Abstract
Background
Papillon-Lefèvre syndrome (PLS; OMIM 245000) is a rare autosomal recessive disorder. Clinically, PLS is characterized by hyperkeratosis involving the palms, soles, elbows and knees which is followed later on by periodontitis, destruction of alveolar bone and loss of primary and permanent teeth. The condition is caused by mutations in the cathepsin C (CTSC) gene.
Methods
We analyzed the DNA of members from 3 consanguineous families for mutations in the CTSC gene by direct sequencing analysis. We then performed haplotype analysis.
Results
We identified an identical recurrent missense mutation, R272P, in all 3 families. Microsatellite marker analysis around the CTSC gene revealed the same haplotype on the mutation-carrying allele in all 3 families.
Conclusion
The presence of this common mutation in families from 2 different geographical areas provides evidence for a founder effect for CTSC mutations in PLS.
Key Words: Cathepsin C, Founder mutation, Palmoplantar keratoderma/hyperkeratosis, Periodontitis
Introduction
Cathepsin C (CTSC), also known as dipeptidyl aminopeptidase I, is a lysosomal proteinase of the papain type that functions in removing dipeptides from the amino terminus of its substrates [1, 2]. It is chemically composed of 4 identical units. Each unit is composed of 3 different polypeptide chains: the heavy chain, the light chain and the exclusion domain [3]. The exclusion domain is noncovalently attached to the heavy and light chains, forming a heterotrimeric structure [4]. CTSC is expressed in many tissues, such as the epithelium including those of the palms, soles, knees and keratinized oral gingiva [5, 6], immune cells, including natural killer cells, polymorphonuclear cells and T lymphocytes [7], and other cells such as fibroblasts and osteoclasts [8]. Thus, disturbances in the function of CTSC will lead to abnormalities especially in the places where it is highly expressed, such as the epithelium and the immune cells [7]. For instance, immune cells such as natural killer cells and T lymphocytes require the presence of activated granzyme A and granzyme B in order for them to be able to exert their cytolytic activity, and these granzymes are activated by the cleavage of their N-terminal peptide that is performed by CTSC. Therefore, mutations in the CTSC gene are expected to lead to an increased frequency of infections [1, 7, 9, 10]. Similarly in the skin, mutations in CTSC might affect the epithelial differentiation process and lead to hyperkeratosis [11].
Papillon-Lefèvre syndrome (PLS; OMIM 245000) is an autosomal recessive disorder occurring at a frequency of 1–4 persons per million with no sex predilection. PLS occurs due to mutations in the CTSC gene on chromosome 11q14 and involves the skin, gingiva and teeth [12]. To date, more than 50 mutations have been reported in the literature. Here, we report 3 families from distinct geographical regions demonstrating the same recurrent mutation in the CTSC gene. Our data provide evidence for a founder effect among CTSC mutations in PLS.
Patients and Methods
Patients
We analyzed 3 families affected with classical features of PLS, 2 from Pakistan and 1 from Lebanon. After obtaining informed consent, we collected peripheral blood samples from members of these families in EDTA-containing tubes (under institutional approval and in adherence to the Declaration of Helsinki principles). Genomic DNA was isolated from the samples according to standard techniques.
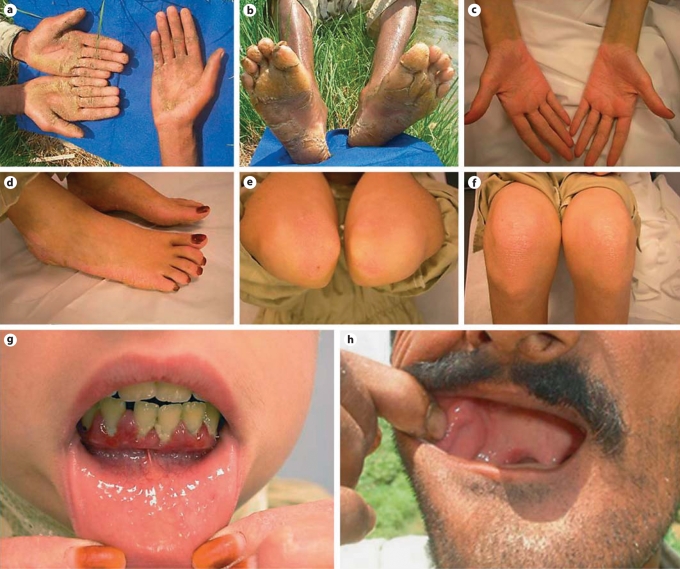
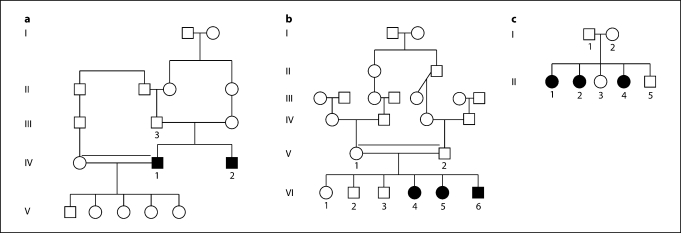
The patients reported that initially they developed palmoplantar keratoderma which later on progressed proximally followed a few years later by gingivitis and loss of teeth. On physical examination, the patients showed palmoplantar hyperkeratosis (fig. 1a, b) with progredient (proximal involvement, extending beyond the hand margins; fig. 1c) and transgredient spread (involvement of the ventral and dorsal surfaces of the hand; fig. 1d). Many of them also had psoriasiform lesions involving the elbows (fig. 1e) and knees (fig. 1f). Young patients had evidence of gingivitis with periodontitis (fig. 1g) while older patients had nearly complete loss of their teeth (fig. 1h). The affected individuals reported recurrent dermatophytic infections. The patients were otherwise healthy. Both males and females were equally affected, suggesting autosomal recessive inheritance (fig. 2a–c).
Fig. 1.
Affected individuals showing palmoplantar hyperkeratosis (a, b) with progredient (c) and transgredient (d) spread. Psoriasiform lesions were present over the elbows (e) and knees (f). g A 12-year-old girl at the start of the second cycle of tooth involvement presenting with gingivitis and periodontitis. h A man in his fourth decade with nearly complete loss of his teeth.
Fig. 2.
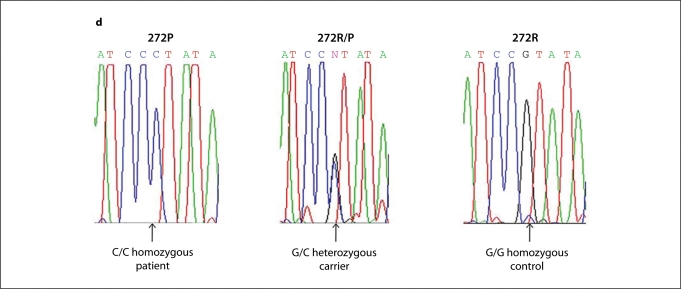
Pedigrees from the 3 families reveal an autosomal recessive pattern of inheritance. Two families are from Pakistan (a, b) and one from Lebanon (c). d Affected individuals from the 3 families showing a homozygous mutation with a G→C change at nucleotide 815 (indicated by arrow), carriers being heterozygotes for the mutation; a control sequence is shown for comparison. Filled circles and squares represent females and males, respectively. Double lines indicate consanguinity.
Mutation Analysis
All exons of the CTSC gene with adjacent sequences of exon-intron borders were amplified by PCR with primers and conditions described previously [13]. The amplified PCR products were directly sequenced in an ABI Prism 310 Automated Sequencer, using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems, Foster City, Calif., USA).
Haplotype Analysis
In order to determine whether the mutation detected was a founder mutation or whether the site was a hot spot for mutations, genomic DNA samples from affected individuals were amplified using the 2 known microsatellite markers CTSC-3′and CTSC-int3 [14] that flank the site of the mutation using the primers listed in table 1. The amplification conditions for each PCR were 94°C for 2 min, followed by 35 cycles of 94°C for 30 s, 55°C for 30 s and 72°C for 30 s, with a final extension at 72°C for 7 min. The amplified PCR products were directly sequenced in an ABI Prism 310 Automated Sequencer, using the ABI Prism Big Dye Terminator Cycle Sequencing Ready Reaction Kit (PE Applied Biosystems). We determined the number of microsatellite repeats by visual inspection, which consisted of AT in the case of CTSC-3′ and GT in the case of CTSC-int3.
Table 1.
Primers used to amplify the microsatellite markers
| Microsatellite markers |
||
|---|---|---|
| CTSC-3′ |
CTSC-int3 |
|
| Forward primers | ACCCCTCATAAGCCCTTCTCTATC | TCTCATGCCACCATTCAGGTAA |
| Reverse primers | CTAAACTCAGGCCCTTCATCATTC | CAATTTATCAACTTTTGCACAGT |
Results
Identification of a Recurrent Mutation in the CTSC Gene
We performed direct sequencing analysis using DNA from affected members in all 3 families. Unexpectedly, we detected a missense mutation c.815G→C which led to a change of the amino acid arginine into proline, R272P, in affected patients from each family (fig. 2d). The uninvolved members were either heterozygous for the mutation or had the wild-type sequence consistent with autosomal recessive inheritance.
Haplotype Analysis
Direct sequencing analysis of both microsatellite markers around the CTSC gene showed a homozygous sequencing pattern and the same number of microsatellite repeats on the mutation-carrying allele in all 3 families, with 21 repeats of AT in the case of CTSC-3′and 14 GT repeats in the case of CTSC-int3.
Discussion
CTSC is a lysosomal enzyme that has been shown to be essential for the activation of other serine proteases such as elastase, cathepsin G or proteinase 3 [15] that are required for the activation of both the innate and the adaptive immune systems. High levels of CTSC in the skin suggest a role for it in epithelial differentiation and desquamation [13]. Mutations in the CTSC gene have been reported to result in 3 closely related conditions, i.e. PLS, Haim-Munk syndrome and prepubertal periodontitis [16]. Each of these conditions is known to exhibit autosomal recessive inheritance, and the complete absence of CTSC activity is required in order to develop the clinical phenotype of PLS [13]. Thus, residual amounts of CTSC may be sufficient to perform its functions.
PLS usually manifests itself in the first 2 years of life. It starts initially with palmoplantar hyperkeratosis and transgredient spread with the soles being more extensively involved [12]. Psoriasiform lesions may also develop over the elbows, knees, and knuckles [13]. This is followed later on by periodontitis and gingivitis with subsequent loss of primary and permanent teeth [12]. In general the development of the primary teeth is uneventful, but at the age of 3 years gingivitis and ulceration of the ginigiva start, leading to complete loss of the primary teeth by the age of 5 years [12, 17, 18]; however, after the loss of the primary teeth, the gingiva returns to normal until about 15 years of age when the same cycle starts again and results in the loss of the permanent teeth [12, 19, 20]. The involvement of the gingivae and the periodontitis are believed to be secondary to the persistence of bacteria and the inability to activate the immune system which requires CTSC to function normally [5, 21]. Some patients may also be predisposed to pyodermas, and sometimes severe and even fatal internal abscesses may occur [22]. It has also been found that Japanese PLS patients are predisposed to develop melanoma at sites of hyperkeratosis [1].
To date, several mutations in the CTSC gene have been reported in patients with PLS, including missense, nonsense, insertion and deletion mutations. The most common mutation is the missense mutation R272P on exon 6, whereas exon 7 contains most of the mutations [16]. There is no genotype-phenotype correlation [21].
Here, we analyze 3 families from 2 different geographical regions, one from Lebanon and the others from Pakistan. Involved members from the 3 families had typical clinical features of PLS with no difference in severity among members of the same or different families. Genetic analysis revealed the same recurrent missense mutation R272P in affected members of the 3 families. Previously 2 families with PLS have been reported from Lebanon with one of them showing the same missense mutation as our families here and the second family having a nonsense mutation R210X [13]. This mutation, R272P, has never been reported in Pakistani patients, although 4 different missense mutations have previously been found in patients from Pakistan [13, 15]. R272P is located within the heavy chain of CTSC [3] in a highly conserved region encoding part of the second turn of the central helix from the carboxy terminus of CTSC. This mutation is likely to cause distortion of the three-dimensional structure of the protein resulting in a dysfunctional protein [23]. Interestingly, the mutation R272P has been reported in populations from different regions in the world including Lebanon, Turkey, France, the Netherlands, Saudi Arabia and Europe [13, 14, 15, 24, 25, 26]. The fact that the mutation R272P is the most common mutation in the CTSC gene might suggest that it is located at a site which is a hot spot for mutation, but our results are consistent with it being a founder mutation. Moreover it has been shown in 4 Saudi Arabian families that R272P is a founder mutation. The close geographical proximity between Lebanon, Saudi Arabia and Pakistan may point toward a common ancestral origin for PLS.
Acknowledgements
We gratefully acknowledge the families for having participated in this study. This study was supported by USPHS NIH grant from NIH/NIAMS RO1 AR44924 (to A.M.C.).
References
- 1.Nakajima K, Nakano H, Takiyoshi N, Rokunohe A, Ikenaga S, Aizu T, Kaneko T, Mitsuhashi Y, Sawamura D. Papillon-Lefèvre syndrome and malignant melanoma: a high incidence of melanoma development in Japanese palmoplantar keratoderma patients. Dermatology. 2008;217:58–62. doi: 10.1159/000124340. [DOI] [PubMed] [Google Scholar]
- 2.Rao NV, Rao GV, Hoidal JR. Human dipeptidyl-peptidase I: gene characterization, localization, and expression. J Biol Chem. 1997;272:10260–10265. doi: 10.1074/jbc.272.15.10260. [DOI] [PubMed] [Google Scholar]
- 3.Mølgaard A, Arnau J, Lauritzen C, Larsen S, Petersen G, Pedersen J. The crystal structure of human dipeptidyl peptidase I (cathepsin C) in complex with the inhibitor Gly-Phe-CHN2. Biochem J. 2007;401:645–650. doi: 10.1042/BJ20061389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Olsen JG, Kadziola A, Lauritzen C, Pedersen J, Larsen S, Dahl SW. Tetrameric dipeptidyl peptidase I directs substrate specificity by use of the residual pro-part domain. FEBS Lett. 2001;506:201–206. doi: 10.1016/s0014-5793(01)02911-8. [DOI] [PubMed] [Google Scholar]
- 5.de Haar SF, Hiemstra PS, van Steenbergen MT, Everts V, Beertsen W. Role of polymorphonuclear leukocyte-derived serine proteinases in defense against Actinobacillus actinomycetemcomitans. Infect Immun. 2006;74:5284–5291. doi: 10.1128/IAI.02016-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hart TC, Hart PS, Bowden DW, Michalec MD, Callison SA, Walker SJ, Zhang Y, Firatli E. Mutations of the cathepsin C gene are responsible for Papillon-Lefèvre syndrome. J Med Genet. 1999;36:881–887. [PMC free article] [PubMed] [Google Scholar]
- 7.Meade JL, de Wynter EA, Brett P, Sharif SM, Woods CG, Markham AF, Cook GP. A family with Papillon-Lefèvre syndrome reveals a requirement for cathepsin C in granzyme B activation and NK cell cytolytic activity. Blood. 2006;107:3665–3668. doi: 10.1182/blood-2005-03-1140. [DOI] [PubMed] [Google Scholar]
- 8.Janjua SA, Iftikhar N, Hussain I, Khachemoune A. Dermatologic, periodontal, and skeletal manifestations of Haim-Munk syndrome in two siblings. J Am Acad Dermatol. 2008;58:339–344. doi: 10.1016/j.jaad.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Smyth MJ, McGuire MJ, Thia KY. Expression of recombinant human granzyme B: a processing and activation role for dipeptidyl peptidase I. J Immunol. 1995;154:6299–6305. [PubMed] [Google Scholar]
- 10.Pham CT, Ley TJ. Dipeptidyl peptidase I is required for the processing and activation of granzymes A and B in vivo. Proc Natl Acad Sci USA. 1999;96:8627–8632. doi: 10.1073/pnas.96.15.8627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pilger U, Hennies HC, Truschnegg A, Aberer E. Late-onset Papillon-Lefèvre syndrome without alteration of the cathepsin C gene. J Am Acad Dermatol. 2003;49(suppl):S240–S243. doi: 10.1016/s0190-9622(03)01558-5. [DOI] [PubMed] [Google Scholar]
- 12.Canger EM, Celenk P, Devrim I, Yenisey M, Gunhan O. Intraoral findings of Papillon-Lefèvre syndrome. J Dent Child (Chic) 2008;75:99–103. [PubMed] [Google Scholar]
- 13.Toomes C, James J, Wood AJ, Wu CL, McCormick D, Lench N, Hewitt C, Moynihan L, Roberts E, Woods CG, Markham A, Wong M, Widmer R, Ghaffar KA, Pemberton M, Hussein IR, Temtamy SA, Davies R, Read AP, Sloan P, Dixon MJ, Thakker NS. Loss-of-function mutations in the cathepsin C gene result in periodontal disease and palmoplantar keratosis. Nat Genet. 1999;23:421–424. doi: 10.1038/70525. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Y, Lundgren T, Renvert S, Tatakis DN, Firatli E, Uygur C, Hart PS, Gorry MC, Marks JJ, Hart TC. Evidence of a founder effect for four cathepsin C gene mutations in Papillon-Lefèvre syndrome patients. J Med Genet. 2001;38:96–101. doi: 10.1136/jmg.38.2.96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de Haar SF, Jansen DC, Schoenmaker T, De Vree H, Everts V, Beertsen W. Loss-of-function mutations in cathepsin C in two families with Papillon-Lefèvre syndrome are associated with deficiency of serine proteinases in PMNs. Hum Mutat. 2004;23:524. doi: 10.1002/humu.9243. [DOI] [PubMed] [Google Scholar]
- 16.Selvaraju V, Markandaya M, Prasad PV, Sathyan P, Sethuraman G, Srivastava SC, Thakker N, Kumar A. Mutation analysis of the cathepsin C gene in Indian families with Papillon-Lefèvre syndrome. BMC Med Genet. 2003;4:5. doi: 10.1186/1471-2350-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lundgren T, Renvert S. Periodontal treatment of patients with Papillon-Lefèvre syndrome: a 3-year follow-up. J Clin Periodontol. 2004;31:933–938. doi: 10.1111/j.1600-051X.2004.00591.x. [DOI] [PubMed] [Google Scholar]
- 18.Firatli E, Gürel N, Efeoglu A, Badur S. Clinical and immunological findings in 2 siblings with Papillon-Lefèvre syndrome. J Periodontol. 1996;67:1210–1215. doi: 10.1902/jop.1996.67.11.1210. [DOI] [PubMed] [Google Scholar]
- 19.Fardal O, Drangsholt E, Olsen I. Palmar plantar keratosis and unusual periodontal findings: observations from a family of 4 members. J Clin Periodontol. 1998;25:181–184. doi: 10.1111/j.1600-051x.1998.tb02425.x. [DOI] [PubMed] [Google Scholar]
- 20.Ullbro C, Brown A, Twetman S. Preventive periodontal regimen in Papillon-Lefèvre syndrome. Pediatr Dent. 2005;27:226–232. [PubMed] [Google Scholar]
- 21.Noack B, Görgens H, Hempel U, Fanghänel J, Hoffmann T, Ziegler A, Schackert HK. Cathepsin C gene variants in aggressive periodontitis. J Dent Res. 2008;87:958–963. doi: 10.1177/154405910808701017. [DOI] [PubMed] [Google Scholar]
- 22.Pham CT, Ivanovich JL, Raptis SZ, Zehnbauer B, Ley TJ. Papillon-Lefèvre syndrome: correlating the molecular, cellular, and clinical consequences of cathepsin C/dipeptidyl peptidase I deficiency in humans. J Immunol. 2004;173:7277–7281. doi: 10.4049/jimmunol.173.12.7277. [DOI] [PubMed] [Google Scholar]
- 23.Turk D, Janjić V, Stern I, Podobnik M, Lamba D, Dahl SW, Lauritzen C, Pedersen J, Turk V, Turk B. Structure of human dipeptidyl peptidase I (cathepsin C): exclusion domain added to an endopeptidase framework creates the machine for activation of granular serine proteases. EMBO J. 2001;20:6570–6582. doi: 10.1093/emboj/20.23.6570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hart TC, Hart PS, Michalec MD, Zhang Y, Marazita ML, Cooper M, Yassin OM, Nusier M, Walker S. Localisation of a gene for prepubertal periodontitis to chromosome 11q14 and identification of a cathepsin C gene mutation. J Med Genet. 2000;37:95–101. doi: 10.1136/jmg.37.2.95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang Y, Hart PS, Moretti AJ, Bouwsma OJ, Fisher EM, Dudlicek L, Pettenati MJ, Hart TC. Biochemical and mutational analyses of the cathepsin C gene (CTSC) in three North American families with Papillon-Lefèvre syndrome. Hum Mutat. 2002;20:75. doi: 10.1002/humu.9040. [DOI] [PubMed] [Google Scholar]
- 26.Lefèvre C, Blanchet-Bardon C, Jobard F, Bouadjar B, Stalder JF, Cure S, Hoffmann A, Prud’Homme JF, Fischer J. Novel point mutations, deletions, and polymorphisms in the cathepsin C gene in nine families from Europe and North Africa with Papillon-Lefèvre syndrome. J Invest Dermatol. 2001;117:1657–1661. doi: 10.1046/j.0022-202x.2001.01595.x. [DOI] [PubMed] [Google Scholar]