Abstract
Significant advances are being made in our understanding of basic pathophyiological and biochemical mechanisms that cause HuntingtonÕs disease (HD). There is increasing reason to believe that pathologic alterations occur in the brain for years before symptoms manifest. The “classic” hallmark of neuropathology in HD is selective neurodegeneration in which vulnerable populations of neurons degenerate while less vulnerable populations are spared. While, the earliest and most striking neuropathologic changes have been found in the neostriatum, neuronal loss has been identified in many other regions of the brain. We report topologically selective, early, and progressive changes in the cortex, striatum, extra-striatal brain structures and white matter throughout the spectrum of disease. Our growing understanding of HD underscores the reality that points to the complexity of HD. A single, well-defined genetic mutation causes a cascade of events whose final result is an aggregate insult of the homeostatic process. We explore possible explanations for the selective vulnerability of the brain in HD.
The ultimate goal in HD is to develop disease-modifying therapies that will prevent the onset of clinical symptoms in those individuals who are at risk and slow the progression of symptoms in those individuals already affected with symptoms. Understanding changes in brain morphometry and their relationship to clinical symptoms may provide important new and important insights into basic pathophysiological mechanisms at play in the disease.
Keywords: Huntington’s disease, cortex, neurodegeneration, pre-manifest HD, sub-cortical atrophy, white matter degeneration, oxidative stress, circuitry
Background
Huntington’s disease (HD) is a fully penetrant autosomal dominant inherited neurodegenerative disorder that is characterized by progressive motor dysfunction, emotional and psychiatric disturbances, cognitive dysfunction, and weight loss. HD occurs worldwide in all races and ethnic groups 1. Its prevalence is 5–10 cases per 100,000, and there is a new mutation rate as high as 1–3% 2. There are about 30,000 affected individuals in North America while another 150,000 have a genetic risk for developing the disease. The average age of clinical onset is about 37 years of age, however the range is from infancy into the 80’s. Affected individuals are rapidly disabled by early functional decline and require multidisciplinary care and supervision for another 15–25 years before succumbing to the effects of severe physical, psychiatric and mental deterioration. There is no therapy proven to delay onset or slow progression, and current medical care focuses on symptom management and maximizing function 3–5. Progressive functional, cognitive, and physical decline leads to increasingly intense levels of care, creating a disproportionate strain on family and healthcare resources
The HD gene, first cloned in 1993 6 codes for a large, highly conserved, protein of unknown function (“huntingtin”) containing 3144 amino acids. In individuals with HD, a polymorphic trinucleotide repeat sequence (CAGn), near the 5′ end of the gene, is expanded beyond the normal repeat range 6, leading to translation of an expanded polyglutamine sequence in the protein. In the normal population the number of CAG repeats varies from 17 to 29. In individuals with HD there are more than 38 repeats; there is reduced penetrance for CAG repeats between 36 and 397. The CAG repeat also exhibits dramatic instability when transmitted to subsequent generations 8; repeats between 27 and 35 are not associated with disease expression but may expand in paternal transmission, resulting in the disease in descendents 9. Once expanded into the pathogenic ranges, there is an inverse relationship between the CAG repeat number and the age of onset 10, while a correlation with progression has not been demonstrated. Even for a specific CAG repeat, there is considerable heterogeneity in the actual “time to onset”; the CAG repeat accounts for only 50–65% of the variance in age of onset 11. Other genetic modifiers are currently being sought.
“The basics”: What is known
The hallmark of neuropathology in HD is selective neurodegeneration in which vulnerable populations of neurons degenerate while less vulnerable populations are spared 12. The earliest and most striking neuropathologic changes are found in the neostriatum 13 but neuronal loss has been identified in may other regions of the brain 14. Proliferative and degenerative changes in vulnerable neurons 14, 15 suggest that the presence of mutant huntingtin leads to both compensatory and degenerative genetic programs in a prolonged process leading to neuronal dysfunction and death 14, 16(20).
Altered synaptic plasticity in the R6/2 mouse model of HD occurs prior to the more overt disease phenotypes 17, indicating early synaptic pathology in the disease. Morphological changes occur in medium spiny neurons of the striatum and in cortical pyramidal cells prior to degeneration, including dendritic remodeling and altered size and number of dendritic spines 14, 18. This suggests that these neurons undergo a period of stress and injury prior to perishing. It is important to consider that cortical degeneration and cell death follows cortical dysfunction 19.
Although many leads have been uncovered, a direct pathway from the genetic mutation to neuronal dysfunction and death has not yet been established. Huntingtin is a widely expressed, predominantly cytoplasmic, protein of unknown function found heterogeneously in neurons throughout the brain 12, 20, 21. In HD, both normal and mutant alleles are expressed and both gain of function alterations in which mutant huntingtin is toxic and loss of function alterations 22, 23 in which suppression of normal huntingtin functions might also be toxic have been identified. Proteolysis of mutant huntingtin, whereby an abnormal and ultimately toxic N-terminus fragment of huntingtin is released, may play a role in causing disease 24–26. This fragment has been shown in humans and in animal and cellular models to form protein aggregates in the nucleus, cytoplasm, and processes of neurons 12, 27. These aggregates induce chaperone expression and become ubiquitinated yet persist, indicating protein misfolding and failed proteolysis 28.
Huntingtin aggregates also sequester a variety of other proteins including chaperones 29 proteasomal proteins 30, normal huntingtin 31, and transcription factors 32–34 35, another means of disturbing protein homeostasis. These aggregates can be readily detected in the brains of individuals at risk for HD who died prior to exhibiting any symptoms and in brains from individuals who died throughout the course of the disease 12 36. While the role of huntingtin aggregates continues to be debated 37, most evidence continues to point to a proximal toxicity residing in mutant huntingtin or its proteolytic fragments and their interactions with other proteins, including huntingtin itself or the dozens of other proteins that have been demonstrated to associate with huntingtin or huntingtin aggregates, including a number of transcription factors which have been implicated in widespread transcriptional alterations that appear to occur in HD. Several recent studies have also suggested that the transcriptional alterations that occur in human brain are variable across cortical regions, providing one more clue to selective vulnerability that requires further investigation 38, 39.
The connection with mitochondrial dysfunction
In postmortem studies, severe deficiencies in mitochondrial complex II and III and sometimes complex IV have been reported in the striatum 40, 41. Inhibition of complex II may impart dysfunction by reducing the levels of complex II/succinate dehydrogenase subunits and result in diminished activity. A decrease in succinate oxidation has also been shown to occur in the striatum 42, 43. Dysfunction of complex I has been reported in human blood in HD 44 and expression of sub-units of mitochondrial complex-I have been found to be reduced in HD brain 45. Elevated lactate levels have also been shown to correlate with CAG repeat length in HD occipital cortex and in striatum 46, 47.
Mutant huntingtin might also exert direct negative effects on mitochondria, through physical interaction with the huntingtin protein or its fragments. Mutant huntingtin has been localized to neuronal mitochondrial membranes and incubation of mutant mitochondria with normal mitochondria results in depolarization of the mitochondrial membrane at lower Ca2+ loads 48, 49. Neuronal dysfunction could subsequently occur secondary to disruption of the mitochondrial membrane potential, excitotoxic induced CA 2+ influx 50 and diminished ATP production. Mitochondrial dysfunction and secondary energy (ATP) depletion could subsequently lead further to unmitigated free radical production and cell death51, 52. There is broad evidence in human HD and in cellular and animal models that free radicals then cause cellular damage and up-regulated anti-oxidant responses in the brain and periphery53, 54.
Mitochondria are a major site of production of free radicals (reactive oxygen species) and primary targets of free radicals. Reactive oxygen species induce a variety of oxidative adducts of DNA and RNA. Hydroxy adducts of guanidine (DNA) or guanosine (RNA) are especially common, stable, and readily measured in tissues and tissue fluids. In DNA, hydroxy adducts induce repair (base excision or nucleotide excision) which is not always complete and is exhaustible because of oxidative injury to repair mechanisms. In situ, DNA adducts are mutagenic, causing substitutions, transcriptional blocks, and deletions which could contribute to transcriptional dysfunction in HD. 8OH2′dG (8-hydroxy 2-deoxyguanosine) is a hydroxy adduct of DNA that has been studied in HD55, 56. Levels of these hydroxy adducts in HD brain and blood are elevated dramatically; in particular, elevated levels of these adducts have been found in mitochondrial DNA isolated from human HD parietal cortex 57. Parenthetically, however, there was no evidence of oxidative damage to mitochondrial DNA from human cerebellum in the same study. This supports the notion that selective vulnerability of in HD, be it in neurons or astrocytes, for example, is directly tied into regionally specific pathophysiologic mechanisms and that further study of what drives this selectivity is extremely important.
Huntingtin is also appears involved in axonal trafficking 58, as shown in in vitro assays, and mutant huntingtin impairs trafficking of vesicles and mitochondria in transgenic mice 59 and in mammalian neurons 60. These changes are present prior to the onset of clinical symptoms, supporting a possible role of altered mitochondrial localization in axons in HD. Reactive oxygen species can also act as signaling molecules for transcription and therefore chronic exposure to free radicals in HD could activate cascades of genes. Peroxisome proliferators-activated receptor gamma co-activator (PGC-1 alpha) is a co-activator of several transcription factors and a potent stimulator of mitochondrial biogenesis and respiration whose altered expression has also been implicated in HD pathogenesis 45, 61.
A window into a complex biology
Modern neuro-imaging methods have been developed that provide a faster more reliable and sensitive method to study the entire brain 62, 63. They allow for a more comprehensive characterization of the progressive changes that occur in the brain in Huntington’s disease. A broader approach to understanding the selective vulnerability of distinct brain regions in HD is critical to understanding basic pathophysiological mechanisms. Much is not known, but it is becoming increasingly clear that the striatum, while severely affected in HD, is only a small piece of the puzzle and cannot fully, in isolation, explain the varied and progressive symptoms of HD.
Alterations in brain anatomy in HD are complex and begin very early. Striatal atrophy is known to begin more than a decade before motor symptoms develop 64; by the time of diagnosis, the striatum may be atrophied by as much as 50% 65. More recently, it has become clear that other brain regions also begin to atrophy during the pre-manifest period, more than a decade before a clinical diagnosis. This may reflect in part the capacity of the brain to compensate during periods of neuronal dysfunction 66–68 by recruiting other brain 69. Neuronal death unfortunately is at present inevitable in HD. Elucidating the specific relationships between selective brain degeneration and clinical symptoms becomes all the more important as targeted therapies are developed.
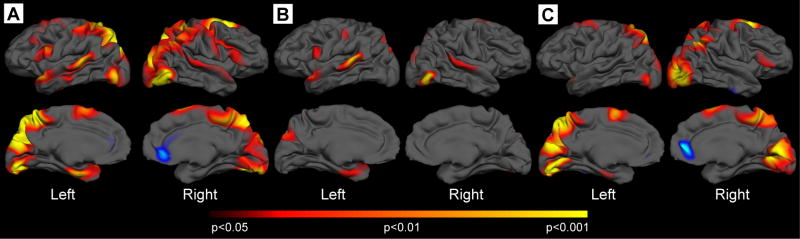
The cortex has been little explored in HD but by end-stage, the entire brain is atrophic. We have found that the cortex begins to atrophy during the pre-manifest period, as shown in Figure 1. It is significant, as compared to age and gender matched controls subjects, occurs in some regions while others appear spared, and corresponds to loss of approximately the cortical thickness of approximately 5 to 10%. The areas that appear the most affected in these pre-manifest subjects correspond to sensori-motor (approximating BA areas 4, 6, and 8), superior parietal (approximating BA7), superior temporal (approximating BA areas 22, 41, 42), middle and inferior temporal, occipital (approximating BA 17, 18, 19) and precuneus (approximating BA 31), and occipital cortical areas 70. In contrast, there was no significant reduction in whole brain volumes in this group (p=0.46); this supports the importance of a fine-grained approach to the study of brain morphometric changes in HD. By looking too globally, we may miss important and clinically relevant changes. When this group of subjects was further stratified according to years from estimated onset (further > 12 years, closer < 12 years) 71, subjects further from onset show thinning in portions of motor, precuneus, superior temporal and occipital. Additional areas were thinned in the group closer to onset including parietal, other regions of occipital and more posterior regions of the superior frontal cortex. Interestingly, the anterior cingulate appeared thicker.
Figure 1.
Topologically selective thinning occurs in Pre-manifest Huntington’s disease. A. Surface maps of cortical thinning were generated by using a general linear model at each vertex across the entire mantle in 31 gene-carriers without symptoms of HD. Significant cortical thinning was present in sensor-motor, parietal, superior temporal, entorhinal, precuneus, occipital and portions of frontal cortex. B. Thickness maps of individuals greater than 12 years to expected onset; thinning is already present over the portions of superior temporal, motor, precuneus and entorhinal cortical regions. C. Thickness maps of individuals less than 12 years to expected onset; thinning is much more extensive, and begins to involve occipital, parietal, cuneus, posterior frontal cortical areas. Maps are presented on a semi-inflated cortical surface of an average brain. The color scale at the bottom represents the significance of the thickness change, transitioning from red (p<0.05) to yellow (p<0.001).
Early symptomatic HD subjects showed thinning of other cortical regions, to encompass more of posterior frontal areas as well as more of the occipital and parietal areas. This suggests a pattern of degeneration, starting with primary cortical regions, areas which subserve simpler functions, including motor, sensory and visual cortical regions, with progression to include uni-modal areas, regions that discriminate, categorize and integrate information within a single modality to form a precept of the same modality, and subsequently of hetero-modal multi-association cortical regions, regions whose central role is in the integration of precepts from various modalities to form complex multi-modal precepts 72. It is of note that more anterior frontal, middle and inferior temporal areas appeared relatively preserved in early HD. It is also interesting the anterior cingulate also appeared thicker in early symptomatic HD subjects; increases in microglial activation have been reported in this region in HD 68. Motor and occipital cortical regions were also more thinned, corresponding to a loss of the cortical mantle of approximately 30% in these regions.
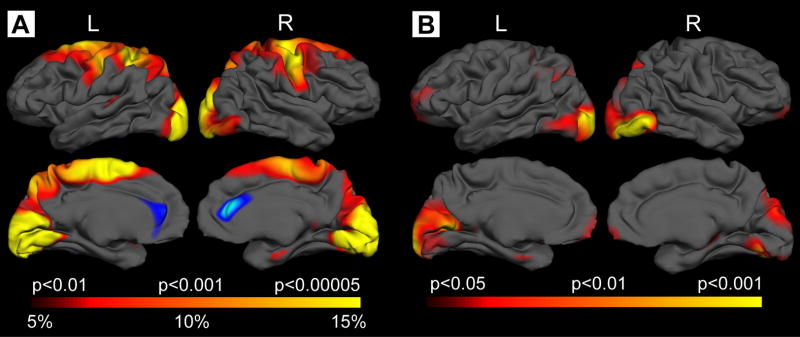
These are very preliminary models that must be validated with longitudinal studies. There are several points on which to comment, however. First, even within gyral regions, specific regions of a gyrus appeared thinned while other regions of the gyrus appeared to remain unaffected. In the motor cortex, for example, in more superior regions thinning was more significant and extensive in the most superior regions, and less so in more inferior regions. Secondly, when we took into account cortical changes that might be related to striatal volume, we found regions of thinning that were independent, as shown in Figure 2. Because the cortex and striatum are so interconnected, it may be difficult to determine if cortical degeneration precedes or follows striatal atrophy, but our findings suggest that distinct mechanisms may also be present. Cortical thinning may reflect not only neuronal loss but also the dysmorphology and loss of dendrites and axons and associated connections that occur 73.
Figure 2.
Topologically selective thinning in Early symptomatic HD: extension of changes seen in Pre-manifest subjects. A. Surface maps of cortical thinning were generated by using a general linear model at each vertex across the entire cortical mantle. In Stage I and Stage II subjects, significant thinning was present in sensor-motor, parietal, posterior superior and middle frontal, enthorhinal, precuneus, cuneus and occipital cortical areas. Maps are presented on a semi-inflated cortical surface of an average brain. The color scale at the bottom represents the significance of the thickness change, transitioning from red (p<0.01) to yellow (p<0.00005). B. After adjusting for striatal volume, significant thinning of the cortex was still present over occipital, parietal, and precuneus, suggesting some degree of independence between striatal and cortical pathology.
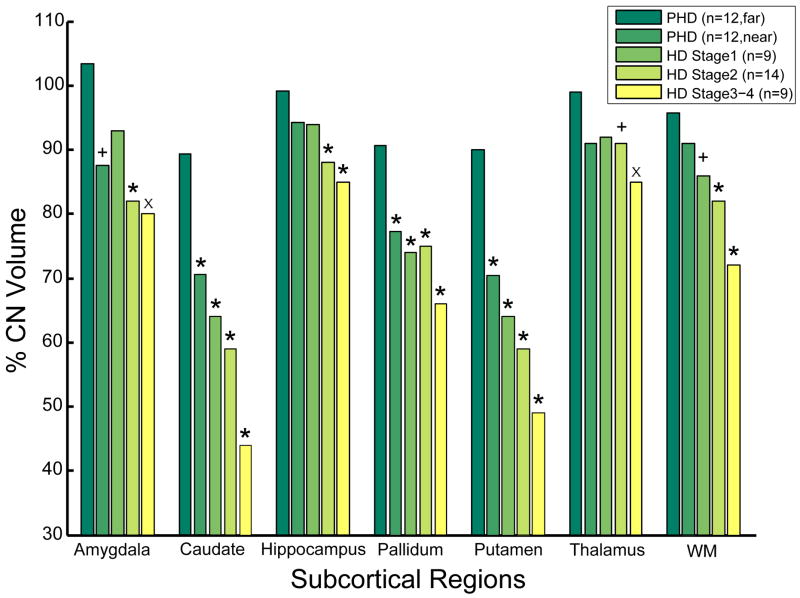
It is also very important to emphasize that it appears that no brain structure is spared in HD and that extensive changes in several other structures are present very early as well. In pre-manifest subjects greater than 12 years to estimated onset, we have found reductions in the volumes of not only the striatum (approximately 20%), but also of the amygdala, thalamus, hippocampus, and white matter. Parenthetically, we and others have shown that white matter changes are also regionally selective, significant and potentially important in HD74–76. As shown in Figure 3, a complex, topologically selective pattern emerges if considers progressive changes that span a continuum from pre-manifest through to advanced HD.
Figure 3.
Bar-graphs demonstrating changes in the volumes of several other brain regions, spanning from pre-manifest individuals greater than 12 years to estimated onset through Stage III HD. A complex pattern of progressive changes emerges that also demonstrates the extensive involvement of brain structures in HD.
The precise relationship between topological alterations in brain structures and clinical symptoms of HD remains to be elucidated. Attempts to ascribe the symptoms of HD to striatal degeneration or to dysfunction of cortico-striatal circuity have been unsatisfying. Striatal degeneration alone, does not adequately explain the clinical symptoms of HD, that are highly variable, individual and neurologically complex. At the time of diagnosis, more than half of the striatum has degenerated, yet symptoms, which encompass multiple clinical domains, continue to progress. A growing body of evidence suggests that the natural history of gradually progressive motor and cognitive dysfunction in HD, which may begin years prior to a clinical diagnosis, based on unequivocal motor signs of HD77, is related to regionally specific changes of many diverse brain regions 65, 70, 78, 79 that include striatum and cortex.
The relationship between cortical atrophy and cognitive dysfunction has only recently and cursorily been investigated 80, 81. Others have focused on neuropsychiatric symptoms 82. We have found distinct relationships between regionally specific cortical degeneration and worsening performance on the cognitive and functional components of the UHDRS 72. Selective degeneration of specific cortical regions may explain some of the clinical heterogeneity. In some patients, psychiatric symptoms are early and debilitating, in others, cognitive dysfunction and in yet others chorea. Motor symptoms can also be quite variable, with some individuals showing the more typical chorea, while in others bradykinesia and dystonia are more prominent. In HD subjects with more prominent Parkinsonism, we have found more significant thinning in frontal cortical regions, specifically affecting portions of pre-motor and supplementary motor cortex 72. Of note, in patients with Parkinson’s disease, a disease characterized by akinesia, bradykinesia and hypokinesia, functional imaging studies have shown hypoactivity of the mesial premotor and prefrontal areas 83.
Summary
What emerges is a greater understanding that a single well-defined genetic expansion results in a complex cascade of biological events that ultimately leads to selective degeneration of the brain. Proximal events mediated by mutant huntingtin in turn trigger cascades of both damaging and compensatory molecular processes and genetic programs in which oxidative injury, and energy depletion appear to play major roles. Much remains to be elucidated. Refocusing attention on the human disease, the natural progression of the disease and how associated brain changes result in clinical symptoms may provide a critical window into basic mechanisms that are involved in Huntington’s disease.
Acknowledgments
We are very grateful to our patients and families, who so generously contributed their time and energy to this work, and without whom it would not have been possible. Thanks to Drs. Anne Young and Nancy Wexler for their continued support and encouragement. Funding for this work was provided by the National Institutes of Health, National Institute for Neurological Disorders and Stroke (NS042861, NS045242, P01NS058793), National Center for Complementary and Alternative Medicine (AT000613), the National Institute on Aging (NIAAG024898) and the High Q Foundation.
Bibliography
- 1.Kremer B, et al. A worldwide study of the Huntington’s disease mutation. The sensitivity and specificity of measuring CAG repeats. N Engl J Med. 1994;330:1401–6. doi: 10.1056/NEJM199405193302001. [DOI] [PubMed] [Google Scholar]
- 2.Myers RH, MacDonald ME, Gusella JF. Discrepancy resolved. Nat Genet. 1993;5:215. doi: 10.1038/ng1193-215b. [DOI] [PubMed] [Google Scholar]
- 3.Anderson KE, et al. Cognitive correlates of obsessive and compulsive symptoms in Huntington’s disease. Am J Psychiatry. 2001;158:799–801. doi: 10.1176/appi.ajp.158.5.799. [DOI] [PubMed] [Google Scholar]
- 4.Rosenblatt A, Leroi I. Neuropsychiatry of Huntington’s disease and other basal ganglia disorders. Psychosomatics. 2000;41:24–30. doi: 10.1016/S0033-3182(00)71170-4. [DOI] [PubMed] [Google Scholar]
- 5.Hersch SM. Huntington’s disease: prospects for neuroprotective therapy 10 years after the discovery of the causative genetic mutation. Curr Opin Neurol. 2003;16:501–6. doi: 10.1097/01.wco.0000084229.82329.03. [DOI] [PubMed] [Google Scholar]
- 6.Group HsDCR. A novel gene containing a trinucleotide repeat that is expanded and unstable on Huntington’s disease chromosomes. The Huntington’s Disease Collaborative Research Group. Cell. 1993;72:971–83. doi: 10.1016/0092-8674(93)90585-e. [DOI] [PubMed] [Google Scholar]
- 7.Quarrell OW, et al. Reduced penetrance alleles for Huntington’s disease: a multi-centre direct observational study. J Med Genet. 2007;44:e68. doi: 10.1136/jmg.2006.045120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wheeler VC, et al. Factors associated with HD CAG repeat instability in Huntington disease. J Med Genet. 2007;44:695–701. doi: 10.1136/jmg.2007.050930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Myers RH. Huntington’s disease genetics. NeuroRx. 2004;1:255–62. doi: 10.1602/neurorx.1.2.255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Andresen JM, et al. The relationship between CAG repeat length and age of onset differs for Huntington’s disease patients with juvenile onset or adult onset. Ann Hum Genet. 2007;71:295–301. doi: 10.1111/j.1469-1809.2006.00335.x. [DOI] [PubMed] [Google Scholar]
- 11.Andrew SE, et al. The relationship between trinucleotide (CAG) repeat length and clinical features of Huntington’s disease. Nat Genet. 1993;4:398–403. doi: 10.1038/ng0893-398. [DOI] [PubMed] [Google Scholar]
- 12.Gutekunst CA, et al. Nuclear and neuropil aggregates in Huntington’s disease: relationship to neuropathology. J Neurosci. 1999;19:2522–34. doi: 10.1523/JNEUROSCI.19-07-02522.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Vonsattel JP, et al. Neuropathological classification of Huntington’s disease. J Neuropathol Exp Neurol. 1985;44:559–77. doi: 10.1097/00005072-198511000-00003. [DOI] [PubMed] [Google Scholar]
- 14.Ferrante RJ, Kowall NW, Richardson EP., Jr Proliferative and degenerative changes in striatal spiny neurons in Huntington’s disease: a combined study using the section-Golgi method and calbindin D28k immunocytochemistry. J Neurosci. 1991;11:3877–87. doi: 10.1523/JNEUROSCI.11-12-03877.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sotrel A, et al. Evidence for neuronal degeneration and dendritic plasticity in cortical pyramidal neurons of Huntington’s disease: a quantitative Golgi study. Neurology. 1993;43:2088–96. doi: 10.1212/wnl.43.10.2088. [DOI] [PubMed] [Google Scholar]
- 16.Milnerwood AJ, Raymond LA. Corticostriatal synaptic function in mouse models of Huntington’s disease: early effects of huntingtin repeat length and protein load. J Physiol. 2007;585:817–31. doi: 10.1113/jphysiol.2007.142448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Murphy KP, et al. Abnormal synaptic plasticity and impaired spatial cognition in mice transgenic for exon 1 of the human Huntington’s disease mutation. J Neurosci. 2000;20:5115–23. doi: 10.1523/JNEUROSCI.20-13-05115.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laforet GA, et al. Changes in cortical and striatal neurons predict behavioral and electrophysiological abnormalities in a transgenic murine model of Huntington’s disease. J Neurosci. 2001;21:9112–23. doi: 10.1523/JNEUROSCI.21-23-09112.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cepeda C, et al. The corticostriatal pathway in Huntington’s disease. Prog Neurobiol. 2007;81:253–71. doi: 10.1016/j.pneurobio.2006.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.DiFiglia M, et al. Huntingtin is a cytoplasmic protein associated with vesicles in human and rat brain neurons. Neuron. 1995;14:1075–81. doi: 10.1016/0896-6273(95)90346-1. [DOI] [PubMed] [Google Scholar]
- 21.Persichetti F, et al. Huntington’s disease CAG trinucleotide repeats in pathologically confirmed post-mortem brains. Neurobiol Dis. 1994;1:159–66. doi: 10.1006/nbdi.1994.0019. [DOI] [PubMed] [Google Scholar]
- 22.Zuccato C, et al. Loss of huntingtin-mediated BDNF gene transcription in Huntington’s disease. Science. 2001;293:493–8. doi: 10.1126/science.1059581. [DOI] [PubMed] [Google Scholar]
- 23.Cattaneo E, et al. Loss of normal huntingtin function: new developments in Huntington’s disease research. Trends Neurosci. 2001;24:182–8. doi: 10.1016/s0166-2236(00)01721-5. [DOI] [PubMed] [Google Scholar]
- 24.Goldberg YP, et al. Cleavage of huntingtin by apopain, a proapoptotic cysteine protease, is modulated by the polyglutamine tract. Nat Genet. 1996;13:442–9. doi: 10.1038/ng0896-442. [DOI] [PubMed] [Google Scholar]
- 25.Wellington CL, et al. Caspase cleavage of gene products associated with triplet expansion disorders generates truncated fragments containing the polyglutamine tract. J Biol Chem. 1998;273:9158–67. doi: 10.1074/jbc.273.15.9158. [DOI] [PubMed] [Google Scholar]
- 26.Wellington CL, Hayden MR. Of molecular interactions, mice and mechanisms: new insights into Huntington’s disease. Curr Opin Neurol. 1997;10:291–8. doi: 10.1097/00019052-199708000-00003. [DOI] [PubMed] [Google Scholar]
- 27.Davies SW, et al. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–48. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- 28.Paulson HL. Protein fate in neurodegenerative proteinopathies: polyglutamine diseases join the (mis)fold. Am J Hum Genet. 1999;64:339–45. doi: 10.1086/302269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jana NR, et al. Polyglutamine length-dependent interaction of Hsp40 and Hsp70 family chaperones with truncated N-terminal huntingtin: their role in suppression of aggregation and cellular toxicity. Hum Mol Genet. 2000;9:2009–18. doi: 10.1093/hmg/9.13.2009. [DOI] [PubMed] [Google Scholar]
- 30.Chai Y, et al. Evidence for proteasome involvement in polyglutamine disease: localization to nuclear inclusions in SCA3/MJD and suppression of polyglutamine aggregation in vitro. Hum Mol Genet. 1999;8:673–82. doi: 10.1093/hmg/8.4.673. [DOI] [PubMed] [Google Scholar]
- 31.Kazantsev A, et al. Insoluble detergent-resistant aggregates form between pathological and nonpathological lengths of polyglutamine in mammalian cells. Proc Natl Acad Sci U S A. 1999;96:11404–9. doi: 10.1073/pnas.96.20.11404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Steffan JS, et al. The Huntington’s disease protein interacts with p53 and CREB-binding protein and represses transcription. Proc Natl Acad Sci U S A. 2000;97:6763–8. doi: 10.1073/pnas.100110097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Steffan JS, et al. Histone deacetylase inhibitors arrest polyglutamine-dependent neurodegeneration in Drosophila. Nature. 2001;413:739–43. doi: 10.1038/35099568. [DOI] [PubMed] [Google Scholar]
- 34.Holbert S, et al. The Gln-Ala repeat transcriptional activator CA150 interacts with huntingtin: neuropathologic and genetic evidence for a role in Huntington’s disease pathogenesis. Proc Natl Acad Sci U S A. 2001;98:1811–6. doi: 10.1073/pnas.041566798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nucifora FC, Jr, et al. Interference by huntingtin and atrophin-1 with cbp-mediated transcription leading to cellular toxicity. Science. 2001;291:2423–8. doi: 10.1126/science.1056784. [DOI] [PubMed] [Google Scholar]
- 36.Gomez-Tortosa E, et al. Quantitative neuropathological changes in presymptomatic Huntington’s disease. Ann Neurol. 2001;49:29–34. [PubMed] [Google Scholar]
- 37.Perutz MF, Windle AH. Cause of neural death in neurodegenerative diseases attributable to expansion of glutamine repeats. Nature. 2001;412:143–4. doi: 10.1038/35084141. [DOI] [PubMed] [Google Scholar]
- 38.Hodges A, et al. Regional and cellular gene expression changes in human Huntington’s disease brain. Hum Mol Genet. 2006;15:965–77. doi: 10.1093/hmg/ddl013. [DOI] [PubMed] [Google Scholar]
- 39.Brochier C, et al. Quantitative gene expression profiling of mouse brain regions reveals differential transcripts conserved in man and affected in disease models. Physiol Genomics. 2008 doi: 10.1152/physiolgenomics.00125.2007. [DOI] [PubMed] [Google Scholar]
- 40.Gu M, et al. Mitochondrial defect in Huntington’s disease caudate nucleus. Ann Neurol. 1996;39:385–9. doi: 10.1002/ana.410390317. [DOI] [PubMed] [Google Scholar]
- 41.Bowling AC, Beal MF. Bioenergetic and oxidative stress in neurodegenerative diseases. Life Sci. 1995;56:1151–71. doi: 10.1016/0024-3205(95)00055-b. [DOI] [PubMed] [Google Scholar]
- 42.Browne SE, Beal MF. Oxidative damage in Huntington’s disease pathogenesis. Antioxid Redox Signal. 2006;8:2061–73. doi: 10.1089/ars.2006.8.2061. [DOI] [PubMed] [Google Scholar]
- 43.Tabrizi SJ, et al. Biochemical abnormalities and excitotoxicity in Huntington’s disease brain. Ann Neurol. 1999;45:25–32. doi: 10.1002/1531-8249(199901)45:1<25::aid-art6>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 44.Parker WD, Jr, et al. Evidence for a defect in NADH: ubiquinone oxidoreductase (complex I) in Huntington’s disease. Neurology. 1990;40:1231–4. doi: 10.1212/wnl.40.8.1231. [DOI] [PubMed] [Google Scholar]
- 45.Weydt P, et al. Thermoregulatory and metabolic defects in Huntington’s disease transgenic mice implicate PGC-1alpha in Huntington’s disease neurodegeneration. Cell Metab. 2006;4:349–62. doi: 10.1016/j.cmet.2006.10.004. [DOI] [PubMed] [Google Scholar]
- 46.Koroshetz WJ, et al. Energy metabolism defects in Huntington’s disease and effects of coenzyme Q10. Ann Neurol. 1997;41:160–5. doi: 10.1002/ana.410410206. [DOI] [PubMed] [Google Scholar]
- 47.Jenkins BG, et al. Effects of CAG repeat length, HTT protein length and protein context on cerebral metabolism measured using magnetic resonance spectroscopy in transgenic mouse models of Huntington’s disease. J Neurochem. 2005;95:553–62. doi: 10.1111/j.1471-4159.2005.03411.x. [DOI] [PubMed] [Google Scholar]
- 48.Fernandes HB, et al. Mitochondrial sensitivity and altered calcium handling underlie enhanced NMDA-induced apoptosis in YAC128 model of Huntington’s disease. J Neurosci. 2007;27:13614–23. doi: 10.1523/JNEUROSCI.3455-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lim D, et al. Calcium homeostasis and mitochondrial dysfunction in striatal neurons of huntington disease. J Biol Chem. 2008;283:5780–9. doi: 10.1074/jbc.M704704200. [DOI] [PubMed] [Google Scholar]
- 50.Oliveira JM, et al. Mitochondrial dysfunction in Huntington’s disease: the bioenergetics of isolated and in situ mitochondria from transgenic mice. J Neurochem. 2007;101:241–9. doi: 10.1111/j.1471-4159.2006.04361.x. [DOI] [PubMed] [Google Scholar]
- 51.Grunewald T, Beal MF. Bioenergetics in Huntington’s disease. Ann N Y Acad Sci. 1999;893:203–13. doi: 10.1111/j.1749-6632.1999.tb07827.x. [DOI] [PubMed] [Google Scholar]
- 52.Brouillet E, et al. Chronic mitochondrial energy impairment produces selective striatal degeneration and abnormal choreiform movements in primates. Proc Natl Acad Sci U S A. 1995;92:7105–9. doi: 10.1073/pnas.92.15.7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beal MF, et al. Kynurenic acid concentrations are reduced in Huntington’s disease cerebral cortex. J Neurol Sci. 1992;108:80–7. doi: 10.1016/0022-510x(92)90191-m. [DOI] [PubMed] [Google Scholar]
- 54.Beal MF. Does impairment of energy metabolism result in excitotoxic neuronal death in neurodegenerative illnesses? Ann Neurol. 1992;31:119–30. doi: 10.1002/ana.410310202. [DOI] [PubMed] [Google Scholar]
- 55.Chen CM, et al. Increased oxidative damage and mitochondrial abnormalities in the peripheral blood of Huntington’s disease patients. Biochem Biophys Res Commun. 2007;359:335–40. doi: 10.1016/j.bbrc.2007.05.093. [DOI] [PubMed] [Google Scholar]
- 56.Hersch SM, et al. Creatine in Huntington disease is safe, tolerable, bioavailable in brain and reduces serum 8OH2′dG. Neurology. 2006;66:250–2. doi: 10.1212/01.wnl.0000194318.74946.b6. [DOI] [PubMed] [Google Scholar]
- 57.Polidori MC, et al. Oxidative damage to mitochondrial DNA in Huntington’s disease parietal cortex. Neurosci Lett. 1999;272:53–6. doi: 10.1016/s0304-3940(99)00578-9. [DOI] [PubMed] [Google Scholar]
- 58.Rong J, et al. Regulation of intracellular trafficking of huntingtin-associated protein-1 is critical for TrkA protein levels and neurite outgrowth. J Neurosci. 2006;26:6019–30. doi: 10.1523/JNEUROSCI.1251-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Orr AL, et al. N-terminal mutant huntingtin associates with mitochondria and impairs mitochondrial trafficking. J Neurosci. 2008;28:2783–92. doi: 10.1523/JNEUROSCI.0106-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Trushina E, et al. Mutant huntingtin impairs axonal trafficking in mammalian neurons in vivo and in vitro. Mol Cell Biol. 2004;24:8195–209. doi: 10.1128/MCB.24.18.8195-8209.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.McGill JK, Beal MF. PGC-1alpha, a new therapeutic target in Huntington’s disease? Cell. 2006;127:465–8. doi: 10.1016/j.cell.2006.10.023. [DOI] [PubMed] [Google Scholar]
- 62.Fischl B, et al. Whole brain segmentation: automated labeling of neuroanatomical structures in the human brain. Neuron. 2002;33:341–55. doi: 10.1016/s0896-6273(02)00569-x. [DOI] [PubMed] [Google Scholar]
- 63.Fischl B, Liu A, Dale AM. Automated manifold surgery: constructing geometrically accurate and topologically correct models of the human cerebral cortex. IEEE Trans Med Imaging. 2001;20:70–80. doi: 10.1109/42.906426. [DOI] [PubMed] [Google Scholar]
- 64.Aylward EH, et al. Reduced basal ganglia volume associated with the gene for Huntington’s disease in asymptomatic at-risk persons. Neurology. 1994;44:823–8. doi: 10.1212/wnl.44.5.823. [DOI] [PubMed] [Google Scholar]
- 65.Aylward EH, et al. Onset and rate of striatal atrophy in preclinical Huntington disease. Neurology. 2004;63:66–72. doi: 10.1212/01.wnl.0000132965.14653.d1. [DOI] [PubMed] [Google Scholar]
- 66.Georgiou-Karistianis N, et al. Increased cortical recruitment in Huntington’s disease using a Simon task. Neuropsychologia. 2007;45:1791–800. doi: 10.1016/j.neuropsychologia.2006.12.023. [DOI] [PubMed] [Google Scholar]
- 67.Saft C, et al. fMRI reveals altered auditory processing in manifest and premanifest Huntington’s disease. Neuropsychologia. 2008;46:1279–89. doi: 10.1016/j.neuropsychologia.2007.12.002. [DOI] [PubMed] [Google Scholar]
- 68.Wolf RC, et al. Dorsolateral prefrontal cortex dysfunction in presymptomatic Huntington’s disease: evidence from event-related fMRI. Brain. 2007;130:2845–57. doi: 10.1093/brain/awm210. [DOI] [PubMed] [Google Scholar]
- 69.Buckner RL, Vincent JL. Unrest at rest: default activity and spontaneous network correlations. Neuroimage. 2007;37:1091–6. doi: 10.1016/j.neuroimage.2007.01.010. discussion 1097–9. [DOI] [PubMed] [Google Scholar]
- 70.Rosas HD, et al. Regional cortical thinning in preclinical Huntington disease and its relationship to cognition. Neurology. 2005;65:745–7. doi: 10.1212/01.wnl.0000174432.87383.87. [DOI] [PubMed] [Google Scholar]
- 71.Langbehn DR, et al. A new model for prediction of the age of onset and penetrance for Huntington’s disease based on CAG length. Clin Genet. 2004;65:267–77. doi: 10.1111/j.1399-0004.2004.00241.x. [DOI] [PubMed] [Google Scholar]
- 72.Rosas HD, et al. Cerebral cortex and the clinical expression of Huntington’s disease: complexity and heterogeneity. Brain. 2008;131:1057–68. doi: 10.1093/brain/awn025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Sapp E, et al. Huntingtin localization in brains of normal and Huntington’s disease patients. Ann Neurol. 1997;42:604–12. doi: 10.1002/ana.410420411. [DOI] [PubMed] [Google Scholar]
- 74.Rosas HD, et al. Diffusion tensor imaging in presymptomatic and early Huntington’s disease: Selective white matter pathology and its relationship to clinical measures. Mov Disord. 2006;21:1317–25. doi: 10.1002/mds.20979. [DOI] [PubMed] [Google Scholar]
- 75.Fennema-Notestine C, et al. In vivo evidence of cerebellar atrophy and cerebral white matter loss in Huntington disease. Neurology. 2004;63:989–95. doi: 10.1212/01.wnl.0000138434.68093.67. [DOI] [PubMed] [Google Scholar]
- 76.de la Monte SM, Vonsattel JP, Richardson EP., Jr Morphometric demonstration of atrophic changes in the cerebral cortex, white matter, and neostriatum in Huntington’s disease. J Neuropathol Exp Neurol. 1988;47:516–25. doi: 10.1097/00005072-198809000-00003. [DOI] [PubMed] [Google Scholar]
- 77.Group HS. Unified Huntington’s Disease Rating Scale: reliability and consistency. Huntington Study Group. Mov Disord. 1996;11:136–42. doi: 10.1002/mds.870110204. [DOI] [PubMed] [Google Scholar]
- 78.Lawrence AD, et al. Visual object and visuospatial cognition in Huntington’s disease: implications for information processing in corticostriatal circuits. Brain. 2000;123(Pt 7):1349–64. doi: 10.1093/brain/123.7.1349. [DOI] [PubMed] [Google Scholar]
- 79.Kipps CM, et al. Progression of structural neuropathology in preclinical Huntington’s disease: a tensor based morphometry study. J Neurol Neurosurg Psychiatry. 2005;76:650–5. doi: 10.1136/jnnp.2004.047993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Kassubek J, et al. Global cerebral atrophy in early stages of Huntington’s disease: quantitative MRI study. Neuroreport. 2004;15:363–5. doi: 10.1097/00001756-200402090-00030. [DOI] [PubMed] [Google Scholar]
- 81.Peinemann A, et al. Executive dysfunction in early stages of Huntington’s disease is associated with striatal and insular atrophy: a neuropsychological and voxel-based morphometric study. J Neurol Sci. 2005;239:11–9. doi: 10.1016/j.jns.2005.07.007. [DOI] [PubMed] [Google Scholar]
- 82.Kipps CM, et al. Disgust and happiness recognition correlate with anteroventral insula and amygdala volume respectively in preclinical Huntington’s disease. J Cogn Neurosci. 2007;19:1206–17. doi: 10.1162/jocn.2007.19.7.1206. [DOI] [PubMed] [Google Scholar]
- 83.Berardelli A, et al. Pathophysiology of bradykinesia in Parkinson’s disease. Brain. 2001;124:2131–46. doi: 10.1093/brain/124.11.2131. [DOI] [PubMed] [Google Scholar]