Abstract
Systems genetics relies on common genetic variants to elucidate biologic networks contributing to complex disease-related phenotypes. Mice are ideal model organisms for such approaches, but linkage analysis has been only modestly successful due to low mapping resolution. Association analysis in mice has the potential of much better resolution, but it is confounded by population structure and inadequate power to map traits that explain less than 10% of the variance, typical of mouse quantitative trait loci (QTL). We report a novel strategy for association mapping that combines classic inbred strains for mapping resolution and recombinant inbred strains for mapping power. Using a mixed model algorithm to correct for population structure, we validate the approach by mapping over 2500 cis-expression QTL with a resolution an order of magnitude narrower than traditional QTL analysis. We also report the fine mapping of metabolic traits such as plasma lipids. This resource, termed the Hybrid Mouse Diversity Panel, makes possible the integration of multiple data sets and should prove useful for systems-based approaches to complex traits and studies of gene-by-environment interactions.
Human complex trait genetics has been revolutionized by the ability to carry out association studies on a genome-wide basis. Such genome-wide association studies (GWAS) have now been applied to numerous complex traits and have resulted in the identification of hundreds of novel genes for traits, such as diabetes, cancer, and various inflammatory diseases (Altshuler et al. 2008; Manolio 2009). This success can be attributed to many factors, including technological developments in acquiring high-throughput genotype data (Matsuzaki et al. 2004; Gunderson et al. 2005), development of catalogs of common human variation, such as the HapMap (The International HapMap Consortium 2005), and development of analytic methodologies for association studies (de Bakker et al. 2005). These have allowed the human genetics community to leverage the increased power and resolution of association studies compared to linkage analyses. Despite these successes, the fraction of the genetic component that is explained by the associated genes in GWA studies has been relatively modest for most traits (Hardy and Singleton 2009). For example, traits such as type 2 diabetes and lipoprotein levels have relatively high heritability, in the range of 50%, and yet the genes discovered by GWAS for these traits explain in aggregate less than 10% of the phenotypic variance. This can likely be attributed to several factors; in particular, the effects of a single common variant on a disease trait tend to be very weak and GWA studies have low power to detect rare variation involved in disease (Cohen et al. 2004; Frikke-Schmidt et al. 2004).
Mouse models have been used effectively for the identification of genes contributing to simple Mendelian traits, but unfortunately there have been few successes for genes contributing to complex, multigenic traits. Traditional genetic analysis in mice involves crossing different inbred strains and mapping the traits of interest using linkage analysis. An important problem has been the lack of resolution in identifying the causal gene(s) from the results of a linkage study. Fine mapping in such cases generally requires the construction of congenic strains, in which the region of interest from one strain is transferred onto the background of the second strain by a series of crosses. But this frequently proves difficult because the alleles contributing to complex traits generally exhibit subtle effects that approach the levels of noise (Flint and Mott 2008), and several closely linked genes may influence the trait at a given locus.
Encouraged by the success of human association studies, several groups have proposed mouse genome-wide association studies. These initial pioneering studies demonstrated the potential of mouse genome-wide association studies with their early successes, but they have also raised some important challenges including complex population stratification among the mouse strains and concerns about the lack of power to detect loci with modest effects (Pletcher et al. 2004; de Bakker et al. 2005; Payseur and Place 2007). In fact, these two issues are intimately related. Population structure inflates the association statistics, both creating spurious associations, as well as artificially increasing the apparent strength of true association signals. The initial mouse genome-wide association studies reported a tremendous number of genome-wide significant signals, some of which overlapped with known loci. This, combined with the knowledge that mouse strains have high heritability for traits, suggested that mouse association studies had sufficient statistical power. However, these initial studies did not adequately correct for population structure, which when taken into account, eliminates the vast majority of predicted associations (Kang et al. 2008). Thus, the inability to correct for population structure in the initial studies led them to severely overestimate their statistical power.
We have explored a wide range of possible designs for mouse association studies using simulations that can accurately measure statistical power after correction for population structure. We assembled a combined set of inbred strains, which we term the “Hybrid Mouse Diversity Panel” (HMDP) that includes 100 commercially available inbred strains consisting of 29 classic inbred (CI) strains and three sets of recombinant inbred (RI) strains. Here, we report that the HMDP has sufficient power to map traits that contribute to 10% of the overall variance. Importantly, the resolution of the panel is an order of magnitude better than linkage analysis. Practical advantages of the HMDP include the elimination of costly genotyping, as these strains have now been genotyped at over 135,000 SNPs (http://mouse.cs.ucla.edu/mouseHapMap/), and the availability of the strains from The Jackson Laboratory. In addition, each strain is renewable and, therefore, diverse molecular and phenotypic data can be collected ad infinitum. Thus, this panel should be useful for the analysis of gene-by-environment interactions where multiple individuals of the same genotype need to be studied. Moreover, the fact that the data involving clinical, expression, proteomic, and metabolic traits are cumulative makes this resource ideal for systems biology.
Results
Strain selection for the Hybrid Mouse Diversity Panel
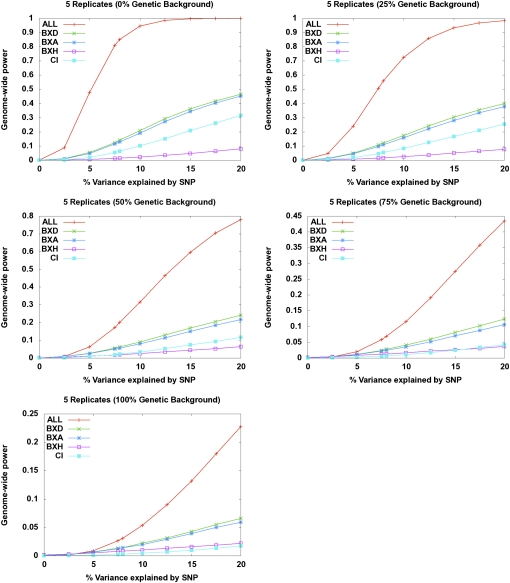
Our goal is to develop a panel of inbred mouse strains for performing association studies with adequate statistical power and resolution for mapping of complex traits. While hundreds of inbred strains have been derived, a relatively small fraction of these are useful for an association panel, and we can use several intuitions to guide our choices for the inbred strains. Certain strains, such as congenics and closely related members of a family of strains (e.g., many members of the C57BL family) are minimally informative because of their largely identical genetic ancestry (Beck et al. 2000). These strains are only informative for the small number of loci that differ, and we include only one representative of each of these strains in our panel. Altogether, we selected 29 CI strains for the panel (Supplemental Table 1). This set of inbred strains is representative of previous mouse association studies that were performed (Pletcher et al. 2004; Liu et al. 2006, 2007; Cervino et al. 2007). We carried out power calculations to estimate the level of SNP effect that could be detected by the inbred panel under various conditions of heritability and P-value cutoff (Fig. 1; Supplemental Table 2). These analyses indicated that 29 strains were not sufficient to detect loci that explain less than 20% of the total trait variance after correcting for population structure.
Figure 1.
Power calculations. We estimated the power for the 29 inbred strains, the individual RI panels (BXD, AXB/BXA, and BXH) and the combined HMDP. Simulations assume five replicates per strain. The x-axis indicates increasing effect size of SNP, and the y-axis is estimated power. Each panel represents simulations performed under different scenarios in which the genetic background (or population structure effect) accounts for an increasing proportion of the total variance of the phenotype.
Our approach differs from previous mouse association studies in that we additionally include in our panel 71 RI strains (Supplemental Table 1). RI strains are derived by crossing a pair of inbred parental strains and then deriving a set of inbred progeny through brother–sister mating for 20 or more generations. These strains consist of roughly 50% genetic contribution from each of the parental strains, such that each allele that is polymorphic among the parents is present in about 50% of the strains in the RI set. The RI mice maximize power to detect associations at loci polymorphic between the parental strains. Power is further increased by combining multiple RI sets, considering that the complex genetic relatedness among the strains is accounted for by the availability of high-density markers. The selected RI sets are derived from crosses between C57BL/6J (B) and either DBA/2J (D), A/J (A), or C3H/HeJ (H) and cover a significant fraction of the SNPs in our panel. Inclusion of RI strains substantially adds to the overall power to detect loci with small effects (Fig. 1; Supplemental Table 2) both because of their genetic structure, as well as increasing the number of total strains in the set. For example, in the HMDP we have 70% power to detect SNPs that contribute ∼10% of the overall variance of a complex trait.
Validating the HMDP resolution using expression quantitative trait loci
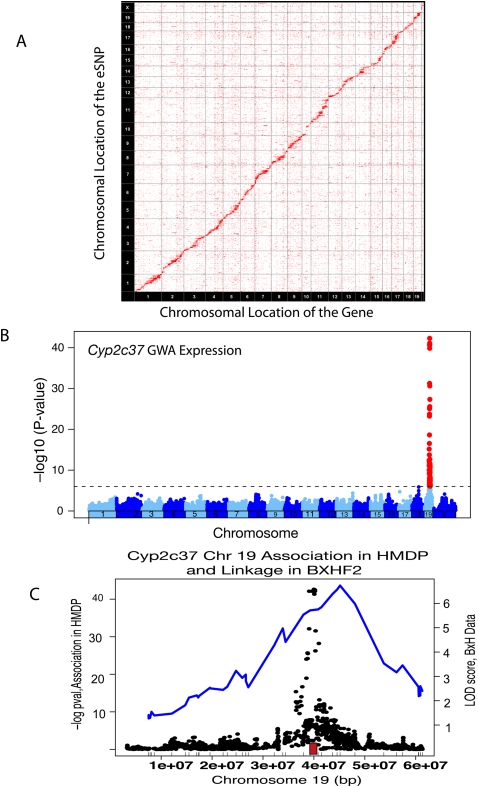
Gene expression traits provide a biologically relevant means to effectively estimate both mapping power and resolution in the HMDP. We have performed hepatic expression array analyses on three individual mice of each strain in the HMDP. This resulted in the identification of 2691 probes with local (commonly called cis-acting) expression quantitative trait loci (eQTL) and 3174 probes with at least one distal (trans-acting) eQTL at a P-value of ≤4.1 × 10−6. Figure 2A shows a plot of the location of each of the genes on the array (y-axis) and the corresponding location of each significant eQTL (x-axis). The local eQTL occur on the diagonal, and the remaining signals represent trans-eQTL signals. The number of cis-eQTL compares favorably to our previous studies of eQTL in inbred crosses of BXH, 2118 cis-eQTL (Wang et al. 2006), and BXD, 1171 cis-eQTL (Doss et al. 2005), and eQTL in outbred mice, 492 cis-eQTL (Ghazalpour et al. 2008).
Figure 2.
Expression SNPs from HMDP. (A) Transcript levels in liver of HMDP mice were profiled and significant associations are plotted according to chromosomal position (x-axis) versus the location of the structural gene (y-axis). The strong diagonal line represents cis-eQTL, whereas the remainder are trans-eQTL signals. (B) Genome-wide association results in the HMDP demonstrating a strong association for Cyp2c37 transcript levels in liver on chromosome 19. (C) Chromosome 19 specifically, with an overlay between the linkage results from the BXHApoe−/− F2 cross and the association from the HMDP panel for the Cyp2c37 cis-eQTL on chromosome 19. (Red box) The location of Cyp2c37. The tick marks on the x-axis are the location of the chromosome 19 markers used in the BXHApoe−/− F2 intercross.
Figure 2B illustrates a typical cis-eQTL for the gene Cyp2c37 on chromosome 19. We have previously carried out expression QTL analysis on several crosses in liver and the LOD score plot from one such cross for Cyp2c37 is shown alongside the association data in Figure 2C (Schadt et al. 2003, 2005; Wang et al. 2006; Yang et al. 2006; Chen et al. 2008). Whereas the linkage peak is quite broad, encompassing many megabases (Mb), the peak association markers map within 500 kb of the Cyp2c37 gene. A list of the top 100 cis-eQTL identified in the HMDP, along with peak SNP markers is presented in Supplemental Table 3. Several cis-eQTL, previously identified in linkage studies, have known mechanisms of altered gene expression and high-resolution mapping of these genes is shown in Supplemental Figure 1 (Schadt et al. 2003; Doss et al. 2005; Aherrahrou et al. 2008).
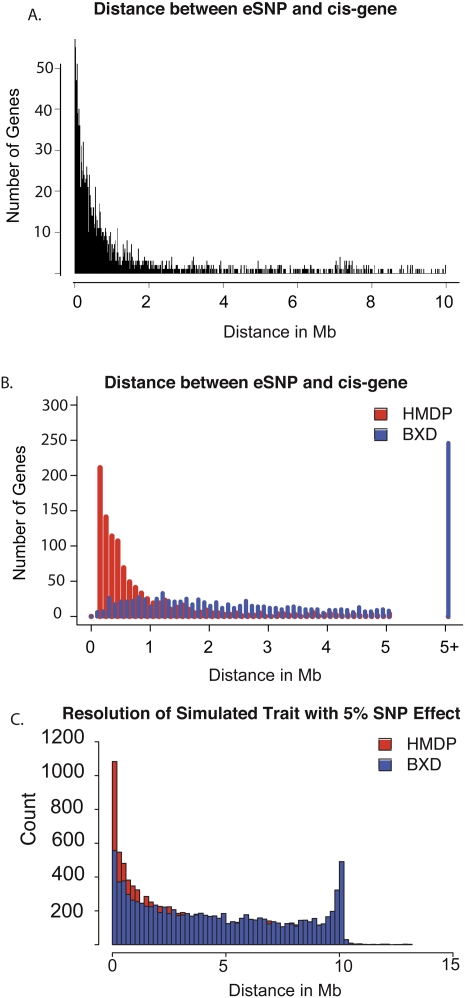
An important criterion for the effectiveness of a mouse association panel is the mapping resolution or the size of the region that we can detect as associated with a trait. Due to many population bottlenecks in the history of the inbred mouse strains, long regions of linkage disequilibrium (LD) are common throughout the mouse genome. RI strains contain even longer regions of LD, since there are a limited number of recombinations that occur when they are being derived. Intuitively, by adding the CI strains to the RI strains, we can improve the mapping resolution. The cis-eQTL provide a convenient measure for the overall resolution of the HMDP as it is reasonable to assume that the majority of causal DNA variations contributing to cis-eQTL would reside within 1 Mb of the gene itself. Thus, mapping the distance between the peak eQTL and the 5′ or 3′ end of the gene provides a measure of the accuracy of our association analysis. The results indicate that the peak SNPs usually occur within 1 Mb of either end of the gene (Fig. 3A). These results contrast with the resolution achieved using RI strains alone, where only 18% map within 1 Mb (Fig. 3B).
Figure 3.
Expression traits demonstrate high resolution of HMDP. (A) Distance between peak cis-eQTL and the transcription start site of the corresponding gene in the HMDP. The majority of cis-eQTL map within 500 kb of the transcription start site of the corresponding gene. (B) Comparison of resolution in the full HMDP (red bars) to BXD recombinant inbred panel (blue bars) showing 1000 cis-eQTL in both populations. (C) Simulated resolution of a SNP with 5% effect in the HMDP (red bars) and BXD recombinant inbred panel (blue bars).
In general, cis-eQTL have a high genetic effect associated with them, and thus we modeled low and medium effect traits using detailed simulations. We found that a SNP, with an effect size of 5%, has a 95% confidence interval of 2.6–2.7 Mb in the HMDP. This compares favorably with the BXD RI panel, which has a 95% confidence interval of 4.5–4.6 Mb (Fig. 3C). As expected the minimum P-values for these simulations did not always reach statistical significance. In the HMDP, 23% of the minimum detected SNPs had a nominal P-value less than 1 × 10−5, while in the BXD RI panel only 2% of the tested SNPs had a P-value less than 1 × 10−5. Simulated resolution of SNPs with effect sizes varying between 2.5% and 17.5% are shown in Supplemental Figure 2.
Application of the HMDP: Mapping high density lipoprotein levels
A major goal of the HMDP is to achieve high-confidence, high-resolution genetic data contributing to complex phenotypes. Of particular interest are phenotypes related to human disease, such as those contributing to metabolic syndrome and atherosclerosis, and we focus on plasma lipids to demonstrate the overall approach. We phenotyped the HMDP strains, using 6–12 males per strain, for a variety of metabolic traits and corrected for population structure using efficient mixed model association (EMMA) (see Methods). Supplemental Figure 3 compares the P-values for uncorrected and EMMA corrected high-density lipoprotein (HDL) data. Supplemental Figure 3B shows the dramatic reduction of P-value inflation following application of EMMA, many of which are false positive signals (Payseur and Place 2007). Using permutation analysis (see Methods), we determined that a P-value of 4.1 × 10−6 was significant at a genome-wide level.
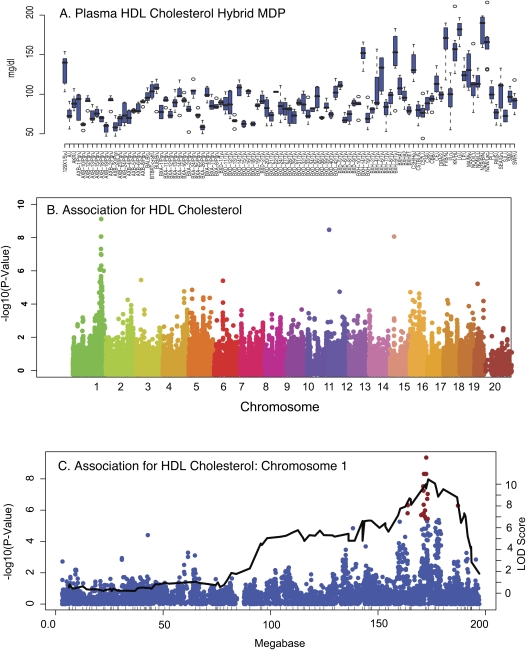
To validate the association approach, we first asked whether we could detect a previously identified common variation among inbred strains affecting HDL-cholesterol levels. We and others have previously shown that variations of the apolipoprotein A-II (Apoa2) gene locus affecting APOA2 protein levels in the plasma commonly occur among inbred strains, and that these significantly influence HDL-cholesterol levels (Doolittle et al. 1990; Warden et al. 1993; Wang et al. 2004; Castellani et al. 2008; Flint and Mott 2008). Figure 4 shows the variation observed for HDL in the HMDP and the corresponding genome-wide association results. Improved mapping resolution of clinical traits is demonstrated by comparing the HMDP results for HDL levels with linkage results from a large cross of C57BL/6J and C3H/HeJ (Fig. 4C). We observed a total of 21 SNPs on distal chromosome 1 associated with HDL-cholesterol at P-value < 4.1 × 10−6. One of the peak SNPs in the region is located 30 kb upstream of the Apoa2 gene at 173.1 Mb. Surprisingly, the peak SNP at 172.4 Mb, within an intron of the gene Nos1ap, is the peak HDL-associated SNP.
Figure 4.
Detection of associations for plasma lipids in HMDP strains coincide with a corresponding QTL in C57BL/6 × C3H/HeJ F2 crosses. Six to ten mice of each strain were examined for the given phenotypes as described in Methods. (A) Plasma HDL levels in the HMDP. (B) GWAS for plasma high-density lipoprotein cholesterol. (C) Comparison of association results with linkage results on chromosome 1. Linkage data from a previously reported F2 cross between C3H/HeJ and C57BL/6J (Wang et al. 2007). These results demonstrate the power of the HMDP to detect associations for QTL observed in the F2 cross, and also highlight the vastly improved resolution of association testing with the HMDP.
Power calculations, presented above, indicate that an approach combining RI and CI strains would have greater power to detect genetic signals. To illustrate this we mapped the HDL signal on distal chromosome 1, for the CI set and each individual RI, a combined RI set (BXN) and the HMDP set independently (Supplemental Fig. 4). The combined RI panel simulates the design of a linkage approach proposed by Williams et al. (2001). In this case, several loci are associated, but they show poor resolution. These analyses highlight the improved power and resolution to map complex traits in the HMDP, compared to the combined RI panel, the individual RI inbred panels of mice, or the improved power to confidently map complex clinical traits compared to the inbred panel alone.
Our second, more global analysis, was to identify the concordance between our association results with the previously reported QTL for plasma HDL levels (Wang and Paigen 2005). We compared our associations to previously reported QTL loci and eight of the 10 associations were within the 95% confidence interval of a previously reported QTL (Supplemental Table 4). Using a binomial probability distribution and assuming 43% of the mouse genome has been mapped with a HDL QTL (Wang and Paigen 2005), there is significant overlap between the HMDP results and previous QTL, eight of 10 possibilities (P < 0.03), which is further evidence that the HMDP identifies true genetic signals.
Linkage disequilibrium patterns in the HMDP
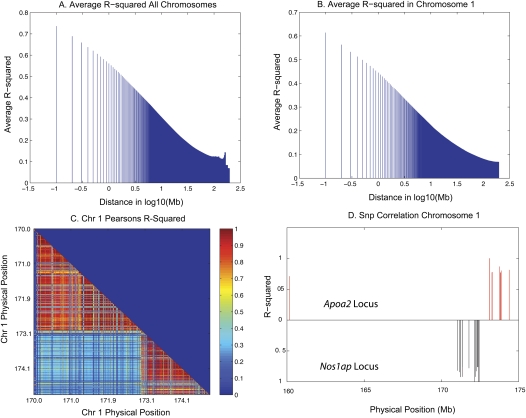
EMMA corrects for background genetic effects at a global level. One additional factor affecting resolution with this approach is the complex linkage disequilibrium (LD) patterns in this population. To calculate the linkage disequilibrium between markers, we calculated the Pearson's pairwise correlation coefficient between all pairs of markers for each chromosome. LD blocks were defined as groups of SNPs with an r2 greater than 0.7. To determine the average correlation between markers for each chromosome, we generated a distribution of mean r2 values for all pairs of informative markers at various distances from each other, using increasing window sizes of 100 kb bins. Figure 5A shows the mean for each window size using all 20 mouse chromosomes to determine the average r2 for each window size across the genome. Thus, on average, blocks showing high LD (r2 > 0.7) extended an average of 500 kb. The average correlation in chromosome 1 for markers 100 kb apart is r2 = 0.7, and for markers 1 Mb apart is r2 = 0.5 (Fig. 5B).
Figure 5.
Linkage disequilibrium in the HMDP. The average r2 for marker pairs was calculated per chromosome and the average of 20 chromosomes is presented here. (A) Average of 20 chromosomes. (B) Average r2 for chromosome 1. The average correlation for markers 100 kb apart is r = 0.7, and for markers 1 Mb apart is r2 = 0.5 (C) Linkage disequilibrium blocks on distal chromosome 1. (D) Long-range LD patterns in the HMDP for these peak SNPs at 172.4 Mb, within coordinates of the Nos1ap gene, and 173.1, 30 kb upstream of the Apoa2 transcription start site on chromosome 1.
The RI strains exhibit extensive LD due to infrequent recombination, and a low level of LD will be observed for many megabases. At individual loci this can have significant implications on actual identification of underlying candidate genes, and we focus on loci associated with HDL on chromosome 1 as an example. Figure 5C shows a plot of the correlation coefficient squared (r2) for the distal region of chromosome 1 containing the Apoa2 and Nos1ap genes. We next calculated the pairwise correlation between the peak SNP on chromosome 1 at 172.4 Mb and all SNPs on chromosome 1. The LD pattern for this SNP is centered around the LD block at 171–172 Mb (Fig. 5D). We repeated this analysis for the SNP closest to Apoa2 at 173.1 Mb. This particular SNP has a complex LD pattern with correlation among SNPs spanning 160–185 Mb (Fig. 5D). Notably, we did not observe correlation above 0.7 (r2) between these two individual SNPs, an indication of distinct genetic signals.
Application of the HMDP: Interrogating novel human GWAS genes
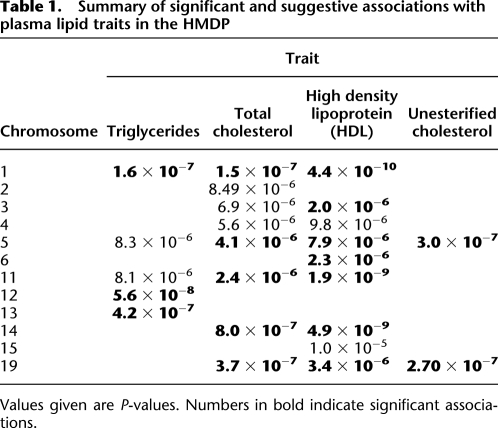
In addition to plasma HDL levels, we found significant associations for total cholesterol (TC), triglycerides (TG), and unesterified cholesterol (UC). The values for these traits are presented in Supplemental Figure 5 and genome-wide mapping results presented in Supplemental Figure 6 and summarized in Table 1. Several of these are of particular interest because they demonstrate how murine studies complement human associations. For example, the signal on chromosome 15 at 58.6 Mb is within 1 Mb of the novel human GWAS plasma lipid genes, Trib1 and Nsmce2 (Willer et al. 2008). A considerable advantage of murine studies is the availability of peripheral tissues for transcriptional, proteomic, and metabolomic profiling. For example, the expression of Trib1 in liver is under cis-regulation (P < 1 × 10−5) and is negatively correlated with TC (r = −0.27), HDL (r = −0.23), and UC (r = −0.30) levels. Conversely Nsmce2 is under distant regulation (P < 1.6 × 10−6) and is also significantly correlated with TC (r = −0.30), HDL (r = −0.30), and UC (r = −0.29) levels.
Table 1.
Summary of significant and suggestive associations with plasma lipid traits in the HMDP
Values given are P-values. Numbers in bold indicate significant associations.
Discussion
Why is a mouse association resource important for the dissection of complex diseases? Mice provide the ability to carry out experimental validation, and unlimited access to tissues. A primary motivation for these studies was increased resolution of murine genetic studies. Less obvious, though equally important, is the systems-based approach that the HMDP enables, as similar to RI panels, the data are cumulative. High-resolution mapping studies in mice should complement human association studies and also make possible the development of coexpression networks allowing functional annotation of the identified genes (Oldham et al. 2006; Lusis et al. 2008; Schadt et al. 2008). From our initial results presented here, and freely available at http://mouse.cs.ucla.edu/emmaserver/, several significant results have emerged that support the use of the HMDP for systems genetic studies. First, we focus on cis-eQTL to demonstrate the robust nature and high-resolution mapping in this population. Second, we map a complex clinically relevant phenotype, plasma HDL levels, to demonstrate concordance with previous mouse studies and, more importantly, to demonstrate how the HMDP can be used to further inform novel human GWAS genes. Third, as with any approach, there are limitations to the HMDP, and alternative approaches, such as the Collaborative Cross and studies of outbred stocks, have their own strengths. Each of these points is discussed below.
A variety of “genetical genomic” studies in humans, rats, mice, and plants have shown that genetic variations influencing gene expression are very common in natural populations (Petretto et al. 2006; Emilsson et al. 2008; Price et al. 2008). Trans-acting loci contributing to transcript levels have proven difficult to validate due to the problem of multiple comparisons, but cis-acting loci provide a relatively straightforward means of examining the power and resolution of our HMDP association approach. We and others have previously validated the cis-acting of the loci by quantifying transcript levels derived from each allele in heterozygous mice using coding polymorphisms (Doss et al. 2005). Using whole genome expression array analysis in livers of the HMDP strains, we identified over 2500 cis-eQTL, comparable to the numbers identified in large crosses with several hundred mice. Moreover, analyses of cis-eQTL provide a convenient measure of the resolution of this approach.
Actual identification and validation of genes contributing to disease phenotypes are of considerable interest, but relatively few genes contributing to common complex variations in mice have been identified and validated. However, one gene that has been shown in a number of studies to contribute to complex metabolic traits is the apolipoprotein A2 gene on distal mouse chromosome 1 (Mehrabian et al. 1993; Warden et al. 1993; Purcell-Huynh et al. 1995). In addition to Apoa2, we have identified a number of additional loci contributing to plasma lipid traits. Two of these loci contain genes identified as novel candidates associated with plasma lipid traits in human GWAS, Trib1 on mouse chromosome 15, and Amac1 on mouse chromosome 11. Therefore, studies in mice may provide additional mechanisms for novel human GWAS genes.
As with any genetic technique there are limitations to the HMDP approach. One potential problem with our association approach is long-range LD. In particular, Petkov et al. (2005) have provided convincing evidence of functional LD both within blocks and also between regions on separate chromosomes. Thus, some association signals could represent such regions of distant LD. We have addressed this concern by testing for the presence of LD between loci identified for HDL.
The HMDP is only one of several recent strategies that attempt to improve mapping resolution in mice. Clearly, association analysis in outbred, heterogeneous stocks of mice can be used (Valdar et al. 2006b; Flint and Mott 2008; Ghazalpour et al. 2008; Farber et al. 2009). One of these studies found hundreds of significant associations for 97 typed traits with an average 95% confidence interval of 2.8 Mb, which is similar to the resolution in the HMDP (Valdar et al. 2006b). A recent analysis focused on eQTL demonstrates that outbred stocks have similar resolution to the HMDP (Huang et al. 2009). Two disadvantages of using outbred stocks are the cost of high-density genotyping and the fact that each mouse is unique and thus can only be studied for a limited number of phenotypes. An advantage of outbred stock strategies over the HMDP is that there is no limit to the number of genetically distinct animals that can be included in the study, while the HMDP is limited to the number of available inbred strains.
There have also been a number of studies that have exploited the mosaic structure of common inbred mouse strains to perform association mapping (Grupe et al. 2001; Klein et al. 2004; Liao et al. 2004; Guo et al. 2006; Liu et al. 2006, 2007; Moran et al. 2006; Cervino et al. 2007; Guo et al. 2007; McClurg et al. 2007). The methods have proved effective for localizing genes with large effects, but not for genes with effect sizes less than 10%, as is usually observed with complex traits (Cervino et al. 2007; Flint and Mott 2008). In addition to the lack of power, population structure is a major problem that can result in false positives (Cervino et al. 2007; Kang et al. 2008). Recently, genes previously identified in murine association mapping have failed to replicate in linkage studies designed to confirm these novel loci (Manenti et al. 2009) underscoring the importance of developing population structure correction methods designed to improve power and reliability of murine association mapping.
Another recently proposed strategy for increasing the resolution and mapping power in mice is through the development of a very large set of RI strains, termed the Collaborative Cross (Churchill et al. 2004). Although it may not have gene-level resolution (Flint and Mott 2008), simulation studies have predicted a power of 94% for a QTL with 5% at a resolution of 1.75 Mb (Valdar et al. 2006a). Additionally, the Collaborative Cross will include many loci that are polymorphic among the wild-derived strains and a genetic structure more representative of the quantity of genetic variation present in human populations when it is completed, estimated to be 2012. Our current strain panel is only capable of mapping SNPs polymorphic in the set of strains within the HMDP and has limited ability to map loci that are polymorphic among the wild-derived strains or have low sequence diversity among the inbreds, since they are poorly represented in the HMDP. Once even a subset of the Collaborative Cross strains are fully backcrossed to homozygosity and genotyped, these mice would complement the methods described here and significantly increase the power to map additional loci.
In summary, we have utilized a “hybrid” strategy for association mapping in mice that combines CI strains, as well as RI strains, to address several key limitations of complex genetic mapping in mice: low resolution of linkage approaches, the high degree of false positive signals found in murine association mapping, and the critical need for permanent resources for systems-based approaches. Our simulated data indicated that such a hybrid population has sufficient power to detect seven out of 10 variations with a modest effect size of 10%. Our results from mapping complex metabolic traits and expression phenotypes support these simulations and validate this approach.
Methods
Animals
Male mice from the hybrid HMDP panel were purchased from the Jackson Labs. Mice were between 6 and 10 wk of age and to ensure adequate acclimatization to a common environment the mice were aged until 16 wk of age. All mice were maintained on a chow diet (Ralston-Purina Co.) until sacrifice at 16 wk of age. A complete list of strains included in the study is listed in Supplemental Table 1. Following a 16-h fast, mice were bled retro-orbitally under isoflurane anesthesia. Plasma lipids were determined as previously described (Mehrabian et al. 1993). Mice were euthanized by cervical dislocation, livers dissected out and flash frozen in liquid nitrogen.
Genotyping
Inbred strains were previously genotyped by the Broad Institute (http://www.broadinstitute.org/mouse/hapmap), and they are combined with the genotypes from Wellcome Trust Center for Human Genetics (WTCHG). Genotypes of RI strains at the Broad SNPs were inferred from WTCHG genotypes by interpolating alleles at polymorphic SNPs among parental strains, calling ambiguous genotypes missing. Details of genotype imputation are in Supplemental Methods. Of the 140,000 SNPs available, 107,145 were informative with an allele frequency greater than 5% and were used for GWAS.
RNA isolation and expression profiling
Flash frozen samples were weighed and homogenized in Qiazol according to the manufacturer's protocol. Following homogenization, livers were isolated in RNeasy 96 columns (Qiagen) using the manufacturer's protocol. The image data were processed using the Affymetrix GCOS algorithm, utilizing quantile normalization or the robust multiarray (RMA) method to determine the specific hybridizing signal for each gene. Expression data can be obtained from Gene Expression Omnibus (GEO) databases for liver (accession no. GSE16780). A detailed protocol of RNA processing is provided in the Supplemental Methods.
Genome-wide association mapping accounting for population structure
We applied the following linear mixed model to account for the population structure and genetic relatedness among strains in the genome-wide association mapping (Kang et al. 2008):
where μ represents mean, x represents SNP effect, u represents random effects due to genetic relatedness with Var(u) = σg2K and Var(e) = σe2, where K represents IBS (identity-by-state) matrix across all genotypes. A restricted maximum likelihood (REML) estimate of σg2 and σe2 are computed using EMMA, and the association mapping is performed based on the estimated variance component with a standard F-test to test β ≠ 0. A potential problem with our approach is that we assume the variance of the phenotype is the same for each strain. Unfortunately, the optimization technique utilized by EMMA, which increases the efficiency by two orders of magnitude over other mixed model implementations, requires this assumption (Kang et al. 2008). We are currently exploring extending the EMMA methodology to allow for multiple variance components that will allow us to incorporate different per-strain variance estimates. We defined an eQTL as local if the peak association signal was within a 10-Mb sliding window of the physical location of the gene(s). We then calculated the average distance between these local eQTL and the transcription start site of the corresponding gene(s) transcription start site.
Estimation of power and mapping resolution
We evaluated the statistical power of the HMDP through simulation studies with various parameters including the variance explained by SNP, variance explained by genetic background, and variance explained by random errors, and the number of repeated measurements per strain. For the comparison of power with single RI set or CI only studies, we selected a subset of the simulated phenotypes for each RI or CI set and evaluated the power in the same way. Since there are eight possibilities of SNPs being polymorphic among three sets of RI strains, the putative causal SNPs are categorized into eight classes and power is evaluated for each class. The significance threshold per each RI set is determined separately using parametric bootstrapping described below. See Supplemental Methods for comparison of the BXD RI set to the full HMDP and simulations.
Genome-wide significance threshold
Genome-wide significance threshold in genome-wide association mapping is determined by the family-wise error rate (FWER) as the probability of observing one or more false positives across all SNPs per phenotype. We ran 100 different sets of permutation tests and parametric bootstrapping of size 1000, and observed that the mean and standard error of the genome-wide significance threshold at FWER of 0.05 were 3.9 × 10−6 ± 0.3 × 10−6 and 4.0 × 10−6 ± 0.3 × 10−6, respectively. This is approximately an order of magnitude larger than the significance threshold obtained by Bonferroni correction (4.6 × 10−7). We also performed parametric bootstrapping under simulated genetic background effect from population structure using EMMA. With 50% and 100% of variance explained by genetic background, the thresholds were determined to be 1.6 × 10−6 ± 0.2 × 10−6 and 1.7 × 10−6 ± 0.2 × 10−6. The reduction in the significance threshold compared to no genetic background effect is due to the fact that inter-SNP correlation due to long-range LD reduces when conditioning on the population structure. A detailed explanation of these analyses is provided in the Supplemental Methods.
Validation of clinical and expression associations
We also compare eQTL and clinical HDL associations in the HMDP to QTL identified in a previously reported F2 cross between C3H/HeJ and C57BL/6J to demonstrate the improved resolution of the approach (Wang et al. 2007).
Acknowledgments
We thank Pingzi Wen, Yi Shou Shi, and Hannah Qi for their expert assistance with mouse care and experiments. This research was supported in part by US National Institute of Health NIH grants HL28481, HL30568, HL079353, and DK072206 and contract N01-ES-45530 to Perlegen Sciences. H.M.K, N.F., E.K., and E.E. were supported by the National Science Foundation grants 0513612, 0731455, and 0729049 and NIH grant 1K25HL080079. B.J.B. was supported by NIH Training grant T32-DK07789. C.R.F was supported by NIH Ruth L. Kirschstein F32 Fellowship 5F32DK074317. A.G. was supported by AHA0825204F from the American Heart Association.
Footnotes
[Supplemental material is available online at http://www.genome.org. The microarray data from this study have been submitted to the NCBI Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/geo) under accession no. GSE16780.]
Article published online before print. Article and publication date are at http://www.genome.org/cgi/doi/10.1101/gr.099234.109.
References
- Aherrahrou Z, Doehring LC, Ehlers EM, Liptau H, Depping R, Linsel-Nitschke P, Kaczmarek PM, Erdmann J, Schunkert H. An alternative splice variant in Abcc6, the gene causing dystrophic calcification, leads to protein deficiency in C3H/He mice. J Biol Chem. 2008;283:7608–7615. doi: 10.1074/jbc.M708290200. [DOI] [PubMed] [Google Scholar]
- Altshuler D, Daly MJ, Lander ES. Genetic mapping in human disease. Science. 2008;322:881–888. doi: 10.1126/science.1156409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beck JA, Lloyd S, Hafezparast M, Lennon-Pierce M, Eppig JT, Festing MF, Fisher EM. Genealogies of mouse inbred strains. Nat Genet. 2000;24:23–25. doi: 10.1038/71641. [DOI] [PubMed] [Google Scholar]
- Castellani LW, Nguyen CN, Charugundla S, Weinstein MM, Doan CX, Blaner WS, Wongsiriroj N, Lusis AJ. Apolipoprotein AII is a regulator of very low density lipoprotein metabolism and insulin resistance. J Biol Chem. 2008;283:11633–11644. doi: 10.1074/jbc.M708995200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervino AC, Darvasi A, Fallahi M, Mader CC, Tsinoremas NF. An integrated in silico gene mapping strategy in inbred mice. Genetics. 2007;175:321–333. doi: 10.1534/genetics.106.065359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Zhu J, Lum PY, Yang X, Pinto S, MacNeil DJ, Zhang C, Lamb J, Edwards S, Sieberts SK, et al. Variations in DNA elucidate molecular networks that cause disease. Nature. 2008;452:429–435. doi: 10.1038/nature06757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Churchill GA, Airey DC, Allayee H, Angel JM, Attie AD, Beatty J, Beavis WD, Belknap JK, Bennett B, Berrettini W, et al. The Collaborative Cross, a community resource for the genetic analysis of complex traits. Nat Genet. 2004;36:1133–1137. doi: 10.1038/ng1104-1133. [DOI] [PubMed] [Google Scholar]
- Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Yelensky R, Pe'er I, Gabriel SB, Daly MJ, Altshuler D. Efficiency and power in genetic association studies. Nat Genet. 2005;37:1217–1223. doi: 10.1038/ng1669. [DOI] [PubMed] [Google Scholar]
- Doolittle MH, LeBoeuf RC, Warden CH, Bee LM, Lusis AJ. A polymorphism affecting apolipoprotein A-II translational efficiency determines high density lipoprotein size and composition. J Biol Chem. 1990;265:16380–16388. [PubMed] [Google Scholar]
- Doss S, Schadt EE, Drake TA, Lusis AJ. Cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emilsson V, Thorleifsson G, Zhang B, Leonardson AS, Zink F, Zhu J, Carlson S, Helgason A, Walters GB, Gunnarsdottir S, et al. Genetics of gene expression and its effect on disease. Nature. 2008;452:423–428. doi: 10.1038/nature06758. [DOI] [PubMed] [Google Scholar]
- Farber CR, van Nas A, Ghazalpour A, Aten JE, Doss S, Sos B, Schadt EE, Ingram-Drake L, Davis RC, Horvath S, et al. An integrative genetics approach to identify candidate genes regulating BMD: Combining linkage, gene expression, and association. J Bone Miner Res. 2009;24:105–116. doi: 10.1359/JBMR.080908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flint J, Mott R. Applying mouse complex-trait resources to behavioural genetics. Nature. 2008;456:724–727. doi: 10.1038/nature07630. [DOI] [PubMed] [Google Scholar]
- Frikke-Schmidt R, Nordestgaard BG, Jensen GB, Tybjaerg-Hansen A. Genetic variation in ABC transporter A1 contributes to HDL cholesterol in the general population. J Clin Invest. 2004;114:1343–1353. doi: 10.1172/JCI20361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghazalpour A, Doss S, Kang H, Farber C, Wen PZ, Brozell A, Castellanos R, Eskin E, Smith DJ, Drake TA, et al. High-resolution mapping of gene expression using association in an outbred mouse stock. PLoS Genet. 2008;4:e1000149. doi: 10.1371/journal.pgen.1000149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grupe A, Germer S, Usuka J, Aud D, Belknap JK, Klein RF, Ahluwalia MK, Higuchi R, Peltz G. In silico mapping of complex disease-related traits in mice. Science. 2001;292:1915–1918. doi: 10.1126/science.1058889. [DOI] [PubMed] [Google Scholar]
- Gunderson KL, Steemers FJ, Lee G, Mendoza LG, Chee MS. A genome-wide scalable SNP genotyping assay using microarray technology. Nat Genet. 2005;37:549–554. doi: 10.1038/ng1547. [DOI] [PubMed] [Google Scholar]
- Guo Y, Weller P, Farrell E, Cheung P, Fitch B, Clark D, Wu SY, Wang J, Liao G, Zhang Z, et al. In silico pharmacogenetics of warfarin metabolism. Nat Biotechnol. 2006;24:531–536. doi: 10.1038/nbt1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y, Lu P, Farrell E, Zhang X, Weller P, Monshouwer M, Wang J, Liao G, Zhang Z, Hu S, et al. In silico and in vitro pharmacogenetic analysis in mice. Proc Natl Acad Sci. 2007;104:17735–17740. doi: 10.1073/pnas.0700724104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009;360:1759–1768. doi: 10.1056/NEJMra0808700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang GJ, Shifman S, Valdar W, Johannesson M, Yalcin B, Taylor MS, Taylor JM, Mott R, Flint J. High resolution mapping of expression QTLs in heterogeneous stock mice in multiple tissues. Genome Res. 2009;19:1133–1140. doi: 10.1101/gr.088120.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- The International HapMap Consortium. A haplotype map of the human genome. Nature. 2005;437:1299–1320. doi: 10.1038/nature04226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang HM, Zaitlen NA, Wade CM, Kirby A, Heckerman D, Daly MJ, Eskin E. Efficient control of population structure in model organism association mapping. Genetics. 2008;178:1709–1723. doi: 10.1534/genetics.107.080101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klein RF, Allard J, Avnur Z, Nikolcheva T, Rotstein D, Carlos AS, Shea M, Waters RV, Belknap JK, Peltz G, et al. Regulation of bone mass in mice by the lipoxygenase gene Alox15. Science. 2004;303:229–232. doi: 10.1126/science.1090985. [DOI] [PubMed] [Google Scholar]
- Liao G, Wang J, Guo J, Allard J, Cheng J, Ng A, Shafer S, Puech A, McPherson JD, Foernzler D, et al. In silico genetics: Identification of a functional element regulating H2-Eα gene expression. Science. 2004;306:690–695. doi: 10.1126/science.1100636. [DOI] [PubMed] [Google Scholar]
- Liu P, Wang Y, Vikis H, Maciag A, Wang D, Lu Y, Liu Y, You M. Candidate lung tumor susceptibility genes identified through whole-genome association analyses in inbred mice. Nat Genet. 2006;38:888–895. doi: 10.1038/ng1849. [DOI] [PubMed] [Google Scholar]
- Liu P, Vikis H, Lu Y, Wang D, You M. Large-scale in silico mapping of complex quantitative traits in inbred mice. PLoS One. 2007;2:e651. doi: 10.1371/journal.pone.0000651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lusis AJ, Attie AD, Reue K. Metabolic syndrome: From epidemiology to systems biology. Nat Rev Genet. 2008;9:819–830. doi: 10.1038/nrg2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manenti G, Galvan A, Pettinicchio A, Trincucci G, Spada E, Zolin A, Milani S, Gonzalez-Neira A, Dragani TA. Mouse genome-wide association mapping needs linkage analysis to avoid false-positive loci. PLoS Genet. 2009;5:e1000331. doi: 10.1371/journal.pgen.1000331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manolio TA. Cohort studies and the genetics of complex disease. Nat Genet. 2009;41:5–6. doi: 10.1038/ng0109-5. [DOI] [PubMed] [Google Scholar]
- Matsuzaki H, Dong S, Loi H, Di X, Liu G, Hubbell E, Law J, Berntsen T, Chadha M, Hui H, et al. Genotyping over 100,000 SNPs on a pair of oligonucleotide arrays. Nat Methods. 2004;1:109–111. doi: 10.1038/nmeth718. [DOI] [PubMed] [Google Scholar]
- McClurg P, Janes J, Wu C, Delano DL, Walker JR, Batalov S, Takahashi JS, Shimomura K, Kohsaka A, Bass J, et al. Genomewide association analysis in diverse inbred mice: power and population structure. Genetics. 2007;176:675–683. doi: 10.1534/genetics.106.066241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehrabian M, Qiao JH, Hyman R, Ruddle D, Laughton C, Lusis AJ. Influence of the apoA-II gene locus on HDL levels and fatty streak development in mice. Arterioscler Thromb. 1993;13:1–10. doi: 10.1161/01.atv.13.1.1. [DOI] [PubMed] [Google Scholar]
- Moran JL, Bolton AD, Tran PV, Brown A, Dwyer ND, Manning DK, Bjork BC, Li C, Montgomery K, Siepka SM, et al. Utilization of a whole genome SNP panel for efficient genetic mapping in the mouse. Genome Res. 2006;16:436–440. doi: 10.1101/gr.4563306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldham MC, Horvath S, Geschwind DH. Conservation and evolution of gene coexpression networks in human and chimpanzee brains. Proc Natl Acad Sci. 2006;103:17973–17978. doi: 10.1073/pnas.0605938103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Place M. Prospects for association mapping in classical inbred mouse strains. Genetics. 2007;175:1999–2008. doi: 10.1534/genetics.106.067868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petkov PM, Graber JH, Churchill GA, DiPetrillo K, King BL, Paigen K. Evidence of a large-scale functional organization of mammalian chromosomes. PLoS Genet. 2005;1:e33. doi: 10.1371/journal.pgen.0010033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petretto E, Mangion J, Pravanec M, Hubner N, Aitman TJ. Integrated gene expression profiling and linkage analysis in the rat. Mamm Genome. 2006;17:480–489. doi: 10.1007/s00335-005-0181-1. [DOI] [PubMed] [Google Scholar]
- Pletcher MT, McClurg P, Batalov S, Su AI, Barnes SW, Lagler E, Korstanje R, Wang X, Nusskern D, Bogue MA, et al. Use of a dense single nucleotide polymorphism map for in silico mapping in the mouse. PLoS Biol. 2004;2:e393. doi: 10.1371/journal.pbio.0020393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price AL, Patterson N, Hancks DC, Myers S, Reich D, Cheung VG, Spielman RS. Effects of cis and trans genetic ancestry on gene expression in African Americans. PLoS Genet. 2008;4:e1000294. doi: 10.1371/journal.pgen.1000294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell-Huynh DA, Weinreb A, Castellani LW, Mehrabian M, Doolittle MH, Lusis AJ. Genetic factors in lipoprotein metabolism. Analysis of a genetic cross between inbred mouse strains NZB/BINJ and SM/J using a complete linkage map approach. J Clin Invest. 1995;96:1845–1858. doi: 10.1172/JCI118230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Monks SA, Drake TA, Lusis AJ, Che N, Colinayo V, Ruff TG, Milligan SB, Lamb JR, Cavet G, et al. Genetics of gene expression surveyed in maize, mouse and man. Nature. 2003;422:297–302. doi: 10.1038/nature01434. [DOI] [PubMed] [Google Scholar]
- Schadt EE, Lamb J, Yang X, Zhu J, Edwards S, Guhathakurta D, Sieberts SK, Monks S, Reitman M, Zhang C, et al. An integrative genomics approach to infer causal associations between gene expression and disease. Nat Genet. 2005;37:710–717. doi: 10.1038/ng1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schadt EE, Molony C, Chudin E, Hao K, Yang X, Lum PY, Kasarskis A, Zhang B, Wang S, Suver C, et al. Mapping the genetic architecture of gene expression in human liver. PLoS Biol. 2008;6:e107. doi: 10.1371/journal.pbio.0060107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Flint J, Mott R. Simulating the collaborative cross: Power of quantitative trait loci detection and mapping resolution in large sets of recombinant inbred strains of mice. Genetics. 2006a;172:1783–1797. doi: 10.1534/genetics.104.039313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdar W, Solberg LC, Gauguier D, Burnett S, Klenerman P, Cookson WO, Taylor MS, Rawlins JN, Mott R, Flint J. Genome-wide genetic association of complex traits in heterogeneous stock mice. Nat Genet. 2006b;38:879–887. doi: 10.1038/ng1840. [DOI] [PubMed] [Google Scholar]
- Wang X, Paigen B. Genetics of variation in HDL cholesterol in humans and mice. Circ Res. 2005;96:27–42. doi: 10.1161/01.RES.0000151332.39871.13. [DOI] [PubMed] [Google Scholar]
- Wang X, Korstanje R, Higgins D, Paigen B. Haplotype analysis in multiple crosses to identify a QTL gene. Genome Res. 2004;14:1767–1772. doi: 10.1101/gr.2668204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Yehya N, Schadt EE, Wang H, Drake TA, Lusis AJ. Genetic and genomic analysis of a fat mass trait with complex inheritance reveals marked sex specificity. PLoS Genet. 2006;2:e15. doi: 10.1371/journal.pgen.0020015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang SS, Schadt EE, Wang H, Wang X, Ingram-Drake L, Shi W, Drake TA, Lusis AJ. Identification of pathways for atherosclerosis in mice: Integration of quantitative trait locus analysis and global gene expression data. Circ Res. 2007;101:e11–e30. doi: 10.1161/CIRCRESAHA.107.152975. [DOI] [PubMed] [Google Scholar]
- Warden CH, Hedrick CC, Qiao JH, Castellani LW, Lusis AJ. Atherosclerosis in transgenic mice overexpressing apolipoprotein A-II. Science. 1993;261:469–472. doi: 10.1126/science.8332912. [DOI] [PubMed] [Google Scholar]
- Willer CJ, Sanna S, Jackson AU, Scuteri A, Bonnycastle LL, Clarke R, Heath SC, Timpson NJ, Najjar SS, Stringham HM, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams RW, Gu J, Qi S, Lu L. The genetic structure of recombinant inbred mice: High-resolution consensus maps for complex trait analysis. Genome Biol. 2001;2:research0046.1–research0046.18. doi: 10.1186/gb-2001-2-11-research0046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang X, Schadt EE, Wang S, Wang H, Arnold AP, Ingram-Drake L, Drake TA, Lusis AJ. Tissue-specific expression and regulation of sexually dimorphic genes in mice. Genome Res. 2006;16:995–1004. doi: 10.1101/gr.5217506. [DOI] [PMC free article] [PubMed] [Google Scholar]