL. Retrotransposons and their recognition of pol II promoters: A comprehensive survey of the transposable elements derived from the complete genome sequence of Schizosaccharomyces pombe. Genome Res. 2003;13:1984–1997. doi: 10.1101/gr.1191603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbon S, Ireland A, Mungall CJ, Shu S, Marshall B, Lewis S. AmiGO: Online access to ontology and annotation data. Bioinformatics. 2009;25:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen DR, Toone WM, Mata J, Lyne R, Burns G, Kivinen K, Brazma A, Jones N, Bahler J. Global transcriptional responses of fission yeast to environmental stress. Mol Biol Cell. 2003;14:214–229. doi: 10.1091/mbc.E02-08-0499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai J, Xie W, Brady TL, Gao J, Voytas DF. Phosphorylation regulates integration of the yeast Ty5 retrotransposon into heterochromatin. Mol Cell. 2007;27:289–299. doi: 10.1016/j.molcel.2007.06.010. [DOI] [PubMed] [Google Scholar]
- Ebina H, Levin HL. Stress management: How cells take control of their transposons. Mol Cell. 2007;27:180–181. doi: 10.1016/j.molcel.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Forsburg SL, Rhind N. Basic methods for fission yeast. Yeast. 2006;23:173–183. doi: 10.1002/yea.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haniford DB. Transpososome dynamics and regulation in Tn10 transposition. Crit Rev Biochem Mol Biol. 2006;41:407–424. doi: 10.1080/10409230600987415. [DOI] [PubMed] [Google Scholar]
- Kim JM, Vanguri S, Boeke JD, Gabriel A, Voytas DF. Transposable elements and genome organization: A comprehensive survey of retrotransposons revealed by the complete Saccharomyces cerevisiae genome sequence. Genome Res. 1998;8:464–478. doi: 10.1101/gr.8.5.464. [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Leem YE, Ripmaster TL, Kelly FD, Ebina H, Heincelman ME, Zhang K, Grewal SIS, Hoffman CS, Levin HL. Retrotransposon Tf1 is targeted to pol II promoters by transcription activators. Mol Cell. 2008;30:98–107. doi: 10.1016/j.molcel.2008.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesage P, Todeschini AL. Happy together: The life and times of Ty retrotransposons and their hosts. Cytogenet Genome Res. 2005;110:70–90. doi: 10.1159/000084940. [DOI] [PubMed] [Google Scholar]
- Levin HL, Weaver DC, Boeke JD. Two related families of retrotransposons from Schizosaccharomyces pombe. Mol Cell Biol. 1990;10:6791–6798. doi: 10.1128/mcb.10.12.6791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Levin HL. A complex structure in the mRNA of Tf1 is recognized and cleaved to generate the primer of reverse transcription. Genes & Dev. 1997;11:270–285. doi: 10.1101/gad.11.2.270. [DOI] [PubMed] [Google Scholar]
- Lin JH, Levin HL. Reverse transcription of a self-primed retrotransposon requires an RNA structure similar to the U5-IR stem-loop of retroviruses. Mol Cell Biol. 1998;18:6859–6869. doi: 10.1128/mcb.18.11.6859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mata J, Lyne R, Burns G, Bahler J. The transcriptional program of meiosis and sporulation in fission yeast. Nat Genet. 2002;32:143–147. doi: 10.1038/ng951. [DOI] [PubMed] [Google Scholar]
- McClintock B. The significance of responses of the genome to challenge. Science. 1984;226:792–801. doi: 10.1126/science.15739260. [DOI] [PubMed] [Google Scholar]
- Mouse Genome Sequencing Consortium. Initial sequencing and comparative analysis of the mouse genome. Nature. 2002;420:520–562. doi: 10.1038/nature01262. [DOI] [PubMed] [Google Scholar]
- Schaak J, Mao J, Soll D. The 5.8S RNA gene sequence and the ribosomal repeat of Schizosaccharomyces pombe. Nucleic Acids Res. 1982;10:2851–2864. doi: 10.1093/nar/10.9.2851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- Sehgal A, Lee CY, Espenshade PJ. SREBP controls oxygen-dependent mobilization of retrotransposons in fission yeast. PLoS Genet. 2007;3:e131. doi: 10.1371/journal.pgen.0030131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singleton TL, Levin HL. A long terminal repeat retrotransposon of fission yeast has strong preferences for specific sites of Insertion. Eukaryot Cell. 2002;1:44–55. doi: 10.1128/EC.01.1.44-55.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang GP, Ciuffi A, Leipzig J, Berry CC, Bushman FD. HIV integration site selection: Analysis by massively parallel pyrosequencing reveals association with epigenetic modifications. Genome Res. 2007;17:1186–1194. doi: 10.1101/gr.6286907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weaver DC, Shpakovski GV, Caputo E, Levin HL, Boeke JD. Sequence analysis of closely related retrotransposon families from fission yeast. Gene. 1993;131:135–139. doi: 10.1016/0378-1119(93)90682-s. [DOI] [PubMed] [Google Scholar]
- Weiner AM. SINEs and LINEs: The art of biting the hand that feeds you. Curr Opin Cell Biol. 2002;14:343–350. doi: 10.1016/s0955-0674(02)00338-1. [DOI] [PubMed] [Google Scholar]
- Wessler SR. Turned on by stress. Plant retrotransposons. Curr Biol. 1996;6:959–961. doi: 10.1016/s0960-9822(02)00638-3. [DOI] [PubMed] [Google Scholar]
- Wilhelm BT, Marguerat S, Watt S, Schubert F, Wood V, Goodhead I, Penkett CJ, Rogers J, Bahler J. Dynamic repertoire of a eukaryotic transcriptome surveyed at single-nucleotide resolution. Nature. 2008;453:1239–1243. doi: 10.1038/nature07002. [DOI] [PubMed] [Google Scholar]
- Wood V, Gwilliam R, Rajandream MA, Lyne M, Lyne R, Stewart A, Sgouros J, Peat N, Hayles J, Baker S, et al. The genome sequence of Schizosaccharomyces pombe. Nature. 2002;415:871–880. doi: 10.1038/nature724. [DOI] [PubMed] [Google Scholar]
- Wu XL, Li Y, Crise B, Burgess SM. Transcription start regions in the human genome are favored targets for MLV integration. Science. 2003;300:1749–1751. doi: 10.1126/science.1083413. [DOI] [PubMed] [Google Scholar]