Abstract
Ubiquitylation describes a process in which ubiquitin, a 76-amino-acid polypeptide, is covalently attached to target proteins. Traditionally, ubiquitin-conjugated proteins are targeted for degradation by the 26S proteasome. However, non-proteolytic roles in histone regulation, DNA repair and signal transduction have been reported. Here, the role of ubiquitylation in the cell death pathway in Drosophila is reviewed. Interestingly, ubiquitylation serves both pro- and anti-apoptotic functions. Although pro-apoptotic ubiquitylation leads to proteolytic degradation, recent evidence suggests that anti-apoptotic ubiquitylation may involve, at least in part, non-proteolytic functions.
Keywords: Drosophila, ubiquitin, cell death, apoptosis, IAP
Ubiquitylation is a complex process requiring the activity of several enzymes. The initial step is the activation of ubiquitin (UB) by an UB-activating (UBA) enzyme or E1 (enzyme 1).1,2 The Drosophila genome contains only one UB-specific E1, termed ‘UBA1’.3 There are additional E1 enzymes such as UBA2 in Drosophila; however, they are involved in the activation of other UB-like molecules such as Sumo, ATG8, ATG12, etc. UBA1 is believed to be required for all UB-dependent reactions.3 In the next step, activated UB is transferred to an UB-conjugating (UBC) enzyme, or E2.1 There are several E2 enzymes encoded in the Drosophila genome. At least UBCD1 (also known as EFFETE) plays an essential role for the control of apoptosis in Drosophila (see below).4,5 The final step is the transfer of the activated UB to the substrate protein. This transfer is mediated by an UB ligase, or E3.1 The E3 ligases confer substrate specificity; hence, the Drosophila genome contains hundreds of genes encoding them. The two major types of E3 UB ligases carry either a Hect or a RING (really interesting new gene) domain as the functional domains for UB ligation. The E3 UB ligases in the cell death pathway are the inhibitor of apoptosis proteins (IAP), which are RING domain ligases.6 The RING domain binds to the E2 and mediates the transfer of UB to the substrate without forming a covalent intermediate, unless it is targeted for ubiquitylation itself (see below).
The UB–substrate bond is an isopeptide bond between the ε-amino group of a lysine residue in the target protein and the C-terminal carboxyl group of the UB moiety.2 Repeated cycles of ubiquitylation lead to formation of poly-UB chains on the target protein. Poly-UB chains also form through isopeptide bonds between the ε-amino group of a lysine residue of the first UB and the C-terminal carboxyl group of the next one. UB contains seven lysine (K) residues (K6, K11, K27, K29, K33, K48 and K63).2 Any of these lysines may be used for poly-UB formation. The lysine used for poly-UB formation determines the fate of the target protein. The best characterized are K48- and K63-linked poly-UB chains. K48-linked poly-UB chains target proteins for degradation by the 26S proteasome. K63-linked chains regulate non-proteolytic events, such as signal transduction in the NF-κB pathway.2,7 Linear (head-to-tail) poly-UB chains have also been reported, which appear to be involved in NF-κB activation.8–10
Polyubiquitylation does not occur in all cases. Monoubiquitylation results from a single ubiquitylation event on a lysine residue. Monoubiquitylation also controls non-proteolytic events such as DNA repair, histone activity and endocytosis.1,7,11
The Cell Death Pathway in Drosophila
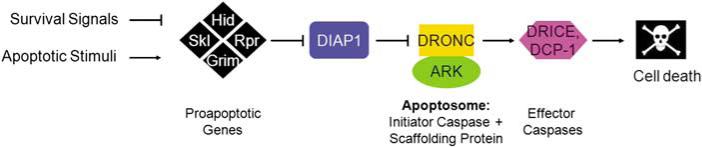
As in vertebrates, caspases are critical for cell death in Drosophila. Of the seven Drosophila caspases, only the initiator caspase, DRONC, and the effector caspases, DRICE and DCP-1, are required for apoptosis (Figure 1).12–19 The remaining four caspases either function in innate immunity (DREDD)20 or their function is unknown. DRONC is most similar to caspase-9,21 and assembles with the adaptor molecule ARK (Apaf1-related killer, also known as Hac-1 and D-Apaf-1)22–25 into the apoptosome (Figure 1). The apoptosome activates DRICE and DCP-1, caspase-3-like proteases that induce apoptosis (Figure 1).
Figure 1.
The apoptotic pathway in Drosophila
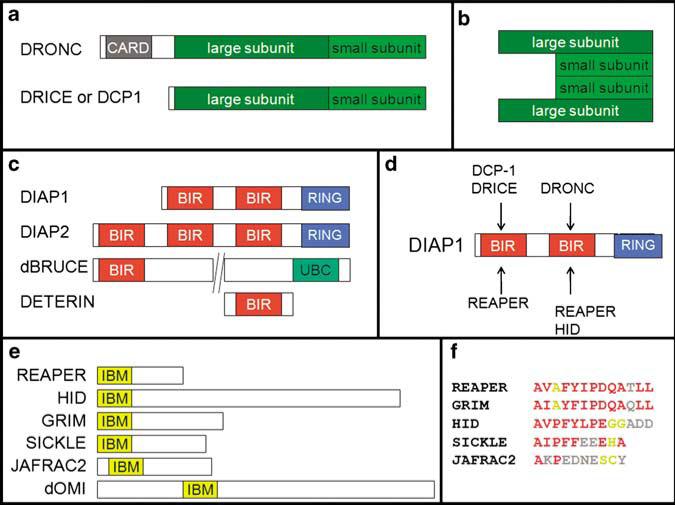
Caspases are produced as inactive zymogen precursors, composed of a prodomain, a large and a small catalytic domain (Figure 2a). Activation of caspases occurs differently between initiator and effector caspases. The initiator caspase, DRONC, is activated through dimerization, which is facilitated by the adaptor molecule, ARK, during apoptosome formation.26,27 The dimerization event is essential for activation of DRONC.26 Cleavage between the large and the small catalytic subunits is neither required nor sufficient for DRONC activation,26,27 although it occurs in dying cells. In contrast, effector caspases are constitutive dimers. Activation of effector caspases requires proteolytic cleavage, cleaving off the prodomain and separating the large and small catalytic subunits. Association of two dimers into a heterotetramer forms the active protease (Figure 2b).15
Figure 2.
Domain structure of apoptotic proteins in Drosophila. Not drawn to scale. (a) Schematic outline of the zymogen form of the initiator caspase, DRONC, and the effector caspases, DRICE and DCP-1. Initiator caspases such as DRONC contain a long prodomain that harbors regulatory motifs such as the caspase activation and recruitment domain (CARD). Effector caspases have only short prodomains. (b) After cleavage of the zymogen form of effector caspases, two large and two small subunits form the active caspase. (c) Schematic outline of IAPs in Drosophila. BIR, Baculovirus IAP repeat; RING, really interesting new gene; UBC, ubiquitin conjugation. (d) Binding preferences of the two BIR domains of DIAP1. (e) The RHG proteins in Drosophila. IBM, IAP-binding motif. (f) Alignment of the IBM of several RHG proteins. Residues in red are identical and in yellow are conserved
In addition, an additional layer of caspase control is imposed by IAPs, which bind directly to caspases and inhibit them (Figure 1). However, paradoxically, IAPs only bind to caspases after they have been cleaved and activated (see below). The only known exception is DRONC, in which case Drosophila IAP1 (DIAP1) binds to the prodomain of the monomeric zymogen.28,29 It is believed that IAP inhibition of activated caspases protects cells against inappropriate caspase activity; hence, IAPs represent the last line of defense for the survival of the cell.
The Drosophila genome contains four IAPs: DIAP1, DIAP2, dBRUCE and DETERIN (Figure 2c).19 Undoubtedly, of these the most important one is DIAP1. Loss-of-function mutations of diap1 are embryonic lethal and the mutant embryos die by massive apoptosis,30–32 suggesting that DIAP1 is absolutely critical for control of apoptosis. diap2 mutations do not have an apoptotic phenotype but affect innate immunity.33,34 It is interesting to note that DIAP2 acts together with the non-apoptotic caspase, DREDD, in the IMD pathway in innate immunity. dbruce mutations cause male sterility,35 but otherwise they do not have a significant apoptotic phenotype. However, dbruce mutants dominantly enhance REAPER- and GRIM-induced cell death in the eye,36 indicating that dBRUCE may also have some function for caspase regulation in somatic cells. The role of DETERIN,37 the SURVIVIN homolog in Drosophila, is unknown due to lack of mutants.
Structurally, IAPs contain between one and three BIR (baculovirus IAP repeat) domains and some have a C-terminally located RING E3 domain. For example, DIAP1 has two BIR and one RING domains; DIAP2 has three BIR and one RING domains (Figure 2c). Interestingly, dBRUCE has an E2 UBC domain instead of a RING domain (Figure 2c). The BIR domains mediate protein–protein interaction with caspases and IAP antagonists (Figure 2d and see below). The presence of several BIR domains increases the flexibility and potency of caspase inhibition. For example, BIR1 of DIAP1 preferentially interacts with DRICE and DCP-1, whereas BIR2 is more specific for DRONC (Figure 2d).38
In cells committed to die, the inhibition of caspases by DIAP1 has to be overcome. This is accomplished by the IAP antagonists REAPER, HID, GRIM, JAFRAC2, SICKLE and dOMI, which are acting upstream in the pathway (Figures 1 and 2e).39–43 Among these, REAPER, HID and GRIM are the best characterized both genetically and biochemically; hence, this protein family is often referred to as RHG proteins. They integrate a large number of apoptotic and survival signals to control apoptosis. For example, activation of the EGFR signaling pathway leads to inactivation of the pro-apoptotic gene HID, and thus to survival.44–46 Other signals, such as steroid hormones, X-ray, developmental defects, unfolded proteins, etc., lead to activation of the RHG proteins and cell death.47–50 The RHG proteins bind directly to DIAP1 and induce its auto-ubiquitylation and degradation.4,5,51,52 The RHG proteins contain an IAP binding motif (IBM) (Figure 2e and f), which binds to the BIR domains of IAPs. The IBM is invariantly located at the N terminus of the proteins (Figure 2e). In those cases where it is not immediately present at the N terminus, proteolytic processing has to occur to present it at the N terminus. There is some specificity as to which RHG motif binds to what BIR domain. The IBM of REAPER and GRIM binds with high affinity to both BIR1 and BIR2 of DIAP1. However, HID, SICKLE and JAFRAC2 have a preference for BIR2 only (Figure 2d).38
Not only IAP antagonists contain an IBM, several caspases also display an IBM after proteolytic processing. This was shown first for mammalian caspase-9,53 and was later also found for the Drosophila caspases DRICE and DCP-1, and mammalian caspase-7.54 In the case of caspase-9, an IBM is exposed at the N terminus of the small catalytic subunit (p12) after proteolytic processing, which can bind to the BIR3 domain of XIAP.53 In contrast, the IBM of DRICE, DCP-1 and Caspase-7 is present at the N terminus of the large catalytic subunit (p20) after proteolytic removal of the prodomain.54 The newly generated IBM of DRICE and DCP-1 can interact with the BIR1 of DIAP1 (Figure 2d). It is generally believed that this IBM/BIR interaction causes inhibition of cleaved caspase-9, DRICE and DCP-1.
However, as will be discussed below IAP-mediated ubiquitylation is also critical for control of caspase activity. All components of the Drosophila pathway, the RHG proteins, IAPs and caspases are subject to ubiquitylation. These ubiquitylation events serve in some cases an anti-apoptotic function and in others a pro-apoptotic one.
Regulation of DIAP1 by Ubiquitylation
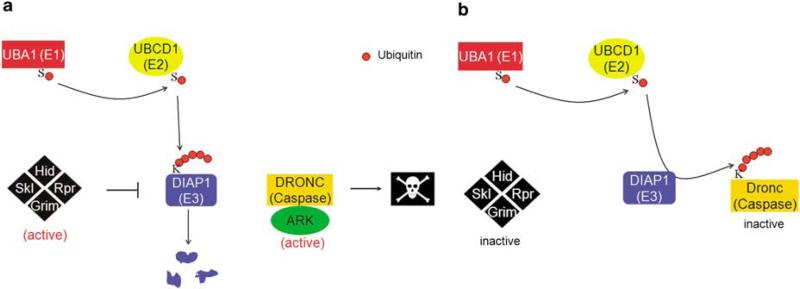
The regulation of DIAP1 by ubiquitylation is very complex and requires at least two, but possibly three, E3 UB ligases. The first E3 ligase identified was DIAP1's own RING domain. In response to apoptotic signals, binding of the IAP antagonists, REAPER, HID and GRIM, triggers or changes the ubiquitylation activity of the RING domain such that it auto-ubiquitylates DIAP1.4,5,51,52 Auto-ubiquitylated DIAP1 is then targeted to the proteasome for degradation (Figure 3a). The E2 enzyme, UBCD1, has been implicated in DIAP1 turnover.4,5 Another factor to be involved appears to be MORGUE, which contains an UB enzyme variant (UEV) domain and a F-box.55,56 The UEV is an UBC domain, but lacks a critical cysteine residue in its catalytic site. F-box-containing proteins are components of Cullin-RING ligases (CRLs). It is unclear how MORGUE functions mechanistically. Because the UEV domain lacks the critical cysteine residue required for ubiquitylation, MORGUE may not be able to perform ubiquitylation events. On the other hand, the F-box motif mediates ubiquitylation by CRLs. These different domain activities may be the reason for the conflicting data that have been reported. morgue mutants accumulate DIAP1 protein, suggesting that it is involved in DIAP1 degradation.55 In contrast, overexpression of MORGUE in S2 cells appears to stabilize DIAP1.55
Figure 3.
Ubiquitylation events mediated by the RING domain of DIAP1. (a) In dying cells, the RHG proteins stimulate the RING domain to auto-ubiquitylate DIAP1, which is subsequently degraded by the proteasome. (b) In living cells, the RING domain of DIAP1 mediates ubiquitylation of the initiator caspase DRONC. This ubiquitylation event inactivates DRONC
RING-mediated ubiquitylation of DIAP1 leads to its proteolytic degradation and thus to release of DIAP1-inhibited caspases, which can now induce apoptosis (Figure 3a). Therefore, in this context, ubiquitylation serves a pro-apoptotic function. Further support for a pro-apoptotic function of ubiquitylation came from the analysis of weak alleles of the only UB-E1 enzyme, Uba1. In weak Uba1 mutants, the levels of activated UB are reduced.3,57 This causes less-efficient ubiquitylation and degradation of short-lived proteins including DIAP1 (with a half-life of 30 min),51 which accumulates in weak Uba1 mutants and hence increases the survival of the affected cells.3,57 However, complete loss of Uba1 and thus complete loss of ubiquitylation causes a strong apoptotic phenotype.3,57
The second ubiquitylation system found to regulate DIAP1 protein levels is the N-end-rule pathway.58,59 According to the N-end-rule, the most N-terminal residue of a protein determines its stability. There are stabilizing residues such as methionine (Met), and destabilizing ones such as asparagine (Asn).58,59 DIAP1 is synthesized as a protein carrying a stabilizing Met residue at its N terminus. However, removal of the first 20 residues through proteolytic cleavage by the effector caspase, DRICE, exposes Asn21 at the N terminus. N-terminally located Asn is converted to aspartate by asparaginase. After further modification with arginine, it is recognized and ubiquitylated by the N-end-rule specific UB-ligase E3α.58,59
Caspase-dependent N-end-rule-induced ubiquitylation of DIAP1 was described in two reports.60,61 However, the outcome of this process was different in these two reports. Yokokura et al.61 found that N-end rule mediated ubiquitylation and degradation of DIAP1 promote cell death. In contrast, Ditzel et al.60 reported that N-end-rule-mediated degradation of DIAP1 actually protects cells from apoptosis, that is, behaves anti-apoptotically. These authors attributed the anti-apoptotic role to simultaneous ubiquitylation and inactivation of associated caspases, which may still be bound to DIAP1 after removal of the N-terminal 20 residues. The reason for the differences in these reports is not clear.
Finally, it has recently been reported that DIAP2 can also ubiquitylate DIAP1 in vitro.62 This ubiquitylation event requires the RING domain of DIAP2. Further analysis in S2 cells indicates that DIAP2-mediated ubiquitylation of DIAP1 leads to K48-linked poly-UB chains, likely targeting DIAP1 for proteasome-mediated degradation. However, homozygous diap2 mutant flies are viable and fertile without an apparent apoptotic phenotype, and DIAP1 protein levels are unchanged in diap2 mutant animals.33,34 Therefore, the in vivo relevance of DIAP2-mediated ubiquitylation of DIAP1 remains unclear.
Regulation of the Initiator Caspase DRONC by Ubiquitylation
As mentioned above, the RING E3 ligase domain of DIAP1 auto-ubiquitylates under apoptotic conditions. In this respect, the RING domain behaves in a pro-apoptotic manner. However, homozygous mutations affecting the RING domain of diap1 (diap1ΔRING) cause apoptosis even in the absence of apoptotic signals.4,31,63 Therefore, the RING domain of DIAP1 also has an important anti-apoptotic function. Furthermore, the apoptotic phenotype of diap1ΔRING is completely suppressed by the caspase inhibitor, P35,63 suggesting that the RING domain plays a critical role in controlling caspase activity. This observation also implies that physical interaction between the BIR domains and caspases is not sufficient for apoptosis inhibition. Indeed, it was shown that DIAP1 can ubiquitylate DRONC in a RING-dependent manner in vitro (Figure 3b).29,64 Following these observations, models have been proposed according to which DIAP1-mediated ubiquitylation of monomeric (i.e., inactive) DRONC induces its degradation.29,65 However, although there has been some evidence in favor of the degradation models, clear genetic proof has not been obtained. Although DRONC protein accumulates in diap1ΔRING mutants, which may confirm the degradation models, this Dronc accumulation was observed in diap1ΔRING mutant cells kept ‘undead’ by P35 expression.63 It is unclear how such cells respond to their ‘undead’ status over an extended period of time. ‘Undead’ cells can induce transcription of growth factors for compensatory proliferation.63,66,67 Therefore, they may also induce the transcription of dronc, which may account for increased DRONC protein levels. Moreover, Wilson et al.,64 showed that DIAP1 can ubiquitylate monomeric DRONC without inducing proteasome-mediated degradation. A recent observation by Herman-Bachinsky et al.62 suggests that the RING domain of DIAP1 mediates non-proteolytic K63 polyubiquitylation. Although this has been shown for auto-ubiquitylation of DIAP1, by extrapolation it may suggest a similar activity toward DRONC (which of course also depends on the E2 partner in this process). In other words, the consequence of ubiquitylation of DRONC by DIAP1 is unclear; proteasome-mediated degradation may not be the outcome.
However, although ubiquitylation of DRONC by DIAP1 may not induce degradation of monomeric Dronc, the situation changes once DRONC becomes incorporated into the apoptosome. In cultured S2 cells, DIAP1 appears to ubiquitylate processed DRONC for degradation.65 More recent work has suggested that the apoptosome components, DRONC and ARK, mutually suppress their protein abundance in a DIAP1-dependent manner.68 Hence, only DRONC present in the apoptosome, but not free, monomeric DRONC, may be degraded by the proteasome. Interestingly, ubiquitylation requires DRONC-induced processing of ARK. Therefore, this mechanism resembles DIAP1 degradation by the N-end-rule pathway after DRICE processing (see above). Degradation of the apoptosome components may be another safeguard mechanism against inappropriate caspase activation, or may help to restrict caspase activity in non-apoptotic processes.
Regulation of the Effector Caspase DRICE by Ubiquitylation
Recent observations have suggested that DIAP1-mediated ubiquitylation does not only regulate DRONC but also the effector caspase DRICE. Here the evidence suggests a degradation-independent mechanism.69 Inhibition of the proteasome by proteasome inhibitors did not cause accumulation of DRICE protein. Similarly, overexpression of DIAP1 in imaginal discs, the precursor structures of the adult animal, does not change the protein levels of DRICE. Even a DRICE mutant in which all nine surface lysine residues are changed to arginine is resistant to DIAP1 inactivation without significant accumulation of the mutant protein.69 Therefore, these observations suggest that DIAP1-mediated ubiquitylation does not target DRICE for degradation. Although it is not degraded, ubiquitylated DRICE is enzymatically inactive. Therefore, it appears that ubiquitylation inhibits DRICE by a non-degradative mechanism. This raises the question how ubiquitylation and poly-UB chains inhibit caspase activation and/or activity. Several possibilities exist. In the case of DRICE, the poly-UB chains may sterically block the access of substrates to the catalytic site of the caspase, as determined by computer modeling.69 Another possibility would be by inhibiting dimerization, which is required for initiator caspase activation, or by inhibiting interaction with scaffolding proteins such as ARK. Finally, ubiquitylated caspases may be recognized by UB-binding domain (UBD)-containing proteins,2 which may inhibit the catalytic activity of caspases or sequester them in a subcellular location in the cell where they do not have access to their substrates.
Nevertheless, although DIAP1-mediated ubiquitylation may not be sufficient for DRICE degradation, there may be other E3 ligases controlling DRICE protein levels. For example, the N-end-rule pathway that degrades DIAP1 after DRICE cleavage (see above) may also degrade DRICE in the process.60 In addition, DIAP2 may also function as an E3 ligase targeting DRICE. diap2 mutants display increased DRICE activity and are sensitive to genotoxic stress.70 In summary, DRICE regulation is complex and may require several E3 ligase systems. More work is needed to dissect all aspects of UB-dependent regulation of DRICE.
Additional Targets of DIAP1 Ubiquitylation: RHG Proteins and dTRAF1
In addition to caspases and DIAP1 itself, RHG proteins can also be ubiquitylated by DIAP1 in a RING-dependent manner. This has been shown for REAPER.71 Owing to the lack of antibodies specific for REAPER, it is not known whether this ubiquitylation event triggers degradation of REAPER. However, a ubiquitylation-resistant mutant of REAPER that lacks all lysine residues is a much more efficient inducer of apoptosis than wild-type REAPER in cultured cells,71 providing evidence that ubiquitylation inhibits REAPER and thus defining another anti-apoptotic function of DIAP1. This puts an interesting twist regarding the control of apoptosis because, as described above, REAPER can induce auto-ubiquitylation and degradation of DIAP1. Thus, it appears as if REAPER and DIAP1 mutually control their activity in a negative manner. It is unclear what tips the balance between life (REAPER inactivation) and death (DIAP1 degradation). Certainly, other signaling pathways and stresses may shift the balance one way or another.
Another target of DIAP1-mediated ubiquitylation and degradation is dTRAF1, a component in the TNF receptor complex.72 dTRAF1 can induce JNK-induced apoptosis; thus, DIAP1 protects cells from JNK-induced apoptosis through degradation of dTRAF1. dtraf1 mutants are dominant suppressors of REAPER-induced cell death. These observations suggest that REAPER-mediated degradation of DIAP1 leads to stabilization of dTRAF1 resulting in JNK activation.72
Role of CRLs for Non-Apoptotic Functions of Caspases
In the past few years, evidence has emerged which also implicates caspases in non-apoptotic functions such as proliferation, differentiation, cell signaling and cell remodeling (reviewed by Galluzzi et al.73). Of particular interest for the topic of this review is the non-apoptotic role of caspases during spermatogenesis in Drosophila. Apoptotic factors including DRONC, ARK and DRICE are used in a non-apoptotic manner in a process termed ‘individualization’ during which 64 interconnected postmeiotic spermatids are separated from each other.74 Interestingly, a similar role of ubiquitylation for the control of caspase activation during apoptosis, in this non-apoptotic process ubiquitylation, is also involved. However, ubiquitylation is not mediated by IAPs, but instead by a CRL complex.35 In CRL complexes, Cullins are large scaffolding proteins that link two functional modules: a catalytic module and a substrate recognition module. The catalytic module is an E3 RING domain protein that recruits an E2 enzyme for ubiquitylation of the substrate. The substrate is recruited into the complex through the substrate recognition module. In the case of spermatid individualization, the CRL is of a Cullin-3 type. In a genetic screen for genes required for sperm individualization, mutants of cullin-3, the small RING protein roc1b, and the BTB-Kelch protein, klhl10, as the substrate recognition factor were identified.35 In these mutants, caspase activity is abolished, suggesting that CRL-mediated ubiquitylation is required for caspase activation. The substrate is currently unknown. The genetics implies that the substrate may be an inhibitor of apoptosis. However, DIAP1 and DIAP2 have been excluded. Another good candidate may be the IAP dBRUCE. dbruce mutants cause male sterility35 and they enhance REAPER-induced cell death,36 suggesting that dBRUCE may have an inhibitory effect on caspase activity in spermatids. Future work is needed to confirm this hypothesis.
Deubiquitylation Enzymes
Although most research effort has focused on ubiquitylation of substrates in the apoptotic pathway, recent findings have also implicated deubiquitylation enzymes (DUBs) in the control of caspase activity. DUBs revert the effects of the UB conjugation system. There are at least 15 DUBs in Drosophila. So far, three of them have been implicated in the control of apoptosis. The gene fat facets (faf) has been identified as a regulator of IAP-controlled apoptosis.4,56 Another DUB is encoded by the echinus gene and is involved in cell death during eye development in flies.75 However, in both cases, it is unclear how these genes control apoptosis. More recently, another DUB termed ‘Emperor's thumb’ (ET, also known as Scrawny) was found to have a pronounced effect on apoptosis.76 Interestingly, ET affects REAPER- and GRIM-induced apoptosis, but not HID-induced apoptosis. Loss of et causes a reduction in DIAP1 levels, suggesting that this DUB may counteract DIAP1 ubiquitylation.76 In summary, these examples show that ubiquitylation and deubiquitylation of apoptotic regulatory proteins is critical for the appropriate decision of the cell to live or to die.
Conclusions
Although this review has focused on UB-mediated regulation of apoptosis in Drosophila, many of the mechanisms are probably conserved in mammalian cells (reviewed in Broemer and Meier77). However, there are several areas where Drosophila research leads the way, and these particular areas have been highlighted in this review. Ubiquitylation can lead to proteasomal degradation and depending on the ubiquitylation target, pro- and anti-apoptotic responses can be observed. However, ubiquitylation does not always lead to proteasome-mediated degradation. This has been best characterized for the ubiquitylation of the effector caspase, DRICE, which leads to non-degradative inactivation. Control of the activity of caspases by ubiquitylation can result in inactivation of caspases, but can also limit caspase activity to non-apoptotic processes. Spermatid individualization during sperm maturation in Drosophila requires this limited caspase activity to occur. Interestingly, nature has evolved a different UB ligation mechanism involving CRLs. Regulation of ubiquitylation-mediated signaling involves the interplay of E3 ligases and DUBs, and a recent study extends this paradigm to the UB-mediated control of apoptosis in Drosophila. The years to come will reveal more information about these complex interactions and will fill the gaps in our understanding.
Acknowledgements
I gratefully acknowledge support of the research conducted in my laboratory by grants from the National Institutes of Health (R01GM068016, R01GM074977, R01GM081543) and from The Robert A Welch Foundation (G-1496).
Abbreviations
- UB
ubiquitin
- UBC
UB-conjugating
- IAP
inhibitor of apoptosis proteins
- DIAP1
Drosophila IAP1
- UEV
UB enzyme variant
References
- 1.Welchman RL, Gordon C, Mayer RJ. Ubiquitin and ubiquitin-like proteins as multifunctional signals. Nat Rev Mol Cell Biol. 2005;6:599–609. doi: 10.1038/nrm1700. [DOI] [PubMed] [Google Scholar]
- 2.Hicke L, Schubert HL, Hill CP. Ubiquitin-binding domains. Nat Rev Mol Cell Biol. 2005;6:610–621. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 3.Lee TV, Ding T, Chen Z, Rajendran V, Scherr H, Lackey M, et al. The E1 ubiquitin-activating enzyme Uba1 in Drosophila controls apoptosis autonomously and tissue growth non-autonomously. Development. 2008;135:43–52. doi: 10.1242/dev.011288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ryoo HD, Bergmann A, Gonen H, Ciechanover A, Steller H. Regulation of Drosophila IAP1 degradation and apoptosis by reaper and ubcD1. Nat Cell Biol. 2002;4:432–438. doi: 10.1038/ncb795. [DOI] [PubMed] [Google Scholar]
- 5.Yoo SJ. Grim stimulates Diap1 poly-ubiquitination by binding to UbcD1. Mol Cells. 2005;20:446–451. [PubMed] [Google Scholar]
- 6.Yang Y, Fang S, Jensen JP, Weissman AM, Ashwell JD. Ubiquitin protein ligase activity of IAPs and their degradation in proteasomes in response to apoptotic stimuli. Science. 2000;288:874–877. doi: 10.1126/science.288.5467.874. [DOI] [PubMed] [Google Scholar]
- 7.Mukhopadhyay D, Riezman H. Proteasome-independent functions of ubiquitin in endocytosis and signaling. Science. 2007;315:201–205. doi: 10.1126/science.1127085. [DOI] [PubMed] [Google Scholar]
- 8.Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, et al. A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J. 2006;25:4877–4887. doi: 10.1038/sj.emboj.7601360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, et al. Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol. 2009;11:123–132. doi: 10.1038/ncb1821. [DOI] [PubMed] [Google Scholar]
- 10.Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, et al. Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell. 2009;136:1098–1109. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 11.Haglund K, Dikic I. Ubiquitylation and cell signaling. EMBO J. 2005;24:3353–3359. doi: 10.1038/sj.emboj.7600808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu D, Li Y, Arcaro M, Lackey M, Bergmann A. The CARD-carrying caspase Dronc is essential for most, but not all, developmental cell death in Drosophila. Development. 2005;132:2125–2134. doi: 10.1242/dev.01790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu D, Wang Y, Willecke R, Chen Z, Ding T, Bergmann A. The effector caspases drICE and dcp-1 have partially overlapping functions in the apoptotic pathway in Drosophila. Cell Death Differ. 2006;13:1697–1706. doi: 10.1038/sj.cdd.4401920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kondo S, Senoo-Matsuda N, Hiromi Y, Miura M. DRONC coordinates cell death and compensatory proliferation. Mol Cell Biol. 2006;26:7258–7268. doi: 10.1128/MCB.00183-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kumar S. Caspase function in programmed cell death. Cell Death Differ. 2007;14:32–43. doi: 10.1038/sj.cdd.4402060. [DOI] [PubMed] [Google Scholar]
- 16.Muro I, Berry DL, Huh JR, Chen CH, Huang H, Yoo SJ, et al. The Drosophila caspase Ice is important for many apoptotic cell deaths and for spermatid individualization, a nonapoptotic process. Development. 2006;133:3305–3315. doi: 10.1242/dev.02495. [DOI] [PubMed] [Google Scholar]
- 17.Chew SK, Akdemir F, Chen P, Lu WJ, Mills K, Daish T, et al. The apical caspase dronc governs programmed and unprogrammed cell death in Drosophila. Dev Cell. 2004;7:897–907. doi: 10.1016/j.devcel.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 18.Daish TJ, Mills K, Kumar S. Drosophila caspase DRONC is required for specific developmental cell death pathways and stress-induced apoptosis. Dev Cell. 2004;7:909–915. doi: 10.1016/j.devcel.2004.09.018. [DOI] [PubMed] [Google Scholar]
- 19.Xu D, Woodfield SE, Lee TV, Fan Y, Antonio C, Bergmann A. Genetic control of programmed cell death (apoptosis) in Drosophila. Fly (Austin) 2009;3:78–90. doi: 10.4161/fly.3.1.7800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stoven S, Silverman N, Junell A, Hedengren-Olcott M, Erturk D, Engstrom Y, et al. Caspase-mediated processing of the Drosophila NF-kappaB factor Relish. Proc Natl Acad Sci USA. 2003;100:5991–5996. doi: 10.1073/pnas.1035902100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dorstyn L, Colussi PA, Quinn LM, Richardson H, Kumar S. DRONC, an ecdysone-inducible Drosophila caspase. Proc Natl Acad Sci USA. 1999;96:4307–4312. doi: 10.1073/pnas.96.8.4307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rodriguez A, Oliver H, Zou H, Chen P, Wang X, Abrams JM. Dark is a Drosophila homologue of Apaf-1/CED-4 and functions in an evolutionarily conserved death pathway. Nat Cell Biol. 1999;1:272–279. doi: 10.1038/12984. [DOI] [PubMed] [Google Scholar]
- 23.Kanuka H, Sawamoto K, Inohara N, Matsuno K, Okano H, Miura M. Control of the cell death pathway by Dapaf-1, a Drosophila Apaf-1/CED-4-related caspase activator. Mol Cell. 1999;4:757–769. doi: 10.1016/s1097-2765(00)80386-x. [DOI] [PubMed] [Google Scholar]
- 24.Zhou L, Song Z, Tittel J, Steller H. HAC-1, a Drosophila homolog of APAF-1 and CED-4 functions in developmental and radiation-induced apoptosis. Mol Cell. 1999;4:745–755. doi: 10.1016/s1097-2765(00)80385-8. [DOI] [PubMed] [Google Scholar]
- 25.Srivastava M, Scherr H, Lackey M, Xu D, Chen Z, Lu J, et al. ARK, the Apaf-1 related killer in Drosophila, requires diverse domains for its apoptotic activity. Cell Death Differ. 2007;14:92–102. doi: 10.1038/sj.cdd.4401931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Snipas SJ, Drag M, Stennicke HR, Salvesen GS. Activation mechanism and substrate specificity of the Drosophila initiator caspase DRONC. Cell Death Differ. 2008;15:938–945. doi: 10.1038/cdd.2008.23. [DOI] [PubMed] [Google Scholar]
- 27.Dorstyn L, Kumar S. A biochemical analysis of the activation of the Drosophila caspase DRONC. Cell Death Differ. 2008;15:461–470. doi: 10.1038/sj.cdd.4402288. [DOI] [PubMed] [Google Scholar]
- 28.Meier P, Silke J, Leevers SJ, Evan GI. The Drosophila caspase DRONC is regulated by DIAP1. EMBO J. 2000;19:598–611. doi: 10.1093/emboj/19.4.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chai J, Yan N, Huh JR, Wu JW, Li W, Hay BA, et al. Molecular mechanism of Reaper-Grim-Hid-mediated suppression of DIAP1-dependent Dronc ubiquitination. Nat Struct Biol. 2003;10:892–898. doi: 10.1038/nsb989. [DOI] [PubMed] [Google Scholar]
- 30.Wang SL, Hawkins CJ, Yoo SJ, Muller HA, Hay BA. The Drosophila caspase inhibitor DIAP1 is essential for cell survival and is negatively regulated by HID. Cell. 1999;98:453–463. doi: 10.1016/s0092-8674(00)81974-1. [DOI] [PubMed] [Google Scholar]
- 31.Lisi S, Mazzon I, White K. Diverse domains of THREAD/DIAP1 are required to inhibit apoptosis induced by REAPER and HID in Drosophila. Genetics. 2000;154:669–678. doi: 10.1093/genetics/154.2.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Goyal L, McCall K, Agapite J, Hartwieg E, Steller H. Induction of apoptosis by Drosophila reaper, hid and grim through inhibition of IAP function. EMBO J. 2000;19:589–597. doi: 10.1093/emboj/19.4.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leulier F, Lhocine N, Lemaitre B, Meier P. The Drosophila inhibitor of apoptosis protein DIAP2 functions in innate immunity and is essential to resist Gram-negative bacterial infection. Mol Cell Biol. 2006;26:7821–7831. doi: 10.1128/MCB.00548-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Huh JR, Foe I, Muro I, Chen CH, Seol JH, Yoo SJ, et al. The Drosophila inhibitor of apoptosis (IAP) DIAP2 is dispensable for cell survival, required for the innate immune response to Gram-negative bacterial infection, and can be negatively regulated by the reaper/hid/grim family of IAP-binding apoptosis inducers. J Biol Chem. 2007;282:2056–2068. doi: 10.1074/jbc.M608051200. [DOI] [PubMed] [Google Scholar]
- 35.Arama E, Bader M, Rieckhof GE, Steller H. A ubiquitin ligase complex regulates caspase activation during sperm differentiation in Drosophila. PLoS Biol. 2007;5:e251. doi: 10.1371/journal.pbio.0050251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vernooy SY, Chow V, Su J, Verbrugghe K, Yang J, Cole S, et al. Drosophila Bruce can potently suppress Rpr- and Grim-dependent but not Hid-dependent cell death. Curr Biol. 2002;12:1164–1168. doi: 10.1016/s0960-9822(02)00935-1. [DOI] [PubMed] [Google Scholar]
- 37.Jones G, Jones D, Zhou L, Steller H, Chu Y. Deterin, a new inhibitor of apoptosis from Drosophila melanogaster. J Biol Chem. 2000;275:22157–22165. doi: 10.1074/jbc.M000369200. [DOI] [PubMed] [Google Scholar]
- 38.Zachariou A, Tenev T, Goyal L, Agapite J, Steller H, Meier P. IAP-antagonists exhibit non-redundant modes of action through differential DIAP1 binding. EMBO J. 2003;22:6642–6652. doi: 10.1093/emboj/cdg617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.White K, Grether ME, Abrams JM, Young L, Farrell K, Steller H. Genetic control of programmed cell death in Drosophila. Science. 1994;264:677–683. doi: 10.1126/science.8171319. [DOI] [PubMed] [Google Scholar]
- 40.Grether ME, Abrams JM, Agapite J, White K, Steller H. The head involution defective gene of Drosophila melanogaster functions in programmed cell death. Genes Dev. 1995;9:1694–1708. doi: 10.1101/gad.9.14.1694. [DOI] [PubMed] [Google Scholar]
- 41.Chen P, Nordstrom W, Gish B, Abrams JM. grim, a novel cell death gene in Drosophila. Genes Dev. 1996;10:1773–1782. doi: 10.1101/gad.10.14.1773. [DOI] [PubMed] [Google Scholar]
- 42.Tenev T, Zachariou A, Wilson R, Paul A, Meier P. Jafrac2 is an IAP antagonist that promotes cell death by liberating Dronc from DIAP1. EMBO J. 2002;21:5118–5129. doi: 10.1093/emboj/cdf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Challa M, Malladi S, Pellock BJ, Dresnek D, Varadarajan S, Yin YW, et al. Drosophila Omi, a mitochondrial-localized IAP antagonist and proapoptotic serine protease. EMBO J. 2007;26:3144–3156. doi: 10.1038/sj.emboj.7601745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang Y, Werz C, Xu D, Chen Z, Li Y, Hafen E, et al. Drosophila cbl is essential for control of cell death and cell differentiation during eye development. PLoS ONE. 2008;3:e1447. doi: 10.1371/journal.pone.0001447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bergmann A, Agapite J, McCall K, Steller H. The Drosophila gene hid is a direct molecular target of Ras-dependent survival signaling. Cell. 1998;95:331–341. doi: 10.1016/s0092-8674(00)81765-1. [DOI] [PubMed] [Google Scholar]
- 46.Kurada P, White K. Ras promotes cell survival in Drosophila by downregulating hid expression. Cell. 1998;95:319–329. doi: 10.1016/s0092-8674(00)81764-x. [DOI] [PubMed] [Google Scholar]
- 47.Jiang C, Lamblin AF, Steller H, Thummel CS. A steroid-triggered transcriptional hierarchy controls salivary gland cell death during Drosophila metamorphosis. Mol Cell. 2000;5:445–455. doi: 10.1016/s1097-2765(00)80439-6. [DOI] [PubMed] [Google Scholar]
- 48.Brodsky MH, Nordstrom W, Tsang G, Kwan E, Rubin GM, Abrams JM. Drosophila p53 binds a damage response element at the reaper locus. Cell. 2000;101:103–113. doi: 10.1016/S0092-8674(00)80627-3. [DOI] [PubMed] [Google Scholar]
- 49.Werz C, Lee TV, Lee PL, Lackey M, Bolduc C, Stein DS, et al. Mis-specified cells die by an active gene-directed process, and inhibition of this death results in cell fate transformation in Drosophila. Development. 2005;132:5343–5352. doi: 10.1242/dev.02150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ryoo HD, Domingos PM, Kang MJ, Steller H. Unfolded protein response in a Drosophila model for retinal degeneration. EMBO J. 2007;26:242–252. doi: 10.1038/sj.emboj.7601477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yoo SJ, Huh JR, Muro I, Yu H, Wang L, Wang SL, et al. Hid, Rpr and Grim negatively regulate DIAP1 levels through distinct mechanisms. Nat Cell Biol. 2002;4:416–424. doi: 10.1038/ncb793. [DOI] [PubMed] [Google Scholar]
- 52.Holley CL, Olson MR, Colon-Ramos DA, Kornbluth S. Reaper eliminates IAP proteins through stimulated IAP degradation and generalized translational inhibition. Nat Cell Biol. 2002;4:439–444. doi: 10.1038/ncb798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Srinivasula SM, Hegde R, Saleh A, Datta P, Shiozaki E, Chai J, et al. A conserved XIAP-interaction motif in caspase-9 and Smac/DIABLO regulates caspase activity and apoptosis. Nature. 2001;410:112–116. doi: 10.1038/35065125. [DOI] [PubMed] [Google Scholar]
- 54.Tenev T, Zachariou A, Wilson R, Ditzel M, Meier P. IAPs are functionally non-equivalent and regulate effector caspases through distinct mechanisms. Nat Cell Biol. 2005;7:70–77. doi: 10.1038/ncb1204. [DOI] [PubMed] [Google Scholar]
- 55.Hays R, Wickline L, Cagan R. Morgue mediates apoptosis in the Drosophila melanogaster retina by promoting degradation of DIAP1. Nat Cell Biol. 2002;4:425–431. doi: 10.1038/ncb794. [DOI] [PubMed] [Google Scholar]
- 56.Wing JP, Schreader BA, Yokokura T, Wang Y, Andrews PS, Huseinovic N, et al. Drosophila Morgue is an F box/ubiquitin conjugase domain protein important for grim-reaper mediated apoptosis. Nat Cell Biol. 2002;4:451–456. doi: 10.1038/ncb800. [DOI] [PubMed] [Google Scholar]
- 57.Pfleger CM, Harvey KF, Yan H, Hariharan IK. Mutation of the gene encoding the ubiquitin activating enzyme Uba1 causes tissue overgrowth in Drosophila. Fly. 2007;1:95–105. doi: 10.4161/fly.4285. [DOI] [PubMed] [Google Scholar]
- 58.Ravid T, Hochstrasser M. Diversity of degradation signals in the ubiquitin–proteasome system. Nat Rev Mol Cell Biol. 2008;9:679–690. doi: 10.1038/nrm2468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Varshavsky A. The N-end rule at atomic resolution. Nat Struct Mol Biol. 2008;15:1238–1240. doi: 10.1038/nsmb1208-1238. [DOI] [PubMed] [Google Scholar]
- 60.Ditzel M, Wilson R, Tenev T, Zachariou A, Paul A, Deas E, et al. Degradation of DIAP1 by the N-end rule pathway is essential for regulating apoptosis. Nat Cell Biol. 2003;5:467–473. doi: 10.1038/ncb984. [DOI] [PubMed] [Google Scholar]
- 61.Yokokura T, Dresnek D, Huseinovic N, Lisi S, Abdelwahid E, Bangs P, et al. Dissection of DIAP1 functional domains via a mutant replacement strategy. J Biol Chem. 2004;279:52603–52612. doi: 10.1074/jbc.M409691200. [DOI] [PubMed] [Google Scholar]
- 62.Herman-Bachinsky Y, Ryoo HD, Ciechanover A, Gonen H. Regulation of the Drosophila ubiquitin ligase DIAP1 is mediated via several distinct ubiquitin system pathways. Cell Death Differ. 2007;14:861–871. doi: 10.1038/sj.cdd.4402079. [DOI] [PubMed] [Google Scholar]
- 63.Ryoo HD, Gorenc T, Steller H. Apoptotic cells can induce compensatory cell proliferation through the JNK and the Wingless signaling pathways. Dev Cell. 2004;7:491–501. doi: 10.1016/j.devcel.2004.08.019. [DOI] [PubMed] [Google Scholar]
- 64.Wilson R, Goyal L, Ditzel M, Zachariou A, Baker DA, Agapite J, et al. The DIAP1 RING finger mediates ubiquitination of Dronc and is indispensable for regulating apoptosis. Nat Cell Biol. 2002;4:445–450. doi: 10.1038/ncb799. [DOI] [PubMed] [Google Scholar]
- 65.Muro I, Hay BA, Clem RJ. The Drosophila DIAP1 protein is required to prevent accumulation of a continuously generated, processed form of the apical caspase DRONC. J Biol Chem. 2002;277:49644–49650. doi: 10.1074/jbc.M203464200. [DOI] [PubMed] [Google Scholar]
- 66.Huh JR, Guo M, Hay BA. Compensatory proliferation induced by cell death in the Drosophila wing disc requires activity of the apical cell death caspase Dronc in a nonapoptotic role. Curr Biol. 2004;14:1262–1266. doi: 10.1016/j.cub.2004.06.015. [DOI] [PubMed] [Google Scholar]
- 67.Perez-Garijo A, Martin FA, Morata G. Caspase inhibition during apoptosis causes abnormal signalling and developmental aberrations in Drosophila. Development. 2004;131:5591–5598. doi: 10.1242/dev.01432. [DOI] [PubMed] [Google Scholar]
- 68.Shapiro PJ, Hsu HH, Jung H, Robbins ES, Ryoo HD. Regulation of the Drosophila apoptosome through feedback inhibition. Nat Cell Biol. 2008;10:1440–1446. doi: 10.1038/ncb1803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Ditzel M, Broemer M, Tenev T, Bolduc C, Lee TV, Rigbolt KTG, et al. Inactivation of effector caspases through non-degradative polyubiquitylation. Mol Cell. 2008;32:540–553. doi: 10.1016/j.molcel.2008.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Ribeiro PS, Kuranaga E, Tenev T, Leulier F, Miura M, Meier P. DIAP2 functions as a mechanism-based regulator of drICE that contributes to the caspase activity threshold in living cells. J Cell Biol. 2007;179:1467–1480. doi: 10.1083/jcb.200706027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Olson MR, Holley CL, Yoo SJ, Huh JR, Hay BA, Kornbluth S. Reaper is regulated by IAP-mediated ubiquitination. J Biol Chem. 2003;278:4028–4034. doi: 10.1074/jbc.M209734200. [DOI] [PubMed] [Google Scholar]
- 72.Kuranaga E, Kanuka H, Igaki T, Sawamoto K, Ichijo H, Okano H, et al. Reaper-mediated inhibition of DIAP1-induced DTRAF1 degradation results in activation of JNK in Drosophila. Nat Cell Biol. 2002;4:705–710. doi: 10.1038/ncb842. [DOI] [PubMed] [Google Scholar]
- 73.Galluzzi L, Joza N, Tasdemir E, Maiuri MC, Hengartner M, Abrams JM, et al. No death without life: vital functions of apoptotic effectors. Cell Death Differ. 2008;15:1113–1123. doi: 10.1038/cdd.2008.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Arama E, Bader M, Srivastava M, Bergmann A, Steller H. The two Drosophila cytochrome C proteins can function in both respiration and caspase activation. EMBO J. 2006;25:232–243. doi: 10.1038/sj.emboj.7600920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Copeland JM, Bosdet I, Freeman JD, Guo M, Gorski SM, Hay BA. echinus, required for interommatidial cell sorting and cell death in the Drosophila pupal retina, encodes a protein with homology to ubiquitin-specific proteases. BMC Dev Biol. 2007;7:82. doi: 10.1186/1471-213X-7-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ribaya JP, Ranmuthu M, Copeland J, Boyarskiy S, Blair AP, Hay B, et al. The deubiquitinase emperor's thumb is a regulator of apoptosis in Drosophila. Dev Biol. 2009;329:25–35. doi: 10.1016/j.ydbio.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 77.Broemer M, Meier P. Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol. 2009;19:130–140. doi: 10.1016/j.tcb.2009.01.004. [DOI] [PubMed] [Google Scholar]