Abstract
Toxoplasma gondii is an apicomplexan of both medical and veterinary importance which is classified as an NIH Category B priority pathogen. It is best known for its ability to cause congenital infection in immune competent hosts and encephalitis in immune compromised hosts. The highly stable and specialized microtubule-based cytoskeleton participates in the invasion process. The genome encodes three isoforms of both α- and β-tubulin and we show that the tubulin is extensively altered by specific post-translational modifications (PTMs) in this paper. T. gondii tubulin PTMs were analyzed by mass spectrometry and immunolabeling using specific antibodies. The PTMs identified on α-tubulin included acetylation of Lys40, removal of the last C-terminal amino acid residue Tyr453 (detyrosinated tubulin) and truncation of the last five amino acid residues. Polyglutamylation was detected on both α- and β-tubulins. An antibody directed against mammalian α-tubulin lacking the last two C-terminal residues (Δ2-tubulin) labeled the apical region of this parasite. Detyrosinated tubulin was diffusely present in subpellicular microtubules and displayed an apparent accumulation at the basal end. Methylation, a PTM not previously described on tubulin, was also detected. Methylated tubulins were not detected in the host cells, human foreskin fibroblasts, suggesting that this may be a modification specific to the Apicomplexa.
Keywords: Toxoplasma gondii, cytoskeleton, tubulin, post-translational modification, proteomics, microtubules, conoid
INTRODUCTION
Toxoplasma gondii (T. gondii), a member of the phylum Apicomplexa, is an obligate intracellular parasite and an important human pathogen. Its ability to be transmitted by water and food has resulted in its being classified as a category B biodefense pathogen. The apicomplexan microtubule cytoskeleton is important for maintenance of cell shape and for host cell invasion.1 It forms unusual specialized sub-membranous and apical structures that are highly stable.2 The stability and the complexity of these structures are likely due to both the accumulation of multiple posttranslational modifications (PTM) on the α- and β-tubulin heterodimers that form microtubules and the presence of specific microtubule associated proteins. Indeed, several lines of evidence strongly indicate that the different combinations of posttranslational modifications of tubulins constitute a code that defines which proteins are recruited onto microtubules.3 Except for acetylation of α-tubulins at lysine 40 4, most posttranslational modifications of both α- and β-tubulin occur at their rather acidic carboxy termini, with polyglutamylation and polyglycylation being reported for both α- and β-tubulin, and detyrosination/tyrosination and removal of the penultimate glutamate residue for α-tubulin.3, 5, 6
Both α- and β-tubulins are usually encoded by multigene families. Although tubulins were initially considered to be single-genes in T. gondii 7, genomic data available at www.toxodb.org suggests that two additional α- and two additional β-tubulin genes exist in this organism (Fig. 1).8, 9 The newly characterized α-tubulin genes are only 68% and 40% identical to the α1 gene (583.m00022), while the β-tubulin genes are as high as 97% identical to β1 gene (57.m00003), with a highly divergent C-terminal tail. These tubulin isotypes may be stage specific, (i.e., only expressed in tachyzoites, bradyzoites, gametes or oocytes) or they may be expressed in specific structures (i.e. the apical polar ring, conoid, or flagella of male gametes).
Figure 1. Alignment of T. gondii α- and β-tubulin sequences.
Labels refer to release-4 gene predictions that are used in the Einstein EPICDB database 8. Tubulin subtypes are shown. Color coding: red: total conservation, identical in both subgroups, both in alpha and beta tubulin; blue: subgroup specific conservation i.e. identical within a subgroup but different from the other subgroup; green: conserved position (at least 7 out of 10 physico-chemical properties are shared by the amino acids occurring in the position) white: positions not conserved. The alignment was prepared by AMAS and CLUSTALW program 26, 27.
The diversification of tubulin isotypes and of their posttranslational modifications may contribute to the shape of the parasites and their unique membrane/cytoskeletal structure. Thus, a detailed analysis of tubulin expression profiles should further expand our understanding of the role of microtubule cytoskeleton in T. gondii cell structure and invasion.
A variety of antibodies have been used to determine PTM of tubulins in different species.10–13 Plessmann et al. described acetylation, detyrosination and polyglutamylation of α-tubulin from T. gondii trachyzoites using such antibodies in combination with mass spectrometry analysis of α-tubulin C-terminal peptides.10 Mass spectrometry-based analysis revealed that side chains of up to three glutamate residues were added on glutamate 445 of its primary sequence and that the five last amino-acids could be removed, presumably by a proteolytic process.10 In the present study, we detected the expression of one α-tubulin and two β-tubulins and their PTMs in T. gondii tachyzoites using both mass spectrometry and antibodies. In addition to previously described PTMs of tubulins, we found that both α- and β-tubulins were methylated on their C-termini. Methylation has not previously been described for tubulins from other organisms and potentially represents a specific tubulin PTM in T. gondii and other apicomplexan parasites.
MATERIALS AND METHODS
Growth of Toxoplasma gondii in vitro
RH strain T. gondii were grown in human foreskin fibroblasts (HFF) in DME media with 10% fetal calf serum and 1% Penicillin Streptomycin (GIBCO-BRL) in a 5% CO2 incubator. Parasites were transferred to fresh cultured human fibroblast monolayers biweekly.
Purification of membrane and cytoskeleton protein fractions from Toxoplasma gondii
1.2×1010 purified Toxoplasma gondii RH strain tachyzoites from tissue culture were resuspended in 20 ml of SMDI buffer (250 mM Sucrose, 10 mM MOPS-KOH, pH 7.2, 2mM DTT, 1X protease inhibitor cocktail) and disrupted by French press at a pressure of 1000 PSI, medium setting. The lysate was centrifuged at 756 × g at 4°C for 10 min to pellet unbroken cells. Intact parasites and large debris were suspended in 10 ml SMDI buffer and disrupted once more by French press at a pressure of 1000 PSI, medium setting. The pooled supernatant was centrifuged at 25,000 × g at 4°C for 20 min. The supernatant was saved for analysis as the cytosolic fraction. The pellet was suspended in 10 ml of 30% Percoll in SMDI buffer. After centrifugation at 75,000 × g in an ultracentrifuge (Rotor TLA 100.3; 30,000 rpm) at 4°C for 25 min, the top band was collected from the self-generated gradient. The band was diluted in SMDI buffer and spun at 100,000 × g for 90min. at 4°C (Rotor TLA 100.3; 40,000 rpm). A band collected between the buffer and resultant Percoll cushion contained the T. gondii ghosts consisting of membranes and cytoskeleton.
To remove the membrane fraction, T. gondii ghosts were suspended in an equal volume of 2% thioglucopyranoside in 40 mM Tris pH 7.6 by pipeting the mixture up and down 10 times (suspension kept on ice) followed by a brief vortex. After centrifugation at 20,000 × g in an Eppendorf centrifuge for 20 min. at 4°C, the supernatant was saved as the membrane fraction (extraction 1). This extraction was repeated twice with 300 μl of 1% thioglucopyranoside (to make extractions 2 and 3). The insoluble material which remained after the membrane fraction extraction was washed twice with 40 mM Tris pH 7.6. Following this washing step the material was solubilized in 500 μl of urea lysis buffer (7.5 M urea, 2.5 M thiourea, 40 mM Tris pH 7.6, 2.5% octyl-β-glucoside, 6.25 mM TCEP, 1.25 × protease inhibitor) followed by homogenization on ice 10 times using a Potter homogenizer. The material was then centrifuged at 8,000 rpm for 10 min at 4°C and the supernatant was collected and saved. This extraction was repeated twice on the remaining pellet using 300 μl of the urea lysis buffer and the supernatants were pooled with the initial extraction to produce the cytoskeleton fraction. Virtually no insoluble material remained following the third extraction. The pooled extractions (i.e., cytoskeleton fraction) were then frozen in liquid nitrogen and stored at −80°C until use for protein analysis.
Isoelectric Focusing
Isoelectric focusing was performed with 24 cm Immobiline DryStrips pH 4.5–5.5, (GE Healthcare). 50 μl of sample in 450 μl of IPG buffer (8M urea, 2M thiourea, 4% CHAPS, 0.5% TX-100, 0.5% ampholytes (pH 4.5–5.5), 20mM DTT and a few grains bromphenol Blue) was incubated for 30 min at room temperature and then spun in an Eppendorf centrifuge at 12,000 rpm for 3 min. The supernatant was loaded on the IPG strip using Ettan IPGphor 2 (Amersham Biosciences) with a focusing time of 28 hrs (96,000 volts-hours total). The focused IPG strips were stored at −20°C in glass tubes.
2D-electrophoresis
Prior to second dimension SDS-PAGE, the focused IPG strips were incubated twice with Tris acetate equilibration buffer (PRC, Inc, 75 mM Tris, 6M urea, 30% glycerol, 0.25% DTT) for 10 minutes. Following this reduction step was alkylation with iodoacetamide (twice of 10 min incubation in 75 mM Tris, 6M urea, 30% glycerol and 2.5% iodoacetamide). The IPG strip was then placed on the top of a 10% Criterion precast gel (Bio-Rad) and run for 2.5 hr at a constant 150V(POWER/PAC 3000 Bio-Rad) in a Criterion Dodeca Cell tank(Bio-Rad) using a standard Tris-glycine SDS running buffer at 12°C. The SDS- PAGE gel was stained with GelCode Blue Stain Reagent (Pierce).
Immunoblotting
SDS-PAGE electrophoresis was performed using 10% precast Tris- HCl Gels for 1D and 10% Criterion Tris-HCl Gels for 2D (Bio-Rad, Hercules, CA) using the Bio-Rad Mini- Protean gel electrophoresis system. Immunoblot analysis was performed by electroblotting SDS-PAGE-resolved proteins onto transfer PVDF Membrane (Millipore) using the Bio-Rad Mini and Midi Polyacrylamide Gel System. All PVDF Membranes were blocked with 3% milk in TBS-Tween pH 7.4 for 1 hour and then incubated with primary antibodies diluted 1:1000 to 1:3000 (see below) in 1% BSA in TBS-Tween for rabbit antibodies and 3% milk solution for mouse antibodies. Following this incubation, membranes were washed for 15 min in TBS-T three times and then incubated with a specific secondary antibody for 45 min (1:3000 anti- mouse IgG-HRP in 3% milk in TBS-T, 1:10,000 anti-rat IgG or 1:2500 anti-rabbit IgG in 1%BSA in TBS-T) followed by three 15 min washes in TBS-T. Bound antibodies were then detected using SuperSignal West Pico Chemiluminescent Substrate (Thermo scientific) and Blue Basic Autorad Film (ISC BioExpess).
Immunoblots were probed with successive antibodies. To remove prior antibody reactions the transfer PVDF Membrane (Millipore, MA) were stripped using Restore Western Blot Stripping Buffer (Thermo Scientific, Waltham, MA) by incubation of the membrane with 15 mL of buffer 10 min at room temperature, following this incubation the membranes were washed twice for 10 min each time with TBS-Tween pH 7.4 and blocked with 3% non-fat dry milk in TBS-Tween for 2 hours. Following stripping membranes were probed with antibodies as described above.
The following primary antibodies were used. Tubulin was detected using the DM1A and DM1B mouse monoclonal antibodies (1:2000 dilution; BioGenex Inc., CA), directed against the C-termini of α- and β-tubulins, respectively. Tubulin PTMs were detected with B3 mouse monoclonal antibody directed against polyglutamylated tubulin (1:1000 dilution; Sigma-Aldrich), with a rabbit polyclonal antibody against detyrosinated tubulin (or glu-tubulin antibody AB3201 1:1000 dilution, Millipore, MA), and with a rabbit polyclonal antibody against Δ2-tubulin (AB3203, 1:2500 dilution, Millipore, MA). Tubulin was also found to react with a polyclonal antibody to tri-methylated K20 histone H4 (AB9053, 1:2000 dilution, Abcam Inc., MA). Horse radish peroxidase-labelled polyclonal antibodies against mouse (1:3000 dilution), rat (1:10000 dilution) and rabbit IgGs (1:3000 dilution) were used as secondary antibodies (GE Healthcare).
Immunofluorescence assay (IFA) analysis
HFF cells were grown to confluence on sterilized coverslips in 24-well plates. Cells were fixed in 3% paraformaldehyde ~20–24 h after infection with tachyzoites expressing YFP-α-tubulin (gift of Dr. Ke Hu, Indiana University) and permeabilized in 0.25% Triton X-100. The following antibody reagents were used: mouse anti-GFP monoclonal antibody conjugated with FITC (Sc-9996, Santa Cruz Biotechnology, CA), rabbit anti-Δ2-TUB (AB3203, Millipore, MA), anti-deTyr-TUB (or anti-Glu-tubulin, AB3201, Millipore, MA) and H4K20me3 (ab9053, Abcam Inc., MA). All antibodies were diluted 1:500 in PBS 1x/3% BSA/Triton-X-100 and detected using either FITC-conjugated goat anti-mouse antibody, or Alexa-568–conjugated goat anti-rabbit antibody (A-11036; Molecular Probes, Eugene, OR). To stain DNA, coverslips were incubated in PBS containing 3 μg/ml Hoechst 33258 stain (Molecular Probes) for 5 min at room temperature followed by several brief washes in PBS. Coverslips were mounted with Antifade (Molecular Probes, Eugene, OR), and images were analyzed by high resolution fluorescence using deconvolution protocols. Microscopy was performed with an inverted microscope (IX81; Olympus) with filter 41001 (excitation 480–440 nm/emission 440 nm) for FITC, filter 41035 (excitation 540–525 nm/emission 620–660 nm) for rhodamine, and filter 31013v2 (excitation 360–340 nm/emission 460–450 nm) for DAPI.
Mass spectrometric analysis
After visualization, the bands containing tubulins (pH 4.5 to 5.5, and molecular weight about 50 kDa) were excised, destained and subjected to in-gel trypsin digestion or CNBr cleavage. Trypsin digestion of dried gel pieces was performed overnight at 37°C with 75 ng/μl trypsin (Sigma-Aldrich, St. Louis, MO) and trypsin Enhancer, ProteasMAX™ surfactant (Promega Corp. Madison, WI). For in-gel CNBr cleavage, the dried gel pieces were treated with 150 mg/mL CNBr in 70% formic acid overnight in the dark. CNBr and formic acid were removed by vacuum centrifugation in Speedvac through 3 cycles of drying and resuspension. The combined tryptic digestion and CNBr cleavage was performed using methods developed by Montfort et. al.14 Briefly, the trypsinized gel pieces were treated with 50 μl of 150 mg/ml CNBr in 70% TFA after washing and dehydration. The supernatant was collected and pooled with the following extractions: the peptides were extracted twice by 5 min sonication in 50 μl of 60% acetonitrile, 1% of TFA and 0.1% octyl-β-glucopyranoside (OBG). The pooled peptide solution was dried in a SpeedVac. The resulting CNBr or/and tryptic peptides were either mixed directly with matrixes (α-cyano-4-hydroxycinnamic acid, for both positive mode negative ion mode) or by LC-MALDI process, briefly, fractioned by reverse phase capillary HPLC (Ultimate 3000, Dionex, CA) followed by mixing with matrix automatically with a robot (Probot, Dionex, CA). Mass spectrometry analysis was carried out using MALDI TOF-TOF mass spectrometer (Applied Biosystems ABI 4800 PROTEOMICS TOF/TOF ANALYZER, Foster City, CA) in both positive and negative ion mode. While most mass spectra were obtained in reflector mode, some of the mass spectra were acquired in linear mode in order to maximize sensitivity at higher m/z range, e.g., figures 5A, 8 and 9. In LC-MALDI experiments, premixed α-cyano-4-hydroxycinnamic acid (G2037A, Agilent Tech.) was used as the matrix since it offered the best result in negative mode mass spectrometry. As previously published a predicted T. gondii proteome (http://toro.aecom.yu.edu/biodefense/), based on genome data available at www.toxodb.org, was used as the protein database 2.
Figure 5. MALDI mass spectrum of peptides of α-tubulins of T. gondii.
A. Positive ion mode MS from α-tubulin (583.m00022) after CNBr cleavage. Identified peptides are labeled with residue numbers from the sequence insert on the figure. The de-tyrosination and glutamylation sites are indicated by double arrows and molecular weight of amino acid residues. Spectra were acquired in linear mode.
B. The C-terminal CNBr and tryptic peptide of α-tubulin shows up to four glutamylations (in the negative mode). The identified peptides from tubulins are labelled using residue numbers. Glutamylation is indicated by double headed arrows. The residue weight of glutamate (129 Da) is indicated above the arrows. 1-4E indicates the number of glutamylations that occurred on each species. Spectra were acquired in reflector mode.
Figure 8. The C-terminal sequence and MALDI mass spectrum of tryptic peptides of α-tubulin (583.m00022) of T. gondii in the positive ion mode.
The peptides from tubulin are labeled using residue numbers. K40-ace 3-60 and K40-ace 1-60 are peptides containing acetylated Lysine 40, as explained in the text. Peaks with m/z of 5561 and 2781 are probably tryptic peptides of other proteins with amolecular weights similar to tubulin. Spectra were acquired in linear mode.
Figure 9. The MALDI mass spectrum of CNBr cleaved peptides of methylated tubulins in the negative ion mode.
The identified peptides from tubulins are labelled using residue numbers and sequence. 1me to 6 me indicates the number of methylations detected on each species identified. The Y axes (relative intensity) are omitted. Spectra were acquired in linear mode.
RESULTS
Tubulins in T. gondii identified by 2D gel and mass spectrometry
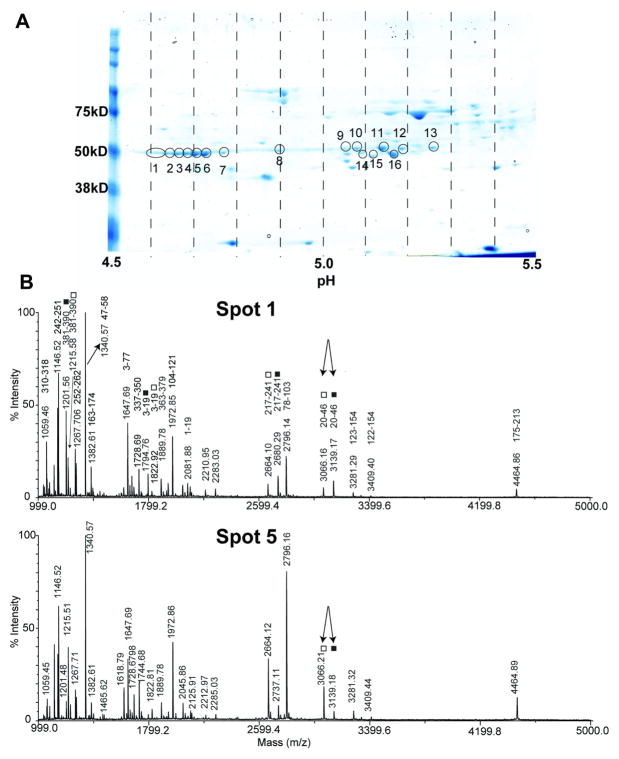
The proteins were separated by 2D gel electrophoresis and subjected to in-gel trypsin digestion and mass spectrometry analysis. Only the spots with an observed molecular mass of 50 kDa and pH of 4.5 to 5.5 in these 2D gels were indentified as tubulins (Fig. 2A). Based on the genome sequence (www.toxodb.org) there are a total of six potential tubulin isotypes in T. gondii. We observed more than six spots in 2D gels and blots, suggesting that there are extensive post-translational modifications of T. gondii tubulins. We identified 2 β-tubulin isotypes and 1 α-tubulin isotype from T. gondii tachyzoites(Table 1). Spots 1, 3, 4, 5, 6 and 7 were identified as β-tubulins with both isotypes 57.m00003 and 41.m00036, while spot 9 through 13 were identified as α-tubulin isotype 583.m00022. This distribution is consistent with the relative pI values of β-tubulins and α-tubulin (Table 1). Spots 2 and 8 contain not only β-tubulins, but also α-tubulin, suggesting that PTMs such as glutamylations occurring on α-tubulin could have shifted it to more acidic pIs (around 4.64 for spot 2 and 4.90 for spot 8, estimated from the pH line).
Figure 2. 2D gel analysis of cytoskeletal tubulins using mass spectrometry.
A. The spots identified as tubulins are circled. Spots 1, 3, 4, 5, 6, 7 are β-tubulin (both isotypes 57.m00003 and 41.m00036), 9 through 13 are α-tubulin (583.m00022), 2 and 8 are mixed α-& β-tubulin. The smaller circled spots 14 through 16 contain only small content of α-tubulin accompanying other protein species. The dotted lines indicate the pH values, each line represents 0.1 pH unit. B. MALDI-TOF mass spectra in the positive mode of spot 1 (top) and spot 5 (bottom) from 2D gel after trypsin digestion on the left panel. Each identified peptide is labelled with residue numbers. Specific peptides for β-tubulin isotypes 57.m00003 and 41.m00036 are indicated with filled square ■ and open square □, respectively. The peptide [20-46] from both isotypes is used to calculate the relative quantity as discussed in the text (arrow). Spectra were acquired in reflector mode.
Table 1.
Tubulins identified by 2D-gel and mass spectrometry in tachyzoites of T. gondii
| Gene | protein | MW | pI | C-terminal peptide sequence and m/z |
|---|---|---|---|---|
| 57.m00003 TgTwinScan _3222 |
β-tubulin | 50073.3 | 4.69 |
416NDLVSEYQQYQDATAEEEGEFDEEEGEh443 * 3236.35 (mono, positive), 3238.20 (avg, positive) 3234. 33 (mono, negative), 3236.19 (Avg, negative) |
| 41.m00036 TgTwinScan _2537 |
β-tubulin | 50037.2 | 4.70 |
416NDLVSEYQQYQDATAEEEGEFDDEDGGh443 * 3136.23(mono, positive), 3138.08(avg, positive) 3134.22 (mono, negative), 3136.07 (avg, negative) |
| 583.m00022 TgTwinScan _5114 |
α-tubulin | 50113.8 | 4.98 |
414EEGEFSEAREDLAALEKDYEEVGIETAEGEG EEEGYGDEY453 4472.87 (mono, positive), 4475.54 (avg, positive) 4470.86 (mono, negative), 4473.53 (avg, negative) 431DYEEVGIETAEGEGEEEGYGDEY453 2568.99 (mono, positive), 2566.97 (mono, negative) |
h represents homoserine lactone converted from methionine during CNBr cleavage.
All of the β-tubulin isotypes were found to overlap and no single isotype was present in any spot. Two isotype-specific peptides were selected to calculate the relative content of each β-tubulin isotype. The sequence of peptide 20FWEVISDEHGIDPTGTYCGDSDLQLER46 (m/z 3139.17 in figure 2) from β-tubulin 57.m00003 is almost identical to the peptide 20FWEVISDEHGVDPTGTYTGDSD LQLER46 (m/z 3066.16 in figure 2) from 41.m00036 except for two amino acid residues (I for V and C for T). It is unlikely that the switch of these two amino acids with similar polarity would significantly change the ionization efficiency of these two peptides; therefore the relative amount of the two peptides should be represented by their relative intensities. In all spots, except spots 2 and 5, the majority of the β-isotype was 57.m00003 which ranged from 1.9 fold to 8 fold of the concentration of the other isotype, 41.m00036, as judged by the relative intensity of isotype-specific peptides (table 2 and Fig. 2B). The exception was spot 5 which contained more of the β-isotype 41.m00036 than 57.m00003 (3.4:1, Fig. 2B). This agrees very well with the theoretical pI of 41.m00036 (4.70). Spots 9 to 13 were identified as α-tubulin isotype 583.m00022 with no trace of other α-tubulin isotypes, suggesting that this isotype undergoes a variety of modifications. The smaller circled spots 14 to 16 in the vicinity of spots 9 to 13 also contain α-tubulin; however, tubulins were not the major component in these spots. It is possible that the tubulin in spots 14–16 is due to contamination from adjacent tubulin spots and due to the low tubulin content in spots 14 –16, these data are not included in table 2. The α-tubulin 44.m02659 has 32 additional amino acids at its carboxy-terminus giving it a calculated molecular weight of 53.8 kDa compared to the molecular weight of 50.1 kDa calculated for α-tubulin 583.m00022 (Fig. 1). This suggests that α-tubulin 44.m02659 would be found a little above the 50 kDa line in the 2D gel (i.e. just above spots 11 and 12 (Fig. 2A). We did not observe this isoform in our 2D gel and mass spectrometry experiment either due to its relatively low content or absence in the tachyzoite life stage.
Table 2.
Content of 2D gel spots determined by mass spectrometry
| gene\spot | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| β57.m00003 | + 1.9* | + 1 | +2.9 | +3.8 | + 1 | + 8 | +3.8 | +3.8 | |||||
| β41.m00036 | + 1 | + 1 | + 1 | + 1 | +3.4 | + 1 | + 1 | + 1 | |||||
| α583.m00022 | + | + | + | + | + |
Posttranslational modifications of tubulins characterized by a combination of antibodies and mass spectrometry
A number of antibodies were used to examine possible posttranslational modifications (PTMs) occurring on T. gondii tubulins, and the PTMs were further confirmed by mass spectrometry. The localization of modified tubulins was also examined using immunofluorescence microscopy.
Truncated tubulin isoforms
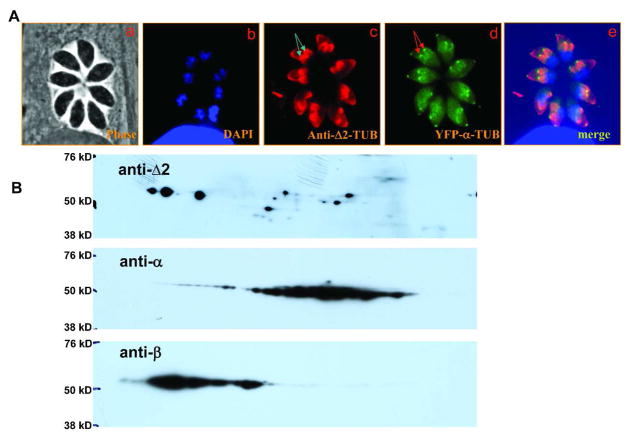
T. gondii tachyzoites were stained with anti-Δ2-tubulin, an antibody specific for α-tubulin lacking the C-terminal tyrosine and the penultimate glutamate residue. Most of the cell was labelled by anti-Δ2-tubulin, except the posterior end (Fig. 3A, panel c). The conoid end of the parasite can be recognized by a bright dot when T.gondii expressing YFP-α-tubulin is stained by anti-GFP (Fig. 3A, panel d).2 When the images of anti-Δ2-tubulin stained cells were superimposed on the cell images stained with YFP-α-tubulin (Fig. 3A, panel e), it can be appreciated that the highest intensity of staining is present in the anterior end, proximal to the conoid, and the intensity of staining gradually diminishes towards the posterior end of the cell. A bright stain is also seen at the apical region of daughter cells developing within the mother cell (Figure 3A, panel c and d, arrows). This suggests that Δ2-tubulin is present in T. gondii and accumulates in the anterior end. To examine which tubulin subunit was deglutamylated, we aligned the 2D-gel immunoblots of the extracted cytoskeleton fraction stained with anti-α- and anti-β-tubulin with that stained with anti-Δ2-tubulin (Fig. 3B). When utilizing mammalian tubulin this anti-Δ2-tubulin is specific for α-tubulin. Surprisingly, anti-Δ2-tubulin demonstrated labelling in both α-tubulin and β-tubulin regions. This may be due to the presence of T. gondii α-tubulin in areas of the gel in which β-tubulin is seen.
Figure 3. Analysis of Δ2-tubulin posttranslational modification of cytoskeletal tubulins using immunofluorescence microscopy and immunoblot.
A. Phase contrast image (a) corresponding to images b through e; immunofluorescence analysis of cells imaged with DAPI for DNA (b), anti-Δ2-tubulin (c), YFP-α-tubulin (d), and the merged image c and d (e). Note the bright staining of anti-Δ2-tubulin around the conoid region of daughter cells (arrows).
B. Immunoblot analysis using anti-Δ2-, anti-α-, and anti-β-tubulin. This demonstrates labelling of both α and β tubulin by anti-Δ2-tubulin. The same immunoblot was used for each antibody. The immunoblot was stripped between antibody incubations using a standard protocol.
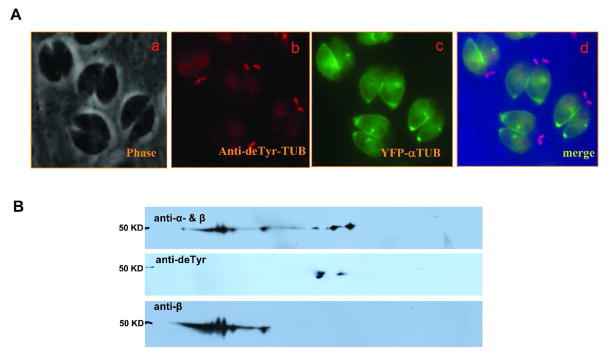
Anti-detyrosinated-tubulin antibody diffusely stained the subpellicular region of the T. gondii tachyzoites (Fig. 4A, panel a-d). This antibody had intense bright staining at the basal end of the cell. This strong labelling by anti-detyrosinated-tubulin antibody in the basal end of T. gondii tachyzoites was also reported in a recent study of α-tubulin.10 This antibody detected several spots of 50k Da in the α-tubulin region (Fig. 4B) of the immunoblot.
Figure 4. Analysis of deTyr-tubulin posttranslational modification of cytoskeletal tubulins using immunofluorescence microscopy and immunoblot.
A. Phase contrast image (a) corresponding to images b through d, immunofluorescence analysis of cells with anti-deTyr-tubulin (b), YFP-α-tubulin (c), and the merge of images of b and c (d). This demonstrates the localization of anti-deTyr-tubulin to the basal region of T. gondii.
B. Immunoblot analysis stained with anti-α- & β-tubulin, anti-deTyr-tubulin and anti-β-tubulin. This demonstrates labelling of α tubulin by anti—deTyr-tubulin. The same immunoblot was used for each antibody. The immunoblot was stripped between antibody incubations using a standard protocol.
Lys392 is the closest trypsin cleavage site from the C-termini of β-tubulins from T. gondii, generating a tryptic peptide with 57 amino acid residues, which is difficult to elute from a gel after in-gel digestion. CNBr cleavage and combined CNBr and trypsin digestion were, therefore, utilized to replace proteolysis, as it generates C-terminal peptides of tubulins in T. gondii which can be eluted from gels and detected by a MALDI TOF-TOF mass spectrometer (Table 1 and Fig. 5). In this work, all the C-terminal peptides of tubulins released by CNBr cleavage or combined CNBr and trypsin digestion were successfully detected by mass spectrometer in both positive and negative ion mode without transferring tubulin to nitrocellulose.
The ion peak corresponding to the C-terminal peptide 414-453 from α-tubulin was observed with an m/z value of 4474.91 (Fig. 5) that is consistent with the calculated average mass [MH+] of 4475.54 for this C-terminal peptide. It should be noted that the average m/z value was used here due to relatively low resolution in the higher m/z region, while at lower m/z region, the monoisotopic m/z value is more appropriate for accurate mass. A portion of this peptide underwent a de-tyrosination event at its C-terminus as evidenced by the peptide showing a mass of 4311.67, which is 163Da less than its tyrosinated form (Fig. 5A). The ion peak with a mass of 3846.83 was identified as the truncated form of the peptide [414-448] lacking the last five residues (Δ5-tubulin Fig. 5A).
Polyglutamylated tubulin isoforms
Three peptides to the right of the C-terminal α-tubulin peptide 414-453 had m/z values of 4604.04, 4733.44 and 4862.90, respectively (Fig. 5A). The difference between these peaks was 129 Da, suggesting that the tyrosinated forms of α-tubulin were polyglutamylated, and a maximum of three glutamylations was observed. The ion peak with a mass of 3846.83 (Δ5-tubulin C-terminal peptide) exhibited the same pattern of glutamylation, showing an addition of 1, 2, or 3 glutamates (Fig. 5A).
In a separate experiment of in-gel CNBr and tryptic digestion of α-tubulin, where an extra separation step by reverse phase HPLC was performed prior to acquisition of MALDI-TOF mass in the negative ion mode (LC-MALDI, see Materials and Methods), up to four glutamylations were detected in the C-terminal tryptic peptide (431-453, 583.m00022, Fig. 5B). The peptide with a mass of 2566.87 corresponded to the tyrosinated C-terminal peptide of α-tubulin, 583.m00022 (calculated mass is 2566.97 in negative mode). Three other peptides showed a mass of 2695.83, 2825.08, 2954.11 and 3082.98, respectively, suggesting that this C-terminal peptide contained isoforms with one, two, three and four glutamylations. The fourth glutamylated isoform was detected due to the increased signal to noise ratio or reduced signal suppression of the mass spectrum after reversed phase HPLC separation.
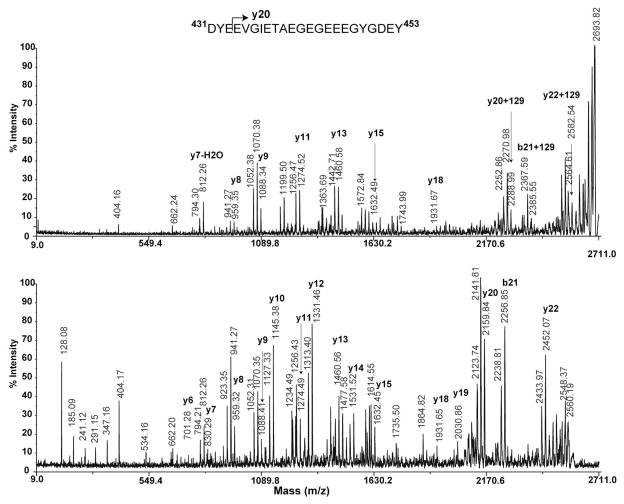
Two ion peaks, 2566.87 and 2695.83, were selected for MS/MS to determine the site of glutamylation on α-tubulin 583.m00002. The MS/MS spectra of both ions in the negative ion mode after collisionally induced dissociation (CID) in the source 2 region of 4800 TOF-TOF mass spectrometer are shown in figure 6. The unmodified C-terminal peptide (Fig. 6, bottom panel) showed almost consecutive y fragment ions from y6 to y22 with a couple of peptides missing. The singly glutamylated peptide (Fig. 6, upper panel) showed identical fragmentation pattern from y7 to y18. The y20 ion peak was shifted to higher mass with an additional 129 Da, indicating glutamylation on either E434 or V435. Since glutamylation occurs on glutamic acid, the glutamylation site in the C-terminal peptide of α-tubulin 583.m00002 should be E434 based on the data. It should be noted that most of the y ions appeared as a cluster in which one y ion was accompanied by two ions with masses of minus 18 and 36. This is due to H2O loss from these y ions when analyzed in the negative ion mode.
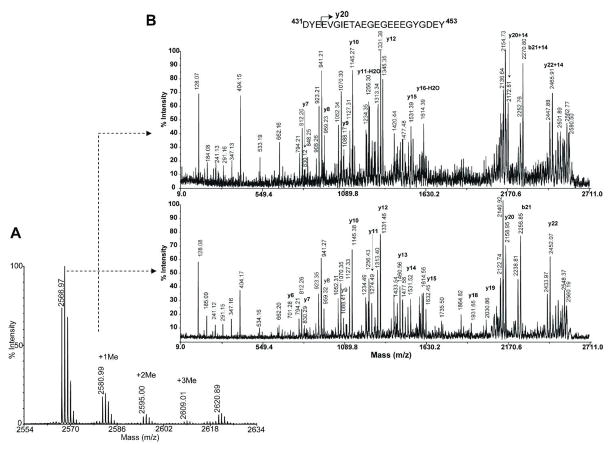
Figure 6. Comparison of fragmentation profiles of the C-terminal peptides of α-tubulin, 583.m00022 from T.gondii to determine the glutamylation site.
Bottom panel, Negative MALDI TOF-TOF MS/MS spectrum of the precursor 2566.87; Upper panel, Negative MALDI TOF-TOF MS/MS spectrum of the precursor 2695.83. The sequence shown is the C-terminal peptide from D431 to Y453 of α-tubulin, 583.m00022. Mainly y ions are shown in the mass spectra. The ions with masses of minus 18 and 36 are y ions having lost one or two H2O molecules. Spectra were acquired in reflector mode.
In β-tubulins, two peptides showing masses of 3134.42 and 3234.41 were identified as C-terminal peptides from 41.m00036 and 57.m00003, respectively (Table 1 and Fig. 7). Remarkably, extensive polyglutamylation was found on both of the β-isotypes 57.m00003 and the α-isotype 41.m00036, suggesting that glutamylation is present in all three of the T.gondii tubulin isotypes we identified. The bottom panel of figure 7 demonstrates up to three glutamylations occurring on β-isotype 41.m00036, represented by peptides with a mass of 3263.43, 3392.51 and 3521.56, respectively. The other β-isotype 57.m00003 showed six glutamylated species with the consecutive addition of 1, 2, 3, 4, 5, 6 glutamates on the C-terminal peptide 416-443 (Fig. 7, upper panel).
Figure 7. MALDI mass spectrum of CNBr cleaved C-terminal peptides of β-tubulins, 57.m00003 and 41.m00036 in the negative ion mode.
Glutamylation is indicated by double headed arrows. The residue weight of glutamate is indicated above each double arrow. 1-6E indicates the number of glutamylations that occurred on each species. The accession number of each C-terminal peptide is shown with each spectrum, and the C-terminal sequences are shown in Table I. The Y axes (relative intensity) are omitted for clarity. Spectra were acquired in reflector mode.
Using an anti-polyGlu-tubulin antibody, both α- and β-tubulins were shown to be polyglutamylated (supplemental material, figure S1).
Acetylation of α-tubulin
Of the three isoforms of α-tubulin in the T.gondii genome only 583.m00022 and 44.m02659 contain an acetylatable lysine residue. Acetylation in lysine 40 of α-tubulin 583.m00022 results in a blocked cleavage at Lys-40, so there should be no corresponding mass of the tryptic peptide [1-40, m/z 4524.21] or [3-40, m/z 4236.82] of α-tubulin. Instead, the tryptic peptide with one extra missed cleavage [1-60] or [3-60] should show up in the mass spectrum of trypsin digestion of α-tubulin (Fig. 8). Indeed, the two peptides corresponding to the modified peptide [1-60] and [3-60] were observed in the mass spectrum of one of the α-tubulin spots in the 2D gel (spot 11, Fig. 2). The average masses of [1-60] and [3-60] alone are 6498.29 and 6210.91, respectively, which become 6540.30 and 6252.92 after acetylation. The two peaks 6540.58 (+40 ppm) and 6253.11 (+30ppm) in the mass spectrum of figure 8 are consistent with acetylation of α-tubulin (583.m00022).
Methylated tubulin isoforms
Methylated tubulin C-terminal peptides were detected in our mass spectrometry experiments using CNBr cleavage of proteins separated on IPG strips. Figures 8a & b show the C-terminal peptides of α-tubulin 583.m00022 acquired in the negative ion mode. The major peptide in figure 9b had a mass of 4471.25, which is close to the monoisotopic mass of C-terminal peptide (i.e., 4471.87) in the negative ion mode (Table 1). Immediately following this peak were two peaks with 14 and 28 Da mass increases, suggesting that methylation occurred on the C-terminal peptide of α-tubulin. The truncated form of this peptide is shown in figure 9a, where a series of four methylated peaks was present in the mass spectrum. A comparable number of methylations were detected in the truncated form and the non-truncated form, suggesting that methylation does not occur on terminal acidic residues Asp451 or Glu452. In β-tubulin, only the peptide originating from isotype 57.m00003 was found to be methylated (Figure 9c). The peptide with a mass of 3812.65 followed by peptides with up to six methylations was assigned as peptide [416-449] (calculated mass 3812.48) generated because of the missed-cleavage of CNBr caused by oxidation of methionine 443. The ion peak with a mass of 3770.62 is very close to the calculated mass of C-terminal peptide [416-449] of β-tubulin 41.m00036 (with one missed-cleavage at M443 calculated mass 3770.44, Figure 9c).
Following HPLC fractionation of an in-gel CNBr and tryptic digest of α-tubulin, up to three methylations were detected in the C-terminal peptide (431-453, 583.m00022, Fig. 10A). The peptide with a mass of 2566.97 corresponds to the C-terminal peptide of α-tubulin, 583.m00022 (calculated mass is 2566.97 in the negative ion mode). Three other peptide ions showed a mass of 2580.99, 2595.00 and 2609.01, respectively, suggesting that this C-terminal peptide contained isoforms with one, two and three methylations. The first two peaks, 2566.97 and 2580.99, were successfully fragmented with CID (Fig. 10B). Similar to the tandem mass spectrum of glutamylated peptide, the methylated peptide started showing divergence from y20 fragment ion with a 14 Da increase, indicating that methylation occurs on E434.
Figure 10. Comparison of fragmentation profiles of the C-terminal peptides of α-tubulin 583.m00022 from T.gondii to determine the methylation site.
A. Negative ion MALDI TOF-TOF MS spectrum of C-terminal peptide of α-tubulin, 583.m00022, containing methylated peptide precursor, Spectra were acquired in reflector mode.
B. bottom panel, negative ion MALDI TOF-TOF MS/MS spectrum of the precursor 2566.97, B. upper panel, negative ion MALDI TOF-TOF MS/MS spectrum of the precursor 2580.99. The sequence shown is the C-terminal peptide from D431 to Y453 of α-tubulin, 583.m00022. Mainly y ions are shown in the mass spectra. The ions with masses of minus 18 and 36 are y ions having lost one or two H2O molecules. Note that figure 10b, bottom panel is the same figure as figure 6, bottom panel. Spectra were acquired in reflector mode.
Interestingly, in a recent immunofluorescence analysis study of histone H4 in our lab, a polyclonal rabbit antibody specific for tri-methylated K20 of histone H4 was noted to react not only with histones in the nucleus (arrow), but also with tubulins in T. gondii especially in the anterior region as shown by immunofluorescence analysis (supplemental material, figure S2). This antibody also reacted with purified T. gondii tubulin (supplemental material and discussion, figure S2).
DISCUSSION
The sequencing of the Toxoplasma genome has led to the identification of multiple α and β tubulins in this organism as well as genes for γ, δ and ε tubulins. In T. gondii, tubulins are found in the subpellicular microtubules and the conoid as well as in standard spindle microtubules and flagella (in male gametes). Subpellicular microtubules are non-dynamic and maintain the characteristic shape and polarity of T. gondii. These microtubules interact with the pellicle consisting of the inner membrane complex and plasma membrane15. Subpellicular microtubules are intimately associated with the cytosolic face of the pellicle and form a spiral occupying the anterior two thirds of the parasite body starting at apical tip and continuing to just bellow the nucleus1. T. gondii has 22 subpellicular microtubules each of which is about 5μm in length16. They are associated with the apical polar ring and a circular microtubule organizing center (MTOC) which is unique to the Apicomplexa16, 17. There is an intimate association of these subpellicular microtubules with the inner membrane complex via the inner membrane particles which contain microtubule associated proteins18. The conoid at the anterior end of the parasite consists of a novel polymer of tubulin attached to the preconoidal rings.19 The regulation, organization and use of tubulin in the Apicomplexa are distinct from other eukaryotic systems. Given the usefulness of drugs which interact with tubulin for the treatment of other parasitic diseases20, a deeper understanding of T. gondii tubulins may facilitate the development of new therapeutic agents.
The posttranslational modifications of tubulins in T.gondii are summarized in table 3. Tubulin α-isotypes undergo a broader set of posttranslational modifications than β-isotypes. These include acetylation of Lys40, a detyrosination and tyrosination cycle of its last C-terminal amino acid residue Y453, truncation of last two and five amino acid residues, polyglutamylation and methylation. Among these modifications, only polyglutamylation and methylation are shared between α- and β-tubulins. While some modified tubulins are found intensively in the anterior region, e.g., methylation and Δ2 tubulin, others are accumulated more in basal region. In a study of α-tubulin in T.gondii by Plessmann et al., extensive acetylation was determined using antibody 6-11 B-1, which is specific for acetylated tubulin.10 Our mass spectrometry data confirms the presence of this modification on α-tubulin. The combination of antibody labelling for localization of tubulin isoform distribution and confirmation of their presence and identity on the sequences of T. gondii tubulins by mass spectrometry provides a powerful tool in the study of these posttranslational modifications.
Table 3.
Posttranslational modifications determined using antibodies and mass spectrometry
| Acetylation | Detyrosination/tyrosination | Δ2a | Δ5b | Polyglutamylation | Methylation | |
|---|---|---|---|---|---|---|
| β 57.m00003 | +d | + 6e | + | |||
| β 41.m00036 | +d | + 3 | ||||
| α 583.m00022 | + | + | + | + | + 4 | + |
| location | N/A | Subpellicular Basalc | anterior | N/A | N/A | anterior |
Δ2 represents the tubulin isotype with the last two amino acid residues truncated.
Δ5 represents the tubulin isotype with the last five amino acid residues truncated.
Possible basal complex
Detected by antibody but not confirmed by mass spectrometry.
The numbers indicate the number of glutamylated species.
We also found a similar glutamylation pattern of the α-tubulin C-terminal peptide to the one obtained by Plessmann et. al. who used Lys-C digestion of α-tubulin, purification of resulting acidic peptides on MonoQ FPLC, followed by reverse phase HPLC for desalting and analysis by MALDI-TOF MS and sequencing by Edman degradation.10 We confirmed by mass spectrometry that α-tubulin was acetylated and that an isoform truncated by 5 amino acid residues was expressed in T.gondii. This Δ5 isoform of α-tubulin has not been described in tubulin from other sources than T.gondii and may represent a modification specific to the Apicomplexa. Production of an antibody recognizing specifically the truncated C-terminus of T. gondii tubulin would be useful to assess if this truncation is genuine and where it is located in the cell. Given the four putative tubulin tyrosine ligase-like (TTLL) enzymes and 6 putative carboxypeptidases that have been identified in the genome of T. gondii (www.ToxoDb.org), it is possible that these carboxypeptidases not only remove the last tyrosine (Tyr453) from α-tubulin, but may also cut at position Tyr449, leading to the C-terminal loss of the last 5 residues. The alignment of α-tubulin C-terminal sequences from T.gondii, P. falciparum and other apicomplexans as well as those from Dinoflagellates demonstrated that they are well conserved (supplemental material, Table 1). Except B. bovis, most of the apicomplexan α-tubulins show identical extreme C-terminal sequences with Glu-Tyr as the last two amino acid residues and a second tyrosine as the fifth residue from the C-terminus. Importantly, the ciliate Tetrahymena α-tubulin has the second tyrosine but lacks the last four residues, identical to those of T.gondii. This type of change may be important in terms of recruiting different proteins on microtubules, thereby creating new organelles.
Anti-Δ2-tubulin labelled both α-tubulin and β-tubulin regions, however, only α-tubulin, but not β-tubulin with this post translational modification, was observed in the mass spectra (Fig. 5A). It is important to note that this antibody (anti-Δ2-tubulin) was raised against the C-terminus of Δ2-tubulin and may be cross-reacting with other tubulins under conditions used for immunological analysis. It is also possible that the spots recognized in β-tubulin region are posttranslationally modified α-tubulins with their pIs shifted by modifications. In fact, we detected some α-tubulins in low pI region (pH 4.5-5) where usually β-tubulins are located in 2D gel and mass spectrometry analysis.
Anti-deTyr-tubulin antibody strongly labelled the basal region, however, only a very weak fluorescence signal was observed at the basal end with YFP-tubulin, indicating that there is not much tubulin at the basal end. In fact, the identified major components of this basal region are TgMORN1 and TgCentrin2.2 While it is possible that anti-deTyr-tubulin antibody cross-reacted with other proteins, this antibody did detect spots in the α-tubulin region (Fig. 4B) of the immunoblot and the presence of the detyrosinated tubulin was confirmed in mass spectra (Fig. 5 A), suggesting that some of the tubulins are detyrosinated in T. gondii.
Anti-polyGlu-tubulin antibody labelled the central zones of the immunoblot (supplemental material, Fig. S1), consistent with a shift of pIs of α-tubulins to the acidic side due to polyglutamylation. Mass spectrometry data also confirmed that α-tubulin, including the truncated Δ5-tubulin, is polyglutamylated. Interestingly, no equivalent shift is seen for the β-tubulins, although mass spectrometry data showed that they are polyglutamylated. The antibody used in this work is B3 mouse monoclonal antibody, which recognizes glutamate side chains containing two or more glutamates, it is possible that only α-tubulin becomes polyglutamylated on one site and that β-tubulin is monoglutamylated on several sites, and therefore, polyglutamylated β-tubulin is not detected by the B3 antibody. We identified a glutamylation site using tandem mass spectrometry on E434 in α-tubulin. This is different than the residue E445 deduced for glutamylation by Plessmann et. al using an Edman sequencing experiment10. Although polyglutamylation is often present on only one particular glutamic acid residue in the C-terminus of both α- and β-tubulin 5, 6, 21, it was found to be involved with multiple sites in Tritrichomonas, i.e., E444, E445, E446 and E447 on α-tubulin.22 It is possible that polyglutamylation of tubulin in T.gondii also involves multiple sites and that both E434 and E445 are glutamylated. Under the conditions in which we carried out MS/MS (CID MS/MS in negative mode), the rich content of glutamic acid residues results in multiple H2O loss. This will not only complicate the mass spectra, but also decrease the signal intensity of all the fragment ions. Due to the low signal to noise ratio obtained from tandem mass spectra of the multiply glutamylated ions, it is not completely resolved whether polyglutamylation was on a single site or involved multiple sites.
The mass spectrometry data were consistent with the immunoblot probed with anti-polyglutamylated-tubulin antibody which suggested that both α- and β-tubulin were polyglutamylated. As discussed earlier, polyglutamylation on glutamate will decrease the pI value of the α-tubulin isotypes, thus shifting their migration toward the β-tubulin region. Although it is not clear if the polyglutamylation occurred on a single or multiple sites of the tubulin isotypes due to the degree of sequence coverage obtained by tandem mass spectrometry. A closer look at divergent part of the C-terminal sequence of the two isotypes revealed that E438, E440 and E442 on 57.m00003 were replaced by G, D and D residues on 41.m00036, resulting in fewer available sites for glutamylation on 41.m00036 (see sequence in Table 1). Assuming that multiple sites of polyglutamylation are possible, it is predicted that 57.m00003 should vary more in this modification.
Methylation has been implicated in transcriptional activation, gene silencing, heterochromatin formation, mitosis, and DNA repair23. Recent studies have suggested that methylation is a frequent posttranslational modification in eukaryotic cells and that methylation may be introduced by sample handling24, 25; however, our control experiment using HFF and bovine brain tubulin suggests that the observed tubulin methylation is not formed exogenously (supplemental material, Fig. S2). Most common methylations occur on nitrogen or oxygen atoms of proteins. Amino acid residues methylated on nitrogen include the guanidine moiety and the side chain amide nitrogens of arginine, the side chain amide nitrogen of asparagine, the nitrogen of the Δ-amino of lysine and the imidazole ring of histidine. Oxygen methylation usually occurs on side chain carboxylates of glutamates and aspartates 24. S-methylation at the thiolate side chain of cysteine and C-methylation of arginine and glutamine side chains 19 have also been reported. Among all types of methylations, only N-methylation can cause multiple methylation events on a single residue, i.e. lysine can be tri-methylated and arginine can undergo up to two methylations. Examination of the amino acid sequences of the CNBr C-terminal peptides of T. gondii tubulins suggests that methylation could occur on lysine, arginine, aspartate and glutamate residues for α-tubulin and on glutamine, aspartate and glutamate residues for β-tubulin. To this end, it should be noted that the H4K20me3 antibody which we found to react with T. gondii tubulin (Figure S3) recognizes the tri-N-methylated lysine at the position 20 in histone 4. Assuming both α- and β-tubulins share the same methylation pathway, it is unlikely that methylation will occur on arginine or lysine since only α-tubulin contains these residues at the C-terminus. Similarly, there will not be C-methylation on glutamine, which only exists in β-tubulins. But the fact that we still observed the methylation patterns on α-tubulin C-terminal tryptic peptides, which lack the glutamine residues, suggests that O-methylation occurs on side chain carboxylates of glutamates and aspartates. By mass spectrometry we located a methylation site on E434 of α-tubulin. This is also consistent with retaining the methylation pattern on Δ5-tubulin C-terminal peptides. Interestingly, E434 was shared as the modification site by both methylation and polyglutamylation. This indicates that the two isoforms of tubulin are mutually exclusive and, therefore, tubulins would be either shifted to more acidic pI when glutamylated or to more basic pI when methylated. Furthermore, we did not detect peptides with mass increments of +143 Da (129+14), suggesting that O-methylation does not occur on the glutamates added posttranslationally via the α carboxyl group. This may explain why the anti-polyGlu-tubulin antibody labelled a central region of the immunoblot of T.gondii tubulins. The mass spectra displaying multiple methylation peaks are consistent with the rich content of glutamate and aspartate in both α- and β-tubulins. Localization of other sites of methylation is not yet complete due to the low concentration of the methylated species, but is ongoing in our laboratory.
While many functions of tubulin and microtubules are conserved in T. gondii (e.g., spindle formation on cell division and flagella in male gametes) other functions such as shape, gliding motility, polarity and invasion are specialized in the Apicomplexa. In addition to microtubule associated proteins, many of these functions may be related to post translational modifications that occur on tubulin in T. gondii and other apicomplexa. These modifications may alter the stability of the microtubules as well as their association with other proteins. In addition to previously described tubulin posttranslational modifications, methylation was identified as a novel tubulin posttranslational modification. If this modification is confirmed in future studies to be specific to the Apicomplexa, the methyl transferase involved would represent a potential new therapeutic target.
Supplementary Material
Figure S1. Immunoblot analysis of Tyr-tubulin and polyGlu tubulin posttranslational modification of cytoskeletal tubulins Immunoblot stained with anti-polyGlu-tubulins (a), anti-β & anti-polyGlu-tubulin (b) and anti-α-tubulin (c). The same immunoblot was used for each antibody. The immunoblot was stripped between antibody incubations using a standard protocol.
Figure S2. Analysis of methylation posttranslational modification of cytoskeletal tubulins using immunofluorescence microscopy and immunoblot. A. Phase contrast image (a) corresponds to images b through e. Phase contrast image (a′) corresponds to images b′ through e′. Immunofluorescence analysis of cells imaged with DAPI for DNA (b, b′), YFP-α-tubulin (c, c′), anti-H4K20me3 (d, d′), and the merge of images of c and d (e, e′). The nucleus is indicated by arrows in figure d′ and the conoid of daughter cells is indicated in arrows in figure e. B. 2D immunoblot analysis using anti-α & β-tubulin (1), anti-α-tubulin (2), and anti-H4K20me3 (3) demonstrating reaction of anti-H4K20me3 (and possible methylation) in both α and β tubulin. The same immunoblot was used for each antibody. The immunoblot was stripped between antibody incubations using a standard protocol.
C. Immunoblot analysis using and anti-H4K20me3 (1) anti- β-tubulin (2) and anti-α-tubulin (3) for control (bovine brain tubulin, lane 1), Human Foreskin Fibroblasts (HFF, lane 2), HFF pellet (lane 3) and T.gondii tubulins (lane 4) in a 1D gel. D. Coomassie blue (1) stain of separated α and β tubulin from T. gondii and Immunoblot analysis with anti-H4k20me3 (2) and anti-β- and anti-α-tubulin (3) after tubulins were separated on a 1D gel.
Supplemental Text for Figure S2 Detection of methylation on T. gondii tubulin using immunofluorescence microscopy and immunoblot. The H4K20me3 antibody labelled the anterior region of T. gondii intensely. This antibody is known to react with the methyl modifications at K20 in H4 (histone). By immunoblot, the H4K20me3 antibody reacted with T. gondii tubulins focusing at pIs more basic than the β-tubulin region (Fig. S2-B). Since T. gondii were grown in human foreskin fibroblasts (HFF), it was necessary to exclude that the reaction of anti-H4K20me3 originated from the HFF.
The soluble fraction of uninfected HFF cells were prepared using the same protocol as cytoskeleton fraction of T.gondii and subjected to 1D SDS PAGE and immunoblot analysis in parallel with the cytoskeleton fraction of T. gondii. Tubulin was also purified using aTaxol assisted pelleting method 28 and subjected to 1D SDS PAGE and immunoblot analysis. Tubulins from bovine brain (served as the control), HFF cells, T. gondii were stained by anti-α & β-tubulins (Fig. S2-C). However, only the tubulins from T. gondii were labeled by anti-H4K20me3 (Fig. S2-C), suggesting that the tubulins from T. gondii, but not HFF, react with this serum and are likely methylated (Fig. S2-C). There is a band in HFF cells reactive with H4K20me3 antibody with a molecular weight of about 40 kDa, but no evidence for the presence of tubulin was found in this 40 kDa band by mass spectrometry. A band near 76 kDa was strongly labeled with H4K20me3, however mass spectrometry analysis revealed that there was no tubulin in this region. In the narrow pH range (pH 4.5-5.5) 2D gel of cytoskeletal tubulin, H4K20me3 antibody stained a number of ~50 kDa spots (Fig. S2-B). The alignment of these spots with those detected by a- or β-tubulin antibodies (Fig. S2-B) suggested that this antibody and possible methylation occurs on both a- and β-tubulins from T. gondii. The H4K20me3 antibody reactive β-tubulins were located at the basic end of the β-tubulin region on the gel, which overlaps with a-tubulin. This is expected, since the pI of these β-tubulins would be shifted to the basic side if methylation occurs on glutamates and aspartates of their C-termini. Figure S3-C, panel 1 also demonstrates that some spots of approximately 60 kDa and 75 kDa were also recognized by H4K20me3 antibody. These spots did not contain tubulin by mass spectrometry analysis. To further confirm the occurrence of methylation on tubulin, a- and β-tubulins were separated on a 10% mini-gel (Bio-Rad, CA) and eluted from gel slices using an Electro-Eluter (Model 422 Electro-Eluter, Bio-Rad, CA), followed by immunoblot analysis. The separated a- and β-tubulins both demonstrated labelling by H4K20me3 antibody at the tubulin bands as indicated by anti-a and β-tubulin (Fig. S2-D), consistent with methylation on both a- and β-tubulins. While it is possible that the anti-H4K20me3 reaction with T. gondii is due to some unrelated cross reaction in this rabbit polyclonal serum, we believe this is unlikely, since staining by IFA and immunoblot clearly demonstrated that this was specific to T. gondii tubulin, limited reactivity was seen with anti-H4K20me3 and other proteins in T. gondii and independent mass spectrometry data confirmed that methylation occurred on T. gondii tubulin. Confirmation of the immunolocalization of T. gondii methyl tubulin antibody will require the production of a T. gondii methyl tubulin specific antiserum.
Table 1. Alignment of α-tubulin C-terminal sequences and relation to classification.
Acknowledgments
With thanks to Mathieu Gissot for his initial observations on the methyl histone antibody reactions with T. gondii. This work was supported by the Biodefense Proteomic Research Program, Contract HHSN266200400054C-02, NIH-NIAID-DMID and by NIH AI39454.
References
- 1.Morrissette NS, Sibley LD. Microbiol Mol Biol Rev. 2002;66:21–38. doi: 10.1128/MMBR.66.1.21-38.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hu K, Johnson J, Florens L, Fraunholz M, Suravajjala S, DiLullo C, Yates J, Roos DS, Murray JM. PLoS Pathog. 2006;2:e13. doi: 10.1371/journal.ppat.0020013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Verdier-Pinard P, Pasquier E, Xiao H, Burd B, Villard C, Lafitte D, Miller LM, Angeletti RH, Horwitz SB, Braguer D. Anal Biochem. 2008 doi: 10.1016/j.ab.2008.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.LeDizet M, Piperno G. Proc Natl Acad Sci U S A. 1987;84:5720–5724. doi: 10.1073/pnas.84.16.5720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 6.Redeker V, Levilliers N, Schmitter JM, Le Caer JP, Rossier J, Adoutte A, Bre MH. Science. 1994;266:1688–1691. doi: 10.1126/science.7992051. [DOI] [PubMed] [Google Scholar]
- 7.Nagel SD, Boothroyd JC. Mol Biochem Parasitol. 1988;29:261–273. doi: 10.1016/0166-6851(88)90081-3. [DOI] [PubMed] [Google Scholar]
- 8.Madrid-Aliste CJ, Dybas JM, Angeletti RH, Weiss LM, Kim K, Simon I, Fiser A. BMC Genomics. 2009;10:38. doi: 10.1186/1471-2164-10-38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu K, Roos DS, Angel SO, Murray JM. J Cell Sci. 2004;117:5697–5705. doi: 10.1242/jcs.01494. [DOI] [PubMed] [Google Scholar]
- 10.Plessmann U, Reiter-Owona I, Lechtreck KF. Parasitol Res. 2004;94:386–389. doi: 10.1007/s00436-004-1220-7. [DOI] [PubMed] [Google Scholar]
- 11.Weber K, Schneider A, Westermann S, Muller N, Plessmann U. FEBS Lett. 1997;419:87–91. doi: 10.1016/s0014-5793(97)01436-1. [DOI] [PubMed] [Google Scholar]
- 12.Fennell BJ, Al-shatr ZA, Bell A. Int J Parasitol. 2008;38:527–539. doi: 10.1016/j.ijpara.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 13.Soucek K, Kamaid A, Phung AD, Kubala L, Bulinski JC, Harper RW, Eiserich JP. Prostate. 2006;66:954–965. doi: 10.1002/pros.20416. [DOI] [PubMed] [Google Scholar]
- 14.van Montfort BA, Doeven MK, Canas B, Veenhoff LM, Poolman B, Robillard GT. Biochim Biophys Acta. 2002;1555:111–115. doi: 10.1016/s0005-2728(02)00264-5. [DOI] [PubMed] [Google Scholar]
- 15.Ajioka JW, Soldati D. Toxoplasma: Molecular And Cellular Biology. Taylor & Francis; 2007. [Google Scholar]
- 16.Nichols BA, Chiappino ML. J Protozool. 1987;34:217–226. doi: 10.1111/j.1550-7408.1987.tb03162.x. [DOI] [PubMed] [Google Scholar]
- 17.Russell DG, Burns RG. J Cell Sci. 1984;65:193–207. doi: 10.1242/jcs.65.1.193. [DOI] [PubMed] [Google Scholar]
- 18.Morrissette NS, Murray JM, Roos DS. J Cell Sci. 1997;110(Pt 1):35–42. doi: 10.1242/jcs.110.1.35. [DOI] [PubMed] [Google Scholar]
- 19.Ermler U, Grabarse W, Shima S, Goubeaud M, Thauer RK. Science. 1997;278:1457–1462. doi: 10.1126/science.278.5342.1457. [DOI] [PubMed] [Google Scholar]
- 20.Traub-Cseko YM, Ramalho-Ortigao JM, Dantas AP, de Castro SL, Barbosa HS, Downing KH. Trends Parasitol. 2001;17:136–141. doi: 10.1016/s1471-4922(00)01834-1. [DOI] [PubMed] [Google Scholar]
- 21.Rudiger M, Plessman U, Kloppel KD, Wehland J, Weber K. FEBS Lett. 1992;308:101–105. doi: 10.1016/0014-5793(92)81061-p. [DOI] [PubMed] [Google Scholar]
- 22.Schneider A, Plessmann U, Felleisen R, Weber K. FEBS Lett. 1998;429:399–402. doi: 10.1016/s0014-5793(98)00644-9. [DOI] [PubMed] [Google Scholar]
- 23.Walsh C. Posttranslational modification of proteins: expanding nature’s inventory. Roberts and Co; 2006. [Google Scholar]
- 24.Sprung R, Chen Y, Zhang K, Cheng D, Zhang T, Peng J, Zhao Y. J Proteome Res. 2008;7:1001–1006. doi: 10.1021/pr0705338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jung SY, Li Y, Wang Y, Chen Y, Liu XD, Eissa NT, Zhao Y, Qin J. Anal Chem. 2008. [Google Scholar]
- 26.Livingstone CD, Barton GJ. Comput Appl Biosci. 1993;9:745–756. doi: 10.1093/bioinformatics/9.6.745. [DOI] [PubMed] [Google Scholar]
- 27.Thompson JD, Higgins DG, Gibson TJ. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miller LM, Menthena A, Chatterjee C, Verdier-Pinard P, Novikoff PM, Horwitz SB, Angeletti RH. Biochemistry. 2008;47:7572–7582. doi: 10.1021/bi8005225. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Immunoblot analysis of Tyr-tubulin and polyGlu tubulin posttranslational modification of cytoskeletal tubulins Immunoblot stained with anti-polyGlu-tubulins (a), anti-β & anti-polyGlu-tubulin (b) and anti-α-tubulin (c). The same immunoblot was used for each antibody. The immunoblot was stripped between antibody incubations using a standard protocol.
Figure S2. Analysis of methylation posttranslational modification of cytoskeletal tubulins using immunofluorescence microscopy and immunoblot. A. Phase contrast image (a) corresponds to images b through e. Phase contrast image (a′) corresponds to images b′ through e′. Immunofluorescence analysis of cells imaged with DAPI for DNA (b, b′), YFP-α-tubulin (c, c′), anti-H4K20me3 (d, d′), and the merge of images of c and d (e, e′). The nucleus is indicated by arrows in figure d′ and the conoid of daughter cells is indicated in arrows in figure e. B. 2D immunoblot analysis using anti-α & β-tubulin (1), anti-α-tubulin (2), and anti-H4K20me3 (3) demonstrating reaction of anti-H4K20me3 (and possible methylation) in both α and β tubulin. The same immunoblot was used for each antibody. The immunoblot was stripped between antibody incubations using a standard protocol.
C. Immunoblot analysis using and anti-H4K20me3 (1) anti- β-tubulin (2) and anti-α-tubulin (3) for control (bovine brain tubulin, lane 1), Human Foreskin Fibroblasts (HFF, lane 2), HFF pellet (lane 3) and T.gondii tubulins (lane 4) in a 1D gel. D. Coomassie blue (1) stain of separated α and β tubulin from T. gondii and Immunoblot analysis with anti-H4k20me3 (2) and anti-β- and anti-α-tubulin (3) after tubulins were separated on a 1D gel.
Supplemental Text for Figure S2 Detection of methylation on T. gondii tubulin using immunofluorescence microscopy and immunoblot. The H4K20me3 antibody labelled the anterior region of T. gondii intensely. This antibody is known to react with the methyl modifications at K20 in H4 (histone). By immunoblot, the H4K20me3 antibody reacted with T. gondii tubulins focusing at pIs more basic than the β-tubulin region (Fig. S2-B). Since T. gondii were grown in human foreskin fibroblasts (HFF), it was necessary to exclude that the reaction of anti-H4K20me3 originated from the HFF.
The soluble fraction of uninfected HFF cells were prepared using the same protocol as cytoskeleton fraction of T.gondii and subjected to 1D SDS PAGE and immunoblot analysis in parallel with the cytoskeleton fraction of T. gondii. Tubulin was also purified using aTaxol assisted pelleting method 28 and subjected to 1D SDS PAGE and immunoblot analysis. Tubulins from bovine brain (served as the control), HFF cells, T. gondii were stained by anti-α & β-tubulins (Fig. S2-C). However, only the tubulins from T. gondii were labeled by anti-H4K20me3 (Fig. S2-C), suggesting that the tubulins from T. gondii, but not HFF, react with this serum and are likely methylated (Fig. S2-C). There is a band in HFF cells reactive with H4K20me3 antibody with a molecular weight of about 40 kDa, but no evidence for the presence of tubulin was found in this 40 kDa band by mass spectrometry. A band near 76 kDa was strongly labeled with H4K20me3, however mass spectrometry analysis revealed that there was no tubulin in this region. In the narrow pH range (pH 4.5-5.5) 2D gel of cytoskeletal tubulin, H4K20me3 antibody stained a number of ~50 kDa spots (Fig. S2-B). The alignment of these spots with those detected by a- or β-tubulin antibodies (Fig. S2-B) suggested that this antibody and possible methylation occurs on both a- and β-tubulins from T. gondii. The H4K20me3 antibody reactive β-tubulins were located at the basic end of the β-tubulin region on the gel, which overlaps with a-tubulin. This is expected, since the pI of these β-tubulins would be shifted to the basic side if methylation occurs on glutamates and aspartates of their C-termini. Figure S3-C, panel 1 also demonstrates that some spots of approximately 60 kDa and 75 kDa were also recognized by H4K20me3 antibody. These spots did not contain tubulin by mass spectrometry analysis. To further confirm the occurrence of methylation on tubulin, a- and β-tubulins were separated on a 10% mini-gel (Bio-Rad, CA) and eluted from gel slices using an Electro-Eluter (Model 422 Electro-Eluter, Bio-Rad, CA), followed by immunoblot analysis. The separated a- and β-tubulins both demonstrated labelling by H4K20me3 antibody at the tubulin bands as indicated by anti-a and β-tubulin (Fig. S2-D), consistent with methylation on both a- and β-tubulins. While it is possible that the anti-H4K20me3 reaction with T. gondii is due to some unrelated cross reaction in this rabbit polyclonal serum, we believe this is unlikely, since staining by IFA and immunoblot clearly demonstrated that this was specific to T. gondii tubulin, limited reactivity was seen with anti-H4K20me3 and other proteins in T. gondii and independent mass spectrometry data confirmed that methylation occurred on T. gondii tubulin. Confirmation of the immunolocalization of T. gondii methyl tubulin antibody will require the production of a T. gondii methyl tubulin specific antiserum.
Table 1. Alignment of α-tubulin C-terminal sequences and relation to classification.