Abstract
Genetic variants underlying the strong heritable component of prostate cancer remain largely unknown. Genome-wide association studies of prostate cancer have yielded several variants that have significantly replicated across studies, predominantly in cases unselected for family history of prostate cancer. Additional candidate gene variants have also been proposed, many evaluated within familial prostate cancer study populations. Such variants hold great potential value for risk stratification, particularly for early onset or aggressive prostate cancer, given co-morbidities associated with current therapies. Here, we investigate a Caucasian study population of 523 independent familial prostate cancer cases and 523 age-matched controls without a personal or family history of prostate cancer. We replicate identified associations at genome-wide association study loci 8q24, 11q13, and 2p15 (P = 2.9 × 10−4 to P = 4.7 × 10−5), demonstrating study population power. We also find evidence to support reported associations at candidate genes RNASEL, EZH2, and NKX3-1 (P = 0.031 to P = 0.0085). We further explore a set of candidate genes related to RNASEL and to its role in retroviral restriction, identifying nominal associations at XPR1 and RBM9. Effects at 8q24 appear more pronounced for those diagnosed at an early age, while at 2p15 and RNASEL effects were more pronounced at a later age. However, these trends did not reach statistical significance. Effects at 2p15 were statistically significantly more pronounced for those diagnosed with aggressive disease.
INTRODUCTION
Among common cancers, prostate cancer has the greatest heritable risk, estimated to be roughly 50% (1, 2). Family history remains the best predictor of risk for prostate cancer. Those with a positive family history are likely to have inherited a greater genetic load for the disease than those with a negative family history. The underlying structure of this heritable risk remains largely unknown. Highly penetrant Mendelian mutations have been widely sought through linkage analysis (3). RNASEL, ELAC2, MSR1, PODXL were identified as candidate prostate cancer genes by this approach, and each has been proposed to harbor common low-penetrance variants contributing to population risk of prostate cancer. Given the established role of RNASEL and MSR1 in innate immunity and viral susceptibility, investigators have also proposed additional candidate genes in these pathways as modifiers of prostate cancer risk. These have included genes encoding the Toll-like receptors and the IL-1 receptor antagonist. Additional candidates have been proposed based upon genes identified through studies of animal models of prostate cancer (NKX3-1), of somatic changes in gene expression in prostate cancer (PPARG, EZH2), of overlapping hereditary cancer syndromes (BRCA2, CDH1), and of salient biochemical pathways (AR, SRD5A2, CDKN1B, TGFB1, CYP17A1, CYP1A1) (4). Among all of these candidates, many of the observed associations have replicated inconsistently, suggesting extensive genetic heterogeneity in prostate cancer predisposition, population-specific findings, or type I errors. Recent genome-wide association studies (GWAS) of prostate cancer have identified additional SNPs that are significantly associated with prostate cancer and, encouragingly, that have replicated broadly across global study populations (5–7).
In this study, we investigated heritable prostate cancer risk in a study population of familial prostate cancer cases, and controls without a personal or family history of prostate cancer. This design was intended to compare two extremes of the distribution of genetic load for prostate cancer. We first sought to replicate the prostate cancer association of several SNPs that have globally replicated within the prostate cancer GWAS literature to assess the relative power of the study population. Second, we conducted a replication study of a series of published genes of less certain significance, including: RNASEL, ELAC2, PODXL, TLR10, TLR1, TLR6, TLR4, IL1RN, NKX3-1, PPARG, EZH2, CDH1, CDKN1B, TGFB1, CYP17A1, and CYP1A1. Third, we further explored potential evidence for the association of prostate cancer with a series of additional genes related to retroviral infection. The impetus for this was the prominence of innate immunity genes among published candidates, as well as the recent intriguing discovery of a novel retrovirus of the xenotropic murine leukemia virus family in prostate adenocarcinomas with the risk RNASEL genotype (8, 9).
A relatively large number of retroviral restriction genes are known, but few reside near published potential hereditary prostate cancer linkage regions. RNASEL was included among the genes of the replication set noted above, and we further explored two potential RNASEL regulatory genes near potential linkage areas: the RNASEL inhibitor RNS4I at 4q31 (10), and RBM9 at 22q12 (11). RBM9 encodes a protein predicted to regulate transcript stability by binding 3’ UTR AU-rich elements, a mechanism that may regulate RNASEL and that is also tied to viral replication (12, 13). RBM9 is located at the peak of the 22q12 hereditary prostate cancer locus of the International Consortium for Prostate Cancer Genetics (11, 14). XPR1 is a cell surface receptor with a direct role in resistance of specific mouse strains to murine leukemia viruses (15). The human ortholog of XPR1 is found 1.6 Mb centromeric to RNASEL within the 1q25 HPC1 prostate cancer locus. Additional retroviral restriction genes near potential linkage regions that were explored include APOBEC3 genes (16) at 22q13 (11), APOBEC4 (17) at 1q25 (18), AICDA (19) at 12p13 (10, 20, 21), PIN1 (22, 23) at 19p13 (20, 24–27), and peptidyl-prolyl isomerases (28) PPID at 4q32 (18) and PPIH at 1p34 (26, 29–31).
MATERIALS AND METHODS
Study population
Study subjects were Americans of Northern European descent, ascertained with informed consent between 2002 and 2008 from Vanderbilt University Medical Center and from the VA Tennessee Valley Healthcare System (adjacent hospitals) with institutional review board oversight. Familial prostate cancer cases were ascertained at the time of treatment for the principal diagnosis of prostate cancer, and controls were ascertained at the time of routine preventative screening for prostate cancer. All prostate cancer probands included in the study were from pedigrees with a family history of prostate cancer (≥ 2 affected), and all control probands were from pedigrees without a family history of prostate cancer. Family history included 1st and 2nd degree relatives. Controls had a screening prostate specific antigen (PSA) test < 4 ng/ml at the time of ascertainment, had no personal history of prostate cancer, no record of a PSA test ≥ 4 ng/ml, and no record of abnormal digital rectal examination. Controls were individually matched to cases on age in a 1:1 ratio (± 2.5 years; age at screen for controls, age at diagnosis for cases). The study included 523 unrelated, independent familial prostate cancer probands and 523 matched control probands. Table 1 provides characteristics of the study population. Stratification analyses of Gleason score preferentially employed final prostatectomy specimen Gleason score (available for 87% of cases) rather than initial diagnostic biopsy Gleason score.
Table 1.
Study population characteristics
| Controls | Cases | ||
|---|---|---|---|
| No. | 523 | 523 | |
| Mean Age*, y | 61.3 | 61.2 | |
| Mean No. Brothers | 1.8 | 1.7 | |
| Median PSA* | 0.9 | 5.6 | |
| Median Gleason Sum | - | 6 | |
| Gleason Sum ≤ 6, No. | - | 275 | |
| Gleason Sum ≥ 7, No. | - | 231 | |
| Affected in Pedigree, No.** | 0 | 523 | - |
| 2 | - | 316 | |
| ≥3 | - | 207 | |
At diagnosis for cases, at screen for controls.
Proband plus 1st and 2nd degree affected relatives
SNP genotyping
DNA was extracted from whole blood on an Autopure LS robot using the Puregene DNA Purification System Standard Protocol (Qiagen, Valencia, CA). DNA was quantified using the PicoGreen dsDNA Quantitation Kit (Invitrogen, Carlsbad, CA), imaged with a Molecular Devices/LJL Analyst HT (Molecular Devices, Union City, CA). SNP genotyping was conducted using the Illumina GoldenGate platform (Illumina, San Diego, CA). SNPs rs10896450, rs6983267, and rs1800470 were exceptions, and were genotyped using the TaqMan platform (Applied Biosystems, Foster City, CA). The rs10896450 assay was kindly provided by Dr. J. Gudmundsson. We obtained 99.7% of the genotypes of SNPs that successfully converted for assay (Figure 1, Table 2 and Table 3, and Supplemental Table 1).
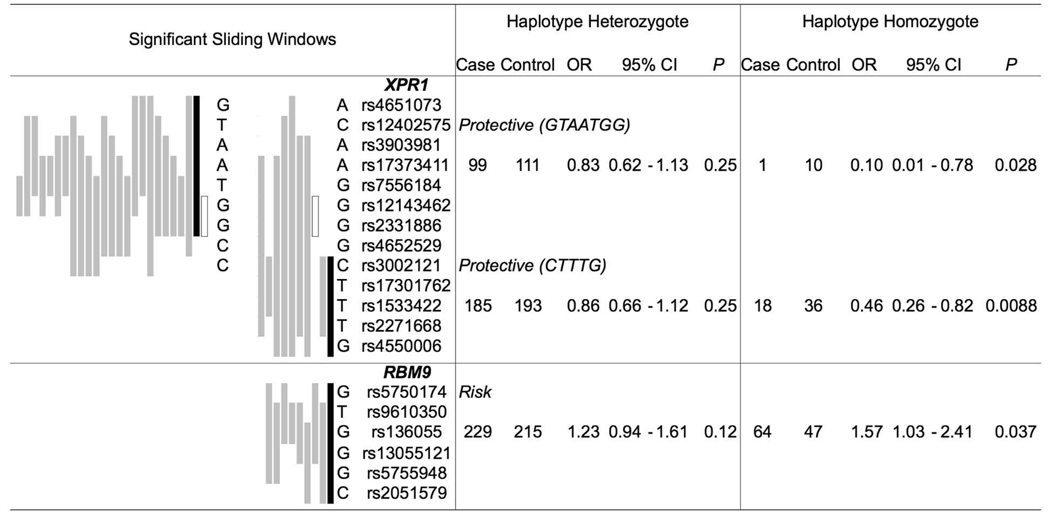
Figure 1.
Haplotype association with prostate cancer. To left: Sliding haplotype windows of P ≤ 0.05 by χ2 analysis, graphically ordered as most to least significant (from black to grey). To right: Logistic regression analysis for haplotypes designated in black (number of subjects 1,046). Among all genes of the study, only RNASEL, XPR1, and RBM9, yielded haplotype windows of P ≤ 0.05. The open boxed haplotype window of XPR1 is common to both protective haplotypes.
Table 2.
Replication evidence for association with prostate cancer among published loci.
| Minor Allele Heterozygote | Minor Allele Homozygote | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Chr | Gene | Variant | Case | Control | OR | 95% CI | P | Case | Control | OR | 95% CI | P | ||
| 2p15 | (GWAS) | rs721048 | T | 176 | 157 | 1.32 | 1.00 – 1.73 | 0.047 | 41 | 11 | 4.06 | 2.07 – 7.98 | 4.7 × 10−5 | |
| 8q24 cen. | (GWAS) | rs6983267 | T | 225 | 252 | 0.65 | 0.49 – 0.87 | 0.0040 | 100 | 138 | 0.49 | 0.34 – 0.70 | 7.5 × 10−5 | |
| 8q24 cen. | POU5F1P1 | rs6998061 | G527E | A | 218 | 236 | 0.75 | 0.58 – 0.98 | 0.032 | 79 | 108 | 0.56 | 0.39 – 0.80 | 0.0015 |
| 8q24 tel. | (GWAS) | rs1447295 | A | 133 | 85 | 1.78 | 1.30 – 2.44 | 2.9 × 10−4 | 7 | 3 | 2.36 | 0.60 – 9.21 | 0.22 | |
| 11q13 | (GWAS) | rs10896450 | A | 247 | 256 | 0.72 | 0.54 – 0.96 | 0.024 | 89 | 128 | 0.51 | 0.35 – 0.73 | 2.2 × 10−4 | |
| 1q25 | RNASEL | rs11807829 | C | 240 | 227 | 1.06 | 0.81 – 1.37 | 0.67 | 49 | 60 | 0.83 | 0.54 – 1.27 | 0.39 | |
| rs533259 | T | 68 | 71 | 0.94 | 0.64 – 1.37 | 0.74 | 3 | 2 | 1.48 | 0.25 – 8.90 | 0.67 | |||
| rs627928 | E541D | T | 262 | 253 | 0.91 | 0.69 – 1.21 | 0.52 | 81 | 110 | 0.64 | 0.44 – 0.92 | 0.018 | ||
| rs486907 | R462Q | A | 259 | 228 | 1.34 | 1.03 – 1.76 | 0.031 | 76 | 73 | 1.22 | 0.84 – 1.78 | 0.29 | ||
| 2q13 | IL1RN | rs2637988 | G | 235 | 248 | 0.89 | 0.68 – 1.16 | 0.37 | 79 | 80 | 0.92 | 0.63 – 1.34 | 0.67 | |
| rs3213448 | A | 94 | 94 | 1.01 | 0.74 – 1.38 | 0.94 | 5 | 11 | 0.47 | 0.16 – 1.35 | 0.16 | |||
| rs3087263 | A | 96 | 93 | 1.03 | 0.75 – 1.41 | 0.87 | 7 | 10 | 0.70 | 0.26 – 1.84 | 0.47 | |||
| rs380092 | T | 208 | 218 | 0.95 | 0.73 – 1.22 | 0.67 | 53 | 48 | 1.10 | 0.72 – 1.67 | 0.67 | |||
| rs315952 | C | 194 | 204 | 0.93 | 0.72 – 1.20 | 0.58 | 44 | 43 | 1.00 | 0.64 – 1.57 | 0.98 | |||
| 3p25 | PPARG* | rs1805192 | P12A | C | 0 | 0 | 0 | 0 | ||||||
| rs1801282 | P12A** | G | 105 | 114 | 0.92 | 0.68 – 1.24 | 0.58 | 8 | 3 | 2.66 | 0.69 – 10.17 | 0.15 | ||
| 4p14 | TLR10* | rs4129009 | I775V | G | 145 | 167 | 0.83 | 0.63 – 1.08 | 0.17 | 22 | 18 | 1.15 | 0.60 – 2.21 | 0.67 |
| rs11466658 | R525W | T | 28 | 26 | 1.08 | 0.62 – 1.88 | 0.79 | 0 | 0 | |||||
| rs11096955 | I369L | C | 214 | 221 | 0.93 | 0.72 – 1.21 | 0.60 | 67 | 71 | 0.90 | 0.62 – 1.32 | 0.60 | ||
| rs11096957 | N241H | C | 214 | 221 | 0.93 | 0.72 – 1.20 | 0.58 | 67 | 71 | 0.88 | 0.60 – 1.30 | 0.53 | ||
| 4p14 | TLR1 | rs3923647 | H305L | T | 30 | 28 | 1.07 | 0.63 – 1.83 | 0.80 | 0 | 0 | |||
| rs4833095 | N248S | C | 181 | 198 | 0.88 | 0.68 – 1.13 | 0.30 | 33 | 31 | 1.00 | 0.59 – 1.67 | 0.98 | ||
| rs5743611 | R80T | C | 69 | 74 | 0.93 | 0.65 – 1.32 | 0.67 | 5 | 7 | 0.80 | 0.25 – 2.56 | 0.71 | ||
| 4p14 | TLR6 | rs5743815 | V427A | C | 21 | 12 | 1.75 | 0.86 – 3.57 | 0.13 | 0 | 0 | |||
| rs5743810 | P249S | T | 264 | 256 | 1.08 | 0.81 – 1.44 | 0.58 | 99 | 101 | 1.02 | 0.71 – 1.45 | 0.91 | ||
| rs1039559 | C | 265 | 253 | 1.14 | 0.86 – 1.53 | 0.36 | 125 | 125 | 1.10 | 0.78 – 1.54 | 0.60 | |||
| 7q32 | PODXL* | rs3735035 | G112S | T | 272 | 271 | 0.90 | 0.67 – 1.21 | 0.48 | 109 | 122 | 0.80 | 0.56 – 1.14 | 0.21 |
| rs3212298 | V358I | A | 45 | 60 | 0.71 | 0.47 – 1.07 | 0.10 | 2 | 2 | 0.94 | 0.13 – 6.84 | 0.95 | ||
| 7q36 | EZH2* | rs2302427 | D185H | C | 69 | 98 | 0.63 | 0.44 – 0.89 | 0.0085 | 4 | 5 | 0.67 | 0.18 – 2.56 | 0.56 |
| 8p21 | NKX3-1 | rs1567669 | T | 201 | 244 | 0.71 | 0.55 – 0.92 | 0.010 | 55 | 51 | 0.93 | 0.61 – 1.42 | 0.74 | |
| 9q33 | TLR4 | rs11536869 | G | 43 | 41 | 1.02 | 0.66 – 1.60 | 0.91 | 0 | 2 | ||||
| rs1927911 | T | 196 | 212 | 0.92 | 0.71 – 1.19 | 0.52 | 47 | 35 | 1.38 | 0.84 – 2.29 | 0.21 | |||
| rs11536879 | G | 45 | 32 | 1.47 | 0.91 – 2.38 | 0.12 | 0 | 1 | ||||||
| rs5030717 | G | 110 | 107 | 1.02 | 0.75 – 1.38 | 0.91 | 3 | 4 | 0.72 | 0.12 – 4.34 | 0.72 | |||
| rs5030728 | A | 225 | 237 | 0.90 | 0.70 – 1.15 | 0.41 | 44 | 47 | 0.86 | 0.55 – 1.36 | 0.53 | |||
| rs4986791 | T399I | T | 53 | 59 | 0.88 | 0.59 – 1.32 | 0.55 | 2 | 1 | 1.74 | 0.15 – 19.63 | 0.65 | ||
| 10q24 | CYP17A1 | rs10883783 | A | 226 | 222 | 1.09 | 0.85 – 1.40 | 0.52 | 55 | 42 | 1.36 | 0.89 – 2.09 | 0.16 | |
| rs3824755 | C | 79 | 95 | 0.81 | 0.58 – 1.12 | 0.19 | 2 | 4 | 0.53 | 0.10 – 2.92 | 0.47 | |||
| rs6163 | A | 249 | 240 | 1.10 | 0.85 – 1.43 | 0.46 | 85 | 83 | 1.08 | 0.75 – 1.56 | 0.67 | |||
| rs2486758 | C | 174 | 161 | 1.13 | 0.87 – 1.49 | 0.36 | 21 | 22 | 1.00 | 0.54 – 1.84 | 1.00 | |||
| 12p13 | CDKN1B | rs34329 | G | 225 | 227 | 0.99 | 0.76 – 1.28 | 0.92 | 48 | 46 | 1.04 | 0.67 – 1.61 | 0.85 | |
| rs3093736 | A | 30 | 34 | 0.89 | 0.54 – 1.46 | 0.65 | 0 | 0 | ||||||
| 16q22 | CDH1* | rs16260 | 5'(−160A) | A | 201 | 211 | 0.97 | 0.75 – 1.25 | 0.81 | 45 | 37 | 1.21 | 0.77 – 1.91 | 0.41 |
| 15q24 | CYP1A1 | rs4646421 | T | 96 | 83 | 1.21 | 0.86 – 1.70 | 0.27 | 3 | 6 | 0.55 | 0.14 – 2.22 | 0.40 | |
| 17p12 | ELAC2* | rs4792311 | S217L | A | 224 | 216 | 1.08 | 0.84 – 1.40 | 0.56 | 46 | 42 | 1.13 | 0.73 – 1.74 | 0.60 |
| 19q13 | TGFB1* | rs1800470 | L10P | C | 242 | 239 | 0.96 | 0.73 – 1.27 | 0.79 | 66 | 78 | 0.76 | 0.51 – 1.12 | 0.16 |
candidate, rather than tagging SNPs evaluated.
different residue variant from an alternative transcript.
Table 3.
Stratification of prostate cancer cases: by age of diagnosis, by Gleason score.
| Minor Allele Heterozygote | Minor Allele Homozygote | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Variant | Case | Control | OR | 95% CI | P | Case | Control | OR | 95% CI | P | ||
| Case Age ≤ 60 (508 Subjects) | ||||||||||||
| 8q24 cen. | rs6983267 | T | 102 | 108 | 0.70 | 0.46 – 1.08 | 0 .11 | 43 | 76 | 0.39 | 0.24 – 0.65 | 2.7 × 10−4 |
| 8q24 tel. | rs1447295 | A | 71 | 43 | 2.11 | 1.34 – 3.32 | 0.0013 | 3 | 0 | |||
| 11q13 | rs10896450 | A | 124 | 132 | 0.69 | 0.45 – 1.04 | 0.08 | 42 | 59 | 0.52 | 0.30 – 0.88 | 0.015 |
| 2p15 | rs721048 | T | 83 | 77 | 1.24 | 0.84 – 1.84 | 0.28 | 19 | 7 | 2.77 | 1.14 – 6.74 | 0.025 |
| RNASEL | rs627928 | T | 129 | 124 | 1.00 | 0.68 – 1.48 | 0.99 | 41 | 50 | 0.79 | 0.47 – 1.33 | 0.37 |
| RNASEL | rs486907 | A | 114 | 113 | 1.08 | 0.74 – 1.58 | 0.67 | 38 | 33 | 1.19 | 0.70 – 2.03 | 0.53 |
| EZH2 | rs2302427 | C | 32 | 39 | 0.76 | 0.44 – 1.29 | 0.31 | 2 | 3 | 0.63 | 0.10 – 3.96 | 0.63 |
| NKX3-1 | rs1567669 | T | 100 | 130 | 0.65 | 0.44 – 0.94 | 0.023 | 31 | 23 | 1.13 | 0.61 – 2.10 | 0.70 |
| Case Age ≥ 61 (538 Subjects) | ||||||||||||
| 8q24 cen. | rs6983267 | T | 123 | 144 | 0.63 | 0.43 – 0.94 | 0.023 | 57 | 62 | 0.62 | 0.37 – 1.05 | 0.08 |
| 8q24 tel. | rs1447295 | A | 62 | 42 | 1.62 | 1.04 – 2.53 | 0.033 | 4 | 3 | 1.43 | 0.32 – 6.46 | 0.64 |
| 11q13 | rs10896450 | A | 123 | 124 | 0.77 | 0.51 – 1.14 | 0.19 | 47 | 69 | 0.51 | 0.31 – 0.83 | 0.0074 |
| 2p15 | rs721048 | T | 93 | 80 | 1.39 | 0.95 – 2.03 | 0.09 | 22 | 4 | 6.21 | 2.11 – 18.29 | 9.3 × 10−4 |
| RNASEL | rs627928 | T | 133 | 129 | 0.84 | 0.56 – 1.24 | 0.37 | 40 | 60 | 0.52 | 0.30 – 0.88 | 0.016 |
| RNASEL | rs486907 | A | 145 | 115 | 1.72 | 1.16 – 2.53 | 0.007 | 38 | 40 | 1.24 | 0.73 – 2.09 | 0.43 |
| EZH2 | rs2302427 | C | 37 | 59 | 0.57 | 0.36 – 0.89 | 0.015 | 2 | 2 | 0.75 | 0.10 – 5.51 | 0.78 |
| NKX3-1 | rs1567669 | T | 101 | 114 | 0.78 | 0.54 – 1.12 | 0.19 | 24 | 28/ | 0.78 | 0.44 – 1.38 | 0.39 |
| Case Gleason ≤ 6 (550 Subjects) | ||||||||||||
| 8q24 cen. | rs6983267 | T | 123 | 127 | 0.72 | 0.49 –1.07 | 0.11 | 50 | 75 | 0.46 | 0.28 – 0.76 | 0.0021 |
| 8q24 tel. | rs1447295 | A | 66 | 47 | 1.57 | 1.02 – 2.42 | 0.043 | 3 | 1 | 2.66 | 0.26 – 26.65 | 0.41 |
| 11q13 | rs10896450 | A | 143 | 128 | 0.93 | 0.63 – 1.36 | 0.70 | 42 | 69 | 0.51 | 0.30 – 0.85 | 0.0093 |
| 2p15 | rs721048 | T | 89 | 82 | 1.22 | 0.85 – 1.75 | 0.29 | 19 | 9 | 2.23 | 0.99 – 5.01 | 0.053 |
| RNASEL | rs627928 | T | 140 | 140 | 0.86 | 0.59 – 1.25 | 0.43 | 37 | 51 | 0.60 | 0.35 – 1.03 | 0.06 |
| RNASEL | rs486907 | A | 139 | 127 | 1.39 | 0.95 – 2.03 | 0.09 | 44 | 35 | 1.59 | 0.94 – 2.67 | 0.08 |
| EZH2 | rs2302427 | C | 40 | 57 | 0.62 | 0.40 – 0.96 | 0.034 | 1 | 4 | 0.23 | 0.03 – 2.06 | 0.19 |
| NKX3-1 | rs1567669 | T | 112 | 133 | 0.72 | 0.50 – 1.02 | 0.06 | 29 | 29 | 0.86 | 0.48 – 1.57 | 0.63 |
| Case Gleason ≥ 7 (462 Subjects) | ||||||||||||
| 8q24 cen. | rs6983267 | T | 92 | 117 | 0.54 | 0.35 – 0.84 | 0.0063 | 45 | 57 | 0.48 | 0.28 – 0.83 | 0.0088 |
| 8q24 tel. | rs1447295 | A | 62 | 34 | 2.23 | 1.37 – 3.63 | 0.0013 | 4 | 2 | 2.24 | 0.40 – 12.50 | 0.36 |
| 11q13 | rs10896450 | A | 98 | 122 | 0.51 | 0.32 – 0.81 | 0.0039 | 43 | 53 | 0.49 | 0.28 – 0.84 | 0.0099 |
| 2p15 | rs721048 | T | 81 | 68 | 1.56 | 1.01 – 2.40 | 0.046 | 21 | 2 | 12.25 | 2.83 – 52.93 | 7.8 × 10−4 |
| RNASEL | rs627928 | T | 111 | 107 | 0.93 | 0.60 – 1.42 | 0.72 | 40 | 52 | 0.68 | 0.40 – 1.16 | 0.16 |
| RNASEL | rs486907 | A | 111 | 97 | 1.26 | 0.85 – 1.88 | 0.25 | 31 | 36 | 0.92 | 0.52 – 1.63 | 0.79 |
| EZH2 | rs2302427 | C | 27 | 38 | 0.67 | 0.38 – 1.19 | 0.17 | 3 | 1 | 2.23 | 0.22 – 22.73 | 0.50 |
| NKX3-1 | rs1567669 | T | 82 | 103 | 0.66 | 0.44 – 1.00 | 0.051 | 25 | 21 | 1.01 | 0.55 – 1.85 | 0.98 |
SNPs within four regions associated with sporadic prostate cancer in published GWAS studies were selected for assay (5–7, 32–34). These included the centromeric 8q24 LD block rs6983267 and rs6998061, telomeric 8q24 LD block rs1447295 (r2 of 1.0 with rs4242382 and rs4242384), 11q13 rs10896450 (r2 of 1.0 with rs10896449, and r2 of 0.98 with rs7931342), and 2p15 rs721048.
Tagging SNPs were selected using LDSelect (35) with an r2 threshold of 0.8 in CEU subject data of HapMap Release 21 of Phase II on the NCBI build 35 assembly, among SNPs amenable to Illumina assay. The minor allele frequency (MAF) threshold was 0.05, as exceeded by any Caucasian study population of dbSNP, including CEU. Tagging SNPs and candidate functional SNPS are designated within Table 2 and Supplementary Table 1. Additional SNPs that failed assay included: rs1927907 (TLR4), rs454078 (IL1RN), rs1985604 (PIN1), rs9998052 (RNS4I (ABCE1)). At PPARG two separate P12A missense SNPs are annotated, rs1805192 in the primary transcript (NM_005037, NM_138711, NM_015869, NM_138712), and rs1801282 in an alternative transcript (NM_015869).
Statistical analyses
We employed a conditional logistic regression model with two beta parameters to estimate odds ratios (OR) and 95% confidence intervals (CI) of heterozygous and homozygous states (Intercooled Stata 10, Stata Corporation, College Station, TX). The model was adjusted for the matching variable age, and for the number of study subject brothers as a potential confounder, since proband selection was based upon family history. We added dichotomous interaction terms to our conditional logistic regression models to assess interaction between genotype and age of diagnosis (≤ 60 or ≥ 61 years of age) on cancer risk. Wald χ2 tests for interaction were performed. A similar approach was used to assess interaction between genotype and prostate cancer aggressiveness (Gleason score ≤ 6 or ≥7).
Individual study subject diplotypes were estimated by PHASE v2.1(36), run for all 1046 subjects as a group. For a given window of consecutive SNPs at a gene, the diplotype may be assigned with evaluation of the specified window, or with evaluation of larger windows encompassing the same markers. The window diplotype of highest probability ≥ 0.9 among them was employed for sliding window tests of association (37, 38). The sliding window approach evaluated a haplotype window of N markers, sliding the window along a gene map in single marker increments. The width of the window was varied from two to the maximum number of SNPs evaluated at a gene. Each N-marker haplotype was compared to the remaining haplotypes of the window as a group among cases and controls. The resulting 2×2 contingency table of frequencies was evaluated by a χ2 test statistic. A nominally significant haplotype was subsequently modeled by conditional logistic regression as described above. All P values were calculated with respect to two-sided alternative hypotheses. Because each locus was investigated to replicate a previously published association, or alternatively to explore a potential association for future replication, P values were unadjusted for multiple comparisons.
RESULTS
Four loci that have been significantly associated with prostate cancer in prior genome wide association studies and that have independently replicated across global study populations were also significantly associated with familial prostate cancer in our study. Table 2 presents odds ratios and significance of each of these, at top. The 2p15 variant rs721048 (32) followed an additive model with a particularly strong effect for homozygous carriers, OR = 4.06 (P = 4.7 × 10−5). The 8q24 variant rs1447295 originally identified in the Icelandic study (5) had a relatively low risk allele frequency (7.1% case, 4.4% control) and greatest effect was observed among heterozygous carriers, OR = 1.78 (P = 2.9 × 10−4). That variant marks a linkage disequilibrium (LD) block that is detected by several additional SNPs that have been observed to be significantly associated with prostate cancer across studies. The adjacent centromeric LD block is independently associated with prostate cancer risk, detected by rs6983267 (6, 7). We evaluated rs6983267 and the adjacent missense variant rs6998061 (G527E) of the POU5F1P1 gene, observing moderate pairwise LD, r2 = 0.69. The association at this LD block appeared to follow an additive model and was best detected by rs6983267, with a homozygote OR = 0.49 (P = 7.5 × 10−5). The T allele frequency was 0.417 among cases and 0.508 among controls. SNP rs10896450 on 11q13 was originally detected through GWAS (personal communication, J. Gudmundsson, and (6, 7)), and also confirmed as significantly associated with prostate cancer in our study. It also followed an additive model with greatest effect among homozygotes, OR = 0.51 (P = 2.2 × 10−4). The minor allele had a frequency of 20.5% among cases and 24.7% among controls. To facilitate comparisons with other publications, results of an additive model for these variants are given in Supplementary Table 2. Each of these associations indicated good power to detect risk-modifying genetic variants among candidate loci within this familial prostate cancer study population.
Of several candidate genes proposed to be associated with prostate cancer in the published literature and evaluated here, three yield additional evidence to support an association with prostate cancer within our study. This data is presented in Table 2 beneath the GWAS replication set. We observed significant associations at RNASEL (39, 40), EZH2 (41), and NKX3-1 (42). Our evaluation of the remaining candidates was not significant in either single allele or haplotype-based analyses. At RNASEL, homozygotes for the minor allele of rs627928 (E541D) had a significantly reduced risk of prostate cancer, OR = 0.64 (P=0.018). That variant marks one of three common haplotypes at the gene on an LD block that includes tested SNPs rs533529, rs627928, and rs486907 (all pairwise D’ = 1.0). Another of the three haplotypes is marked by the minor allele of rs486907 (R462Q); heterozygotes for this minor allele had a significantly increased risk of prostate cancer, OR = 1.34 (P = 0.031). At EZH2, rs2302427 (D185H) had a MAF of 3.7% among cases and 5.2% among controls, conferring significantly reduced risk of prostate cancer among heterozygotes, OR = 0.63 (P = 0.0085). Tagging SNP rs1567669 at NKX3-1 also yielded evidence to support an association with prostate cancer among heterozygotes, OR = 0.71 (P = 0.010).
A third set of candidate loci was also evaluated in this study. These were genes related to RNASEL and retroviral restriction, and near genomic regions supported by prior linkage evidence. The impetus for this exploration was the recent discovery of the XMRV gammaretrovirus in prostate cancer tissue, apparently more commonly present among evaluated RNASEL 462Q/Q homozygotes (8, 9). Results at these genes are presented in Supplementary Table 1. Only three SNPs yielded nominal evidence of an association with prostate cancer, one each at PPIH, XPR1, and AICDA. Sliding window haplotype analysis often redundantly identifies an association, but no significant haplotype profile differences between cases and controls were observed at PPIH or AICDA. Haplotype-based sliding window analyses did, however, yield further evidence to support a protective association at XPR1 (Figure 1). XPR1 is encoded by 15 exons that reside within a region of strong LD among study subjects. Homozygotes of each of two haplotypes at XPR1 had a nominally significantly reduced risk of prostate cancer (OR = 0.10 (P = 0.028) and OR = 0.46 (P = 0.009)). These two protective haplotypes share alleles that distinguish them from other common XPR1 haplotypes: rs17373411 A, rs2331886 G, rs3002121 C, rs1533422 T, and rs2271668 T. Haplotype A-G-C-T-T was protective of prostate cancer among homozygotes, OR = 0.31 (P = 5.4 × 10−4). Haplotype analysis additionally identified a risk association at RBM9, which also resides within a region of strong LD. Among the seven common haplotypes across RBM9 in the study population, one was nominally associated with excess risk of prostate cancer among homozygotes, OR = 1.57 (P = 0.037). XPR1 and RBM9 may thus be reasonable candidates for further evaluation in independent study populations.
Finally, we investigated two clinical facets of prostate cancer of importance for prevention and prognosis: age of diagnosis, and Gleason score as an index of disease aggressiveness (Table 3). We first stratified the study population according to cancer diagnosis by age 60 or after age 61 (two roughly comparably sized groups), and evaluated variants of Table 2 that significantly modified prostate cancer risk. Each of the two independent 8q24 variants had apparently stronger association among those with a younger age of diagnosis (rs1447295 heterozygote OR = 2.11, P = 0.0013; rs6983267 homozygote OR = 0.39, P = 2.7 × 10−4). In contrast, other variants had apparently greater effects at a later age of diagnosis. 2p15 rs721048 homozygotes had an OR = 6.21 (P = 9.3 × 10−4) among cases diagnosed ≥ age 61, relative to an OR = 2.77 (P = 0.025) among those diagnosed ≤ age 60. Our results suggest that heritable variation may modify risk of both early and late onset prostate cancer. However, tests of interaction between genotype and age stratum on cancer risk were not significant for any of the SNPs of Table 3. Study power to establish different effects for these SNPs across age strata was limited.
We separately stratified the study population according to an index of disease aggressiveness, Gleason sum of ≤ 6 (well- or moderately-differentiated and relatively indolent), and ≥ 7 (poorly differentiated and relatively aggressive). Again, this resulted in the division of the study population into two roughly comparably sized groups. More pronounced effects were observed among those with more aggressive cancer (Gleason ≥ 7) relative to those with less aggressive cancer (Gleason ≤ 6) for variants of 8q24 (rs1447295) and of 2p15 (rs721048). However, among the SNPs of Table 3, the test for interaction between genotype and Gleason score stratum on cancer risk was significant only for 2p15 rs721048 (homozygote interaction P = 0.024). A further test of heterogeneity across age of diagnosis and Gleason score revealed that these facets are not independent among the familial cases. Those of the high Gleason score group tended to have an older mean age of diagnosis (on average 2.3 years older, t test P = 0.0016).
DISCUSSION
Our study strongly replicated observed associations between prostate cancer and genetic variants that have been recently uncovered by three genome wide association studies. For all four GWAS loci that we investigated in our familial prostate cancer study, we observed greater effect sizes than have been observed in prior studies unselected for family history. A study design sampling case and control probands based upon family history may improve power to detect disease loci. SNPs detecting the association of adjacent linkage disequilibrium blocks of 8q24 have validated broadly (43). The risk-modifying allele of the telomeric LD block at rs1447295 nicely replicated within our study. The minor risk allele was sufficiently infrequent that nearly all carriers were heterozygotes. Our stratification results were concordant with published findings. We observed greater risk among those with aggressive prostate cancer or with an early age of diagnosis. Amundadottir et al. (5), Helfand et al. (44), Wang et al. (45), Schumacher et al. (46), Severi et al. (47), and Suuriniemi et al. (48) each observed a greater effect at rs1447295 in more aggressive disease. The GWAS of Thomas et al. (7) was also concordant at rs4242382 (r2 of 1.0 with rs1447295), with a greater effect among aggressive cases. Schumacher et al. additionally evaluated age of diagnosis, finding a more pronounced effect among those with an early age of diagnosis. The observation of a stronger association for rs1447295 with aggressive disease and early age of diagnosis is particularly notable, given that our familial cases with more aggressive disease had a significantly later rather than a younger age of diagnosis (by 2.3 years, on average).
Our investigation of the adjacent centromeric 8q24 block included a candidate SNP in moderate LD with the published SNP rs6983267. POU5F1P1 at that locus is a retrotransposed pseudogene, with a fully intact open reading frame that has been demonstrated to be expressed in cancerous tissue (49). In contrast, the parent gene POU5F1 (OCT4) on 6p21 is not expressed in some transformed cells (50). The SNP that we selected for assay is the missense variant G527E (rs6998061). We demonstrate that it too detects an association with prostate cancer, with a protective minor allele that acts in an additive fashion. However, the missense SNP did not improve upon the strong association signal of rs6983267, and thus does not supplant it as the best candidate of the LD block.
Three separate GWAS studies have identified the association of 11q13 with prostate cancer, though at different SNPs. These SNPs are rs7931342 of Thomas et al. (7), rs10896449 of Eeles et al. (6), and rs10896450 of the Icelandic GWAS (personal communication, J. Gudmundsson). All three SNPs are in LD and equivalently detect the association. Within our data, the minor allele of 11q13 rs10896450 was significantly protective and acted additively. Our stratified analyses at 11q13 also indicated a slightly greater effect among aggressive cases relative to indolent cases, but similar effects within older and younger age of diagnosis groups. A trend toward a greater effect among those with aggressive disease was not evident in data of Thomas et al. However, only 11.6% of cases of the initial phase of that study had a family history of prostate cancer, and the proportion of replication phase subjects with a family history was not reported. Eeles et al. had noted that the association at 11q13 was stronger among those with a positive family history.
A fourth variant originally detected in the Icelandic GWAS on 2p15 and subsequently validated across multiple study populations (32), including an overlapping subject group of this study, was re-investigated here to enable cross-comparisons with other variants. Among the original replication study groups, the two with the greatest proportion of familial cases had yielded strongest evidence for the association. In our study of familial cases, homozygotes for the minor risk allele had a four-fold increased risk for prostate cancer. The association at 2p15 appeared stronger among both older and more aggressive cases, sub-groups that were significantly correlated. The Icelandic study of the 2p15 variant had also noted a greater effect among more aggressive cases.
Among candidate genes that have been proposed to be associated with risk of prostate cancer within the published literature, our investigation replicated associations at RNASEL, NKX3-1, and EZH2. Our study failed to support an association of prostate cancer with variants of PPARG, TLR10, PODXL, CDH1, ELAC2, IL1RN, TLR1, TLR6, TLR4, CYP17A1, CDKN1B, and CYP1A1.
Association results across published studies have been inconsistent at RNASEL. Our result for E541D is consistent with the meta-analysis result of Li et al., who demonstrated a significant odds ratio of 1.37 for major allele (Glu) homozygotes or heterozygotes relative to minor allele (Asp) homozygotes among familial Caucasian cases (39). Our investigation concordantly found that minor allele (Asp) homozygotes had a significant odds ratio of 0.64 relative to major allele homozygotes. Converting our analysis of the full study population to the more comparable major allele as dominant model, we observe an odds ratio of 1.47 (P = 0.018). This effect was most prominent among subjects diagnosed beyond age 61. The meta-analysis of R462Q among Caucasians by Rennert et al. found that minor allele (Gln) heterozygotes and homozygotes each had a significantly elevated risk of prostate cancer (40). Our data for all cases supports a risk effect, and as at E541D, it was most apparent in those diagnosed beyond age 61. Caucasian study populations segregate three major haplotypes for the RNASEL LD block harboring these risk-modifying variants. Study subjects may inherit minor allele diplotypes: null/null, null/risk (462Q), null/protective (541D), protective/protective, protective/risk, and risk/risk. Estimated effects of RNASEL variants on prostate cancer risk generally appear modest.
Linkage, association, and mouse model evidence has been published supporting a role for NKX3-1 in prostate cancer. Zheng et al. identified a peak heterogeneity lod score of 2.04 at NKX3-1 (51). Gelmann et al. found that the minor allele of rs2228013 (C154T, R52C, MAF 0.04) was associated with prostate cancer risk among subjects of the Physician Health Study with more aggressive disease (stage C or D, or Gleason score ≥ 7) (42). Three SNPs at the 4kb NKX3-1 gene were genotyped within HapMap Phase II, and did not include rs2228013. Among them, rs1567669 has a pairwise r2 of 0.57 with rs4872176 and of 0.69 with rs11781886 in CEU subjects. Only rs1567669 within the 3’ UTR successfully converted for genotyping assay in our study. We observed a significant protective effect among heterozygotes for the minor allele of rs1567669. Collectively, linkage and association results support further investigation of the role of NKX3-1 in familial prostate cancer.
Only a single prior study has investigated the potential role of EZH2 in prostate cancer predisposition, despite its established role in the progression of aggressive prostate cancer. Bachmann et al. conducted a mutation screen at EZH2 among hereditary prostate cancer pedigrees linked to 7q35, and evaluated the discovered variants for evidence of association with sporadic and familial prostate cancer (41). They observed eight haplotypes of frequency ≥ 0.01, one of which was significantly less frequent among familial cases than among controls. That haplotype was distinguished from all other haplotypes by the minor allele of rs2302427 (185H). The MAF was 0.10 among controls, and 0.06 among familial cases. Concordantly, we observed a MAF of 0.10 among controls, and 0.07 among familial cases, a significant difference within our study. Additional investigation of the role of this gene in familial prostate cancer appears to be warranted.
Our further exploration of several genes related to RNASEL and viral restriction identifies several nominally significant associations with familial prostate cancer. Given the number of multiple comparisons made across these novel candidates, none remain significant upon Bonferroni correction. However, candidates XPR1 and RBM9 may remain of interest for subsequent replication efforts. Haplotype-based sliding window tests at these two genes redundantly identified nominal associations with familial prostate cancer in our study. XPR1 is within the HPC1 linkage interval 1.6 Mb centromeric to RNASEL, and RBM9 is within the 882 kb one recombinant consensus region of the ICPCG hereditary prostate cancer locus on 22q12.3 (11, 14, 52). Within HapMap CEU data, each gene resides alone within an LD block, suggesting that an association is unlikely to be attributable to an adjacent gene in LD with either candidate.
In summary, our investigation has strongly replicated associations at 8q24, 11q13, and 2p15 in a familial prostate cancer study population. Odds ratios that we observed at these loci were generally stronger than described within study populations unselected for family history of prostate cancer, suggesting that greater power for discovery may be afforded by study of familial cases. Our data are also concordant with prior published prostate cancer associations at RNASEL, NKX3-1, and EZH2. Overall, the level of significance that we observed for these was not as great as that observed for the GWAS replication set. In subsequent stratification analyses of age of diagnosis, our data further suggests that the heritable component of prostate cancer may predispose to late- as well as to early-onset disease. This observation has potentially important implications for GWAS strategies. We failed to support the association of a set of additional genes with familial prostate cancer. This latter group included several genes associated with prostate cancer within published literature, and additional candidates potentially related to retroviral restriction and located near hereditary prostate cancer linkage regions. Haplotype-based analysis supported an association at only two of these candidates, XPR1 and RBM9. Both of these are intriguing candidates, based upon genomic position and function, and may be worthy of further investigation.
Supplementary Material
ACKNOWLEDGMENTS
We extend particular thanks to the study participants and to Drs Sam Chang, Peter Clark, Michael Cookson, Rodney Davis, S. Duke Herrell, Richard Hock, William Maynard, Douglas Milam, Jason Pereira, and Joseph Smith. This work was supported by an award from the V Foundation, by a MERIT grant from the US Department of Veterans Affairs, by grant W81XWH-06-1-0057 from the Department of the Army, and by the Cancer Center Support Grant P30 CA068485.
Footnotes
Published in Cancer Epidemiol Biomarkers Prev: http://cebp.aacrjournals.org/content/18/7/2137.long
REFERENCES
- 1.Lichtenstein P, Holm NV, Verkasalo PK, et al. Environmental and heritable factors in the causation of cancer--analyses of cohorts of twins from Sweden, Denmark, and Finland. N Engl J Med. 2000;343:78–85. doi: 10.1056/NEJM200007133430201. [DOI] [PubMed] [Google Scholar]
- 2.Page WF, Braun MM, Partin AW, Caporaso N, Walsh P. Heredity and prostate cancer: a study of World War II veteran twins. Prostate. 1997;33:240–245. doi: 10.1002/(sici)1097-0045(19971201)33:4<240::aid-pros3>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 3.Langeberg WJ, Isaacs WB, Stanford JL. Genetic etiology of hereditary prostate cancer. Front Biosci. 2007;12:4101–4110. doi: 10.2741/2374. [DOI] [PubMed] [Google Scholar]
- 4.Naylor SL. SNPs associated with prostate cancer risk and prognosis. Front Biosci. 2007;12:4111–4131. doi: 10.2741/2375. [DOI] [PubMed] [Google Scholar]
- 5.Amundadottir LT, Sulem P, Gudmundsson J, et al. A common variant associated with prostate cancer in European and African populations. Nat Genet. 2006;38:652–658. doi: 10.1038/ng1808. [DOI] [PubMed] [Google Scholar]
- 6.Eeles RA, Kote-Jarai Z, Giles GG, et al. Multiple newly identified loci associated with prostate cancer susceptibility. Nat Genet. 2008;40:316–321. doi: 10.1038/ng.90. [DOI] [PubMed] [Google Scholar]
- 7.Thomas G, Jacobs KB, Yeager M, et al. Multiple loci identified in a genome-wide association study of prostate cancer. Nat Genet. 2008;40:310–315. doi: 10.1038/ng.91. [DOI] [PubMed] [Google Scholar]
- 8.Dong B, Kim S, Hong S, et al. An infectious retrovirus susceptible to an IFN antiviral pathway from human prostate tumors. Proc Natl Acad Sci U S A. 2007;104:1655–1660. doi: 10.1073/pnas.0610291104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Urisman A, Molinaro RJ, Fischer N, et al. Identification of a novel Gammaretrovirus in prostate tumors of patients homozygous for R462Q RNASEL variant. PLoS Pathog. 2006;2:e25. doi: 10.1371/journal.ppat.0020025. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Goddard KA, Witte JS, Suarez BK, Catalona WJ, Olson JM. Model-free linkage analysis with covariates confirms linkage of prostate cancer to chromosomes 1 and 4. Am J Hum Genet. 2001;68:1197–1206. doi: 10.1086/320103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Camp NJ, Cannon-Albright LA, Farnham JM, et al. Compelling evidence for a prostate cancer gene at 22q12.3 by the International Consortium for Prostate Cancer Genetics. Hum Mol Genet. 2007;16:1271–1278. doi: 10.1093/hmg/ddm075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lemay J, Maidou-Peindara P, Bader T, et al. HuR interacts with human immunodeficiency virus type 1 reverse transcriptase, and modulates reverse transcription in infected cells. Retrovirology. 2008;5:47. doi: 10.1186/1742-4690-5-47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li XL, Andersen JB, Ezelle HJ, Wilson GM, Hassel BA. Post-transcriptional regulation of RNase-L expression is mediated by the 3'-untranslated region of its mRNA. J Biol Chem. 2007;282:7950–7960. doi: 10.1074/jbc.M607939200. [DOI] [PubMed] [Google Scholar]
- 14.Johanneson B, McDonnell SK, Karyadi DM, et al. Fine mapping of familial prostate cancer families narrows the interval for a susceptibility locus on chromosome 22q12.3 to 1.36 Mb. Hum Genet. 2008;123:65–75. doi: 10.1007/s00439-007-0451-y. [DOI] [PubMed] [Google Scholar]
- 15.Levy JA. Xenotropism: the elusive viral receptor finally uncovered. Proc Natl Acad Sci U S A. 1999;96:802–804. doi: 10.1073/pnas.96.3.802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Santiago ML, Montano M, Benitez R, et al. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321:1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santa-Marta M, Aires da Silva F, Fonseca AM, Rato S, Goncalves J. HIV-1 Vif protein blocks the cytidine deaminase activity of B-cell specific AID in E. coli by a similar mechanism of action. Mol Immunol. 2007;44:583–590. doi: 10.1016/j.molimm.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Smith JR, Freije D, Carpten JD, et al. Major susceptibility locus for prostate cancer on chromosome 1 suggested by a genome-wide search. Science. 1996;274:1371–1374. doi: 10.1126/science.274.5291.1371. [DOI] [PubMed] [Google Scholar]
- 19.Rosenberg BR, Papavasiliou FN. Beyond SHM and CSR: AID and related cytidine deaminases in the host response to viral infection. Adv Immunol. 2007;94:215–244. doi: 10.1016/S0065-2776(06)94007-3. [DOI] [PubMed] [Google Scholar]
- 20.Dumur CI, Dechsukhum C, Ware JL, et al. Genome-wide detection of LOH in prostate cancer using human SNP microarray technology. Genomics. 2003;81:260–269. doi: 10.1016/s0888-7543(03)00020-x. [DOI] [PubMed] [Google Scholar]
- 21.Stanford JL, McDonnell SK, Friedrichsen DM, et al. Prostate cancer and genetic susceptibility: a genome scan incorporating disease aggressiveness. Prostate. 2006;66:317–325. doi: 10.1002/pros.20349. [DOI] [PubMed] [Google Scholar]
- 22.Saitoh T, Tun-Kyi A, Ryo A, et al. Negative regulation of interferon-regulatory factor 3-dependent innate antiviral response by the prolyl isomerase Pin1. Nat Immunol. 2006;7:598–605. doi: 10.1038/ni1347. [DOI] [PubMed] [Google Scholar]
- 23.Watashi K, Khan M, Yedavalli VR, Yeung ML, Strebel K, Jeang KT. Human immunodeficiency virus-1 replication and regulation of APOBEC3G by peptidyl prolyl isomerase Pin1. J Virol. 2008 doi: 10.1128/JVI.01017-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Neville PJ, Conti DV, Krumroy LM, et al. Prostate cancer aggressiveness locus on chromosome segment 19q12-q13.1 identified by linkage and allelic imbalance studies. Genes Chromosomes Cancer. 2003;36:332–339. doi: 10.1002/gcc.10165. [DOI] [PubMed] [Google Scholar]
- 25.Ryo A, Uemura H, Ishiguro H, et al. Stable suppression of tumorigenicity by Pin1-targeted RNA interference in prostate cancer. Clin Cancer Res. 2005;11:7523–7531. doi: 10.1158/1078-0432.CCR-05-0457. [DOI] [PubMed] [Google Scholar]
- 26.Wiklund F, Gillanders EM, Albertus JA, et al. Genome-wide scan of Swedish families with hereditary prostate cancer: suggestive evidence of linkage at 5q11.2 and 19p13.3. Prostate. 2003;57:290–297. doi: 10.1002/pros.10303. [DOI] [PubMed] [Google Scholar]
- 27.Witte JS, Suarez BK, Thiel B, et al. Genome-wide scan of brothers: replication and fine mapping of prostate cancer susceptibility and aggressiveness loci. Prostate. 2003;57:298–308. doi: 10.1002/pros.10304. [DOI] [PubMed] [Google Scholar]
- 28.Braaten D, Luban J. Cyclophilin A regulates HIV-1 infectivity, as demonstrated by gene targeting in human T cells. EMBO J. 2001;20:1300–1309. doi: 10.1093/emboj/20.6.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Christensen GB, Camp NJ, Farnham JM, Cannon-Albright LA. Genome-wide linkage analysis for aggressive prostate cancer in Utah high-risk pedigrees. Prostate. 2007;67:605–613. doi: 10.1002/pros.20554. [DOI] [PubMed] [Google Scholar]
- 30.Suarez BK, Lin J, Burmester JK, et al. A genome screen of multiplex sibships with prostate cancer. Am J Hum Genet. 2000;66:933–944. doi: 10.1086/302818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xu J, Gillanders EM, Isaacs SD, et al. Genome-wide scan for prostate cancer susceptibility genes in the Johns Hopkins hereditary prostate cancer families. Prostate. 2003;57:320–325. doi: 10.1002/pros.10306. [DOI] [PubMed] [Google Scholar]
- 32.Gudmundsson J, Sulem P, Rafnar T, et al. Common sequence variants on 2p15 and Xp11.22 confer susceptibility to prostate cancer. Nat Genet. 2008;40:281–283. doi: 10.1038/ng.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Haiman CA, Patterson N, Freedman ML, et al. Multiple regions within 8q24 independently affect risk for prostate cancer. Nat Genet. 2007;39:638–644. doi: 10.1038/ng2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yeager M, Orr N, Hayes RB, et al. Genome-wide association study of prostate cancer identifies a second risk locus at 8q24. Nat Genet. 2007;39:645–649. doi: 10.1038/ng2022. [DOI] [PubMed] [Google Scholar]
- 35.Carlson CS, Eberle MA, Rieder MJ, Yi Q, Kruglyak L, Nickerson DA. Selecting a maximally informative set of single-nucleotide polymorphisms for association analyses using linkage disequilibrium. Am J Hum Genet. 2004;74:106–120. doi: 10.1086/381000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Stephens M, Smith NJ, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fallin D, Cohen A, Essioux L, et al. Genetic analysis of case/control data using estimated haplotype frequencies: application to APOE locus variation and Alzheimer's disease. Genome Res. 2001;11:143–151. doi: 10.1101/gr.148401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yaspan BL, McReynolds KM, Elmore JB, Breyer JP, Bradley KM, Smith JR. A haplotype at chromosome Xq27.2 confers susceptibility to prostate cancer. Hum Genet. 2008;123:379–386. doi: 10.1007/s00439-008-0486-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Li H, Tai BC. RNASEL gene polymorphisms and the risk of prostate cancer: a meta-analysis. Clin Cancer Res. 2006;12:5713–5719. doi: 10.1158/1078-0432.CCR-05-2799. [DOI] [PubMed] [Google Scholar]
- 40.Rennert H, Zeigler-Johnson CM, Addya K, et al. Association of susceptibility alleles in ELAC2/HPC2, RNASEL/HPC1, and MSR1 with prostate cancer severity in European American and African American men. Cancer Epidemiol Biomarkers Prev. 2005;14:949–957. doi: 10.1158/1055-9965.EPI-04-0637. [DOI] [PubMed] [Google Scholar]
- 41.Bachmann N, Hoegel J, Haeusler J, et al. Mutation screen and association study of EZH2 as a susceptibility gene for aggressive prostate cancer. Prostate. 2005;65:252–259. doi: 10.1002/pros.20296. [DOI] [PubMed] [Google Scholar]
- 42.Gelmann EP, Steadman DJ, Ma J, et al. Occurrence of NKX3.1 C154T polymorphism in men with and without prostate cancer and studies of its effect on protein function. Cancer Res. 2002;62:2654–2659. [PubMed] [Google Scholar]
- 43.Cheng I, Plummer SJ, Jorgenson E, et al. 8q24 and prostate cancer: association with advanced disease and meta-analysis. Eur J Hum Genet. 2008;16:496–505. doi: 10.1038/sj.ejhg.5201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Helfand BT, Loeb S, Cashy J, et al. Tumor characteristics of carriers and noncarriers of the deCODE 8q24 prostate cancer susceptibility alleles. J Urol. 2008;179:2197–2201. doi: 10.1016/j.juro.2008.01.110. discussion 2202. [DOI] [PubMed] [Google Scholar]
- 45.Wang L, McDonnell SK, Slusser JP, et al. Two common chromosome 8q24 variants are associated with increased risk for prostate cancer. Cancer Res. 2007;67:2944–2950. doi: 10.1158/0008-5472.CAN-06-3186. [DOI] [PubMed] [Google Scholar]
- 46.Schumacher FR, Feigelson HS, Cox DG, et al. A common 8q24 variant in prostate and breast cancer from a large nested case-control study. Cancer Res. 2007;67:2951–2956. doi: 10.1158/0008-5472.CAN-06-3591. [DOI] [PubMed] [Google Scholar]
- 47.Severi G, Hayes VM, Padilla EJ, et al. The common variant rs1447295 on chromosome 8q24 and prostate cancer risk: results from an Australian population-based case-control study. Cancer Epidemiol Biomarkers Prev. 2007;16:610–612. doi: 10.1158/1055-9965.EPI-06-0872. [DOI] [PubMed] [Google Scholar]
- 48.Suuriniemi M, Agalliu I, Schaid DJ, et al. Confirmation of a positive association between prostate cancer risk and a locus at chromosome 8q24. Cancer Epidemiol Biomarkers Prev. 2007;16:809–814. doi: 10.1158/1055-9965.EPI-06-1049. [DOI] [PubMed] [Google Scholar]
- 49.Suo G, Han J, Wang X, Zhang J, Zhao Y, Dai J. Oct4 pseudogenes are transcribed in cancers. Biochem Biophys Res Commun. 2005;337:1047–1051. doi: 10.1016/j.bbrc.2005.09.157. [DOI] [PubMed] [Google Scholar]
- 50.Cantz T, Key G, Bleidissel M, et al. Absence of OCT4 expression in somatic tumor cell lines. Stem Cells. 2008;26:692–697. doi: 10.1634/stemcells.2007-0657. [DOI] [PubMed] [Google Scholar]
- 51.Zheng SL, Ju JH, Chang BL, et al. Germ-line mutation of NKX3.1 cosegregates with hereditary prostate cancer and alters the homeodomain structure and function. Cancer Res. 2006;66:69–77. doi: 10.1158/0008-5472.CAN-05-1550. [DOI] [PubMed] [Google Scholar]
- 52.Camp NJ, Farnham JM, Cannon-Albright LA. Localization of a prostate cancer predisposition gene to an 880-kb region on chromosome 22q12.3 in Utah high-risk pedigrees. Cancer Res. 2006;66:10205–10212. doi: 10.1158/0008-5472.CAN-06-1233. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.