Abstract
Here, we map a quantitative trait locus (QTL) with a large effect on predisposition to barbiturate (pentobarbital) withdrawal to a 0.44 Mb interval of mouse chromosome 1 syntenic with human 1q23.2. We report a detailed analysis of the genes within this interval and show that it contains 15 known and predicted genes, 12 of which demonstrate validated genotype-dependent transcript expression and/or nonsynonymous coding sequence variation that may underlie the influence of the QTL on withdrawal. These candidates are involved in diverse cellular functions including intracellular trafficking, potassium conductance and spatial buffering, and multimolecular complex dynamics, and indicate both established and novel aspects of neurobiological response to sedative-hypnotics. This work represents a substantial advancement toward identification of the gene(s) that underlie the phenotypic effects of the QTL. We identify Kcnj9 as a particularly promising candidate and report the development of a Kcnj9-null mutant model that exhibits significantly less severe withdrawal from pentobarbital as well as other sedative-hypnotics (zolpidem and ethanol) versus wild-type littermates. Reduced expression of Kcnj9, which encodes GIRK3 (Kir3.3), is associated with less severe sedative-hypnotic withdrawal. A multitude of QTLs for a variety of complex traits, including diverse responses to sedative-hypnotics, have been detected on distal chromosome 1 in mice, and as many as four QTLs on human chromosome 1q have been implicated in human studies of alcohol dependence. Thus, our results will be primary to additional efforts to identify genes involved in a wide variety of behavioral responses to sedative-hypnotics and may directly facilitate progress in human genetics.
Introduction
Sedative-hypnotic drugs depress CNS function and are commonly prescribed to treat anxiety and insomnia. These medications include benzodiazepines and barbiturates [e.g., pentobarbital (PB)]. Abuse of prescription and other sedative-hypnotics (including ethanol) is among the top five health problems in the United States (Office of National Drug Control Policy, 2004) and is one of the most highly heritable addictive disorders (Goldman et al., 2005). Withdrawal is a hallmark of sedative-hypnotic physiological dependence and constitutes a motivational force that perpetuates drug use/abuse (Little et al., 2005). Unfortunately, current understanding of the gene networks that contribute to sedative-hypnotic withdrawal is limited, and this has hindered treatment and resulted in a lack of alternatives for dependent individuals.
It is well established that there is common genetic influence on withdrawal from a variety of sedative-hypnotics in mice (Belknap et al., 1987, 1988, 1989; Crabbe et al., 1991; Buck et al., 1999; Metten and Crabbe, 1999; Kliethermes et al., 2004; Shirley et al., 2004; Hood et al., 2006; Metten et al., 2007). Although no animal model duplicates clinically defined sedative-hypnotic dependence, models for specific factors, including the withdrawal syndrome, are useful for identifying potential genetic determinants of liability in humans. Although there are more common signs of sedative-hypnotic withdrawal in humans, a genetic contribution to individual differences in withdrawal convulsions is apparent in humans and animal models (Goldstein, 1973; Metten and Crabbe, 1999; Lutz et al., 2006). The handling-induced convulsion (HIC) is a robust measure of sedative-hypnotic withdrawal hyperexcitability in mice after acute and chronic exposure to short- and long-acting sedative-hypnotics (Goldstein and Pal, 1971; Belknap, 1978; Kosobud and Crabbe, 1986; Belknap et al., 1988, 1989; Chan et al., 1989; Crabbe et al., 1991; Metten et al., 2007). However, some withdrawal signs are genetically correlated with HIC severity (i.e., tremors, hypoactivity, emotionality) (Kosobud and Crabbe, 1986; Belknap et al., 1987; Feller et al., 1994), whereas others are not (i.e., tail stiffness) (Kosobud and Crabbe, 1986), so it should be kept in mind that assessment of withdrawal convulsions can inform analyses for correlated withdrawal signs but represents only part of the complex syndrome of sedative-hypnotic withdrawal.
Previously, we mapped a quantitative trait locus (QTL), with a large effect on PB withdrawal, to a large region of chromosome 1 containing hundreds of genes, any one (or more) of which could underlie the QTL association (Buck et al., 1999). One of the most powerful strategies to precisely map a QTL uses interval-specific congenic strains (Darvasi, 1997). Here, using this approach, we precisely map this QTL to a 0.44 Mb interval containing 15 genes and identify QTL candidates. Among these, we identify Kcnj9 as a particularly promising candidate and report the development of a Kcnj9-null mutant model that shows significantly less severe withdrawal from PB as well as other abused sedative-hypnotics (zolpidem and ethanol).
Materials and Methods
Animals.
C57BL/6J (B6), DBA/2J (D2), and B6.D2-Mtv congenic (Taylor and Frankel, 1993) strain mice were purchased from The Jackson Laboratory and bred in our colony at the Veterinary Medical Unit of the Portland Veterans Affairs Medical Center. Recombinant interval-specific congenic strains (R6 and R9) derived from the B6.D2-Mtv (B6.D2) congenic strain and the D2.B6-D1Mit206 (D2.B6) congenic strain and were developed in our colony. R4, R7, R8, and R12 congenic breeders were generously provided by Dr. Aimee Mayeda at the Indianapolis Veterans Affairs Medical Center (Indianapolis, IN).
Development of Kcnj9-null mutants on a D2 genetic background used an existing mutant (B6 background) (Torrecilla et al., 2002) and involved transfer of the Kcnj9-null mutation to the D2 background by repeated backcrosses to D2 strain mice. After six generations of breeding, the genetic background of the resulting mice was estimated to be >98% D2. Mice from backcross generations 7–10 were tested for sedative-hypnotic withdrawal behavior.
All animals used for behavioral testing were bred in our colony. A total of 1443 mice were tested, including 719 congenic mice, 38 D2 and 167 B6 strain mice, and 519 Kcnj9-null mutant, heterozygote, and wild-type mice. Mice were group-housed two to five per cage by strain and sex. Mouse chow (Purina LabDiet 5001; Purina Mills International) and water were available ad libitum. Procedure and colony rooms were kept at a temperature of 21 ± 1°C. Lights were on in the colony from 6:00 A.M. to 6:00 P.M., and behavioral testing was initiated between 7:00 A.M. and 8:00 A.M. All procedures were approved by the Veterans Affairs and Oregon Health & Science University Institutional Animal Care and Use Committees in accordance with United States Department of Agriculture and United States Public Health Service guidelines.
PB withdrawal.
Withdrawal seizures are one of the primary characteristics of the barbiturate withdrawal syndrome (Ho and Harris, 1981) and are a useful index of withdrawal in humans and in animal models. To focus on CNS mechanisms of physiological dependence, withdrawal was assessed after an acute injection of PB, because chronic treatment induces hepatic metabolizing enzymes resulting in metabolic tolerance (Flint and Ho, 1980). Genetic variation in PB withdrawal severity was examined by monitoring HICs (Goldstein and Pal, 1971), a sensitive index of acute barbiturate withdrawal severity (Crabbe et al., 1991). Details of the HIC seven-point scoring system have been published previously (Crabbe et al., 1991; Metten et al., 1998). Individual mice and different inbred strains can differ in baseline (predrug) HIC scores. Therefore, to assess PB withdrawal severity, mice were scored twice for baseline (predrug) HICs 20 min apart, followed by a single sedative-hypnotic dose of PB (60 mg/kg, i.p., 6 mg/ml in saline; Sigma-Aldrich) and were scored hourly up to 10 h after drug. To create an index of PB withdrawal that is independent of individual differences in baseline HIC scores and that reflects differences in withdrawal convulsion severity, postdrug HIC scores were corrected for the individual's average predrug (baseline) HIC score as in previous studies by Metten et al. (1998) and Buck et al. (1999). PB withdrawal severity scores were calculated as the area under the curve (AUC), which was calculated as a sum of corrected postdrug HIC scores as in previous work by Buck et al. (1999). Peak withdrawal scores were also calculated as previously described (Metten and Crabbe, 1999).
Development of interval-specific congenic strains for high-resolution QTL mapping.
The B6.D2 congenic strain was used as our point of departure to generate a series of interval-specific congenic strains for high-resolution QTL mapping. B6.D2 congenic mice were crossed to B6 mice to yield F1(B6.D2 × B6) animals, which were backcrossed to B6 mice. Individual progeny were genotyped using D1Mit and single-nucleotide polymorphism (SNP) markers within or flanking the chromosome 1 QTL affecting PB withdrawal (referred to as Pbw1) (Buck et al., 1999) to identify recombinant mice and define the boundaries of the introgressed intervals. Individual recombinant mice were backcrossed to B6 mice, resulting in multiple offspring with the same recombination. At the same time that recombinations in the previous generation were being replicated, additional recombinants were sought in subsequent backcross generations to develop smaller introgressed intervals and replicated as needed. For six lines, final intercrosses were performed to isolate the donor homozygotes, which constituted the six finished interval-specific congenic strains (i.e., R4, R6, R7, R8, R9, and R12) that were used in our phenotypic analyses of PB withdrawal. We tested for QTL capture by phenotypic comparisons of congenic and background strain mice.
Zolpidem and ethanol withdrawal.
McQuarrie and Fingl (1958) first demonstrated a state of withdrawal CNS hyperexcitability after acute ethanol administration. Acute zolpidem withdrawal is also apparent after a single hypnotic dose (Metten et al., 1998). Kcnj9 knock-out, heterozygote, and wild-type littermates were compared for their acute zolpidem and ethanol withdrawal severities. Adult mice were scored twice for baseline HICs immediately before administration of zolpidem (20 mg/kg, i.p., 2 mg/ml in saline containing 0.1% Tween 80; Tocris Bioscience) and then 15, 30, 45, 60, 75, 90, 120, 150, 180, and 360 min after zolpidem administration as in previous work by Kliethermes et al. (2004). A different group of mice were scored twice for baseline HICs immediately before administration of ethanol (4 g/kg, 20% v/v in saline, i.p.; Aaper Alcohol and Chemical), and then hourly between 2 and 12 h after ethanol administration as in previous work by Buck et al. (1997). To create an index of drug withdrawal that is independent of individual differences in baseline HIC scores and reflects differences in withdrawal convulsion severity, postdrug HIC scores were corrected for the individual's average baseline (predrug) HIC score (described above), and drug withdrawal was indexed as the AUC, calculated as a sum of corrected postdrug HIC scores over the time course as in previous work by Buck et al. (1997), Metten et al. (1998), and Kliethermes et al. (2004).
Genotypic analysis.
DNA was extracted from tail biopsy or ear punch tissue using the Puregene DNA isolation kit (Gentra Biosystems) according to the manufacturer's instructions. PCR amplification and gel electrophoresis was performed as in previous work by Fehr et al. (2002) using SNP and simple sequence length polymorphism markers from the D1Mit series for mouse chromosome 1 (www.informatics.jax.org). Kcnj9-null mutant, heterozygote, and wild-type littermates were differentiated using a PCR-based assay with a common forward primer (G3com) and two reverse primers (G3WT and C3KO). Null mutant and wild-type animals produce 500 and 645 bp PCR products, respectively. A heterozygote produces both PCR products. All PCRs are performed using QIAGEN HotStar under standard conditions with a 55°C annealing temperature. The primer sequences are as follows: G3com (GATACTAGACTAGCGTAACTCTGGAT), G3WT (GATAAAGAGCACAGACTGGGTGTCG), and G3KO (CAAAGCTGAGACATCTCTTTGGCTCTG).
Candidate genes.
Using several databases, we identified as many known and predicted coding and noncoding transcripts as possible within the maximal QTL interval. Databases used included Ensembl [www.ensembl.org; National Center for Biotechnology Information (NCBI) Build 37], miRBase (http://microrna.sanger.ac.uk/), GenBank (http://www.ncbi.nlm.nih.gov/), and University of California, Santa Cruz, Genome Browser (www.genome.ucsc.edu; mm9). Ensembl mouse transcript and NCBI RefSeq sequences were used as consensus sequences in subsequent gene and probe set alignments. The Unigene (NCBI; www.ncbi.nlm.nih.gov) and Allen Brain Atlas (ABA) (www.brainatlas.org) databases were searched to obtain brain expression information for each known and predicted gene within the QTL interval.
SNP annotation.
We compiled several public SNP datasets to annotate all known SNPs within the maximal Pbw1 interval between the B6 and D2 progenitor strains as recently described (Walter et al., 2007). This compilation includes D2 strain sequence data from the National Institute of Environmental Health Sciences/Perlegen Mouse Resequencing Project, NCBI dbSNP (www.ncbi.nlm.nih.gov/SNP/), the Sanger resequencing SNPs from Ensembl (www.ensembl.org), and additional SNP datasets found in the Mouse Phenome SNP Database (http://www.jax.org/phenome/snp.html). Additionally, recent next-generation sequencing of this entire region of chromosome 1 for the D2 strain (Walter et al., 2009), confirmed most known SNPs and detected an abundance of novel SNPs. Together, our analyses provide a sound and unbiased evaluation of all genes within the QTL interval for potential quantitative trait gene (QTG) candidacy based on sequence variation criteria.
Quantitative real-time PCR.
Naive R4 congenic and B6 background strain mice (males 60–90 d of age) were killed by cervical dislocation (n = 12 per strain), the brains were cut in half sagitally, flash-frozen in liquid nitrogen, and stored at −80°C. The genetic composition of the R4 congenic strain is estimated to be >99.5% B6, except for an introgressed 5 Mb donor interval (172.97–177.97 Mb) spanning the QTL derived from the D2 strain. Additionally, a separate set of inbred B6 and D2 progenitor strain mice (n = 10 per strain) were killed by cervical dislocation, and four brain regions (frontal cortex, hippocampus, midbrain, cerebellum) were dissected, flash-frozen in liquid nitrogen, and stored at −80°C. Total RNA was isolated from individual mice using TRIzol reagent (Invitrogen) in a one-step guanidine isothiocyanate procedure as in previous work by Daniels and Buck (2002). Total RNA was reverse transcribed using random hexamers as per protocol (TaqMan Reverse Transcription kit; Applied Biosystems) for all sample sets. Before first-strand cDNA synthesis with High Capacity cDNA Archive kit (Applied Biosystems), a subset of aliquots of mRNA were treated with DNase (Promega) at 37°C for 30 min to eliminate potential contaminating genomic DNA. For other samples, DNase was not used, but subsequent quantitative real-time PCR (Qrt-PCR) probes span introns eliminating the need for DNA removal.
Reference genes were selected using geNorm (Vandesompele et al., 2002), which is an algorithm used to select the most stable reference genes from a set of tested candidate reference genes for accurate normalization of Qrt-PCR data. Ten potential reference genes were analyzed in geNorm, and all 10 were determined to be acceptable based on a gene expression normalization factor (M) (Vandesompele et al., 2002). We chose Reep5 as our primary reference gene because its M value (M = 0.138) identified it as one of the two most stable genes tested. In addition, Reep5 is highly expressed throughout the brain (www.brain-map.org; normalized expression level and density of 100 for all 17 brain regions summarized in coronal image series), making it suitable for both whole-brain and brain regional expression studies for genes with mid- to high-level expression. To assess QTG candidates with low brain expression, we used an alternate reference gene (Gusb). Gusb also passed geNorm tests for our sample set but has ∼32-fold lower expression than Reep5.
Because target probe sequence information for TaqMan assays is proprietary, amplicon sequences were approximated using the Applied Biosystems coordinates (appliedbiosystems.com), which correspond to the center nucleotide of the amplicon and takes into account the provided amplicon length. All primer and target probe sequences were aligned with SNP annotation. Alternative gene-specific TaqMan assays were used when B6/D2 SNPs were detected in the target sequence of the initial probe. Only expression results obtained using probes free of known SNPs are reported.
For each candidate, relative expression was measured using validated gene-specific TaqMan assays, most of which span an intron as an additional control against contaminating genomic DNA. Additionally, custom TaqMan probes (Applied Biosystems) were used for Kcnj9 (forward primer, GTCATTCTCGAGGGCATGGT; reverse primer, CACCAGGTACGAGCTTCGA; reporter sequence, CCACGGGAATGACG) and Atp1a2 (forward primer, ATTGAGGTCTCCCTGAGTAGGTATC; reverse, CACCTCAGTGCACAGTGTCT; reporter, CTGCCCACCACATGCA). Reactions (20 μl) were performed in an ABI Prism7500 thermal cycler using Two-Step PCR Master Mix. Crossing point values (Ct) for target gene expression levels were determined by the standard TaqMan software package and normalized to a reference gene (Reep5; Mm00492230_m1; or Gusb, Mm00446953_m1. The comparative (ΔΔCt) method (Livak and Schmittgen, 2001) was used for relative quantification, which corrects for run-to-run technical variability (e.g., pipetting errors, cDNA concentration, or quality differences) by normalization of sample target and reference gene expression to expression levels of a calibrator sample (cDNA pooled from all samples) included on each run.
cis-Regulation.
In the case of cis-regulation, the expression levels of a transcript map to the structural gene (i.e., DNA variations of a gene directly influence transcript levels of that gene) (Doss et al., 2005). Evidence for cis-regulation was determined by surveying genome-wide microarray gene expression data sets for additional B6D2-derived populations [i.e., BXD recombinant inbred strains and an F2 intercross; www.genenetwork.org/; UCHSC BXD RI Whole Brain M430 2.0 (Nov06) RMA and OHSU/VA B6D2F2 Brain mRNA M430 (Aug05) RMA datasets]. All probe sets interrogating known exons (based on Ensembl) of the genes in the QTL interval were surveyed, except for Atp1a2, in which only one probe set was assessed as the other three harbor excessive SNP-based bias for B6 and D2 allele expression values (Walter et al., 2007). The probe sets surveyed are given in Table 1. QTL candidate genes with expression significantly associated with genotype at a marker within the R4 congenic introgressed interval exhibit apparent cis-regulation.
Table 1.
QTG candidate assessment for allelic structural and expression differences in whole brain
| Gene | TaqMan probe | Gene description | Total D2 versus B6 coding SNPs (Δ amino acid) | Whole-brain mRNA expression: R4 congenic/B6background ratio |
|---|---|---|---|---|
| Novel | ND | Novel retrotransposed gene | ND | ND |
| Wdr42aa | Mm00463652_m1 | WD repeat domain 42A | 6 (0) | 0.86 (p = 1.5 × 10−4)* |
| Pea15aa,b | Mm00440716_g1 | Phosphoprotein enriched in astrocytes 15 | 2 (0) | 1.02 (NS) |
| Casq1a | Mm00486725_m1 | Calsequestrin 1 | 6 (0) | 5.16 (p = 8.0 × 10−13)* |
| Atp1a4a,b | Mm01290854_g1 | ATPase, Na+/K+ transporting, α4 polypeptide | 16 (3) M546Tc, N476S, I74V | 20.67 (p = 4.0 × 10−16)* |
| Igsf8a,b | Mm00712984_m1 | Ig superfamily, member 8 | 12 (2) H221R, T489S | 0.84 (p = 5.3 × 10−4)* |
| Atp1a2a,b,d | Mm00617899_m1 | ATPase, Na+/K+ transporting, α2 polypeptide | 15 (0) | 1.00 (NS) |
| Atp1a2a,b,d | Custom | ATPase, Na+/K+ transporting, α2 polypeptide | 15 (0) | 4.13 (p = 1.3 × 10−20)* |
| Potassium inwardly rectifying channel, subfamily | ||||
| Kcnj9a,b | Mm01290202_m1 | J, member 9 | 6 (0) | 1.06 (NS) |
| Potassium inwardly rectifying channel, subfamily | ||||
| Kcnj9a,d | Mm00434622_m1 | J, member 9 | 6 (0) | 1.06 (NS) |
| Potassium inwardly rectifying channel, subfamily | ||||
| Kcnj9a,d | Custom | J, member 9 | 6 (0) | 1.17 (p = 3.4 × 10−5)* |
| Potassium inwardly rectifying channel, subfamily | ||||
| Kcnj10a,b,d | Mm00445028_m1 | J, member 10 | 5 (1) T262S | 1.31 (p = 1.7 × 10−6)* |
| Pigma | Mm00452712_s1 | Phosphatidylinositol glycan, class M | 5 (1) Q206Hc | 1.07 (NS) |
| Slamf9a | Mm00504048_m1 | SLAM family member 9 | 5 (3) V29I, I204V, L224P | 0.93 (NS) |
| Igsf9a,b | Mm00459672_m1 | Ig superfamily, member 9 | 11 (4) H49R, G284Rc, N388D, Q411Rc | 1.33 (p = 1.0 × 10−5)* |
| Tagln2a,b | Mm00724259_m1 | Transgelin 2 | 1 (0) | 0.97 (NS) |
| Ccdc19a | Mm01283396_g1 | Coiled-coil domain containing 19 | 5 (0) | 0.93 (NS) |
| Vsig8a | Mm00624907_m1 | V-set and Ig domain containing 8 | 0 (0) | 6.07 (p = 1.0 × 10−3)* |
Of the 15 genes within the maximal QTL interval, 11 demonstrate validated genotype-dependent (R4 congenic vs B6 background strain) transcript expression in whole brain and/or nonsynonymous coding sequence variation (B6 vs D2) that may underlie the influence of the QTL on withdrawal. All known coding region SNPs are annotated here. ND, Not determined; NS, not significant.
aNonconservative amino acid difference.
bHuman homolog for this gene exists on syntenic region of human chromosome 1q23.2.
cNonsynonymous SNP(s) exist in human homolog.
dSignificant exclusive cis-regulation (eQTL) is also evident based on whole-brain microarray data for additional B6D2-derived mapping populations (i.e., an F2 intercross and recombinant inbred strains) (www.genenetwork.org: UCHSC BXD Whole Brain M430 2.0 Nov06 RMA and OHSU/VA B6D2F2 Brain mRNA M430 Aug05 RMA). Probe sets surveyed were as follows: 1434134_at, 1416407_at, 1416406_at, 1422598_at, 1437321_at, 1460675_at, 1455136_at, 1428602_at, 1450712_at, 1426115_a_at, 1419601_at, 1418964_at, 1440978_at, 1419315_at, 1420518_a_at, 1426529_a_at, 1429930_at, 1418459_at, and 1436671_at.
*p < 0.0029 [significant after correction for multiple comparisons (i.e., p < 0.05/17 TaqMan probes tested for whole-brain analyses)].
Data analysis.
For most of the behavioral comparisons, the data were not normally distributed; therefore, the data were analyzed using a Kruskal–Wallis ANOVA on ranks. This generates a Mann–Whitney U (two groups) or Kruskal–Wallis H (more than two groups) statistic and corresponding p value. When significance was indicated (p < 0.05), this was followed up with a Kolmogorov–Smirnov two-sample test (SyStat11; Systat Systems). For Affymetrix microarray and Qrt-PCR analyses, the data were analyzed for strain-dependent expression (B6 vs D2) using a two-tailed t test.
The percentages of the total variance attributable to strain (heritability values, h2) for PB withdrawal were calculated for congenic versus B6 background strain comparisons based on R2 values from a one-way ANOVA by strain, or SSbetween strains/SStotal (Belknap et al., 1996). To determine whether the R2 statistics for Kcnj9−/− and congenic analyses were significantly different, a Fisher's r-to-z transformation of the R2 values was made to obtain a normally distributed statistic, followed by a t test of the transformed z values (Sokal and Rohlf, 1995).
For all statistical comparisons, the significance level was set at α < 0.05, except for comparisons of gene expression using Qrt-PCR, in which the significance level was corrected for multiple comparisons based on the number of TaqMan probes tested (i.e., p = 0.0029 = 0.05/17 and p = 0.036 = 0.05/14 for whole-brain and brain regional analyses, respectively, in which a total of 17 and 14 probes were tested).
Results
QTL capture in chromosome 1 congenic strains
We began by testing B6.D2 and D2.B6 congenics and appropriate background strain mice for acute PB withdrawal indexed using the HIC. Genotypic analysis delimited their minimal (and maximal) introgressed intervals as follows: B6.D2, 172.9–188.0 Mb (maximal, 172.3–189.3 Mb); and D2.B6, 152.2–176.5 Mb (maximal, 151.6–177.5 Mb). Figure 1A illustrates the HIC time course associated with PB withdrawal for D2.B6 and B6.D2 congenics and background strain mice. Figure 1B summarizes these data as the corrected AUC and illustrates that D2.B6 congenic mice exhibited significantly less severe PB withdrawal than D2 background strain animals (corrected AUC, 4.2 ± 0.8 and 8.4 ± 1.1, respectively; U(1,49) = 165; p = 0.007) and that the B6.D2 congenic strain had more severe ethanol withdrawal than B6 background strain animals (corrected AUC, 2.7 ± 0.3 and 0.8 ± 0.1, respectively; U(1,152) = 1335; p = 1.7 × 10−8). This emphasizes that the QTL is evident on both genetic backgrounds. Additionally, our results demonstrate that the QTL is evident in both males and females (males, U(1,80) = 431, p = 0.002; females, U(1,72) = 226, p = 1.2 × 10−6, B6.D2 vs background strain comparison). Therefore, in all subsequent analyses, data for males and females were collapsed in a single dataset. Together with our previous work (Buck et al., 1999), these congenic data confirm a highly significant QTL [combined LOD (logarithm of the odds), 17.8; p = 1.5 × 10−18], which exceeds the guidelines recommended by Lander and Kruglyak (1995) for highly significant linkage.
Figure 1.
A, PB withdrawal was indexed using the HIC in D2.B6 congenic strain (gray circles) and background strain (D2) mice (white circles), and in B6.D2 congenic strain (gray squares) and background strain (B6) mice (black squares). The mice were scored twice for baseline (predrug) HICs immediately before administration of 60 mg/kg PB (the arrow marks PB injection at time 0), and hourly up to 10 h after PB administration. After 2–3 h, convulsion scores increase above baseline indicating a state of withdrawal hyperexcitability, which peaks ∼4–6 h after PB administration. B, PB withdrawal severity is indexed as the corrected AUC (mean ± SEM) as in previous work by Buck et al. (1999). The D2.B6 congenic mice exhibited significantly less severe PB withdrawal than D2 background strain animals (p = 0.007) and the B6.D2 congenic strain had more severe ethanol withdrawal than B6 background strain animals (p = 1.7 × 10−8). AUC data represent the strain mean ± SEM (n = 64, 88, 24, and 25 mice per strain, respectively). ***p < 0.005.
Although the data suggest the possibility of a shift in the withdrawal time course in congenic versus background strain animals, statistical analyses did not indicate a statistical difference between D2.B6 congenic and D2 background strain animals (mean time of peak HICs, 5.5 ± 0.4 and 5.1 ± 0.2 h, respectively; U(1,49) = 254; p = 0.07), although a trend was detected for association of the B6 donor region with later onset of withdrawal. The mean time of peak HICs differed between B6.D2 congenic and B6 background strain animals (5.1 ± 0.1 vs 4.2 ± 0.3 h, respectively; U(1,113) = 927; p = 0.003), but in this comparison the D2 donor region was associated with later onset of withdrawal.
QTL fine mapping using interval-specific congenic strains
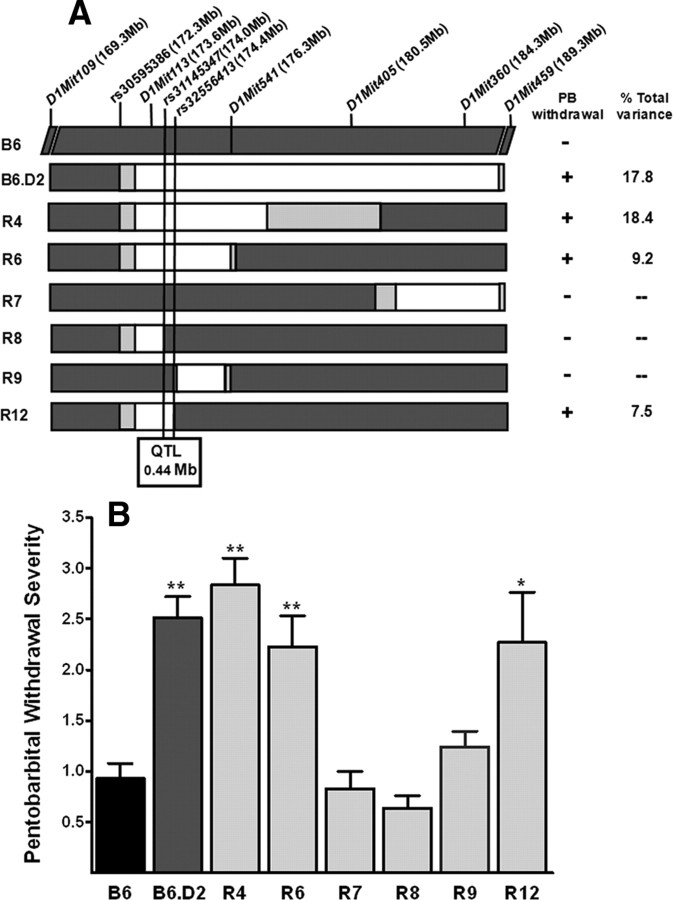
Because the B6.D2 congenic had the smaller introgressed region, it was used as our point of departure to generate a series of interval-specific congenic strains for high-resolution QTL mapping (Fig. 2A). The results of phenotypic comparisons of standard congenic (B6.D2), recombinant (interval-specific) congenics (R4, R6, R7, R8, R9, R12), and B6 background strain mice are shown in Figure 2B; progeny from at least 12 breeder pairs per congenic strain were tested for these analyses. PB withdrawal severity was significantly different between congenic (B6.D2, R4, R6, R7, R8, and R9; AUC = 2.5 ± 0.2, 2.8 ± 0.3, 2.2 ± 0.3, 0.8 ± 0.2, 0.6 ± 0.1, and 1.2 ± 0.2, respectively) and B6 background strain animals (AUC = 0.9 ± 0.2; H(6,687) = 107; p = 1 × 10−15). There was no main effect of sex (p = 0.7) or strain by sex interaction (p = 0.9), so data were collapsed across both sexes. The QTL effect was evident in R6 and R4 congenic animals (p = 0.008 and p = 6.0 × 10−8, respectively; Kolmogorov–Smirnov two-sample test). In subsequent experiments, R12 animals were directly compared with a separate group of B6 background strain animals (AUC, 2.3 ± 0.5 and 0.8 ± 0.2, respectively) and their PB withdrawal severities were also significantly different (U(1,114) = 13.3; p < 0.05). In contrast, R7 and R8 congenics did not differ from B6 background strain mice in their PB withdrawal convulsion severities (both p > 0.9), nor did R9. This narrows the QTL to a maximal 0.44 Mb interval (174.06–174.50 Mb; minimal 0.43 Mb interval, 174.07–174.50 Mb), which accounts for 7.5% of the total variance between the R12 congenic and background strains. Both R6 and R12 showed less severe PB withdrawal than the R4 (p = 0.02 and p = 4.6 × 10−4) or B6.D2 (p = 0.03 and p = 8.1 × 10−6) congenic strains, suggesting that an additional more distal QTL(s) also influences PB withdrawal severity. If two closely linked QTLs are captured in the R4 and B6.D2 congenic intervals, while the R6 and R12 congenic intervals capture only the more proximal QTL, then the QTL effect size, expressed as R2, would be expected to be significantly larger for the first pair of congenics (vs B6 background strain) than for the second pair (also vs B6). Additional analyses revealed that the effect size (R2) in a comparison of R4 and B6.D2 (also vs B6; pooled R2 = 0.181) with R6 and R12 congenics (vs B6; pooled R2 = 0.084) was significantly different (p = 0.025; one-tailed) using a Fisher's r-to-z conversion of pooled R2 values followed by a t test of the z values.
Figure 2.
A, In addition to the starting B6.D2 congenic strain, a panel of six interval specific congenic strains was developed and tested to attain fine mapping of the QTL. The genetic markers examined to establish the congenic interval boundaries are indicated and their locations are given (in megabases) at the top. For each congenic, the donor segment is shown in white and is D2D2 homozygous. Chromosomal regions homozygous for the background (B6) allele are shown in dark gray, and the boundaries between the B6 and D2 regions are shown in light gray. PB withdrawal severity was significantly more severe in R4, R6, and R12 mice compared with B6 background strain mice, whereas R7, R8, and R9 mice did not differ from B6 background strain mice in PB withdrawal liability. Our results narrow the QTL to a minimal 0.43 Mb (maximal, 0.44 Mb) interval that is contained within the R12 introgressed interval but excluded by the R8 and R9 congenic intervals. B, The B6.D2 congenic and six recombinant interval specific congenic strains were tested for their PB withdrawal severities (corrected AUC; mean ± SEM) and compared with B6 background strain mice. PB withdrawal was significantly more severe in B6.D2 (p = 1.2 × 10−7; n = 122), R4 (p = 6.0 × 10−8; n = 105), R6 (p = 0.008; n = 90) mice than in B6 background strain mice (n = 98). Withdrawal was also significantly more severe in R12 (p = 0.049; n = 53) than in B6 background strain mice (n = 61). In contrast, PB withdrawal severity does not differ from B6 background strain mice for R7 (p = 0.94; n = 73), R8 (p = 0.92; n = 59), or R9 (p = 0.07; n = 140). *p < 0.05; **p < 0.01. The estimated percentage of the total variance accounted for in each congenic strain (vs the B6 background strain) is also given.
QTL candidate genes
Using mouse genome databases (Ensembl and NCBI), we identified as many known and predicted transcripts as possible within the QTL interval. As shown in Table 1, the maximal 0.44 Mb QTL interval contains a total of 15 known and predicted genes (i.e., 14 known coding genes and one novel retrotransposed gene predicted by Ensembl), with no noncoding RNA genes known or predicted. These results distinguish this interval as remarkably gene dense.
We used evidence for brain expression as an initial filter in prioritizing potential candidates because convulsions are centrally mediated and HICs are an index of sedative-hypnotic withdrawal severity for which pharmacokinetic factors are not crucially important (Crabbe et al., 1983; Metten and Crabbe, 1994). Using the Unigene and ABA databases and our Qrt-PCR data, we confirmed expression in mouse brain at varying levels for the 14 confirmed genes in the QTL interval and identified these as primary candidates (Table 1). We were unable to assess the novel retrotransposed gene, as it is represented multiple times in the genome, and a unique query based on sequence is not possible.
Candidate gene expression variation
To ensure that all brain regions potentially relevant to the behavioral expression of withdrawal were included in the analyses, whole-brain samples were obtained from R4 congenic and B6 background strains. We nominated candidates based on inherent genotype-dependent expression for three reasons: (1) Many more genes are differentially expressed between naive B6 and D2 mice than after a single injection of a sedative-hypnotic drug (Daniels and Buck, 2002; Kerns et al., 2005) (K. J. Buck, unpublished results); (2) most differences in gene expression between sedative-hypnotic-treated B6 and D2 animals are already apparent before drug exposure (Kerns et al., 2005); and (3) the time frames during which withdrawal-enhanced HICs are evident (e.g., ∼3–8 h after PB and ∼30–210 min after zolpidem) are shorter than generally thought to be required for the majority of gene expression changes to show the corresponding changes in protein critical for behavioral expression of differential withdrawal severity, increasing the likelihood that the relevant gene expression disparity (or disparities) is preexisting. We identified differential expression between R4 congenic and B6 background strain mice for Kcnj9, Kcnj10, Atp1a2, Atp1a4, Wdr42a, Casq1, Igsf8, Igsf9, and Vsig8 (Table 1). Seven genes showed evidence of increased expression in R4 versus background strain mice, whereas only two genes (Wdr42a and Igsf8) showed decreased expression in R4 versus background strain animals (Table 1). Comparison of gene expression between congenic and background strains affords substantially increased confidence that expression differences detected for transcripts physically mapped within the QTL interval are cis-mediated, as would be expected for a true QTG.
In independent tests for cis-regulation, three QTG candidates (Kcnj9, Kcnj10, and Atp1a2) showed exclusive significant cis-regulation in B6D2 F2 and BXD recombinant inbred strain datasets for whole brain (Table 1); all three also showed significant differential expression in the R4 congenic versus background strain comparison. Two genes showed both significant cis- and trans-regulation (Wdr42a and Pigm), but only the former also showed differential expression between R4 congenic and background strains in whole brain. Two genes showed trans-regulation in one dataset only (Pea15a and Ccdc19), and, as expected, neither was differentially expressed between R4 congenic and background strains. These analyses showed no significant cis- or trans-regulation for the remaining seven genes. Consistent with these results, two of these genes were not differentially expressed between R4 congenic and background strains (Slamf9 and Tagln2). However, for the remaining five genes, Qrt-PCR analyses detect differential expression between R4 congenic and background strains. For three of these genes (Atp1a4, Casq1, and Vsig8), this discrepancy may be related to their low expression in brain. For the remaining two genes (Igsf8 and Igsf9), the discrepancy could be attributable to small fold changes not being replicated in microarray data or attributable to variation of where the microarray probe sets and QrtPCR probes hybridize within the genes.
Only two of the genes that showed evidence of differential expression in previous microarray analyses comparing the B6 and D2 progenitor strains (Walter et al., 2007) did not show confirmation using predesigned TaqMan probes (i.e., Kcnj9 and Atp1a2). Both genes have potential alternative transcripts identified in the public databases (NCBI), so this lack of confirmation using Qrt-PCR was potentially related to interrogation of alternative transcripts by microarray versus QrtPCR analyses. Therefore, for these two genes, we designed and tested custom TaqMan probes to specifically interrogate the transcripts implicated in microarray analyses. For Atp1a2, the predesigned TaqMan probe (Mm00617899_m1) targeting the coding region failed to detect differential expression, whereas the custom probe interrogating the extended 3′-untranslated region (3′-UTR) detected significantly greater expression in R4 congenic versus background strain animals for an extended transcript that is detected only in the brain (Table 1). This is consistent with microarray and Qrt-PCR analyses comparing Atp1a2 expression in D2 versus B6 progenitor strains (Walter et al., 2007). Interestingly, the extended 3′-UTR is a potential target for microRNAs (Gaidatzis et al., 2007). For Kcnj9, two predesigned TaqMan probes spanning exons 1–2 (Mm01290202_m1) and 2–3 (Mm00434622_m1) failed to detect differential expression, whereas the custom probe interrogating exons 3–4 identified significantly greater expression in R4 congenic versus background strain animals (Table 1). Thus, our results identify differential Kcnj9 expression for transcripts that extend through exons 3 and 4, but not for potential alternative transcripts that eliminate exon 4 and truncate the C terminus. Both Kcnj9 transcripts are expressed (GenBank accession IDs: NM_008429 and AK015907), but the potential role of the truncated transcript in brain is unknown.
We also assessed genotype-dependent expression of the 14 primary candidates in four brain regions (frontal cortex, hippocampus, midbrain, and cerebellum) using the B6 and D2 progenitor strains (Fig. 3). Only one additional candidate, Ccdc19, was identified based on differential expression in a discrete brain region(s), but not detected using whole brain. Three transcripts (Atp1a2, Casq1, and Atp1a4) showed evidence of higher expression in D2 versus B6 mice in all four regions surveyed, and one gene (Wdr42a) was more highly expressed in B6 versus D2 mice in all four regions surveyed. Four genes (Kcnj9, Kcnj10, Ccdc19, Vsig8) showed evidence of brain regionally specific differential expression. Differential expression in midbrain was apparent for seven genes (Kcnj9, Kcnj10, Atp1a2, Atp1a4, Vsig8, Wdr42a, Casq1) and is of particular interest because withdrawal-associated neuronal activation differs between chromosome 1 congenic and background strains in midbrain (G. Chen and K. J. Buck, unpublished data) and because focused lesions of the substantia nigra pars reticulata significantly attenuate withdrawal from PB and withdrawal after acute and repeated ethanol exposure (Chen et al., 2008) (G. Chen and K. J. Buck, unpublished data).
Figure 3.
Brain regional genotype-dependent expression of QTL candidates in B6 and D2 progenitor strain mice. Qrt-PCR results are based on validated TaqMan assays and illustrate D2 expression relative to B6 expression as a percentage. Positive values indicate D2>B6 expression, and negative values indicate B6>D2 RNA expression. TaqMan assays were as follows: Wdr42a, Mm00463652_m1; Pea15a, Mm00440716_g1; Casq1, Mm00486725_m1; Atp1a4, Mm01290854_g1; Igsf8, Mm00712984_m1; Atp1a2, custom; Kcnj9, custom; Kcnj10, Mm00445028_m1; Pigm, Mm00452712_s1; Slamf9, Mm00504048_m1; Igsf9, Mm00459672_m1; Tagln2, Mm00724259_m1; Ccdc19, Mm01283396_g1; Vsig8, Mm00624907_m1. Reep5 (Mm00492230_m1) was used as the housekeeping gene for relative quantification for most candidate genes. aGusb (Mm00446953_m1) was used as the housekeeping gene for the candidates more lowly expressed in brain. Four genes showed evidence of differential expression in all four brain regions, and three showed evidence of regionally specific differential expression. Only one gene (Casq1) was identified as a QTL candidate based on brain regionally specific differential expression that was not also detected in whole-brain analyses. *p < 0.0036 [significant after correction for multiple comparisons (i.e., p < 0.05/14 probes tested in the brain regional analyses)].
Together with the congenic versus background whole-brain data, this is an unusually high number of genes for such a small region showing strain-specific expression, particularly since they are apparently unrelated. This corroborates previous findings of disproportionately frequent expression differences between the B6 and D2 strains for genes in this region of chromosome 1, even after correction for the increased gene density (Kerns et al., 2005).
Candidate gene sequence variation
We began by searching the public databases to systematically assess which genes in the QTL interval harbor SNPs that change predicted protein sequence between the B6 and D2 progenitor strains. However, even between these two well annotated strains, the majority of SNPs are still unknown or cryptic (Walter et al., 2009). Therefore, to complete this comparison, we performed next-generation sequencing of the QTL interval for the D2 strain (Walter et al., 2009). Together, these analyses detect and confirm nonsynonymous sequence variation for six genes in the QTL interval (i.e., Atp1a4, Igsf8, Kcnj10, Pigm, Slamf9, and Igsf9) (Table 1), four of which also display genotype-dependent expression. These experiments provide a sound and unbiased evaluation of all genes within the QTL interval for potential QTG candidacy based on either the expression or sequence variation criteria.
Kcnj9 mutant analyses
Kcnj9 encodes GIRK3, a subunit member of a family of G-protein-dependent inwardly rectifying K+ channels that primarily mediate postsynaptic inhibitory effects of Gi/o-coupled receptors (Dascal, 1997; Lüscher et al., 1997). We identified it as a particularly promising candidate for this QTL for several reasons, although it should be kept in mind that additional candidates remain in the QTL interval that may also influence withdrawal and warrant additional attention in future studies. First, Kcnj9 exhibited significantly higher expression in D2 versus B6 progenitor strains, and R4 congenic versus background strain mice. Although this differential expression is modest (e.g., 30% in midbrain), it is comparable with a confirmed QTG (Mpdz) for PB and ethanol withdrawal on chromosome 4 (Shirley et al., 2004). Second, channels containing GIRK2 (with which GIRK3 primarily associates) have been implicated in ethanol withdrawal as well as other behavioral responses to this sedative-hypnotic (Blednov et al., 2001; Hill et al., 2003). Third, GIRK channel activation by sedative-hypnotics such as ethanol and nitrous oxide appears to be a property of most types of GIRK channels (Kobayashi et al., 1999; Lewohl et al., 1999), and recent studies (Aryal et al., 2009) identify GIRK channels as direct targets of ethanol. These studies either did not test PB or zolpidem, or found no effect of acute PB exposure (Yamakura et al., 2001), but GIRK3-containing channels were not examined, nor has GIRK channel function in withdrawn animals been assessed. Fourth, a plausible mechanism emerges. GIRK3 interacts with a sorting nexin (SNX27) that promotes endosomal movement of channels and reduced surface expression (Lunn et al., 2007). In this context, reduced GIRK3 expression would be expected to increase GIRK currents resulting in more neuronal inhibition. This is consistent with our finding that lower Kcnj9 expression is associated with less PB withdrawal convulsivity (and also less severe ethanol and zolpidem withdrawal) in Kcnj9 knock-out versus wild-type littermates, as well as less PB withdrawal convulsivity in B6 background strain versus R4 congenic mice. Moreover, our results identify differential Kcnj9 expression for transcripts that extend through exons 3 and 4, but not for potential alternative transcripts that eliminate exon 4 and truncate the C terminus containing the four amino acid residues that associate with SNX27 (Lunn et al., 2007).
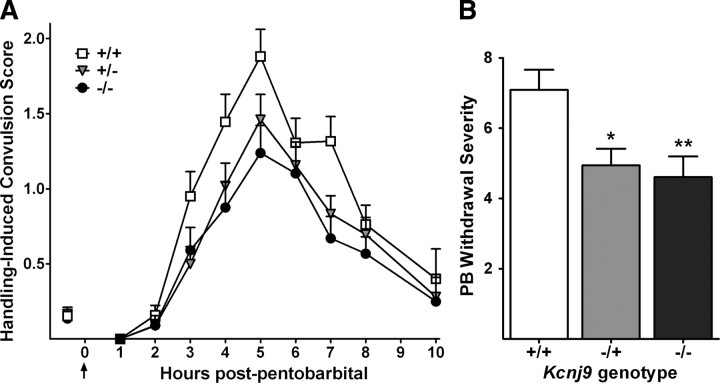
Because our results showed that less severe PB withdrawal is associated with lower Kcnj9 mRNA expression, we predicted that Kcnj9-null mutation would result in significantly less severe PB withdrawal compared with wild-type littermates. The B6 strain exhibits little or no acute sedative-hypnotic withdrawal, and, as expected, we were unable to detect any additional decrease in PB withdrawal severity in Kcnj9−/− mice on this genetic background (data not shown). Testing our hypothesis therefore required that we transfer the Kcnj9−/− mutation onto a genetic background associated with sufficiently severe PB withdrawal. After seven generations of repeated backcrosses to D2 strain mice (which show severe PB withdrawal), the finished null mutant was estimated to be >98% D2. Backcross generations 7–10 were tested. Null mutant, heterozygote, and wild-type littermates had significantly different PB withdrawal severities (H(2,291) = 14.5; p = 0.0007) (Fig. 4). As predicted, Kcnj9−/− mice showed less severe PB withdrawal versus wild-type littermates (p = 0.0033, one-tailed; mean AUC scores ± SEM, 4.6 ± 0.6 and 7.1 ± 0.6, respectively). Kcnj9 heterozygotes also showed less severe PB withdrawal (AUC, 4.9 ± 0.5) versus wild-type littermates (p = 0.037, one-tailed). These data are consistent with Kcnj9 being a QTG that influences PB withdrawal severity. Furthermore, the total variability accounted for in the comparison of R12 congenic (with the smallest introgressed interval) versus background strain animals is consistent with that accounted for in the comparison of Kcni9−/− and wild-type littermates (7.5 and 4.7%, respectively; p = 0.65, NS; using Fisher's r-to-z transformation of the R2 values, followed by a t test of the z values).
Figure 4.
A, PB withdrawal was indexed using the HIC in D2 background Kcnj9-null mutants (black circles), heterozygotes (gray triangles), and wild-type (open squares) littermates. The mice were scored twice for baseline (predrug) HICs immediately before administration of 60 mg/kg PB (the arrow marks PB injection at time 0), and hourly up to 10 h after PB administration. The data represent mean HIC score values ± SEM. PB administration initially lowers convulsion scores (0–1 h). Later, as PB is metabolized, convulsion scores increase above baseline, indicating a state of withdrawal hyperexcitability, which peaks ∼5 h after PB administration. B, PB withdrawal severity is indexed as the corrected AUC as in previous work by Buck et al. (1999), and the data represent Kcnj9-null mutant, heterozygote, and wild-type littermate mean corrected AUC values ± SEM (n = 88, 101, and 102, respectively). Kcnj9-null mutants and heterozygotes exhibited significantly less severe PB withdrawal than wild-type littermates. *p < 0.05; **p < 0.01.
Additionally, we tested 172 null mutant, heterozygote, and wild-type littermates from generations 9–10 for zolpidem withdrawal, and determined that they also differed in their zolpidem withdrawal severities (H(1,152) = 13.5; p = 0.0012) (Fig. 5). Kcnj9-null mutants and heterozygotes exhibited significantly less severe zolpidem withdrawal than wild-type littermates (p = 0.0043 and p = 0.035, respectively; mean AUC scores ± SEM, 11.3 ± 1.4, 15.7 ± 1.3, and 18.7 ± 1.1, respectively). Kcnj9 mutants also showed less severe ethanol withdrawal than wild-type littermates (data not shown).
Figure 5.
A, Zolpidem withdrawal was indexed using the HIC in Kcnj9-null mutants (black circles), heterozygotes (gray triangles), and wild-type (open squares) littermates. The mice were scored twice for baseline (predrug) HICs immediately before administration of 20 mg/kg zolpidem (the arrow marks zolpidem injection at time 0), and at 13 time points after zolpidem administration. The data represent mean HIC score values ± SEM. Approximately 30 min after zolpidem administration, convulsion scores increase above baseline indicating a state of withdrawal hyperexcitability, which peaks ∼1 to 1.5 h after zolpidem administration. B, Zolpidem withdrawal severity is indexed as corrected AUC as in previous work by Kliethermes et al. (2004), and data represent Kcnj9-null mutants, heterozygotes, and wild-type littermate mean corrected AUC values ± SEM (n = 38, 59, and 55, respectively). Kcnj9-null mutant and heterozygote mice exhibited significantly less severe zolpidem withdrawal than wild-type littermates. *p < 0.05; **p < 0.01.
Finally, we compared expression of the other confirmed QTG candidates in Kcnj9−/− and wild-type littermates. Only Kcnj9 and one other gene (Vsig8) differed in whole-brain expression between Kcnj9−/− and wild-type littermates, and only Kcnj9 showed differential expression consistent with the phenotype in both congenic (R4)/background (B6) and wild-type/Kcnj9 mutant comparisons (i.e., higher expression associated with more severe sedative-hypnotic withdrawal) (supplemental Table 1, available at www.jneurosci.org as supplemental material). Notably, Kcnj10, Atp1a2, and Atp1a4, which showed marked differential expression in whole brain between R4 congenic and B6 background strain mice did not differ in expression between Kcnj9−/− and wild-type littermates (expression ratios, 0.98, 0.92, and 1.13, respectively; all nonsignificant). This mitigates the potential confounding effect of linked embryonic stem (ES) cell-derived genes on the Kcnj9−/− phenotype and supports a causal role for Kcnj9. Furthermore, only one nonsynonymous polymorphism was found in both 129Sv/J versus D2 and B6 versus D2 comparisons (rs31557967 in Igsf9, R284G).
Discussion
Our results provide confirmation and fine mapping of a sedative-hypnotic withdrawal QTL (originally referred to as Pbw1) (Buck et al., 1999) on chromosome 1 using congenic animals. Detailed molecular analyses of the 14 confirmed candidates in the QTL interval identified 5 with genotype-dependent brain expression between R4 congenic and B6 background strains, 1 with regionally specific differential expression, 2 with B6/D2 missense variation, and 4 exhibiting both features. Additionally, our studies demonstrate that Kcnj9−/− mice show reduced withdrawal from sedative-hypnotics, including PB, zolpidem, and ethanol.
The most commonly used models of sedative-hypnotic withdrawal are acute models, in which withdrawal is monitored after a single hypnotic dose (Buck et al., 1997, 1999; Metten et al., 2007), and chronic models in which withdrawal is monitored after drug exposure for several days or weeks (Levental and Tabakoff, 1980; Sivam et al., 1982; Belknap et al., 1988, 1989; Terdal and Crabbe, 1994; Skelton et al., 2000). Here, we used acute models, which have the advantage that sedative-hypnotic concentrations in blood and brain vary little across genotype and thus focus on CNS mechanisms of withdrawal. Chronic models result in more intense withdrawal but induce metabolizing enzymes resulting in metabolic tolerance (Flint and Ho, 1980; Lieber, 1999; Hoen et al., 2001).
Our analyses identify Kcnj9 (GIRK3) as a promising QTL candidate. Currently little is known about GIRK3 function in brain and the mechanism by which it may affect sedative-hypnotic withdrawal. GIRK3 is widely expressed in brain in which it contributes to heteromeric GIRK2/3 channels (Torrecilla et al., 2002; Koyrakh et al., 2005; Labouèbe et al., 2007). One plausible mechanism by which GIRK3 may influence sedative-hypnotic withdrawal is via its role in GABAB receptor signaling. Baclofen suppresses withdrawal symptoms in ethanol-dependent rats (Colombo et al., 2000; Knapp et al., 2007) and human alcoholics (Addolorato et al., 2006). Kcnj9−/− (B6 background) mice show significantly increased GIRK–GABAB receptor coupling efficiency in ventral tegmental area dopamine neurons compared with wild-type mice (Labouèbe et al., 2007). Ethanol enhances GABAB receptor-mediated inhibitory postsynaptic transmission in midbrain neurons by facilitating GIRK currents (McDaid et al., 2008; Federici et al., 2009) and enhances baclofen-evoked GIRK currents in cerebellar granule cells in culture and expression systems (Kobayashi et al., 1999; Lewohl et al., 1999) but, in the hippocampus, does not affect GABAB-IPSPs (Morrisett and Swartzwelder, 1993) or postsynaptic response to baclofen (Frye and Fincher, 1996). This discrepancy may reside in the different types of neurons in the different brain regions examined, in the subunits forming GIRK channels in these regions (Karschin et al., 1996), and in the different animal strains examined. Kcnj9−/− mice (B6 background) also exhibit blunted behavioral response to morphine (Marker et al., 2002) (but see Smith et al., 2008), WIN55,212-2 (R-(+)-(2,3-dihydro-5-methyl-3-[(4-morpholinyl)methyl]pyrol[1,2,3-de]-1,4-benzoxazin-6-yl)(1-naphthalenyl)methanone monomethanesulfonate), and clonidine (cannabinoid and α2 adrenergic receptor agonists) (Smith et al., 2008), suggesting the potential involvement of additional Gi/o-coupled receptors in mediating GIRK3 effects on sedative-hypnotic response. Future studies will be needed to assess GIRK coupling to GABAB and other Gi/o-coupled receptors in sedative-hypnotic-dependent and withdrawn animals.
Our analyses also implicate three genes (Kcnj10, Atp1a4, Atp1a2) that potentially influence withdrawal via spatial buffering/siphoning (Orkand et al., 1966; Newman et al., 1984, 1995) in which K+ is taken up into glia at a site of neuronal activity and released distal to the active neurons (Kalsi et al., 2004). Atp1a2 and Atp1a4 encode Na+,K+-ATPase α2 and α4 subunits and show markedly higher expression in R4 congenic versus background strains. Atp1a2 is widely expressed during development but becomes more restricted in adult brain (Herrera et al., 1994), remaining high in astrocytes and meningeal tissues (McGrail et al., 1991; Watts et al., 1991; Peng et al., 1997). Atp1a2−/− mice show neuronal hyperactivity (Ikeda et al., 2003), so if it were the QTG, one might expect lower Atp1a2 expression in D2 versus B6 and R4 congenic versus background strains. However, the opposite was apparent, at least for an extended 3′-UTR transcript, whereas alternative transcripts showed no differential expression. Atp1a4 is known almost entirely for its role in sperm, although our results and the Allen Brain Atlas (http://www.brain-map.org/) confirm widespread expression in brain. Based on Atp1a2, one might expect lower Atp1a4 expression in congenic versus background and/or D2 versus B6 strains, but the opposite was observed. However, nonsynonymous variation could also affect ATPase function. Kcnj10 encodes a potassium channel (Kir4.1) found predominantly (Takumi et al., 1995) but not exclusively (Li et al., 2001) in glia. Kir4.1 and related channels set the resting membrane potential of glia near the equilibrium potential for K+ (Newman et al., 1995). Higher Kcnj10 expression would therefore be expected to reduce CNS excitability, but the opposite was observed in D2 versus B6 and R4 congenic versus background strains. However, nonsynonymous variation could affect Kir4.1 function, and missense variation in human KCNJ10 is related to seizures in association studies (Buono et al., 2004).
Other candidates suggest previously indicated and entirely novel aspects of neurobiology as potentially related to withdrawal. The Casq1 product, calsequestrin-1, is a Ca2+-binding protein known almost entirely for its role in diabetes and calcium sequestration in muscle, in which it plays a dual role in excitation–contraction coupling (Paolini et al., 2007). Higher Casq1 might therefore be expected to decrease neuronal excitability, but the opposite was observed. Vsig8 encodes an uncharacterized protein with V-set and Ig domains, and shows marked genotype-dependent expression warranting additional exploration. Igsf8 and Igsf9 encode centrally expressed members of an Ig superfamily (Doudney et al., 2002; Murdoch et al., 2003). IGSF8 binds tetraspanin molecules that promote the formation/stabilization of multimolecular complexes involving many proteins including protein kinase C isoforms (Zhang et al., 2001) that influence sedative-hypnotic actions (Qi et al., 2007). Igsf9 encodes protein dendrite arborization and synapse maturation 1 (Dasm-1), which is involved in dendrite arborization at excitatory synapses (Shi et al 2004; Mishra et al 2008). Pigm is involved in glycosylphosphatidylinositol synthesis, and hypomorphic promoter mutation in human PIGM causes a syndrome with seizures as a prominent characteristic (Almeida et al., 2006). Wdr42a encodes a member of a family of proteins containing tryptophan–aspartic acid dipeptide (WD) repeats. This family has diverse biological functions (Li and Roberts, 2001), but WDR42A function is unknown.
Our novel Kcnj9−/− and congenic models will be invaluable to assess potential pleiotropic effects of Kcnj9 and/or linked genes on behaviors for which QTLs are detected on chromosome 1, including other sedative-hypnotic responses [e.g., conditioned aversion (Risinger and Cunningham, 1998), preference (Tarantino et al., 1998), sensitivity to locomotor activation (Demarest et al., 1999), and hypothermia (Crabbe et al., 1994)]. The possibility that the QTL gene(s) play an important role in such diverse responses makes it an important target. Moreover, several studies identify markers on human 1q associated with alcoholism (Ehlers et al., 2010) that, although localized to large regions, carry the potential to be syntenic. Therefore, detailed analyses of QTG candidates may inform developing models for genetic influences on human sedative-hypnotic dependence.
QTLs for additional CNS hyperexcitability states are detected on chromosome 1. The mechanisms underlying withdrawal and other hyperexcitability states remain obscure, and influential genes may indeed have more general effects on hyperexcitability. Ferraro et al. (2007) localized an electroshock seizure locus to the Atp1a4–Atp1a2–Igsf8–Kcnj9–Kcnj10–Pigm region. A QTL for kainate-induced seizures is also detected but is a suggestive association mapped to a large region (Ferraro et al., 1997). A QTL for pentylenetetrazole-induced seizures is detected in some populations (Ferraro et al., 1999) (but see Martin et al., 1995). However, analyses for audiogenic seizures did not detect a chromosome 1 QTL (Neumann and Collins, 1991; Frankel et al., 1995), nor did analyses for β-carboline or glycine-induced seizures (Martin et al., 1995) or using polygenic models of epilepsy (Frankel et al., 1995).
Our studies contribute to progress in understanding the genetic determination of sedative-hypnotic withdrawal, but there are some limitations. First, our analyses are primarily based on one measure of sedative-hypnotic withdrawal (convulsivity). However, preliminary analyses indicate less severe PB and ethanol withdrawal-associated depression-like behavior in Kcnj9−/− versus wild-type littermates, consistent with a broader role for Kcnj9 beyond withdrawal convulsivity (L. C. Milner and K. J. Buck, unpublished data). Additionally, Kcnj9−/− and wild-type littermates both exhibit, but do not differ for, PB-induced locomotor sedation and hypothermia, indicating that Kcnj9 does not generally effect sedative-hypnotic acute effects. Second, candidates were nominated based on gene expression comparisons in naive animals. Although future studies using withdrawn animals may identify additional candidates, previous work indicates that most expression differences between D2 and B6 strains (for genes in QTL regions) are apparent at baseline (Kerns et al., 2005). Third, no direct relationships are established to delineate how differential gene expression translates through protein levels to behavior. Finally, one must be cautious in interpreting knock-out results. Embryonic stem cell-derived genes that flank the targeted gene and developmental adaptations may influence results. Future studies (e.g., transgenic rescue) will be needed to disentangle Kcnj9 actions from potential effects of 129-derived genes, and RNA interference or conditional knock-outs will be important to address the latter. Because additional genes in the QTL interval may also influence withdrawal, Kcnj9−/− transcriptome analyses will be important to elucidate the flanking region and allelic sequence/expression variation that might also contribute to phenotypic differences.
Footnotes
This work was supported by Public Health Service Grants DA05228, AA011114, DA011806, AA01731, AA10760, and MH61933, and a Veterans Affairs Merit Award. We gratefully acknowledge Drs. Aimee Mayeda and John Hofstetter for providing some breeder stock; Drs. John Belknap, John Crabbe, and Pamela Metten for helpful discussions on this project; and Gregory Auger for his technical assistance.
References
- Addolorato G, Leggio L, Abenavoli L, Agabio R, Caputo F, Capristo E, Colombo G, Gessa GL, Gasbarrini G. Baclofen in the treatment of alcohol withdrawal syndrome: a comparative study vs. diazepam. Am J Med. 2006;119:276, e213, e278. doi: 10.1016/j.amjmed.2005.08.042. [DOI] [PubMed] [Google Scholar]
- Almeida AM, Murakami Y, Layton DM, Hillmen P, Sellick GS, Maeda Y, Richards S, Patterson S, Kotsianidis I, Mollica L, Crawford DH, Baker A, Ferguson M, Roberts I, Houlston R, Kinoshita T, Karadimitris A. Hypomorphic promoter mutation in PIGM causes inherited glycosylphosphatidylinositol deficiency. Nat Med. 2006;12:846–851. doi: 10.1038/nm1410. [DOI] [PubMed] [Google Scholar]
- Aryal P, Dvir H, Choe S, Slesinger PA. A discrete alcohol pocket involved in GIRK channel activation. Nat Neurosci. 2009;12:988–995. doi: 10.1038/nn.2358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belknap JK. Barbiturate physical dependence in mice: effects of neuroleptics and diazepam on the withdrawal syndrome. Clin Toxicol. 1978;12:427–434. doi: 10.3109/15563657809150013. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Laursen SE, Crabbe JC. Ethanol and nitrous oxide produce withdrawal-induced convulsions by similar mechanisms in mice. Life Sci. 1987;41:2033–2040. doi: 10.1016/0024-3205(87)90477-2. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Danielson PW, Lame M, Crabbe JC. Ethanol and barbiturate withdrawal convulsions are extensively codetermined in mice. Alcohol. 1988;5:167–171. doi: 10.1016/0741-8329(88)90015-8. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Crabbe JC, Laursen SE. Ethanol and diazepam withdrawal convulsions are extensively codetermined in WSP and WSR mice. Life Sci. 1989;44:2075–2080. doi: 10.1016/0024-3205(89)90354-8. [DOI] [PubMed] [Google Scholar]
- Belknap JK, Mitchell SR, O'Toole LA, Helms ML, Crabbe JC. Type I and type II error rates for quantitative trait loci (QTL) mapping studies using recombinant inbred mouse strains. Behav Genet. 1996;26:149–160. doi: 10.1007/BF02359892. [DOI] [PubMed] [Google Scholar]
- Blednov YA, Stoffel M, Chang SR, Harris RA. Potassium channels as targets for ethanol: studies of G-protein-coupled inwardly rectifying potassium channel 2 (GIRK2) null mutant mice. J Pharmacol Exp Ther. 2001;298:521–530. [PubMed] [Google Scholar]
- Buck K, Metten P, Belknap J, Crabbe J. Quantitative trait loci affecting risk for pentobarbital withdrawal map near alcohol withdrawal loci on mouse chromosomes 1, 4, and 11. Mamm Genome. 1999;10:431–437. doi: 10.1007/s003359901018. [DOI] [PubMed] [Google Scholar]
- Buck KJ, Metten P, Belknap JK, Crabbe JC. QTL involved in genetic predisposition to acute alcohol withdrawal in mice. J Neurosci. 1997;17:3946–3955. doi: 10.1523/JNEUROSCI.17-10-03946.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buono RJ, Lohoff FW, Sander T, Sperling MR, O'Connor MJ, Dlugos DJ, Ryan SG, Golden GT, Zhao H, Scattergood TM, Berrettini WH, Ferraro TN. Association between variation in the human KCNJ10 potassium ion channel gene and seizure susceptibility. Epilepsy Res. 2004;58:175–183. doi: 10.1016/j.eplepsyres.2004.02.003. [DOI] [PubMed] [Google Scholar]
- Chan AW, Leong FW, Schanley DL, Langan MC, Penetrante ML. A liquid diet model of chlordiazepoxide dependence in mice. Pharmacol Biochem Behav. 1989;34:839–845. doi: 10.1016/0091-3057(89)90283-9. [DOI] [PubMed] [Google Scholar]
- Chen G, Kozell LB, Hitzemann R, Buck KJ. Involvement of the limbic basal ganglia in ethanol withdrawal convulsivity in mice is influenced by a chromosome 4 locus. J Neurosci. 2008;28:9840–9849. doi: 10.1523/JNEUROSCI.1713-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colombo G, Agabio R, Carai MA, Lobina C, Pani M, Reali R, Addolorato G, Gessa GL. Ability of baclofen in reducing alcohol intake and withdrawal severity: I—Preclinical evidence. Alcohol Clin Exp Res. 2000;24:58–66. [PubMed] [Google Scholar]
- Crabbe JC, Kosobud A, Young ER. Genetic selection for ethanol withdrawal severity: differences in replicate mouse lines. Life Sci. 1983;33:955–962. doi: 10.1016/0024-3205(83)90751-8. [DOI] [PubMed] [Google Scholar]
- Crabbe JC, Merrill C, Belknap JK. Acute dependence on depressant drugs is determined by common genes in mice. J Pharmacol Exp Ther. 1991;257:663–667. [PubMed] [Google Scholar]
- Crabbe JC, Belknap JK, Mitchell SR, Crawshaw LI. Quantitative trait loci mapping of genes that influence the sensitivity and tolerance to ethanol-induced hypothermia in BXD recombinant inbred mice. J Pharmacol Exp Ther. 1994;269:184–192. [PubMed] [Google Scholar]
- Daniels GM, Buck KJ. Expression profiling identifies strain-specific changes associated with ethanol withdrawal in mice. Genes Brain Behav. 2002;1:35–45. doi: 10.1046/j.1601-1848.2001.00008.x. [DOI] [PubMed] [Google Scholar]
- Darvasi A. Interval-specific congenic strains (ISCS): an experimental design for mapping a QTL into a 1-centimorgan interval. Mamm Genome. 1997;8:163–167. doi: 10.1007/s003359900382. [DOI] [PubMed] [Google Scholar]
- Dascal N. Signalling via the G protein-activated K+ channels. Cell Signal. 1997;9:551–573. doi: 10.1016/s0898-6568(97)00095-8. [DOI] [PubMed] [Google Scholar]
- Demarest K, McCaughran J, Jr, Mahjubi E, Cipp L, Hitzemann R. Identification of an acute ethanol response quantitative trait locus on mouse chromosome 2. J Neurosci. 1999;19:549–561. doi: 10.1523/JNEUROSCI.19-02-00549.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doss S, Schadt EE, Drake TA, Lusis AJ. cis-acting expression quantitative trait loci in mice. Genome Res. 2005;15:681–691. doi: 10.1101/gr.3216905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doudney K, Murdoch JN, Braybrook C, Paternotte C, Bentley L, Copp AJ, Stanier P. Cloning and characterization of Igsf9 in mouse and human: a new member of the immunoglobulin superfamily expressed in the developing nervous system. Genomics. 2002;79:663–670. doi: 10.1006/geno.2002.6757. [DOI] [PubMed] [Google Scholar]
- Ehlers C, Walter N, Dick D, Buck K, Crabbe J. A comparison of selected quantitative trait loci associated with alcohol use phenotypes in humans and mouse models. Addict Biol. 2010 doi: 10.1111/j.1369-1600.2009.00195.x. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Federici M, Nisticò R, Giustizieri M, Bernardi G, Mercuri NB. Ethanol enhances GABAB-mediated inhibitory postsynaptic transmission on rat midbrain dopaminergic neurons by facilitating GIRK currents. Eur J Neurosci. 2009;29:1369–1377. doi: 10.1111/j.1460-9568.2009.06700.x. [DOI] [PubMed] [Google Scholar]
- Fehr C, Shirley RL, Belknap JK, Crabbe JC, Buck KJ. Congenic mapping of alcohol and pentobarbital withdrawal liability loci to a <1 cM interval of murine chromosome 4: identification of Mpdz as a candidate gene. J Neurosci. 2002;22:3730–3738. doi: 10.1523/JNEUROSCI.22-09-03730.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feller DJ, Bassir JM, Crabbe JC, Le Fevre CA. Audiogenic seizure susceptibility in WSP and WSR mice. Epilepsia. 1994;35:861–867. doi: 10.1111/j.1528-1157.1994.tb02524.x. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, Schork NJ, St Jean P, Ballas C, Choi H, Berrettini WH. Mapping murine loci for seizure response to kainic acid. Mamm Genome. 1997;8:200–208. doi: 10.1007/s003359900389. [DOI] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Smith GG, St Jean P, Schork NJ, Mulholland N, Ballas C, Schill J, Buono RJ, Berrettini WH. Mapping loci for pentylenetetrazol-induced seizure susceptibility in mice. J Neurosci. 1999;19:6733–6739. doi: 10.1523/JNEUROSCI.19-16-06733.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferraro TN, Golden GT, Dahl JP, Smith GG, Schwebel CL, Macdonald R, Lohoff FW, Berrettini WH, Buono RJ. Analysis of a quantitative trait locus for seizure susceptibility in mice using bacterial artificial chromosome-mediated gene transfer. Epilepsia. 2007;48:1667–1677. doi: 10.1111/j.1528-1167.2007.01126.x. [DOI] [PubMed] [Google Scholar]
- Flint BA, Ho IK. Assessment of tolerance to and physical dependence on pentobarbital, induced by multiple pellet implantation. Eur J Pharmacol. 1980;65:355–363. doi: 10.1016/0014-2999(80)90339-8. [DOI] [PubMed] [Google Scholar]
- Frankel WN, Johnson EW, Lutz CM. Congenic strains reveal effects of the epilepsy quantitative trait locus, El2, separate from other El loci. Mamm Genome. 1995;6:839–843. doi: 10.1007/BF00292432. [DOI] [PubMed] [Google Scholar]
- Frye GD, Fincher A. Sensitivity of postsynaptic GABAB receptors on hippocampal CA1 and CA3 pyramidal neurons to ethanol. Brain Res. 1996;735:239–248. doi: 10.1016/0006-8993(96)00579-3. [DOI] [PubMed] [Google Scholar]
- Gaidatzis D, van Nimwegen E, Hausser J, Zavolan M. Inference of miRNA targets using evolutionary conservation and pathway analysis. BMC Bioinformatics. 2007;8:69. doi: 10.1186/1471-2105-8-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldman D, Oroszi G, O'Malley S, Anton R. COMBINE genetics study: the pharmacogenetics of alcoholism treatment response: genes and mechanisms. J Stud Alcohol. 2005;(Suppl):56–64. doi: 10.15288/jsas.2005.s15.56. discussion 33. [DOI] [PubMed] [Google Scholar]
- Goldstein DB. Alcohol withdrawal reactions in mice: effects of drugs that modify neurotransmission. J Pharmacol Exp Ther. 1973;186:1–9. [PubMed] [Google Scholar]
- Goldstein DB, Pal N. Alcohol dependence produced in mice by inhalation of ethanol: grading the withdrawal reaction. Science. 1971;172:288–290. doi: 10.1126/science.172.3980.288. [DOI] [PubMed] [Google Scholar]
- Herrera VL, Cova T, Sassoon D, Ruiz-Opazo N. Developmental cell-specific regulation of Na+-K+-ATPase α1-, α2-, and α3-isoform gene expression. Am J Physiol. 1994;266:C1301–C1312. doi: 10.1152/ajpcell.1994.266.5.C1301. [DOI] [PubMed] [Google Scholar]
- Hill KG, Alva H, Blednov YA, Cunningham CL. Reduced ethanol-induced conditioned taste aversion and conditioned place preference in GIRK2 null mutant mice. Psychopharmacology (Berl) 2003;169:108–114. doi: 10.1007/s00213-003-1472-4. [DOI] [PubMed] [Google Scholar]
- Ho IK, Harris RA. Mechanism of action of barbiturates. Annu Rev Pharmacol Toxicol. 1981;21:83–111. doi: 10.1146/annurev.pa.21.040181.000503. [DOI] [PubMed] [Google Scholar]
- Hoen PA, Bijsterbosch MK, van Berkel TJ, Vermeulen NP, Commandeur JN. Midazolam is a phenobarbital-like cytochrome p450 inducer in rats. J Pharmacol Exp Ther. 2001;299:921–927. [PubMed] [Google Scholar]
- Hood HM, Metten P, Crabbe JC, Buck KJ. Fine mapping of a sedative-hypnotic drug withdrawal locus on mouse chromosome 11. Genes Brain Behav. 2006;5:1–10. doi: 10.1111/j.1601-183X.2005.00122.x. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Onaka T, Yamakado M, Nakai J, Ishikawa TO, Taketo MM, Kawakami K. Degeneration of the amygdala/piriform cortex and enhanced fear/anxiety behaviors in sodium pump α2 subunit (Atp1a2)-deficient mice. J Neurosci. 2003;23:4667–4676. doi: 10.1523/JNEUROSCI.23-11-04667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalsi AS, Greenwood K, Wilkin G, Butt AM. Kir4.1 expression by astrocytes and oligodendrocytes in CNS white matter: a developmental study in the rat optic nerve. J Anat. 2004;204:475–485. doi: 10.1111/j.0021-8782.2004.00288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karschin C, Dissmann E, Stühmer W, Karschin A. IRK(1-3) and GIRK(1-4) inwardly rectifying K+ channel mRNAs are differentially expressed in the adult rat brain. J Neurosci. 1996;16:3559–3570. doi: 10.1523/JNEUROSCI.16-11-03559.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerns RT, Ravindranathan A, Hassan S, Cage MP, York T, Sikela JM, Williams RW, Miles MF. Ethanol-responsive brain region expression networks: implications for behavioral responses to acute ethanol in DBA/2J versus C57BL/6J mice. J Neurosci. 2005;25:2255–2266. doi: 10.1523/JNEUROSCI.4372-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kliethermes CL, Metten P, Belknap JK, Buck KJ, Crabbe JC. Selection for pentobarbital withdrawal severity: correlated differences in withdrawal from other sedative drugs. Brain Res. 2004;1009:17–25. doi: 10.1016/j.brainres.2004.02.040. [DOI] [PubMed] [Google Scholar]
- Knapp DJ, Overstreet DH, Breese GR. Baclofen blocks expression and sensitization of anxiety-like behavior in an animal model of repeated stress and ethanol withdrawal. Alcohol Clin Exp Res. 2007;31:582–595. doi: 10.1111/j.1530-0277.2007.00342.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi T, Ikeda K, Kojima H, Niki H, Yano R, Yoshioka T, Kumanishi T. Ethanol opens G-protein-activated inwardly rectifying K+ channels. Nat Neurosci. 1999;2:1091–1097. doi: 10.1038/16019. [DOI] [PubMed] [Google Scholar]
- Kosobud A, Crabbe JC. Ethanol withdrawal in mice bred to be genetically prone or resistant to ethanol withdrawal seizures. J Pharmacol Exp Ther. 1986;238:170–177. [PubMed] [Google Scholar]
- Koyrakh L, Luján R, Colón J, Karschin C, Kurachi Y, Karschin A, Wickman K. Molecular and cellular diversity of neuronal G-protein-gated potassium channels. J Neurosci. 2005;25:11468–11478. doi: 10.1523/JNEUROSCI.3484-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labouèbe G, Lomazzi M, Cruz HG, Creton C, Luján R, Li M, Yanagawa Y, Obata K, Watanabe M, Wickman K, Boyer SB, Slesinger PA, Lüscher C. RGS2 modulates coupling between GABAB receptors and GIRK channels in dopamine neurons of the ventral tegmental area. Nat Neurosci. 2007;10:1559–1568. doi: 10.1038/nn2006. [DOI] [PubMed] [Google Scholar]
- Lander E, Kruglyak L. Genetic dissection of complex traits. Nat Genet. 1995;11:241–247. doi: 10.1038/ng1195-241. [DOI] [PubMed] [Google Scholar]
- Levental M, Tabakoff B. Sodium-potassium-activated adenosine triphosphatase activity as a measure of neuronal membrane characteristics in ethanol-tolerant mice. J Pharmacol Exp Ther. 1980;212:315–319. [PubMed] [Google Scholar]
- Lewohl JM, Wilson WR, Mayfield RD, Brozowski SJ, Morrisett RA, Harris RA. G-protein-coupled inwardly rectifying potassium channels are targets of alcohol action. Nat Neurosci. 1999;2:1084–1090. doi: 10.1038/16012. [DOI] [PubMed] [Google Scholar]
- Li D, Roberts R. WD-repeat proteins: structure characteristics, biological function, and their involvement in human diseases. Cell Mol Life Sci. 2001;58:2085–2097. doi: 10.1007/PL00000838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Head V, Timpe LC. Identification of an inward rectifier potassium channel gene expressed in mouse cortical astrocytes. Glia. 2001;33:57–71. doi: 10.1002/1098-1136(20010101)33:1<57::aid-glia1006>3.0.co;2-0. [DOI] [PubMed] [Google Scholar]
- Lieber CS. Microsomal ethanol-oxidizing system (MEOS): the first 30 years (1968–1998)—a review. Alcohol Clin Exp Res. 1999;23:991–1007. [PubMed] [Google Scholar]
- Little HJ, Stephens DN, Ripley TL, Borlikova G, Duka T, Schubert M, Albrecht D, Becker HC, Lopez MF, Weiss F, Drummond C, Peoples M, Cunningham C. Alcohol withdrawal and conditioning. Alcohol Clin Exp Res. 2005;29:453–464. doi: 10.1097/01.alc.0000156737.56425.e3. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lunn ML, Nassirpour R, Arrabit C, Tan J, McLeod I, Arias CM, Sawchenko PE, Yates JR, 3rd, Slesinger PA. A unique sorting nexin regulates trafficking of potassium channels via a PDZ domain interaction. Nat Neurosci. 2007;10:1249–1259. doi: 10.1038/nn1953. [DOI] [PubMed] [Google Scholar]
- Lüscher C, Jan LY, Stoffel M, Malenka RC, Nicoll RA. G protein-coupled inwardly rectifying K+ channels (GIRKs) mediate postsynaptic but not presynaptic transmitter actions in hippocampal neurons. Neuron. 1997;19:687–695. doi: 10.1016/s0896-6273(00)80381-5. [DOI] [PubMed] [Google Scholar]
- Lutz UC, Batra A, Kolb W, Machicao F, Maurer S, Köhnke MD. Methylenetetrahydrofolate reductase C677T-polymorphism and its association with alcohol withdrawal seizure. Alcohol Clin Exp Res. 2006;30:1966–1971. doi: 10.1111/j.1530-0277.2006.00242.x. [DOI] [PubMed] [Google Scholar]
- Marker CL, Cintora SC, Roman MI, Stoffel M, Wickman K. Hyperalgesia and blunted morphine analgesia in G protein-gated potassium channel subunit knockout mice. Neuroreport. 2002;13:2509–2513. doi: 10.1097/00001756-200212200-00026. [DOI] [PubMed] [Google Scholar]
- Martin B, Clément Y, Venault P, Chapouthier G. Mouse chromosomes 4 and 13 are involved in β-carboline-induced seizures. J Hered. 1995;86:274–279. doi: 10.1093/oxfordjournals.jhered.a111581. [DOI] [PubMed] [Google Scholar]
- McDaid J, McElvain MA, Brodie MS. Ethanol effects on dopaminergic ventral tegmental area neurons during block of Ih: involvement of barium-sensitive potassium currents. J Neurophysiol. 2008;100:1202–1210. doi: 10.1152/jn.00994.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGrail KM, Phillips JM, Sweadner KJ. Immunofluorescent localization of three Na,K-ATPase isozymes in the rat central nervous system: both neurons and glia can express more than one Na,K-ATPase. J Neurosci. 1991;11:381–391. doi: 10.1523/JNEUROSCI.11-02-00381.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McQuarrie DG, Fingl E. Effects of single doses and chronic administration of ethanol on experimental seizures in mice. J Pharmacol Exp Ther. 1958;124:264–271. [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Common genetic determinants of severity of acute withdrawal from ethanol, pentobarbital and diazepam in inbred mice. Behav Pharmacol. 1994;5:533–547. doi: 10.1097/00008877-199408000-00014. [DOI] [PubMed] [Google Scholar]
- Metten P, Crabbe JC. Genetic determinants of severity of acute withdrawal from diazepam in mice: commonality with ethanol and pentobarbital. Pharmacol Biochem Behav. 1999;63:473–479. doi: 10.1016/s0091-3057(99)00017-9. [DOI] [PubMed] [Google Scholar]
- Metten P, Belknap JK, Crabbe JC. Drug withdrawal convulsions and susceptibility to convulsants after short-term selective breeding for acute ethanol withdrawal. Behav Brain Res. 1998;95:113–122. doi: 10.1016/s0166-4328(97)00216-7. [DOI] [PubMed] [Google Scholar]
- Metten P, Buck KJ, Merrill CM, Roberts AJ, Yu CH, Crabbe JC. Use of a novel mouse genotype to model acute benzodiazepine withdrawal. Behav Genet. 2007;37:160–170. doi: 10.1007/s10519-006-9094-3. [DOI] [PubMed] [Google Scholar]
- Mishra A, Knerr B, Paixão S, Kramer ER, Klein R. The protein dendrite arborization and synapse maturation 1 (Dasm-1) is dispensable for dendrite arborization. Mol Cell Biol. 2008;28:2782–2791. doi: 10.1128/MCB.02102-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrisett RA, Swartzwelder HS. Attenuation of hippocampal long-term potentiation by ethanol: a patch-clamp analysis of glutamatergic and GABAergic mechanisms. J Neurosci. 1993;13:2264–2272. doi: 10.1523/JNEUROSCI.13-05-02264.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murdoch JN, Doudney K, Gerrelli D, Wortham N, Paternotte C, Stanier P, Copp AJ. Genomic organization and embryonic expression of Igsf8, an immunoglobulin superfamily member implicated in development of the nervous system and organ epithelia. Mol Cell Neurosci. 2003;22:62–74. doi: 10.1016/s1044-7431(02)00021-0. [DOI] [PubMed] [Google Scholar]
- Neumann PE, Collins RL. Genetic dissection of susceptibility to audiogenic seizures in inbred mice. Proc Natl Acad Sci U S A. 1991;88:5408–5412. doi: 10.1073/pnas.88.12.5408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman EA, Frambach DA, Odette LL. Control of extracellular potassium levels by retinal glial cell K+ siphoning. Science. 1984;225:1174–1175. doi: 10.1126/science.6474173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Newman SL, Weikle AA, Neuberger TJ, Bigbee JW. Myelinogenic potential of an immortalized oligodendrocyte cell line. J Neurosci Res. 1995;40:680–693. doi: 10.1002/jnr.490400514. [DOI] [PubMed] [Google Scholar]
- Office of National Drug Control Policy. Washington, DC: Executive Office of the President; 2004. The economic costs of drug abuse in the United States, 1992–2002. (Publication No. 207303) [Google Scholar]
- Orkand RK, Nicholls JG, Kuffler SW. Effect of nerve impulses on the membrane potential of glial cells in the central nervous system of amphibia. J Neurophysiol. 1966;29:788–806. doi: 10.1152/jn.1966.29.4.788. [DOI] [PubMed] [Google Scholar]
- Paolini C, Quarta M, Nori A, Boncompagni S, Canato M, Volpe P, Allen PD, Reggiani C, Protasi F. Reorganized stores and impaired calcium handling in skeletal muscle of mice lacking calsequestrin-1. J Physiol. 2007;583:767–784. doi: 10.1113/jphysiol.2007.138024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng L, Martin-Vasallo P, Sweadner KJ. Isoforms of Na,K-ATPase α and β subunits in the rat cerebellum and in granule cell cultures. J Neurosci. 1997;17:3488–3502. doi: 10.1523/JNEUROSCI.17-10-03488.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi ZH, Song M, Wallace MJ, Wang D, Newton PM, McMahon T, Chou WH, Zhang C, Shokat KM, Messing RO. Protein kinase C epsilon regulates γ-aminobutyrate type A receptor sensitivity to ethanol and benzodiazepines through phosphorylation of γ2 subunits. J Biol Chem. 2007;282:33052–33063. doi: 10.1074/jbc.M707233200. [DOI] [PubMed] [Google Scholar]
- Risinger FO, Cunningham CL. Ethanol-induced conditioned taste aversion in BXD recombinant inbred mice. Alcohol Clin Exp Res. 1998;22:1234–1244. [PubMed] [Google Scholar]
- Shi SH, Cox DN, Wang D, Jan LY, Jan YN. Control of dendrite arborization by an Ig family member, dendrite arborization and synapse maturation 1 (Dasm-1) Proc Natl Acad Sci U S A. 2004;101:13341–13345. doi: 10.1073/pnas.0405370101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shirley RL, Walter NA, Reilly MT, Fehr C, Buck KJ. Mpdz is a quantitative trait gene for drug withdrawal seizures. Nat Neurosci. 2004;7:699–700. doi: 10.1038/nn1271. [DOI] [PubMed] [Google Scholar]
- Sivam SP, Nabeshima T, Ho IK. Acute and chronic effects of pentobarbital in relation to postsynaptic GABA receptors: a study with muscimol. J Neurosci Res. 1982;7:37–47. doi: 10.1002/jnr.490070105. [DOI] [PubMed] [Google Scholar]
- Skelton KH, Nemeroff CB, Owens MJ. A comparison of plasma alprazolam concentrations following different routes of chronic administration in the Sprague-Dawley rat: implications for psychotropic drug research. Psychopharmacology (Berl) 2000;151:72–76. doi: 10.1007/s002130000469. [DOI] [PubMed] [Google Scholar]
- Smith SB, Marker CL, Perry C, Liao G, Sotocinal SG, Austin JS, Melmed K, Clark JD, Peltz G, Wickman K, Mogil JS. Quantitative trait locus and computational mapping identifies Kcnj9 (GIRK3) as a candidate gene affecting analgesia from multiple drug classes. Pharmacogenet Gen. 2008;18:231–241. doi: 10.1097/FPC.0b013e3282f55ab2. [DOI] [PubMed] [Google Scholar]
- Sokal RR, Rohlf FJ. Ed 3. New York: Freeman; 1995. Biometry. [Google Scholar]
- Takumi T, Ishii T, Horio Y, Morishige K, Takahashi N, Yamada M, Yamashita T, Kiyama H, Sohmiya K, Nakanishi S, Kurachi Y. A novel ATP-dependent inward rectifier potassium channel expressed predominantly in glial cells. J Biol Chem. 1995;270:16339–16346. doi: 10.1074/jbc.270.27.16339. [DOI] [PubMed] [Google Scholar]
- Tarantino LM, McClearn GE, Rodriguez LA, Plomin R. Confirmation of quantitative trait loci for alcohol preference in mice. Alcohol Clin Exp Res. 1998;22:1099–1105. [PubMed] [Google Scholar]
- Taylor BA, Frankel WN. A new strain congenic for the Mtv-7/Mls-1 locus of mouse chromosome 1. Immunogenetics. 1993;38:235–237. doi: 10.1007/BF00211523. [DOI] [PubMed] [Google Scholar]
- Terdal ES, Crabbe JC. Indexing withdrawal in mice: matching genotypes for exposure in studies using ethanol vapor inhalation. Alcohol Clin Exp Res. 1994;18:542–547. doi: 10.1111/j.1530-0277.1994.tb00907.x. [DOI] [PubMed] [Google Scholar]
- Torrecilla M, Marker CL, Cintora SC, Stoffel M, Williams JT, Wickman K. G-protein-gated potassium channels containing Kir3.2 and Kir3.3 subunits mediate the acute inhibitory effects of opioids on locus ceruleus neurons. J Neurosci. 2002;22:4328–4334. doi: 10.1523/JNEUROSCI.22-11-04328.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandesompele J, De Preter K, Pattyn F, Poppe B, Van Roy N, De Paepe A, Speleman F. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 2002;3:RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter N, Bottomly D, Laderas T, Mooney M, Darakjian P, Searles R, Harrington CA, McWeeney SK, Hitzemann R, Buck KJ. High throughput sequencing in mice: a platform comparison identifies a preponderance of cryptic SNPs. BMC Genomics. 2009;10:379. doi: 10.1186/1471-2164-10-379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter NA, McWeeney SK, Peters ST, Belknap JK, Hitzemann R, Buck KJ. SNPs matter: impact on detection of differential expression. Nat Methods. 2007;4:679–680. doi: 10.1038/nmeth0907-679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watts AG, Sanchez-Watts G, Emanuel JR, Levenson R. Cell-specific expression of mRNAs encoding Na+,K+ATPase α and β subunit isoforms within the rat central nervous system. Proc Natl Acad Sci U S A. 1991;88:7425–7429. doi: 10.1073/pnas.88.16.7425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakura T, Lewohl JM, Harris RA. Differential effects of general anesthetics on G protein-coupled inwardly rectifying and other potassium channels. Anesthesiology. 2001;95:144–153. doi: 10.1097/00000542-200107000-00025. [DOI] [PubMed] [Google Scholar]
- Zhang XA, Bontrager AL, Hemler ME. Transmembrane-4 superfamily proteins associate with activated protein kinase C (PKC) and link PKC to specific β1 integrins. J Biol Chem. 2001;276:25005–25013. doi: 10.1074/jbc.M102156200. [DOI] [PubMed] [Google Scholar]