Abstract
Mice deficient for the transcription factor NFATc1 fail to form pulmonary and aortic valves, a defect reminiscent of some types of congenital human heart disease. We examined the mechanisms by which NFATc1 is activated and translocated to the nucleus in human pulmonary valve endothelial cells to gain a better understanding of its potential role(s) in post-natal valvular repair as well as valve development. Herein we demonstrate that activation of NFATc1 in human pulmonary valve endothelial cells is specific to vascular endothelial growth factor (VEGF) signaling through VEGF receptor 2. VEGF-induced NFATc1 nuclear translocation was inhibited by either cyclosporin A or a calcineurin-specific peptide inhibitor; these findings suggest that VEGF stimulates NFATc1 nuclear import in human pulmonary valve endothelial cells by a calcineurin-dependent mechanism. Importantly, both cyclosporin A and the calcineurin-specific peptide inhibitor reduced VEGF-induced human pulmonary valve endothelial cell proliferation, indicating a functional role for NFATc1 in endothelial growth. In contrast, VEGF-induced proliferation of human dermal microvascular and human umbilical vein endothelial cells was not sensitive to cyclosporin A. Finally, NFATc1 was detected in the endothelium of human pulmonary valve leaflets by immunohistochemistry. These results suggest VEGF-induced NFATc1 activation may be an important mechanism in cardiac valve maintenance and function by enhancing endothelial proliferation.
Defects in heart development are the most common congenital anomaly, occurring in 1% of all live births (1, 2). Some of the defects, especially those that involve the aortic and pulmonary valves, require surgical intervention, which can include replacement of a defective valve. In the past decade, researchers have elucidated some of the basic signaling interactions necessary for heart development. In addition to initiating heart development, many of these pathways are also required for optimal heart function in post-natal life (3). However, comparatively little is known about the interactions that control more specific events, for example, induction of cardiac valve formation (4). In the mouse, primordial heart valves known as cardiac cushions begin to form by embryonic day 9.5. These cushions appear as swellings in the atrioventricular junction and outflow tract. As development proceeds, the cushions contribute to chamber septation and ultimately result in development of four adult valves.
Initiation of endocardial cushion formation involves a distinct subset of endothelial cells in the cushion-forming area that undergo endothelial to mesenchymal transdifferentiation (EMT).1 It is believed that the underlying myocardium sends inductive signals to the endocardial cells, beginning the process of EMT (5). These newly formed mesenchymal cells migrate into the underlying extracellular matrix, where further remodeling transforms the cushions into fibrous leaflets. Studies have demonstrated that transforming growth factor-β (TGF-β) signaling (6) and the type III TGF-β receptor are required for EMT in avian models (7). Indeed, TGF-β-mediated EMT has been shown to occur in post-natal ovine aortic valve endothelial cells (8). More recently, the phenotypes of mice and zebrafish in which specific genes have been “knocked out” or mutated have provided new insight into the genes required for normal valve development.
For example, targeted gene deletion has demonstrated a specific requirement for NFATc1, also known as NFAT2, in valve development. Knockout of NFATc1 in the mouse leads to defective aortic and pulmonary valve development with subsequent death at embryonic days (E) 14–15 due to congestive heart failure (9, 10). The cardiac cushions in the outflow tract of these mice are hypoplastic, suggesting that lack of NFATc1 leads to dysregulation of an early step in cushion formation. Knockout of other NFAT proteins, such as NFATc2, c3, and c4, has no effect on valve development (11). Members of the NFAT family function as mediators of the CsA-sensitive calcineurin-NFAT signaling pathway. First discovered in the pathway of interleukin-2-mediated T-cell activation and proliferation (12), NFAT (nuclear factor in activated T cells) signaling has since been shown to be crucial for neuronal guidance, skeletal and cardiac muscle hypertrophy, and, as cited above, cardiac valve development (13–15).
The upstream signaling events regulating NFATc1 activity in the valve endothelium are unknown, although all known NFAT proteins are dependent upon cytosolic Ca2+ flux for nuclear translocation. Increased cytosolic Ca2+ leads to activation of calmodulin and ultimately calcineurin, a serine/threonine phosphatase. When activated, calcineurin dephosphorylates residues in the conserved N-terminal region of various NFAT isoforms (16, 17). This dephosphorylation prompts a conformational change in NFAT that exposes a previously inaccessible nuclear localization sequence. NFAT is then shuttled into the nucleus, where it interacts with other transcription factors, including AP-1 and NF-κB, to alter gene transcription (18). Although Ca2+ flux across the membrane is a ubiquitous signaling mechanism, specificity can be accomplished by differential expression of NFAT isoforms or use of specific ligand-receptor complexes that alter the pattern of calcium flux. Inhibitors of NFAT activation include the pharmacological agents CsA, FK506, and the synthetic peptide VIVIT. CsA and FK506 are immunosuppressant drugs that, in combination with immunophilins, bind to the catalytic subunit of calcineurin and inhibit its enzymatic activity (17). VIVIT is a hydrophobic 16-amino acid peptide that mimics the calcineurin-docking motif of NFAT proteins and thereby inhibits calcineurin-mediated activation of NFATc1 (20).
Recent studies have demonstrated that VEGF, an endothelial mitogen known to induce calcium mobilization upon receptor activation, may regulate NFATp activity in endothelial cells (21). VEGF is a potent stimulator of angiogenesis and vascular permeability with the ability to induce endothelial proliferation, migration, and survival in vitro (22). In human umbilical vein endothelial cells (HUVECs), VEGF has been shown to induce nuclear localization of NFATc2, also known as NFATp, by a CsA-sensitive mechanism, which resulted in increased tissue factor expression (23). In a separate study, administration of CsA prohibited VEGF from activating expression of cylcooxygenase-2 and inhibited angiogenesis in a corneal neovascularization model, again suggesting an important role for NFAT in vascular endothelial cells (24).
Studies of VEGF expression in transgenic mice provide evidence for its role in cardiac valve development. LacZ-tagged VEGF knock-in mice were found to express VEGF at E 9.0 in endocardial cells along the entire heart tube (25). By E 9.5, VEGF expression was restricted to endothelial cells of the outflow tract and atrioventricular valve area. This expression pattern coincides precisely with the timing and location of NFATc1 expression in the embryonic valve (9, 10). In a different study, transgenic mice that overexpress VEGF in the embryonic myocardium at E 10.5 demonstrated decreased EMT in the endocardial cushions (26, 27). Although these studies suggest a role for VEGF in heart valve development, little is known regarding the signaling cascade downstream of VEGF in valvular endothelial cells. To study the role of VEGF in NFATc1 signaling in endothelial cells, we performed experiments on human pulmonary valve endothelial cells (HPVEC). Our results demonstrate a unique pathway for VEGF-mediated proliferation of these cells involving an NFATc1-dependent mechanism.
EXPERIMENTAL PROCEDURES
Cell Culture Conditions
HPVEC were isolated from human pulmonary valve leaflets obtained from patients undergoing cardiovascular surgical procedures at Children’s Hospital, Boston under an institutional review board-approved protocol. Removal of the pulmonary valve was a planned part of each procedure. Patients’ ages ranged from 5 months to 20 years. The time interval from surgical excision to cell isolation was less than 1 h. Endothelial cells in primary cultures of human pulmonic valve leaflets were isolated using Ulex europaeus I-coated Dynabeads (28) as previously described (8). HPVECs were cultured on 1% gelatin-coated tissue culture plates in endothelial cell basal medium-2 (EBM-2, Clonetics) supplemented with endothelial cell growth media-2 SingleQuots (human VEGF, EGF, bFGF, insulin-like growth factor-1, ascorbic acid, gentamicin, and heparin; Clonetics), 20% heat-inactivated FBS, and glutamine/penicillin/streptomycin (GPS, Invitrogen). This culture media will be referred to as EBM-2. Cells were maintained in 5% CO2 at 37 °C. Human umbilical vein endothelial cells (HUVECs) were generously provided by Dr. F. W. Luscinskas at Brigham and Women’s Hospital, Boston. Human dermal microvascular endothelial cells (HDMEC) were isolated and cultured as described previously (29).
Endothelial Cell Characterization
Immunofluorescence was performed as previously described (8). Briefly, cells plated onto gelatin-coated glass coverslips were fixed with −20 °C methanol and incubated with primary antibody diluted 1:1000 followed by FITC- or Texas Red-conjugated secondary antibody at 5 µg/ml. von Willebrand factor (vWF) was detected using rabbit anti-human vWF (Dako) and Texas Red anti-rabbit IgG. CD31 (also known as platelet endothelial cell adhesion molecule, PECAM-1) was detected using goat anti-human CD31 (Santa Cruz Biotechnology) and Texas Red anti-goat IgG. To detect lipopolysaccharide (LPS)-induced E-selectin, cells were treated with 1 µg/ml LPS for 3 h prior to staining with mouse anti-human E-selectin (29) and FITC-anti-mouse IgG.
Growth Factor Stimulation
HPVEC were plated onto gelatin-coated glass cover slips for 1–3 days and then switched from EBM-2 medium to endothelial cell basal medium (EBM) containing 10% FBS (without the additive growth factors described above) for at least 24 h. Cells were then stimulated for 30 min with either 50 ng/ml VEGF165, bFGF (R&D Systems), or 1 µm calcium ionophore A23187 (Sigma). 5 µm CsA or 1 µm FK506 (Sigma) was added 2 h before VEGF stimulation. Cells were also stimulated with receptor-selective variants of VEGF: KDR-sel2 was shown to have wild-type affinity for KDR/VEGF-R2 but 2000-fold reduced binding to Flt-1, whereas Flt-1-sel was shown to have wild-type affinity for Flt-1 but 470-fold reduced binding to KDR/ VEGF-R2 (30). Drs. Bing Li and Abraham M. de Vos (Genentech, Inc., South San Francisco, CA) kindly provided these receptor-selective variants, as well as comparably produced wild-type VEGF (VEGFwt, amino acids 1–109). NFATc1 expression was analyzed by fixing cells in 4% paraformaldehyde, permeabilizing with 0.5% Triton X-100, and incubating with mouse anti-human NFATc1 mAb (7A6 from Santa Cruz Biotechnology) diluted 1:500 followed by FITC-conjugated anti-mouse IgG. An aliquot of the anti-human NFATc1 mAb for preliminary experiments was kindly provided by Dr. Gerald Crabtree (Stanford University). Cellular localizations of NFATc2, NFATc3, and NFATc4 were analyzed as described above using monoclonal antibodies (mAbs) purchased from Santa Cruz Biotechnology.
Western Blotting
Cell lysates were prepared as described previously (8) and subjected to SDS-PAGE using commercially available gradient gels. Gels were transferred to nitrocellulose membrane (Millipore). The membrane was rinsed in TBS-T solution (0.1% Tween 20 in TBS, pH 7.5) and incubated in blocking buffer (5% skim milk in TBS-T) at the room temperature. The membranes were incubated with the mouse anti-human NFATc1 (7A6 from Santa Cruz Biotechnology), mouse anti-human NFATc2 (G1-D10 from Santa Cruz Biotechnology), mouse anti-human NFATc3 (F-1 from Santa Cruz Biotechnology), or goat-anti-human NFATc4 (C-20 from Santa Cruz Biotechnology) antibodies diluted 1:500 for overnight at 4 °C, followed by horseradish peroxidase-labeled secondary antibodies diluted 1:2000 for 1 h at room temperature. The immunoreactive bands were visualized using chemiluminescent reagent (LumiGLO, KPL).
MTT Assay
1 × 104 cells/well were plated onto gelatin-coated 48-well plates. One day later, cells were washed with PBS and incubated with CsA at 1, 5, 10, and 20 µm in low serum (2% FBS) or high serum (20% FBS) containing EBM-2 media for 48 h. Five hours before the end of the incubation, 50 µl of MTT solution (5 mg/ml) was added into each well. Media was removed, and 500 µl of Me2SO was added to each well. Dissolved crystals were transferred into 96-well plates, and absorbance at 550 nm was measured.
Cell Proliferation Assay
Cell proliferation was assayed by [3H]thymidine incorporation (31). Cells were plated onto gelatin-coated 48-well plates at a density of 1 × 104 cells/well. The next day, cells were washed with PBS and incubated in thymidine-free endothelial basal labeling medium (Clonetics) with 2% FBS and GPS (starvation medium) for 24 h. The quiescent cells were stimulated with fresh starvation medium containing 10 ng/ml VEGF, KDR-sel2, or Flt-sel, with or without CsA. Approximately 16–24 h after growth factor stimulation, 0.7 µCi of [3H]thymidine (6.7 Ci/mmol, PerkinElmer Life Sciences) was added to each well, and incubations were continued for another 4 h. [3H]Thymidine DNA incorporation in the cell lysates was quantitated in a scintillation counter. Each condition was tested in triplicate, and the mean ± S.D. of [3H]thymidine DNA incorporation was calculated. Statistical significance was determined by Student’s t test, with a p value of <0.05 considered statistically significant. The -fold induction compared with cells with no VEGF stimulation was also determined. Proliferation assays on retrovirally transduced HPVEC were performed as above after the GFP-positive cells were sorted by FACS.
Northern Blot Analysis
Total RNA was isolated from human endothelial cells using an RNeasy kit (Qiagen). Blots were hybridized with 32P-labeled cDNA probes for KDR/VEGFR2, Flt-1/VEGF-R1, and neuropilin-1.
Retroviral Transduction
The retroviral expression vectors, retro-EGFP, and retro-EGFP-VIVIT (contains an oligonucleotide coding for MAGPHPVIVITGPHEE at the N terminus of the green fluorescent protein (GFP)), were kindly provided by Dr. A. Blauvelt at NCI, National Institutes of Health and have been previously described (32). For viral production, 293 cells at 70% confluence were transfected in Dulbecco’s modified Eagle’s medium with 2 µg of retroviral expression vector and 2 µg of the packaging vector, pCL-10A1 (33), using the Effectene transfection reagent (Qiagen) according to the manufacturer’s directions for a 100-mm dish. After 6 h, an equal volume of Dulbecco’s modified Eagle’s medium containing 20% FBS, 1% GPS was added for a final concentration of 10% FBS. After 24 h, medium was replaced with 6 ml of EBM containing 10% FBS, 1% GPS, and, after an additional 24 h, supernatants were collected and filtered (0.45 µm). For retroviral infection, HPVEC at 50% confluence were incubated twice for 4 h with 5 ml of the viral supernatant containing 8 µg/ml Polybrene. 5 ml of EBM-2 was added after the second infection, and, after 24 h, the cells were washed and recovered in EBM-2. Cells positive for high levels of GFP expression (fluorescence signal > 102) were sorted by FACS and immediately plated onto a 48-well tissue culture dish for cell proliferation assays as described above. Similar transduction efficiencies of 40–50% were obtained using either retro-EGFP or retro-EGFP-VIVIT. To confirm that VIVIT was effectively inhibiting NFATc1 nuclear translocation, GFP-positive HPVEC were also plated in parallel onto gelatin-coated glass coverslips, cultured in EBM supplemented with 10% FBS for at least 24 h, and then stimulated with 50 ng/ml VEGF for 30 min. NFATc1 cellular localization was detected with immunofluorescence using mouse anti-human NFATc1 at 1:500 dilution followed by biotinylated anti-mouse IgG (H+L) and Texas Red Avidin D, both at 5 µg/ml.
Immunohistochemistry
Normal human pulmonary valve leaflets (n = 3) were obtained from autopsies with post-mortem intervals ranging from 5 to 20 h according to a protocol approved by the Human Research Committee at the Brigham and Women’s Hospital, Boston, MA. The mean age of the patients was 46.8 years. Formalin-fixed paraffin-embedded human pulmonary valve tissues were sectioned in 5-µm slices. After antigen retrieval, sections were preincubated with 0.3% hydrogen peroxide to inhibit endogenous peroxidase activity and then incubated with primary anti-human CD31 mAb, anti-human NFATc1 mAb, or mouse IgG1 control mAb diluted in phosphate-buffered saline supplemented with 4% of the species-respective normal serum. After washing with phosphate-buffered saline, species-appropriate biotinylated secondary antibodies were applied, followed by avidin-peroxidase complex (Vectastain ABC kit, Vector Laboratories). The reaction was visualized with 3-amino-9-ethyl carbazole substrate (Sigma Chemical Co). Sections were counterstained with Gill’s hematoxylin solution (Sigma) and mounted.
RESULTS
Isolation and Characterization of HPVEC
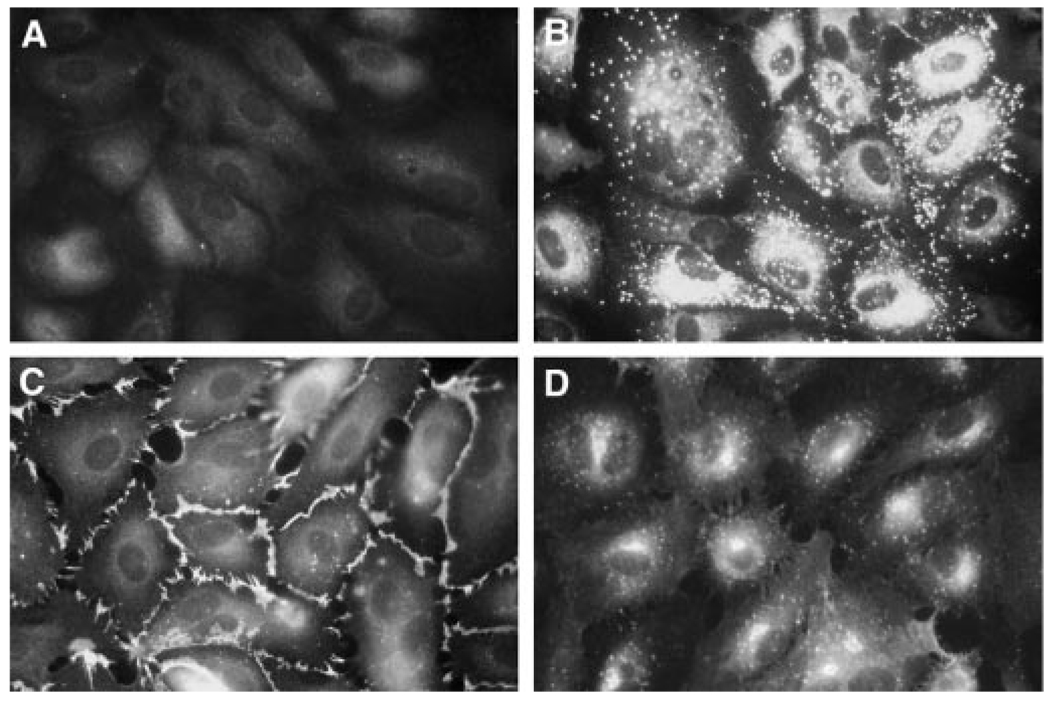
The function of NFATc1 in valvular endothelium was studied using HPVEC isolated from human pulmonary valve leaflets. The cells uniformly expressed the endothelial-specific markers von Willebrand factor (vWF), CD31/PECAM-1, and LPS-inducible Eselectin, demonstrating that HPVECs retain expression of endothelial-specific markers in culture (Fig. 1).
FIG. 1. HPVECs express endothelial-specific markers.
HPVEC were stained by indirect immunofluorescence using rabbit control IgG (A), rabbit anti-human von Willebrand Factor (vWF) (B), goat anti-human CD31/PECAM-1 (C), or mouse anti-human E-selectin (D). In panel D, HPVEC were stimulated with 1 µg/ml LPS for 3 h to induce E-selectin expression. Photographs were taken at 630× magnification.
VEGF Stimulates NFATc1 Nuclear Translocation by a KDR/VEGF-R2-specific and Calcineurin-dependent Mechanism
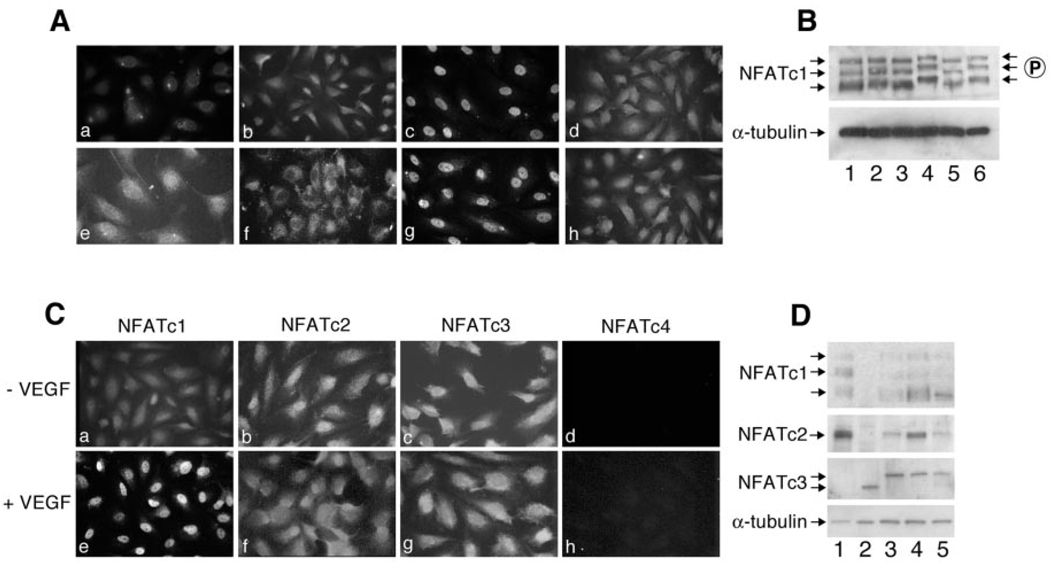
To identify extracellular ligands that stimulate NFATc1 activation in valvular endothelial cells, growth factors were tested for their ability to induce NFATc1 nuclear import in HPVEC. HPVEC were stimulated with 50 ng/ml VEGF, KDR-sel2, Flt-sel, or bFGF and then immunostained with an antibody specific for NFATc1. VEGF (50 ng/ml) was shown previously to be sufficient for stimulating NFATc2 (also known as NFATp) nuclear localization (23). VEGF stimulated the nuclear translocation of NFATc1 (Fig. 2A, panel c) compared with untreated HPVECs (Fig. 2A, panel b), at concentrations of VEGF as low as 10 ng/ml (data not shown). The pharmacological agents CsA (Fig. 2A, panel d) and FK506 (data not shown) inhibited the VEGF-induced nuclear translocation, suggesting a calcineurin-dependent mechanism. As expected, the calcium ionophore A23187 also stimulated NFATc1 nuclear import (data not shown). KDR-sel2 stimulated NFATc1 nuclear translocation (Fig. 2A, panel g), and, as observed with VEGF, the translocation was inhibited when CsA was included (Fig. 2A, panel h). In contrast, Flt-1-sel did not induce translocation (Fig. 2A, panel f), indicating that VEGF-induced NFATc1 translocation is mediated by KDR/VEGF-R2 and not Flt-1/VEGF-R1. When cells were treated with other endothelial mitogens such as bFGF, NFATc1 remained in the cytosol, as evident by a diffuse pattern of immunofluorescence (Fig. 2A, panel e). Other growth factors, such as insulin-like growth factor-1, EGF, and heparin-binding EGF, and the endothelial factors angiopoietin-1 and -2, had no effect on NFATc1 localization (data not shown). These results demonstrate that the mechanism of NFATc1 activation and nuclear translocation in HPVEC is specific to VEGF signaling events mediated by KDR/VEGF-R2.
FIG. 2. VEGF stimulates NFATc1 nuclear translocation by a calcineurin-dependent mechanism.
A, HPVEC were stimulated for 30 min with no growth factor (a and b), 50 ng/ml VEGF (c and d), 50 ng/ml bFGF (e), or 50 ng/ml Flt-1sel (f) or 50 ng/ml KDR-sel-2 (g and h). HPVECs were incubated with a mouse isotyped-matched control IgG1 (a) or with mouse anti-human NFATc1 (b–h), followed by anti-mouse-conjugated FITC. In panels d and h, HPVEC were preincubated with 5 µm CsA for 2 h prior to addition of VEGF and KDR-sel-2, respectively. Photographs were taken at 630× magnification. In B, cell lysates from HPVEC in EBM-2 with growth factors (lane 1), EBM-2 without growth factors for 24 h (lanes 2–6) were analyzed for expression of NFATc1 by Western blot. Cells were stimulated with VEGF for 15 min (lanes 3 and 4) or 24 h (lanes 5 and 6) in the absence (lanes 3 and 5) or presence (lanes 4 and 6) of 5 µm CsA. α-Tubulin levels in each cell lysate are shown as a control. C, expression of NFATc1, NFATc2, NFATc3, and NFATc4 in HPVECs treated without (panels a–d) or with (panels d–h) VEGF was analyzed as described in A using family member-specific mAbs. D, cell lysates from human lymphoma cell lines (lanes 1 and 2), HPVEC (lane 3), HUVEC (lane 4), and HDMEC (lane 5) were analyzed by Western blot using family member-specific mAbs against NFATc1, NFATc2, and NFATc3. α-Tubulin levels in the cell lysates are shown as a control.
NFATc1 activation was further examined by Western blot (Fig. 2B). Dephosphorylation of NFATc1 results in increased mobility on SDS-PAGE (20). As seen in Fig. 2B, three variants of NFATc1, with apparent molecular masses between 90 and 140 kDa, were detected with anti-human NFATc1 mAb (Fig. 2B, lane 1). Removal of growth factors from the culture medium for 24 h had no effect on migration of the NFATc1 isoforms (lane 2). Treatment with VEGF for 15 min (lane 3) or for 24 h (lane 5) resulted in an increase in electrophoretic mobility of all three isoforms. This apparent dephosphorylation at 15 min and 24 h was inhibited in the presence of 5 µm CsA (lanes 4 and 6). Tubulin expression in each cell lysate is shown as a control (Fig. 2B, lower panel).
NFATc2 and NFATc3 Are Expressed in HPVEC but Not Translocated into the Nucleus in Response to VEGF
VEGF-induced translocation of NFATc2, NFATc3, and NFATc4 was examined as described in Fig. 2A to determine which NFAT family members are activated by VEGF. NFAT family members c1, c2, and c3, but not c4, were detected in the cytoplasm of HPVEC in non-stimulated cells by indirect immunofluorescence using isoform-specific mAbs (Fig. 2C, panels a–d). However, only NFATc1 was translocated into the nucleus when HPVEC were treated with 50 ng/ml VEGF for 30 min (Fig. 2C, panels e–h). In Fig. 2D, protein expression of NFATc1, c2, and c3 was analyzed by Western blot of HPVEC (lane 3), HUVEC (lane 4), and HDMEC (lane 5) cell lysates. Human lymphoma cell lines served as positive controls (lanes 1 and 2). As seen in Fig. 2D, NFATc1, c2, and c3 were detected in all three human endothelial cultures. NFATc4 was not detected by Western blot (data not shown). NFATc2 expression was highest in HUVEC, compared with NFATc1 and NFATc3, consistent with previous studies showing NFATc2 (i.e. NFATp) in HUVECs. Also consistent with Armesilla et al. (23), NFATc2 was translocated into the nucleus of HUVECs in response to VEGF (data not shown). These results suggest that VEGF specifically induces translocation of NFATc1, but not NFATc2 or NFATc3, in valvular endothelial cells.
VEGF Induces Proliferation by a CsA-sensitive Mechanism in HPVEC but Not in HDMEC or HUVEC
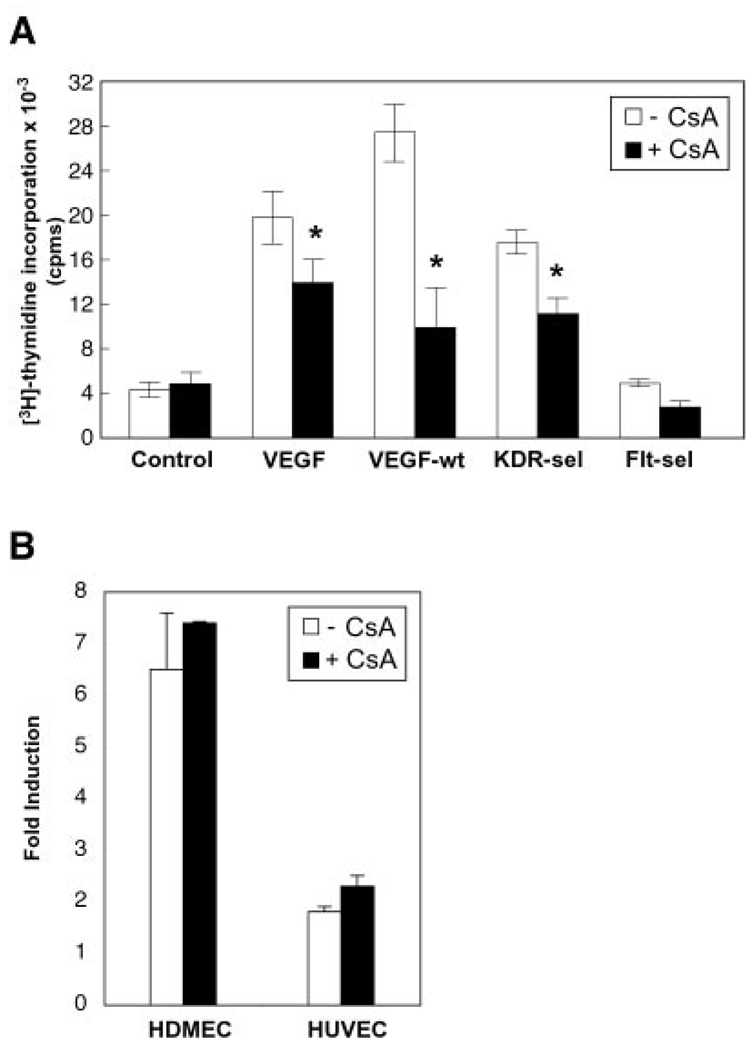
We next sought to investigate the cellular effects of preventing VEGF-induced NFATc1 nuclear import. Because VEGF is a mitogen of endothelial cells (21) and NFAT signaling can increase proliferation in other cell types (16), we used CsA to test whether VEGF induced valvular endothelial proliferation by an NFATc1-dependent mechanism. First, the potential cytotoxicity of CsA (34) was addressed. MTT assays were performed on HPVEC, HDMEC, and HUVEC at 1, 5, 10, and 20 µm CsA in the presence of 2 and 20% FBS (2% FBS was used in the growth factor-induced proliferation assays). Cytotoxicity was not detected at 1, 5, or 10 µm CsA in these three human endothelial cell cultures in either 2% or 20% FBS (data not shown). Quiescent HPVEC were stimulated with VEGF, VEGFwt, KDR-sel2, or Flt-sel (30), in the absence or presence of 5 µm CsA (Fig. 3A). Cell proliferation was assayed by [3H]thymidine incorporation. We found that VEGF, VEGFwt, and KDR-sel 2 induced HPVEC proliferation by 4- to 6-fold in the absence of CsA (Fig. 3A, open bars), but the proliferation was attenuated significantly by CsA (Fig. 3A, black bars). CsA inhibited VEGF-induced proliferation by 30% (p = 0.032), VEGFwt-induced proliferation by 64% (p = 0.031), and KDR-sel2-induced proliferation by 37% (p = 0.004). A similar inhibitory effect was observed with FK506 (data not shown). Flt-sel did not induce proliferation compared with control cells. Thus, VEGF stimulates HPVEC proliferation, via KDR/VEGF-R2, by a calcineurin-dependent mechanism that may be dependent upon the dephosphorylation and nuclear translocation of NFATc1. This result was observed in cultures of HPVEC isolated from the pulmonary valve leaflets of three different patients.
FIG. 3. VEGF induces proliferation by a cyclosporin-sensitive mechanism in HPVEC but not in HDMEC or HUVEC.
A, quiescent HPVEC were stimulated with 10 ng/ml VEGF or with 10 ng/ml of the VEGF receptor-selective variants, VEGFwt, KDR-sel-2, or Flt-sel, in the absence (open bars) or presence (black bars) of 5 µm CsA. Cell proliferation was assayed by [3H]thymidine incorporation. Data represent mean ± S.D. of a representative experiment (n = 3), each performed in triplicate. Asterisks denote a statistically significant (p < 0.05) difference. B, HDMEC and HUVEC were stimulated with 10 ng/ml VEGF in the absence (open bars) or presence (black bars) of 5 µm CsA. Data are plotted as -fold induction. The change in VEGF-mediated proliferation of these cell types after addition of CsA was not statistically significant.
The CsA-mediated inhibition of VEGF-induced proliferation appears to be specific to the valve endothelium, because proliferation of HDMEC and HUVEC was not inhibited by CsA-mediated NFATc1 inactivation (Fig. 3B). 5 µm CsA was sufficient to inhibit VEGF-induced NFATc1 nuclear translocation in HDMEC and HUVEC (data not shown). It is unlikely that this cell-specific CsA effect is due to differences in VEGF receptor expression, because mRNA expression levels for KDR/VEGF-R2, neuropilin-1, and Flt-1/VEGF-R1 did not vary more than 2-fold among the three types of human endothelial cells tested (data not shown). It is possible that the interaction of NFATc1 with valve-specific nuclear factors accounts for the CsA-sensitive proliferation response to VEGF observed in HPVECs.
The NFAT-specific Inhibitor, VIVIT, Inhibits VEGF-induced Proliferation of HPVEC
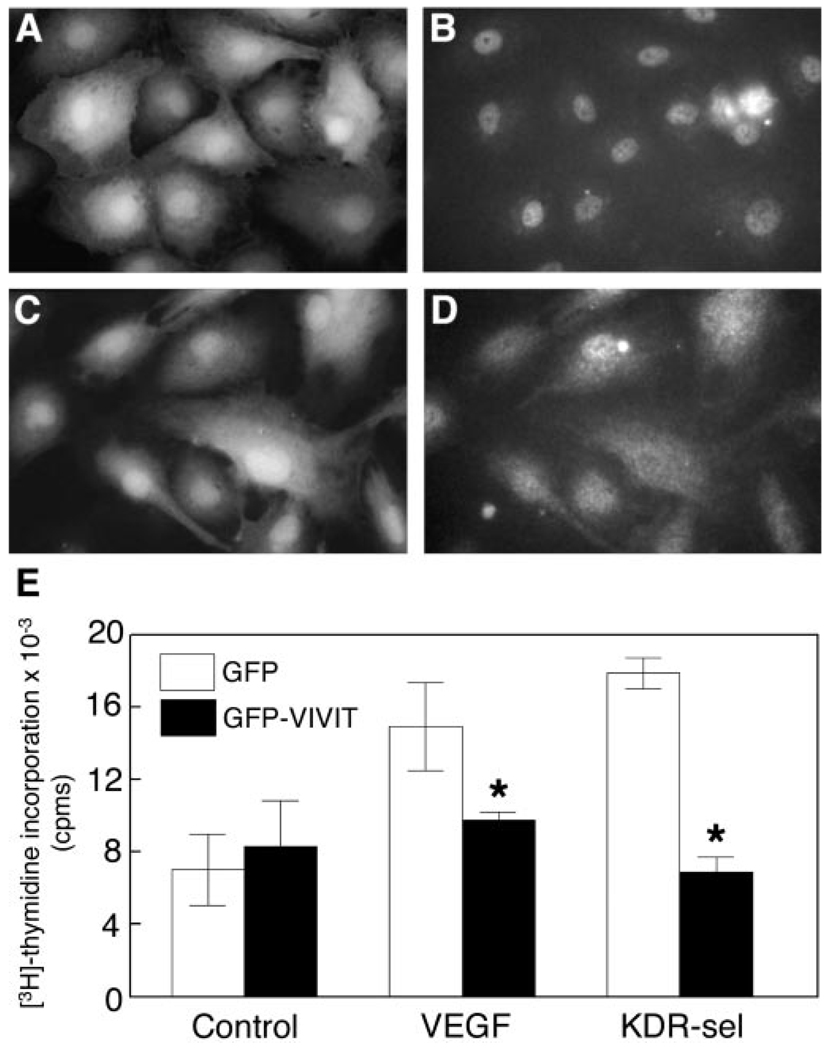
Because CsA and FK506 can potentially disrupt calcineurin-dependent pathways other than the activation of NFAT, we used the synthetic peptide VIVIT to inhibit NFAT activation selectively without disrupting other calcineurin-dependent pathways (20). We demonstrated that VIVIT inhibits VEGF-induced NFATc1 nuclear translocation and that this results in inhibition of VEGF-mediated HPVEC proliferation (Fig. 4). HPVEC retrovirally transduced with GFP-VIVIT (an oligonucleotide coding for VIVIT-containing peptide fused to the N terminus of the green fluorescent protein) did not import NFATc1 to the nucleus upon VEGF stimulation (Fig. 4, C and D). HPVEC expressing GFP alone did show VEGF-induced NFATc1 nuclear localization (Fig. 4, A and B), indicating that the inhibition of NFATc1 nuclear import is specific to the VIVIT peptide and that NFATc1 nuclear shuttling was not disrupted by retroviral transduction. VEGF-induced proliferation was inhibited by 35% (p = 0.022), whereas KDR-sel2-induced proliferation was inhibited 62% (p = 0.001) (Fig. 4E). Although VIVIT peptide could potentially inhibit NFATc2 and c3, our data show that these two family members are not translocated to the nucleus in response to VEGF (Fig. 2C) and therefore would not be functionally disrupted in this experiment. These data provide strong evidence that nuclear translocation of NFATc1 is required for maximal VEGF-induced proliferation of HPVEC. The smaller -fold induction observed in this experiment compared with Fig. 3 was likely due to the sequential retroviral transduction, FACS sorting, and 24 h serum starvation prior to measuring VEGF-induced proliferation.
FIG. 4. VIVIT inhibits VEGF-induced proliferation of HPVEC.
HPVEC were retrovirally transduced with GFP or GFP-VIVIT. The brightest GFP-positive cells (fluorescence signal > 102) were sorted for analysis of NFATc1 localization in GFP-positive (A and B) and GFP-VIVIT-positive (C and D) cells and for proliferation assays (E). GFP-positive and GFP-VIVIT-positive sorted cells are shown in A and C, respectively. For indirect immunofluorescence, the same cells were stimulated with 10 ng/ml VEGF for 30 min followed by staining with anti-human NFATc1 mAb, followed by a Texas Red-conjugated anti-mouse IgG (B and D). Photographs were taken at 630× magnification. For proliferation assays (E), quiescent HPVEC were stimulated with 10 ng/ml VEGF or KDR-sel-2 for 24 h and assayed for [3H]thymidine incorporation. Data represent mean ± S.D. of a representative experiment (n = 3), each performed in triplicate. Asterisks denote a statistically significant (p < 0.05) difference.
NFATc1 Expression in Adult Human Pulmonary Valve Leaflets
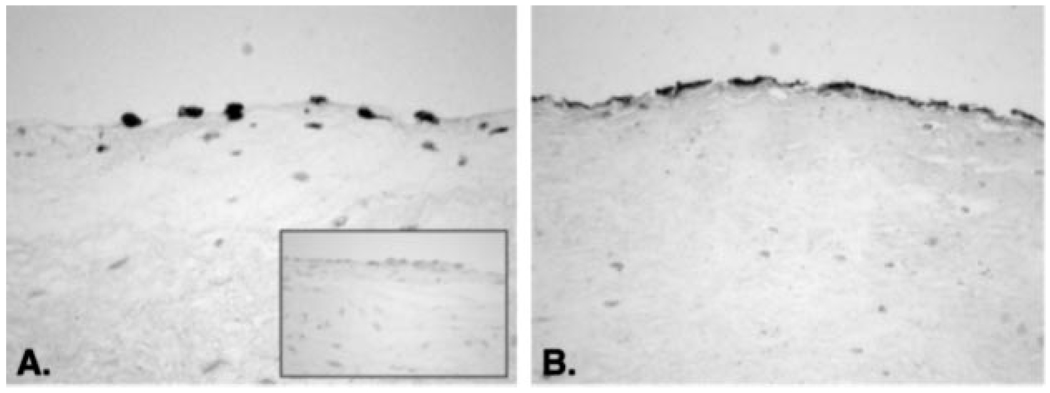
In the mouse, expression patterns of NFATc1 in the developing heart have been examined thoroughly to gain insights into its role in valvulogenesis. NFATc1 can be detected in developing murine hearts beginning at embryonic day E7.5, becomes increasingly restricted to nascent valvular structures by E11.5, but is then undetectable by either reverse transcription-PCR or immunohistochemistry, after E13.5 (9, 10). NFATc1 has been reported undetectable in newborn or adult murine valves as well (9). Despite the lack of reported evidence for NFATc1 expression in post-natal murine valves, we examined NFATc1 protein expression in human adult pulmonary valve leaflets, because our experiments in cultured HPVEC show that NFATc1 is expressed in adult HPVECs and suggest that NFATc1 may play an important physiological role in post-natal valve leaflets. Paraffin sections from human pulmonary and aortic valve leaflets were stained with anti-human NFATc1 or anti-human CD31/PECAM-1 mAbs. Fig. 5 shows representative immunohistochemical staining seen in human pulmonary valve leaflet tissue (n = 3). NFATc1 was detected in a nuclear-localized pattern along the surface of the leaflets (Fig. 5A), coinciding with CD31-positive endothelial cells in an adjacent tissue section (Fig. 5B). The inset in Fig. 5A shows staining with mouse IgG1 as a negative control. Regions of NFATc1-positive endothelial cells were seen intermittently along the leaflet surface, suggesting NFATc1 is expressed in a subset of endothelial cells in adult valves. This observation, combined with the expression of NFATc1 in HPVEC isolated from post-natal pulmonary valve leaflets, obtained from patients ranging in age from 5 months to 20 years of age, indicates that NFATc1 can be expressed focally along the endothelium of post-natal valves.
FIG. 5. Expression and localization of NFATc1 in human pulmonary valve leaflets.
Serial sections of formalin-fixed paraffin-embedded sections human pulmonic valve leaflets were immunostained with anti-human NFATc1 mAb (A) or with anti-human CD31 mAb (B), and endothelial marker. The inset in A shows a section stained with an isotype-matched control mouse IgG1.
DISCUSSION
We show here that HPVEC require nuclear translocation of NFATc1 for maximal VEGF-induced proliferation. Furthermore, NFATc1 nuclear translocation and resulting proliferation is mediated by KDR/VEGF-R2. These results are specific to HPVEC, because addition of CsA did not affect VEGF-induced proliferation of HUVEC and HDMEC (Fig. 3). The expression of other NFAT family members, specifically NFATc2 and NFATc3, was detected in HPVEC, but these proteins were not translocated into the nucleus in response to VEGF (Fig. 2, C and D). This specificity suggests that VEGF and NFATc1 signaling interact to activate a proliferative pathway unique to cardiac valvular endothelial cells.
Our results show a functional relationship between VEGF and NFATc1 in post-natal valvular endothelial cells. Several reports in the literature suggest that a similar pathway may also operate in endocardial cells during cardiac valve formation in the developing mouse. During embryogenesis, NFATc1 is initially expressed in the endocardium by day E8.5. By E11.5–E13.5, NFATc1 is expressed in the lining of the pulmonic, aortic, and atrioventricular valves (9, 10). Similarly, VEGF is expressed in the endocardial cells lining the endocardial cushions at day E8.5 as well as in the myocardium from E9.5 to E13.5 (25). Interestingly, endocardial cells that have undergone transdifferentiation to mesenchymal cells are negative for both NFATc1 and VEGF expression (9, 10, 25), suggesting that NFATc1 and VEGF play pivotal roles both spatially and temporally in development of the endocardial cushion.
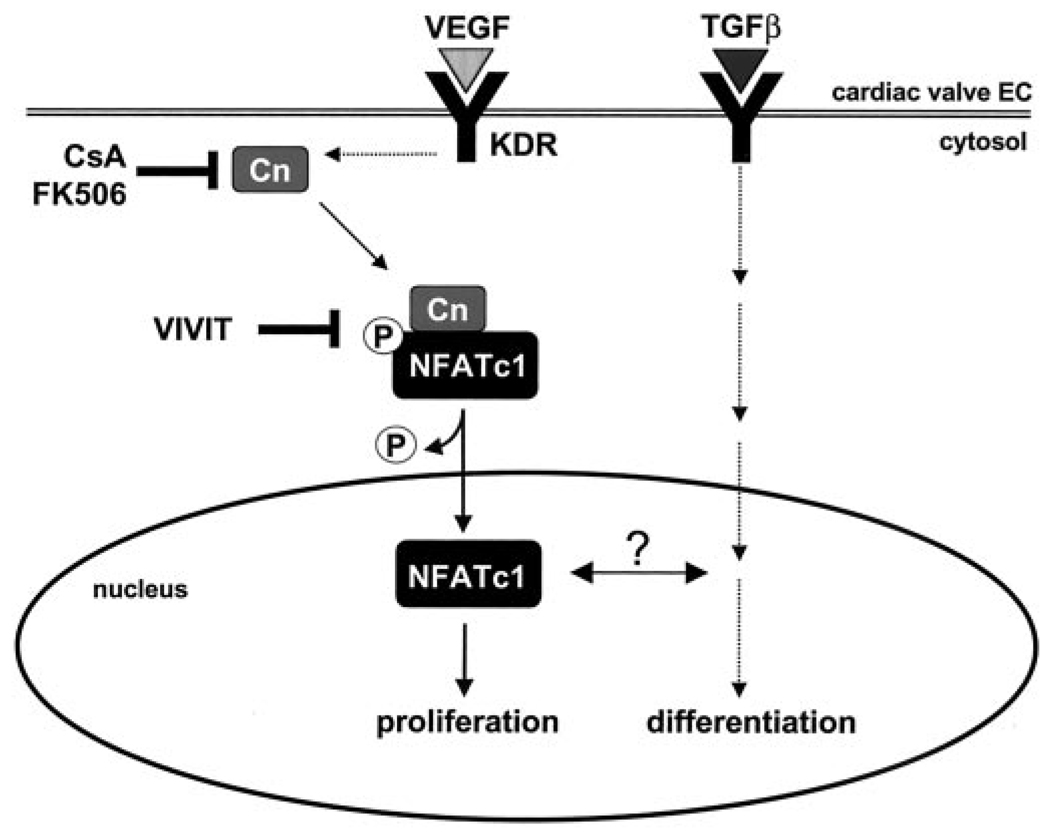
Given our results that VEGF stimulation leads to strong HPVEC proliferation in vitro, one might hypothesize that overexpression of VEGF would result in hyperplastic cardiac valves. However, the opposite result is observed in mice. Premature induction of VEGF expression in developing mouse embryos results in decreased endocardial cushion formation (27). This is phenotypically the same result observed with the NFATc1 murine knockout (9, 10). To explain these findings, we propose that VEGF-mediated NFAT signaling influences a cascade of events, including proliferation, migration, and differentiation, that is critical for valve development and post-natal valvular endothelial function (Fig. 6). In this model, VEGF induces proliferation of valvular endothelial cells and at the same time may influence the TGF-β-induced differentiation to a mesenchymal phenotype. Inhibition of VEGF signaling, either by genetic ablation of NFATc1 or by addition of CsA, would reduce the number of endothelial cells available to undergo EMT.
FIG. 6. Model for VEGF-induced proliferation in cardiac valve endothelium.
These results and previous studies (8) demonstrate that two pathways present during valvulogenesis are also active in post-natal heart valves. In this study, we show that a VEGF-mediated pathway, signaling through KDR/VEGF-R2, results in intranuclear NFATc1 and valvular endothelial cell proliferation. A TGF-β-mediated pathway induces differentiation to a mesenchymal phenotype (8). Whether or not cross-talk between these two pathways, as has been shown in the mouse (27), occurs in post-natal valves and affects post-natal valve function or repair warrants investigation.
We postulate that this model may apply to heart valve regeneration and repair. Because our experiments were carried out in postnatal cells, we hypothesize that the signaling pathways used during development are re-established in mature valve leaflets to replenish endothelial and interstitial cells as needed throughout adult life. The focal expression of NFATc1 in mature valve leaflets (Fig. 5) is consistent with this hypothesis. The source of VEGF that would activate NFATc1 expressed in the leaflet endothelium is unknown. Possible sources of VEGF include the endothelial cells, the mesenchymal interstitial cells, or release of VEGF from circulating cells in the blood. Also consistent with the repair concept, Paranya et al. (8) demonstrated that clonal populations of valve endothelial cells isolated from post-natal valves can undergo EMT in a manner similar to what occurs in fetal valve development. Therefore, TGF-β-mediated EMT could be a common mechanism in both developing and adult heart valves, whereas VEGF-mediated proliferation may be a mechanism for repopulation of the valvular endothelium. VEGF may also elicit other endothelial responses in the valve. Further studies will be required to determine the full spectrum of VEGF-induced events in cardiac valve endothelium.
Our results demonstrate a unique signaling pathway for endothelial cell proliferation. Previous studies have demonstrated that VEGF signal transduction in endothelial cells is dominated by inositol 3,4,5-phosphate/diacylglycerol and extracellular-related kinase (ERK)/mitogen-activated proliferation kinase (MAPK) pathways (35), the latter being dependent on Ras activation (36). Activation of inositol 3,4,5-phosphate leads to mobilization of cytosolic Ca2+, which may ultimately stimulate calcineurin to dephosphorylate NFATc1. Because [3H]thymidine incorporation in HPVECs was reduced ~30–60% after administration of CsA (Fig. 3), calcineurin appears to be an important mediator for VEGF stimulation of HPVEC proliferation. We used the VIVIT peptide to dissect this signaling pathway further and to circumvent potential endothelial toxicity that has been observed with CsA (34). This approach revealed that expression of VIVIT in valvular endothelial cells reduced HPVEC proliferation by 30–60%. A role for ERK and MAPK signaling in HPVECs remains to be defined but may represent the remaining proliferative activity of HPVECs after addition of CsA or VIVIT peptide.
In conclusion, our data provide evidence that VEGF and the receptor KDR/VEGF-R2 are upstream mediators of NFATc1 activation and nuclear translocation in HPVEC, which may lead to the expression of endothelial-specific genes required for valvular endothelial cell proliferation. To elucidate fully the function of NFATc1 in heart valve formation and function, it will be essential to identify additional upstream regulators of NFATc1 activity and downstream valve-specific target genes whose expression is mediated by NFATc1. We speculate that expression of NFATc1 in subsets of endothelium along the native valve leaflets from human adult pulmonary and aortic valve specimens may be a repair mechanism for replacing damaged endothelial cells. Although we have demonstrated an important role for NFATc1 in mediating valvular endothelial proliferation in cultured HPVEC, its functional role in mature heart valves in vivo must be investigated. Understanding the signaling mechanisms controlling valve endothelium proliferation and differentiation will provide insights on heart valve disease and for creating tissue-engineered valves that mimic normal valve function.
Acknowledgments
We thank Drs. Bing Li and Abraham M. de Vos at Genentech Inc., South San Francisco, CA for generously providing the receptor-selective variants of VEGF. We also thank Dr. Andrew. Blauvelt at NCI, National Institutes of Health, for the retroviral expression vectors and Drs. D. Friedman and S. Soker at Children’s Hospital, Boston, for providing the retroviral packaging vector and cDNA probes, respectively. We also thank Dr. Gerald Crabtree at Stanford University for providing an aliquot of the anti-human NFATc1 (clone 7A6) for pilot experiments. Finally, we thank Alan Flint in the Genetics Division at Children’s Hospital for FACS of GFP-positive cells.
Footnotes
This work was supported by the National Institute of Health (Grant RO1-HL60490 to J. B.), a gift from the Harvey-Wolff family, and a stipend from the Academic Societies for Student Research at Harvard Medical School (to E. J.).
The abbreviations used are: EMT, endothelial to mesenchymal transdifferentiation; TGF-β, transforming growth factor-β; CsA, cyclosporin A; HUVEC, human umbilical vein endothelial cell; HPVEC, human pulmonary valve endothelial cell; HDMEC, human dermal microvascular endothelial cell; mAbs, monoclonal antibodies; GFP, green fluorescent protein; EGFP, enhanced GFP; E, embryonic day; EGF, epidermal growth factor; VEGF, vascular endothelial growth factor; EBM-2, endothelial cell basal medium-2; FBS, fetal bovine serum; bFGF, basic fibroblast growth factor; GPS, glutamine/penicillin/streptomycin; FITC, fluorescein isothiocyanate; vWF, von Willebrand factor; PECAM-1, platelet endothelial cell adhesion molecule 1; LPS, lipopolysaccharide; wt, wild-type; MTT, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide; FACS, fluorescence-activated cell sorting; ERK, extracellular-related kinase; MAPK, mitogen-activated proliferation kinase; KDR, kinase domain region.
REFERENCES
- 1.Ferencz C, Rubin JD, McCarter RJ, Brenner JI, Neill CA, Perry LW, Hepner SI, Downing JW. Am. J. Epidemiol. 1985;121:31–36. doi: 10.1093/oxfordjournals.aje.a113979. [DOI] [PubMed] [Google Scholar]
- 2.Hoffman JI. Pediatr. Cardiol. 1995;16:155–165. doi: 10.1007/BF00794186. [DOI] [PubMed] [Google Scholar]
- 3.Chien KR, Olson EN. Cell. 2002;110:153–162. doi: 10.1016/s0092-8674(02)00834-6. [DOI] [PubMed] [Google Scholar]
- 4.Srivasta D, Olson EN. Nature. 2000;407:221–226. doi: 10.1038/35025190. [DOI] [PubMed] [Google Scholar]
- 5.Nakajima Y, Yamagishi T, Hokari S, Nakamura H. Anat. Rec. 2000;258:119–127. doi: 10.1002/(SICI)1097-0185(20000201)258:2<119::AID-AR1>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 6.Ramsdell AF, Markwald RR. Dev. Biol. 1997;188:64–74. doi: 10.1006/dbio.1997.8637. [DOI] [PubMed] [Google Scholar]
- 7.Brown CB, Boyer AS, Runyan RB, Barnett JV. Science. 1999;283:2080–2082. doi: 10.1126/science.283.5410.2080. [DOI] [PubMed] [Google Scholar]
- 8.Paranya G, Vineberg S, Dvorin E, Kaushal S, Roth SJ, Rabkin E, Schoen FJ, Bischoff J. Am. J. Pathol. 2001;159:1335–1343. doi: 10.1016/s0002-9440(10)62520-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de la Pompa JL, Timmerman LA, Takimoto H, Yoshida H, Elia AJ, Samper E, Pooter J, Wakeham A, Marengere L, Langille BL, Crabtree GR, Mak TW. Nature. 1998;392:182–186. doi: 10.1038/32419. [DOI] [PubMed] [Google Scholar]
- 10.Ranger AM, Grusby MJ, Hodge MR, Gravallese EM, de la Brousse FC, Hoey T, Mickanin C, Baldwin HS, Glimcher LH. Nature. 1998;392:186–190. doi: 10.1038/32426. [DOI] [PubMed] [Google Scholar]
- 11.Crabtree GR, Olson EN. Cell. 2002;109:S67–S79. doi: 10.1016/s0092-8674(02)00699-2. [DOI] [PubMed] [Google Scholar]
- 12.Shaw JP, Utz PJ, Durand DB, Toole JJ, Emmel EA, Crabtree GR. Science. 1988;241:202–205. [PubMed] [Google Scholar]
- 13.Chang HY, Takei K, Sydor AM, Born T, Rusnak F, Jay DG. Nature. 1995;376:686–690. doi: 10.1038/376686a0. [DOI] [PubMed] [Google Scholar]
- 14.Musaro A, McCullagh KJA, Naya FJ, Olson EN, Rosenthal N. Nature. 1999;400:581–585. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- 15.Molkentin JD, Lu JR, Antos CL, Markham B, Richardson J, Robbins J, Grant SR, Olson EN. Cell. 1998;93:215–228. doi: 10.1016/s0092-8674(00)81573-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rao A, Luo C, Hogan PG. Annu. Rev. Immunol. 1997;15:707–747. doi: 10.1146/annurev.immunol.15.1.707. [DOI] [PubMed] [Google Scholar]
- 17.Masuda ES, Imamura R, Amasaki Y, Arai K, Arai N. Cell. Signal. 1998;10:599–611. doi: 10.1016/s0898-6568(98)00019-9. [DOI] [PubMed] [Google Scholar]
- 18.Crabtree GR. J. Biol. Chem. 2001;276:2313–2316. doi: 10.1074/jbc.R000024200. [DOI] [PubMed] [Google Scholar]
- 19.McKinsey TA, Olson EN. Curr. Opin. Genet. Dev. 1999;9:267–274. doi: 10.1016/s0959-437x(99)80040-9. [DOI] [PubMed] [Google Scholar]
- 20.Aramburu J, Yaffe MB, Lopez-Rodriguez C, Cantley LC, Hogan PG, Rao A. Science. 1999;285:2129–2133. doi: 10.1126/science.285.5436.2129. [DOI] [PubMed] [Google Scholar]
- 21.Kroll J, Waltenberger J. J. Biol. Chem. 1997;272:32521–32527. doi: 10.1074/jbc.272.51.32521. [DOI] [PubMed] [Google Scholar]
- 22.Ferrara N, Davis ST. Endocrine Rev. 1997;18:4–25. doi: 10.1210/edrv.18.1.0287. [DOI] [PubMed] [Google Scholar]
- 23.Armesilla AL, Lorenzo E, del Arco PG, Martinez-Martinez S, Alfranca A, Redondo JM. Mol. Cell. Biol. 1999;19:2032–2043. doi: 10.1128/mcb.19.3.2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hernandez GL, Volpert OV, Iniguez MA, Lorenzo E, Martinez-Martinez S, Grau R, Fresno M, Redondo JM. J. Exp. Med. 2001;193:607–620. doi: 10.1084/jem.193.5.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Miquerol L, Gertsenstein M, Harpal K, Rossant J, Nagy A. Dev. Biol. 1999;121:307–322. doi: 10.1006/dbio.1999.9355. [DOI] [PubMed] [Google Scholar]
- 26.Miquerol L, Langille BL, Nagy A. Development. 2000;127:3941–3946. doi: 10.1242/dev.127.18.3941. [DOI] [PubMed] [Google Scholar]
- 27.Dor Y, Camenisch TD, Itin A, Fishman GI, McDonald JA, Carmeliet P, Keshet E. Development. 2001;128:1531–1538. doi: 10.1242/dev.128.9.1531. [DOI] [PubMed] [Google Scholar]
- 28.Jackson CJ, Garbett PK, Nissen B, Schieber L. J. Cell Sci. 1990;96:257–262. doi: 10.1242/jcs.96.2.257. [DOI] [PubMed] [Google Scholar]
- 29.Kraling BM, Bischoff J. In Vitro Cell Dev. Biol. Anim. 1998;34:308–315. doi: 10.1007/s11626-998-0007-z. [DOI] [PubMed] [Google Scholar]
- 30.Li B, Fuh G, Meng G, Xin X, Gerritsen ME, Cunningham B, de Vos AM. J. Biol. Chem. 2000;275:29823–29828. doi: 10.1074/jbc.M002015200. [DOI] [PubMed] [Google Scholar]
- 31.Shing Y, Davidson M, Klagsbrun M. Methods Enzymol. 1987;146:42–48. doi: 10.1016/s0076-6879(87)46007-2. [DOI] [PubMed] [Google Scholar]
- 32.Zoeteweij JP, Moses AV, Rinderknecht AS, Davis DA, Overwijk WW, Yarchaon JM, Blauvelt A. Blood. 2001;97:2374–2380. doi: 10.1182/blood.v97.8.2374. [DOI] [PubMed] [Google Scholar]
- 33.Naviaux RK, Costanzi E, Haas M, Verma IM. J. Virol. 1996;70:5701–5705. doi: 10.1128/jvi.70.8.5701-5705.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Alvarez-Arroyo MV, Yague S, Wenger RM, Pereira D, Jimenez S, Gonzalez-Pacheco FR, Castilla MA, Deudero JJP, Caramelo C. Circ. Res. 2002;91:202–209. doi: 10.1161/01.res.0000027562.91075.56. [DOI] [PubMed] [Google Scholar]
- 35.Meadows KN, Bryant P, Pumiglia K. J. Biol. Chem. 2001;276:49289–49298. doi: 10.1074/jbc.M108069200. [DOI] [PubMed] [Google Scholar]
- 36.Zachary I, Glicki G. Cardiovasc. Res. 2001;49:568–581. doi: 10.1016/s0008-6363(00)00268-6. [DOI] [PubMed] [Google Scholar]