Abstract
Introduction
Fluorescence-guided resection (FGR) of brain tumors is an intuitive, practical and emerging technology for visually delineating neoplastic tissue exposed intraoperatively. Image guidance is the standard technique for producing 3-dimensional spatially coregistered information for surgical decision making. Both technologies together are synergistic: the former detects surface fluorescence as a biomarker of the current surgical margin while the latter shows coregistered volumetric neuroanatomy but can be degraded by intraoperative brain shift. We present the implementation of deformation modeling for brain shift compensation in protoporphyrin IX FGR, integrating these two sources of information for maximum surgical benefit.
Methods
Two patients underwent FGR coregistered with conventional image guidance. Histopathological analysis, intraoperative fluorescence and image space coordinates were recorded for biopsy specimens acquired during surgery. A biomechanical brain deformation model driven by intraoperative ultrasound data was used to generate updated MR images.
Results
Combined use of fluorescence signatures and updated MR image information showed substantially improved accuracy compared to fluorescence or the original (i.e., nonupdated) MR images, detecting only true positives and true negatives, and no instances of false positives or false negatives.
Conclusion
Implementation of brain deformation modeling in FGR shows promise for increasing the accuracy of neurosurgical guidance in the delineation and resection of brain tumors.
Key Words: Fluorescence-guided resection, Brain shift, Protoporphyrin IX, Aminolevulinic acid, Deformation modeling, Brain tumor
Introduction
Fluorescence-guided resection (FGR) for intraoperative, real-time delineation of tumors has been gaining wider acceptance within the neurosurgical community [1, 2, 3, 4, 5, 6, 7, 8, 9]. Prior to surgery, δ-aminolevulinic acid (ALA) is administered orally, leading to accumulation of protoporphyrin IX (PpIX) in neoplastic tissue. When excited with blue light, tumor tissue accumulating sufficient levels of PpIX displays a red fluorescence [2, 3, 4]. FGR provides the neurosurgeon with a surface guidance technology that is not susceptible to intraoperative brain shift, in contrast to conventional image guidance systems where navigational accuracy is degraded [10, 11, 12, 13, 14]. However, not all neoplastic tissue fluoresces to levels observable intraoperatively, leaving some tumor tissue undetected [7, 9, 15, 16] and subsurface levels of fluorescent tissue are not visible with current surgical microscope systems, requiring the need for complementary 3-dimensional volumetric guidance in locating abnormal tissue.
To account for image-guided registration degradation secondary to brain shift, biomechanical models that compensate for brain deformation are currently under development [10, 11, 17]. In one form, intraoperative data drive a biomechanical model to estimate the 3-dimensional displacement field and subsequently deform the preoperative MR (pMR) images used for navigation to produce updated MR (uMR) images. The uMR provides the neurosurgeon with more accurate MR image correspondence with the current surgical field. A dual-modality, neurosurgical guidance system that incorporates both FGR and uMR images would offer the neurosurgeon a biomarker of the surgical margin as currently exposed along with coregistered volumetric neuroanatomy that is not degraded by intraoperative brain shift. In this study, we present the implementation of a biomechanical deformation model for brain shift compensation in the setting of PpIX FGR of brain tumors.
Patients and Methods
Patients
Two patients, one with a gliosarcoma (GS) and one with a glioblastoma multiforme (GBM), provided written informed consent for this investigational study and were enrolled in an institutional review board-approved study of coregistered fluorescence-enhanced tumor resection. pMR, T1-weighted images (matrix: 256 × 256, 1.5 mm slice thickness) with gadolinium enhancement (0.2 ml/kg gadolinium-diethylenetriamine pentaacetic acid) and scalp-based registration fiducials were acquired for both patients and used for image guidance. Three hours prior to induction of anesthesia, both patients received a 20 mg/kg body weight oral dose of ALA (DUSA Pharmaceuticals, Tarrytown, N.Y., USA) dissolved in 100 ml of water.
Coregistered FGR Procedure
For FGR, a commercial image guidance system (Treon® StealthStation®; Medtronic, Louisville, Colo., USA) was coregistered with a surgical microscope offering fluorescence capabilities (Zeiss OPMI Pentero®; Carl Zeiss Surgical GmbH, Oberkochen, Germany). The surgical microscope excites with blue light and collects the filtered red fluorescence emission of PpIX. During the procedure, the surgeon intermittently switched from the white light to the blue light illumination mode (fig. 1). Eighteen biopsy specimens were collected (9 from patient 1: GS; 9 from patient 2: GBM) at the beginning, middle, and end of resection in regions within the bulk of the tumor and tumor margins, and were divided into 3 groups: one was placed in 10% buffered formalin, one was frozen in a cryogenic vial, and one was frozen in mounting medium (OCT). The navigation system was used to determine the image space coordinates of each specimen, which was subsequently matched to its corresponding white and blue light digital images.
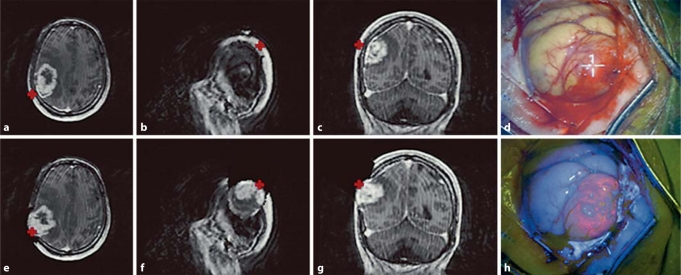
Fig. 1.
Preoperative, updated, and intraoperative images of patient 1. The top row from left to right (a–c) shows preoperative T1-weighted postdurotomy MR images without compensation for brain shift (pMR). The bottom row from left to right (e–g) shows the updated T1-weighted MR images immediately after durotomy with compensation for brain shift (uMR). The surgical microscope was coregistered and tracked by the navigational system. The white light (d) and blue light (h) images corresponding to the tracked coordinates of the microscope focal point in image space (crosses) are shown. A bulging, highly fluorescent tumor tissue confirmed by pathology as neoplastic tissue corresponds to an area of contrast enhancement in the uMR. Compare this area of fluorescent brain tissue to the pMR, which inaccurately points to the scalp region.
Intraoperative Brain Deformation Procedure
Prior to the start of surgery, pMR images were rigidly registered to the patient's head in the physical space coordinate system through a fiducial-based registration. At the time of surgery and before craniotomy, the locations of the fiducial markers were digitized using a stylus probe whose location was identified by a 3-dimensional tracking system (Polaris System Northern Digital Inc., London, Ont., Canada) coupled to a workstation dedicated to the deformation modeling. The registration process was used to match the fiducials’ physical space coordinates with the corresponding image space coordinates. The stylus probe was then placed at two vertical locations to estimate the direction of gravity that was used in the model computations.
After craniotomy, a Philips (Philips Medical Systems, Bothell, Wash., USA) iU22 3-dimensional ultrasound system (US) was used to collect predurotomy and postdurotomy images. The US transducer was tracked by the Polaris System continuously through a rigidly attached infrared light-emitting tracker to allow for the 3-dimensional US images to be registered to the pMR stack [18, 19].
Tissue Processing and Analysis
Formalin-fixed paraffin-embedded tissue was processed for hematoxylin and eosin staining and analyzed by a single neuropathologist (B.T.H.). The neuropathologist was blinded to the pathological diagnoses from the main surgical specimen and from the intraoperative fluorescence or imaging data. Each biopsy tissue section was analyzed for the presence or absence of tumor cells and was evaluated based on the WHO classification schema [20].
Deformation Model
The biomechanical brain deformation model implemented simulates the amount and direction of brain shift and deformation (i.e., displacement) that occurs intraoperatively, by incorporating standard surgical conditions (e.g. direction of gravity) and intraoperative data. Predurotomy and immediate postdurotomy US images were used to measure the displacement that occurs in a subvolume of the brain at the beginning of surgery. They provided sufficient patient-specific intraoperative data, in combination with standard surgical conditions, to drive the biomechanical brain deformation model to estimate displacement for the whole-brain volume which was used to deform the pMR images and generate uMR images. Additional details on the brain deformation model are provided in the Appendix.
Data Analysis
Two-by-two tables were used to calculate the following values and statistical measures for FGR alone, image guidance with (IGW) and without brain shift compensation (IGWO) alone, and FGR coupled with brain shift compensated image guidance (FGR-IGW): (1) true negatives; (2) false negatives; (3) true positives; (4) false positives; (5) sensitivities; (6) specificities; (7) negative predictive values, and (8) positive predictive values. To calculate these measures for FGR, intraoperative red fluorescence was considered a positive test result, whereas absence of fluorescence was recorded as a negative test result. Similarly, to calculate the same values and measures for image guidance, tissue judged radiologically abnormal from T1-weighted pMR images (i.e., preoperative MRI without brain shift compensation) or uMR images (i.e., preoperative MRI with brain shift compensation) was considered a positive test result, whereas tissue judged radiologically normal from pMR or uMR images was scored as a negative test result.
Results
The 2 patients in this study displayed distinct patterns of PpIX fluorescence and different degrees of brain shift (table 1). The GS and GBM (both WHO grade IV) cases displayed areas of strong fluorescence as well as gadolinium-enhancing regions on MRI. The GS surgery (brain shift displacement values: mean = 2.9 mm, max = 16.0 mm) had the largest degree of brain shift (table 2), whereas the GBM surgery (brain shift displacement values: mean = 0.1 mm, max = 1.4 mm) showed much less brain shift. The deformation model was run on a Linux computer (2.33 GHz, 8 GB RAM) with 500 iterations for patient 1 (GS) and 51 iterations for patient 2 (GBM). The computational cost was less than 30 min for patient 1 and less than 6 min for patient 2. Figure 1 shows representative pMR, uMR, and white and blue light images of the coregistered focal point of the operating microscope after dural opening during the surgery of patient 1 from which it is evident that the fluorescing tumor distended. The uMR views correctly compensate for the tumor movement after dural opening whereas their pMR counterparts clearly do not.
Table 1.
Overview of FGR and brain shift for the whole brain
| Patient | Age years | Diagnosis | Tumor location | Tumor volume | Intraoperative fluorescence | Brain shift displacement, mm |
|
|---|---|---|---|---|---|---|---|
| mean | max | ||||||
| 1 | 70 | GS | right parietal | 65.95 | yes | 2.9 | 16.0 |
| 2 | 76 | GBM | left temporal | 4.96 | yes | 0.1 | 1.4 |
Mean and maximum displacements represent the mean and maximum magnitude of brain shift estimated by the biomechanical model.
Table 2.
Statistical measures
| Statistical measures | FGR | IGWO | IGW | FGR-IGW |
|---|---|---|---|---|
| Sensitivity | 0.50 | 0.69 | 0.94 | 1.00 |
| Specificity | 1.00 | 1.00 | 1.00 | 1.00 |
| NPV | 0.20 | 0.29 | 0.67 | 1.00 |
| PPV | 1.00 | 1.00 | 1.00 | 1.00 |
Sensitivities, specificities, negative predictive values (NPV), and positive predictive values (PPV) were calculated for FGR, IGWO, IGW, and FGR-IGW. n = 18.
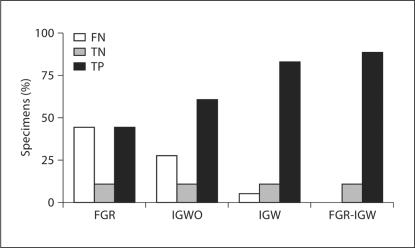
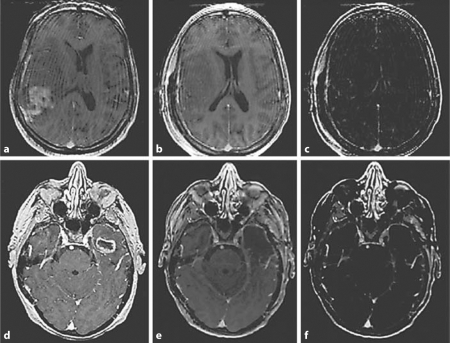
Sensitivity, specificity, negative predictive values, and positive predictive values for both cases are shown in table 2. An increase in the sensitivity and negative predictive value of image guidance was noted after brain shift compensation (sensitivity: IGWO = 0.69 vs. IGW = 0.94, negative predictive value: IGWO = 0.29 vs. IGW = 0.67). FGR-IGW showed the best statistical measures for accurately identifying abnormal tissue (sensitivity = 1.00, negative predictive value = 1.00, specificity = 1.00, positive predictive value = 1.00). True negative, false negative, and true positive values from both cases are graphically displayed in figure 2. No false positives were recorded in this study and two biopsies were true negatives (nonenhancing, nonfluorescent tissue without the presence of tumor cells on histology). The percentage of true positive samples increased (IGWO = 61.1% vs. FGR-IGW = 88.9%) and false negatives decreased (IGWO = 27.8% vs. FGR-IGW = 0.00%) in IGWO versus FGR-IGW. Figure 3 shows representative pre- and postoperative gadolinium-enhanced, T1-weighted images of both cases.
Fig. 2.
Test results for biopsy specimens. Percentage of biopsy specimens (n = 18) for FGR, IGWO, IGW, and FGR-IGW that were true negative (TN), false negative (FN), and true positive (TP) are shown for both cases. No false positives were recorded in this study.
Fig. 3.
Gadolinium-enhanced preoperative and postoperative T1-weighted MR images. Preoperative images for patients 1 (a) and 2 (d). Postoperative images for patients 1 (b) and 2 (e). Postoperative subtraction images for patients 1 (c) and 2 (f).
Discussion
With evidence that the extent of tumor resection correlates with patient survival and quality of life [5, 6, 8, 21, 22, 23], intraoperative brain shift poses a major challenge in brain tumor resections using conventional image guidance technologies. FGR based on ALA-PpIX has recently gained acceptance as a safe and easy way to aid the neurosurgeon in delineating neoplastic tissues during tumor resection. FGR is not limited by degradation of registration accuracy due to the intraoperative brain shift that is known to compromise conventional image guidance systems. However, FGR only provides the neurosurgeon with surface information on the location of neoplastic tissue, and as such, accurate 3-dimensional navigation that accounts for intraoperative brain shift is still needed to complement FGR.
Here, we present the implementation of a brain deformation biomechanical model to compensate for intraoperative brain shift in PpIX FGR of brain tumors. Two surgical cases with different levels of PpIX fluorescence and intraoperative brain shift were studied. FGR-delineated tumor tissue appeared in both the GBM and GS instances. In both surgeries, observable intraoperative fluorescence was mapped only to areas of contrast-enhancing tissue on the uMR images, whereas the pMR images incorrectly mapped fluorescing tissue to radiologically normal regions (e.g. scalp), which on pathological analysis was demonstrated to be neoplastic. Some biopsy specimens of highly enhancing tissue in patient 1 (n = 3) did not display observable fluorescence. In these specimens, a heavy infiltration of mostly the sarcomatous element of the tumor was observed. Meanwhile, tissue sections containing mostly astrocytic elements of the tumor showed intraoperative fluorescence and contrast enhancement on uMR images, suggesting that high-grade glioma tumor cells accumulate more PpIX than high-grade sarcomas.
The uMR images provided more accurate anatomical information increasing both the sensitivity (pMR = 0.69 vs. uMR = 0.94) and negative predictive value (pMR = 0.29 vs. uMR = 0.67) of T1-weighted images for surgical correspondence with histologically abnormal tissue (i.e., tissue with the presence of tumor cells). The dual-modality approach using fluorescence signatures with uMR offers a complementary approach that increased the accurate determination of abnormal tissue in the two cases (fig. 2). Image guidance provides the neurosurgeon with accurate 3-dimensional volumetric information for navigation based on MR-specific image signatures that are widely used to identify abnormal and resectable tissue. PpIX FGR provides the neurosurgeon with an intuitive guidance tool that delineates tumor tissue in real time from which the current resection margin can be evaluated. Thus, the dual-modality approach described here takes advantage of pathophysiological changes in tumor cell metabolism (e.g. increased PpIX production and accumulation), blood-brain barrier breakdown (e.g. gadolinium enhancement on MRI), and tumor-specific biological signatures (e.g. astrocytic tumors) to improve the accuracy of tumor identification in the operating room.
Current technologies for intraoperative updating during brain surgery include intraoperative MRI, CT, and US (table 3). These technologies provide intraoperative feedback to the neurosurgeon, counteracting error in navigational accuracy due to brain deformation [24, 25, 26, 27, 28, 29, 30, 31, 32]. Each technology has pros and cons with respect to its costs, limitations and capabilities for tissue contrast, and effects on surgical procedures and workflow that remain to be fully understood and evaluated in the operative setting. In this study, FGR-IGW combines intraoperative feedback from surface fluorescence with a biomechanical model enabled through intraoperative US to update the pMR images, providing the neurosurgeon with real-time feedback on tumor fluorescence coregistered with periodically compensated 3-dimensional uMR views for enhanced neuronavigation.
Table 3.
Technologies for intraoperative updating during brain surgery
| FGR-IGW | iMRI | iCT | iUS |
|---|---|---|---|
| Pros | |||
| High soft-tissue contrast | High spatial resolution [24, 30] | Mobility of iCT system [29, 31] | Real-time feedback [25, 27, 30, 32] |
| 3-D image guidance using MR image features with increased intraoperative accuracy | High contrast resolution [24, 30] | Capability of monitoring intraoperative complications [29, 31] | Easy-to-use instruments [27] |
| Mobility of microscope with minimal OR modifications | Better differentiation of normal from abnormal tissue [24] | High soft-tissue contrast for contrast enhancing tissue [31, 32] | Relatively inexpensive costs [27] |
| Minimal disruption to surgical workflow | High soft-tissue contrast for noncontrast enhancing tissue [32] | Minor modifications in OR [31, 32] | |
| Real-time feedback of fluores- cence characteristics | Use of same instruments and equipment [32] |
||
| Fewer limitations in patient positioning [32] | |||
| Cons | |||
| Detection of only surface fluo- rescence | No real-time feedback [30] | No real-time feedback [29] | Low contrast resolution [25, 28, 30] |
| Limits in fluorescing tumors using -ALA | Interrupts surgical flow [30] | Low contrast resolution [30, 32] | Low spatial resolution [24, 25, 28] |
| Model updating dependent on sparse data from US | Prolongs time of surgery [30] | Low spatial resolution [32] | Restricted field of view due to probe placement [24] |
| Administration of -ALA with minimal drug side effects | Special shielded room [28, 31, 32] | Increased radiation dose [30, 32] | Difficulty interpreting results, requiring specialized user [25, 27, 30, 32] |
| 5–20 min for uMR generation | Specialized instruments and equipments [32] |
Interrupts surgical flow [29] |
Artifacts due to blood, air, and instruments [25, 27] |
| Specialized personnel in the OR [32] |
Low soft-tissue contrast for noncontrast enhancing tissue [31, 32] |
||
| Limitations in patient positioning [32] |
Artifacts due to patient positioning devices [32] |
||
| High costs [28, 31, 32] |
Prolongs time of surgery [32] |
||
| Limited access to operating field [28] | |||
iMRI = Intraoperative MRI; iCT = intraoperative CT; iUS = intraoperative US.
Although this dual-modality system provides a platform for integrating two powerful guidance technologies, some limitations remain. FGR currently detects only the visually apparent surface levels of fluorescence, since thin layers of blood and/or intervening tissue can obscure subsurface fluorescence emissions. As noted with the GS case, not all tumor tissue produces observable PpIX fluorescence (i.e., sarcomatous element of the tumor). As such, the optimal brain tumor population for efficient use of PpIX FGR needs to be determined. We are currently studying various brain tumor histologies in patients undergoing FGR to determine the most appropriate and relevant biologies for efficient accumulation of PpIX to observable levels. In addition, our group is developing intraoperative fluorescent probes to target non-PpIX-fluorescent tumors. We continue our ongoing efforts to develop a software platform to execute the deformation model intraoperatively as efficiently as possible. Our approach exploits intraoperative US to create a sparse data set to drive the model where one US image is acquired before durotomy, followed by a series of intraoperative US images at different stages during the surgery in order to generate corresponding uMR images [33]. We have also utilized a stereovision system that can provide surface sparse data to improve the generation of uMR images with minimal disruption to the surgical workflow [10, 11].
Conclusion
We present an implementation of a deformation model guided by intraoperatively acquired data in the setting of PpIX FGR of brain tumors. FGR was used to delineate and guide the resection of fluorescent tissue visually evident within the surgical field. A biomechanical brain deformation model provided uMR images by assimilating intraoperative data acquired with 3-dimensional intraoperative US that improved the correspondence between the volumetric representation of tumor and the fluorescing biomarker of tumor associated with the surgeon's visual field. Postoperative analysis of fluorescence signatures and the uMR images were complementary in more accurately identifying tumor tissue confirmed histopathologically from resected specimens. We believe that this dual-modality approach which uses a deformation model that compensates for brain shift coregistered with fluorescence imaging can provide the neurosurgeon with an accurate, intuitive platform that improves intraoperative guidance during tumor resection.
Acknowledgements
This research was supported by National Institutes of Health grants R01NS052274-02 and R01EB002082-13. We acknowledge DUSA Pharmaceuticals (Tarrytown, N.Y., USA), Carl Zeiss (Carl Zeiss Surgical GmbH, Oberkochen, Germany), Medtronic Navigation (Louisville, Colo., USA), and Phillips Medical Systems (Bothell, Wash., USA) for the supply of ALA, and the use of the OPMI Pentero® surgical microscope, StealthStation® Treon® navigation system, and iU22 3D ultrasound system, respectively.
Appendix
A biomechanical brain deformation model computed with the finite element method was used to estimate displacement vectors within the whole-brain volume. This model can be represented by the following coupled equations, where u is the displacement, p is the pore fluid pressure, and the other parameters symbolize the tissue mechanical properties:
| (1) |
| (2) |
These equations are discretized into a matrix form, Kx = b, where K is the stiffness matrix, x is the displacement and pore fluid pressure to be computed, and b contains the forcing conditions.
The patient's parenchymal volume was isolated within the pMR images using a level set segmentation algorithm [34]. A triangular surface mesh and its corresponding tetrahedral volume mesh were generated using the segmented brain. After patient registration, boundary conditions were assigned to different types of surface nodes as follows: (1) craniotomy nodes were identified using the contour line drawn by the surgeon and were allowed to move freely; (2) brainstem nodes were also allowed to move unconstrained; (3) fluid drainage was defined by a plane passing through the lowest craniotomy node which was perpendicular to the direction of gravity, with elements below and above being assigned with different parameters (e.g. saturated with fluid or not); (4) a second plane was determined by moving the fluid plane along the direction of gravity by 20 mm; all boundary nodes above this plane except the craniotomy nodes were assigned as contact nodes that are free to move towards or away from the inner surface of the skull and were constrained to motion tangential to the skull, if and when, they moved into direct contact during the displacement computations; (5) other boundary nodes were assigned as fixed, meaning that they were only allowed to move tangentially with respect to the skull.
Subsequently, a master surface was generated by projecting the brain boundary nodes along the average nodal normal by a specified distance to simulate the inner surface of the skull [17]. The data used to drive the model estimates were the displacements between predurotomy and immediately postdurotomy ultrasound images. A mutual information-based rigid registration followed by a B-spline nonrigid registration was performed to align the predurotomy and postdurotomy ultrasound images and compute displacement vectors. Figure 4 shows typical US images from patient 1 before and after durotomy as well as their un- and re-registered overlays which were used to extract intraoperative displacement data. Displacement vectors were mapped from US coordinates to pMR coordinates through a series of transformations and used as data to drive the biomechanical model. To generate a displacement map for model assimilation, the tumor does not have to be well defined on US per se as long as features exist in both US and MR image volumes which correspond sufficiently well to allow the mutual information registration to occur successfully. Once the displacement map has been generated, the model will assimilate the measured data to produce whole-brain deformation in order to create the model uMR images.
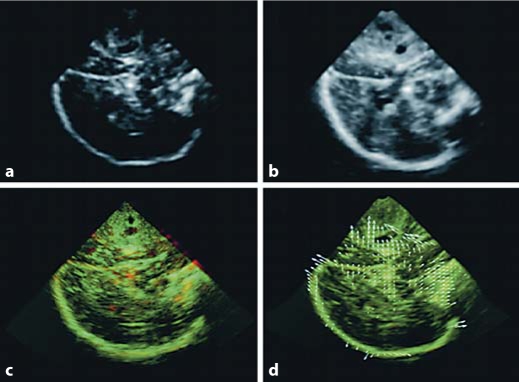
Fig. 4.
Cross-sectional images of intraoperative 3-dimensional US from patient 1. US image before (a) and after (b) dura opening. Overlays of US images before (red) and after (green) dura opening (regions appearing in yellow align) are shown in c and d. Apparent misalignment due to brain shift is evident in c; after both rigid and nonrigid registration with computed displacement vectors (white arrows) is presented in d.
A generalized least-squares inverse method was used to solve the estimation problem by minimizing the difference between measured data, d, and the model estimate, x [13]. The model constraint was embedded in the objective function through Lagrange multipliers to form an augmented quadratic expression
| (3) |
where A is the sampling matrix which computes the model estimate at locations where the measurements were made. W∊ is the inverse of the covariance matrix of the misfit, ∊, between measured data and model estimates, and Wb is the inverse of the covariance of forcing conditions, b.
The objective function is minimized when all derivatives are zero, and the resulting set of equations was solved using the steepest gradient descent algorithm with displacement vectors throughout the whole-brain volume as output files [17, 35]. The pMR image was then deformed using these displacement vectors and uMR images were generated. A schema of the biomechanical brain deformation modeling process is shown in figure 5.
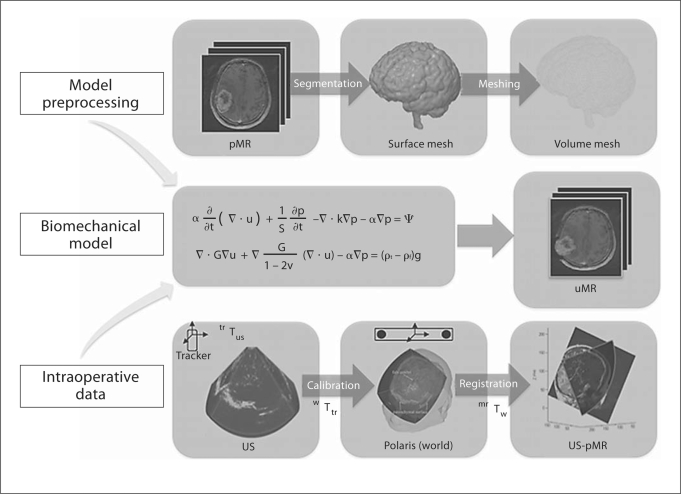
Fig. 5.
Schema of the brain deformation model. Model preprocessing: the brain is segmented from the pMR image to generate triangular surface and tetrahedral volumetric meshes, and appropriate boundary conditions are assigned. All preprocessing steps are performed prior to surgery. Intraoperative data: tracked 3-dimensional US images are acquired before and after durotomy, and are spatially merged with the patient's head and then re-registered with the pMR image. Displacement data are computed from registered pre- and postdurotomy US images to drive a biomechanical model. Biomechanical model: whole-brain deformation is computed based on boundary conditions assigned and intraoperative sparse data provided to generate an uMR image volume.
Footnotes
This study was presented in part at the 77th Annual Meeting of the American Association of Neurological Surgeons, San Diego, Calif., USA, May 5, 2009.
Conflict of Interest
Dr. David W. Roberts serves on the data monitoring committee for a Medtronic deep brain stimulation study.
References
- 1.Utsuki S, Miyoshi N, Oka H, Miyajima Y, Shimizu S, Suzuki S, Fujii K. Fluorescence-guided resection of metastatic brain tumors using a 5-aminolevulinic acid-induced protoporphyrin IX: pathological study. Brain Tumor Pathol. 2007;24:53–55. doi: 10.1007/s10014-007-0223-3. [DOI] [PubMed] [Google Scholar]
- 2.Stummer W, Stocker S, Wagner S, Stepp H, Fritsch C, Goetz C, Goetz AE, Kiefmann R, Reulen HJ. Intraoperative detection of malignant gliomas by 5-aminolevulinic acid-induced porphyrin fluorescence. Neurosurgery. 1998;42:518–525. doi: 10.1097/00006123-199803000-00017. [DOI] [PubMed] [Google Scholar]
- 3.Stummer W, Stepp H, Moller G, Ehrhardt A, Leonhard M, Reulen HJ. Technical principles for protoporphyrin-IX-fluorescence guided microsurgical resection of malignant glioma tissue. Acta Neurochir (Wien) 1998;140:995–1000. doi: 10.1007/s007010050206. [DOI] [PubMed] [Google Scholar]
- 4.Stummer W, Reulen HJ, Novotny A, Stepp H, Tonn JC. Fluorescence-guided resections of malignant gliomas – An overview. Acta Neurochir Suppl. 2003;88:9–12. doi: 10.1007/978-3-7091-6090-9_3. [DOI] [PubMed] [Google Scholar]
- 5.Stummer W, Reulen HJ, Meinel T, Pichlmeier U, Schumacher W, Tonn JC, Rohde V, Oppel F, Turowski B, Woiciechowsky C, Franz K, Pietsch T. Extent of resection and survival in glioblastoma multiforme: identification of and adjustment for bias. Neurosurgery. 2008;62:564–576. doi: 10.1227/01.neu.0000317304.31579.17. [DOI] [PubMed] [Google Scholar]
- 6.Stummer W, Pichlmeier U, Meinel T, Wiestler OD, Zanella F, Reulen HJ. Fluorescence-guided surgery with 5-aminolevulinic acid for resection of malignant glioma: a randomised controlled multicentre phase III trial. Lancet Oncol. 2006;7:392–401. doi: 10.1016/S1470-2045(06)70665-9. [DOI] [PubMed] [Google Scholar]
- 7.Stummer W, Novotny A, Stepp H, Goetz C, Bise K, Reulen HJ. Fluorescence-guided resection of glioblastoma multiforme by using 5-aminolevulinic acid-induced porphyrins: a prospective study in 52 consecutive patients. J Neurosurg. 2000;93:1003–1013. doi: 10.3171/jns.2000.93.6.1003. [DOI] [PubMed] [Google Scholar]
- 8.Pichlmeier U, Bink A, Schackert G, Stummer W. Resection and survival in glioblastoma multiforme: an RTOG recursive partitioning analysis of ALA study patients. Neuro Oncol. 2008;10:1025–1034. doi: 10.1215/15228517-2008-052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hefti M, von Campe G, Moschopulos M, Siegner A, Looser H, Landolt H. 5-Aminolevulinic acid induced protoporphyrin IX fluorescence in high-grade glioma surgery: a one-year experience at a single institution. Swiss Med Wkly. 2008;138:180–185. doi: 10.4414/smw.2008.12077. [DOI] [PubMed] [Google Scholar]
- 10.Sun H, Roberts DW, Farid H, Wu Z, Hartov A, Paulsen KD. Cortical surface tracking using a stereoscopic operating microscope. Neurosurgery. 2005;56:86–97. doi: 10.1227/01.neu.0000146263.98583.cc. [DOI] [PubMed] [Google Scholar]
- 11.Sun H, Lunn KE, Farid H, Wu Z, Roberts DW, Hartov A, Paulsen KD. Stereopsis-guided brain shift compensation. IEEE Trans Med Imaging. 2005;24:1039–1052. doi: 10.1109/TMI.2005.852075. [DOI] [PubMed] [Google Scholar]
- 12.Nimsky C, Ganslandt O, Cerny S, Hastreiter P, Greiner G, Fahlbusch R. Quantification of, visualization of, and compensation for brain shift using intraoperative magnetic resonance imaging. Neurosurgery. 2000;47:1070–1079. doi: 10.1097/00006123-200011000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Lunn KE, Paulsen KD, Liu F, Kennedy FE, Hartov A, Roberts DW. Data-guided brain deformation modeling: evaluation of a 3-D adjoint inversion method in porcine studies. IEEE Trans Biomed Eng. 2006;53:1893–1900. doi: 10.1109/TBME.2006.881771. [DOI] [PubMed] [Google Scholar]
- 14.Hill DL, Maurer CR, Jr, Maciunas RJ, Barwise JA, Fitzpatrick JM, Wang MY. Measurement of intraoperative brain surface deformation under a craniotomy. Neurosurgery. 1998;43:514–526. doi: 10.1097/00006123-199809000-00066. [DOI] [PubMed] [Google Scholar]
- 15.Utsuki S, Oka H, Sato S, Shimizu S, Suzuki S, Tanizaki Y, Kondo K, Miyajima Y, Fujii K. Histological examination of false positive tissue resection using 5-aminolevulinic acid-induced fluorescence guidance. Neurol Med Chir (Tokyo) 2007;47:210–213. doi: 10.2176/nmc.47.210. [DOI] [PubMed] [Google Scholar]
- 16.Toms SA, Lin WC, Weil RJ, Johnson MD, Jansen ED, Mahadevan-Jansen A. Intraoperative optical spectroscopy identifies infiltrating glioma margins with high sensitivity. Neurosurgery. 2005;57:382–391. doi: 10.1227/01.neu.000176855.39826.2d. [DOI] [PubMed] [Google Scholar]
- 17.Ji S, Roberts DW, Hartov A, Paulsen KD. Brain-skull contact boundary conditions in an inverse computational deformation model. Med Image Anal. 2009;13:659–672. doi: 10.1016/j.media.2009.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pallatroni H, Hartov A, McInerney J, Platenik LA, Miga MI, Kennedy FE, Paulsen KD, Roberts DW. Coregistered ultrasound as a neurosurgical guide. Stereotact Funct Neurosurg. 1999;73:143–147. doi: 10.1159/000029775. [DOI] [PubMed] [Google Scholar]
- 19.Hartov A, Roberts DW, Paulsen KD. A comparative analysis of coregistered ultrasound and magnetic resonance imaging in neurosurgery. Neurosurgery. 2008;62:91–99. doi: 10.1227/01.neu.0000317377.15196.45. [DOI] [PubMed] [Google Scholar]
- 20.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A, Scheithauer BW, Kleihues P. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114:97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lacroix M, Abi-Said D, Fourney DR, Gokaslan ZL, Shi W, DeMonte F, Lang FF, McCutcheon IE, Hassenbusch SJ, Holland E, Hess K, Michael C, Miller D, Sawaya R. A multivariate analysis of 416 patients with glioblastoma multiforme: prognosis, extent of resection, and survival. J Neurosurg. 2001;95:190–198. doi: 10.3171/jns.2001.95.2.0190. [DOI] [PubMed] [Google Scholar]
- 22.Barker FG, 2nd, Prados MD, Chang SM, Gutin PH, Lamborn KR, Larson DA, Malec MK, McDermott MW, Sneed PK, Wara WM, Wilson CB. Radiation response and survival time in patients with glioblastoma multiforme. J Neurosurg. 1996;84:442–448. doi: 10.3171/jns.1996.84.3.0442. [DOI] [PubMed] [Google Scholar]
- 23.Albert FK, Forsting M, Sartor K, Adams HP, Kunze S. Early postoperative magnetic resonance imaging after resection of malignant glioma: objective evaluation of residual tumor and its influence on regrowth and prognosis. Neurosurgery. 1994;34:45–60. doi: 10.1097/00006123-199401000-00008. [DOI] [PubMed] [Google Scholar]
- 24.Black PM, Moriarty T, Alexander E, 3rd, Stieg P, Woodard EJ, Gleason PL, Martin CH, Kikinis R, Schwartz RB, Jolesz FA. Development and implementation of intraoperative magnetic resonance imaging and its neurosurgical applications. Neurosurgery. 1997;41:831–842. doi: 10.1097/00006123-199710000-00013. [DOI] [PubMed] [Google Scholar]
- 25.Gronningsaeter A, Kleven A, Ommedal S, Aarseth TE, Lie T, Lindseth F, Lango T, Unsgard G. Sonowand, an ultrasound based neuronavigation system. Neurosurgery. 2000;47:1373–1379. [PubMed] [Google Scholar]
- 26.Grunert P, Muller-Forell W, Darabi K, Reisch R, Busert C, Hopf N, Perneczky A. Basic principles and clinical applications of neuronavigation and intraoperative computed tomography. Comput Aided Surg. 1998;3:166–173. doi: 10.1002/(SICI)1097-0150(1998)3:4<166::AID-IGS6>3.0.CO;2-E. [DOI] [PubMed] [Google Scholar]
- 27.Keles GE, Lamborn KR, Berger MS. Coregistration accuracy and detection of brain shift using intraoperative sononavigation during resection of hemispheric tumors. Neurosurgery. 2003;53:556–562. doi: 10.1227/01.neu.0000080949.44837.4c. [DOI] [PubMed] [Google Scholar]
- 28.Letteboer MM, Willems PW, Viergever MA, Niessen WJ. Brain shift estimation in image-guided neurosurgery using 3-D ultrasound. IEEE Trans Biomed Eng. 2005;52:268–276. doi: 10.1109/TBME.2004.840186. [DOI] [PubMed] [Google Scholar]
- 29.Matula C, Rossler K, Reddy M, Schindler E, Koos WT. Intraoperative computed tomography guided neuronavigation: concepts, efficiency, and work flow. Comput Aided Surg. 1998;3:174–182. doi: 10.1002/(SICI)1097-0150(1998)3:4<174::AID-IGS7>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 30.Nabavi A, Black PM, Gering DT, Westin CF, Mehta V, Pergolizzi RS, Jr, Ferrant M, Warfield SK, Hata N, Schwartz RB, Wells WM, 3rd, Kikinis R, Jolesz FA. Serial intraoperative magnetic resonance imaging of brain shift. Neurosurgery. 2001;48:787–797. doi: 10.1097/00006123-200104000-00019. [DOI] [PubMed] [Google Scholar]
- 31.Nakao N, Nakai K, Itakura T. Updating of neuronavigation based on images intraoperatively acquired with a mobile computerized tomographic scanner: technical note. Minim Invasive Neurosurg. 2003;46:117–120. doi: 10.1055/s-2003-39344. [DOI] [PubMed] [Google Scholar]
- 32.Uhl E, Zausinger S, Morhard D, Heigl T, Scheder B, Rachinger W, Schichor C, Tonn JC. Intraoperative computed tomography with integrated navigation system in a multidisciplinary operating suite. Neurosurgery. 2009;64:231–239. doi: 10.1227/01.NEU.0000340785.51492.B5. [DOI] [PubMed] [Google Scholar]
- 33.Roberts DW, Miga MI, Hartov A, Eisner S, Lemery JM, Kennedy FE, Paulsen KD. Intraoperatively updated neuroimaging using brain modeling and sparse data. Neurosurgery. 1999;45:1199–1206. [PubMed] [Google Scholar]
- 34.Wu Z, Paulsen KD, Sullivan JM., Jr Adaptive model initialization and deformation for automatic segmentation of T1-weighted brain MRI data. IEEE Trans Biomed Eng. 2005;52:1128–1131. doi: 10.1109/TBME.2005.846709. [DOI] [PubMed] [Google Scholar]
- 35.Ji S, Hartov A, Roberts DW, Paulsen KD. Data assimilation using a gradient descent method for estimation of intraoperative brain deformation. Med Image Anal. 2009;13:744–756. doi: 10.1016/j.media.2009.07.002. [DOI] [PMC free article] [PubMed] [Google Scholar]