Abstract
We report a neocentromere event on maize chromosome 3 that occurred due to chromosome breakage. The neocentromere lies on a fragment of the short arm that lacks the primary centromere DNA elements, CentC and CRM. It is transmitted in the genomic background of oat via a new centromere (and kinetochore), as shown by immunolocalization of the oat CENH3 protein. Despite normal transmission of the maize fragment in most progeny, neocentromeres appear to vary in size within the same tissue, as shown by fluorescent measurements. A secondary truncation in one line lowered mitotic transmission to 3% and precipitously reduced the size of the chromosome. The results support the view that neocentromere formation is generally associated with major genomic disturbances such as wide species crosses or deletion of an existing centromere. The data further suggest that new centromeres may undergo a period of instability that is corrected over a period of several generations.
Key Words: CENH3, Centromere, Epigenetic, Kinetochore, Maize, Neocentromere, Oat
Centromeres must be positionally stable – otherwise homologous chromosomes will not pair correctly and can be lost at high frequencies during meiosis. The stability is perplexing because centromeres are not determined in a strictly genetic sense [Dawe and Henikoff, 2006]. A special histone called centromeric histone H3 (CENH3) replaces standard histone H3 and creates an environment that demarcates centromeres from surrounding chromatin. Several other proteins are required to build the kinetochore over a centromere, but CENH3 is widely considered the ‘centromere identifier’ [Warburton et al., 1997; Malik and Henikoff, 2001; Van Hooser et al., 2001]. Although centromere chromatin typically forms on tandem arrayed satellite repeats or special retrotransposon families, there are no common constraints on the DNA sequences CENH3 may interact with [Jiang et al., 2003]. Other data support and extend the idea that centromeres are propagated by epigenetic means. Some of the best evidence comes from chromosome addition lines where alien centromeres are maintained by centromere proteins of the host species [Jin et al., 2004], and rare cases in which the centromere chromatin forms at a new locus, creating a neocentromere [Maggert and Karpen, 2001; Nasuda et al., 2005; Alonso et al., 2007; Marshall et al., 2008].
Neocentromerization has been proposed to underlie key evolutionary events such as rapid centromere evolution, hybrid incompatibility, and speciation [Ventura et al., 2001; O’Neill et al., 2004; Marshall et al., 2008]. Extensive work in human cell lines suggests that neocentromeres have different DNA sequences but structurally similar kinetochores [Saffery et al., 2000; Alonso et al., 2003, 2007]. In contrast, ‘classical’ neocentromeres form at terminal heterochromatic knob repeats of some plants and do not share any protein similarity to the normal centromere/kinetochore [Dawe and Hiatt, 2004]. Despite its presumed importance, the process of new centromere formation is not well understood.
One mode of neocentromere formation appears to involve the direct transfer of kinetochore proteins to a new locus. Such a kinetochore transfer mechanism was first suggested by a study in Drosophila, where chromosomes were broken next to a centromere and the centromere/kinetochore moved laterally to neighboring DNA sequences [Maggert and Karpen, 2001]. In wheat, a similar neocentromere event occurred on a barley chromosome that had been introduced by a wide cross. A spontaneous breakage produced an isochromosome with no known centromere repeats and two mirror images of the chromosome 7 short arm. The newly formed centromere was transmitted normally, although some chromosome variants were produced; for example some had one or part of one of the 2 original arms. Immunolocalization of CENH3 and other key proteins confirmed that a neocentromere had formed close to the position of the original barley centromere [Nasuda et al., 2005]. A common feature of most neocentromeres is their association with genome rearrangements that cause a loss of the original centromere. It seems likely that neocentromeres arise as a natural means of stabilizing broken genomes.
We report the characterization of a second plant neocentromere. An unstable chromosome lacking a visible centromere was reported by Muehlbauer et al. [1999] as a derivative of maize chromosome 3 in an oat genomic background. We analyzed progeny and show that the unusual chromosome is a fragment of the chromosome 3 short arm (ch3S) and lacks known maize centromere DNA sequences. In contrast to the highly unstable derivative originally reported, this neocentromere now reliably transmits the maize chromosome in somatic tissue. We cloned the oat CENH3 gene and raised an antibody to the predicted protein in order to demonstrate the location of the new centromere. In apparent contrast to the neocentromeres formed in Drosophila and barley, the maize neocentromere formed at a site distant from the original, making a templating event in cis unlikely. We also observed wide variance in the size of stably transmitting neocentromeres, suggesting to us that newly formed centromeres undergo a period of epigenetic flux before (presumably) stabilizing.
Materials and Methods
Mapping of the Maize Chromosome 3 Breakage Site
The following conditions were used to PCR amplify markers from each material: 50 ng genomic DNA, 0.2 units HotStarTaq (Qiagen), 0.5 μM of each primer forward and reverse, 2.67 μM dNTPs, and 1× supplied buffer in a 15 μl total volume. The cycling program began with a 15 min incubation at 95°C to activate the enzyme followed by 36 cycles of 94°C for 30 s, 57°C or 52°C for 30 s and 72°C for 75 s, and finally 2 min at 72°C. PCR primers were either downloaded from www.maizegdb.org or designed using the Primer3 program with an optimal annealing temperature of 63°C [Rozen and Skaletsky, 2000]. Southern blots were performed using standard conditions.
Quantitative PCR of Neocentromere Material
qPCR was performed on an Eppendorf Realplex machine using the following conditions: ∼100 ng genomic DNA, 0.375 units AmpliTaq Gold (Applied Biosystems), 3 mM MgCl2, 0.3 μM of each primer forward and reverse, 200 μM dNTPs, 5% DMSO, 1× SYBR Green (Invitrogen), and 1× supplied buffer in a 15 μl total volume. Primers pairs are as follows: CentC F-(GATTGGGCATGTTCGTTGTG), R-(CACTACTTTAGGTCGAAAAC); CRM F-(CTCGTGCTCGTCAACTCAA), R-(ACCGTCACAAGTTGGTGTT); OPIE F-(GATTCCTCGCAAACGGGAW), R-(CTTGCCTACTCCACGTTGT); 5S rDNA F-(GATGCGATCATACCAGCACTA), R-(GAATGCAACACGAGGACTT). The cycling program began with a 5 min incubation at 95°C to activate the enzyme followed by 35 cycles of 95°C for 15 s, 53°C for 15 s and 72°C for 30 s, and finally 1 min at 72°C. Fold depletion was calculated by the 2–ΔΔCt method [Livak and Schmittgen, 2001] using 5S rDNA as the reference standard. Each reaction was performed in triplicate wells and averaged, excluding replicates in which the variability exceeded one cycle.
Fluorescence in situ Hybridization
Root tip preparation and FISH were performed essentially as described, except the digestion was 80 min [Lamb et al., 2007]. PCR products were amplified from maize cultivar Seneca 60, using the aforementioned primers for CentC, OPIE, CRM, or as reported for CentA [Mroczek and Dawe, 2003]
Identification of Oat CENH3 (Avena sativa CENH3)
To generate genomic clones of the 5′ region of the oatCENH3 gene, primers F2 (GCACCMGGCSGTGAGGAA) and R1 (TTCCTGATCTCCCGCAGYGC) were used. From the genomic DNA sequence (GenBank accession FJ155067), new primers were used to screen a cDNA preparation from ‘Carolina Oat’. Primers were oatCH3F2 (ACGCCCAAGAAGCAGCTCAAG) and oatrace2 (GCGACCGTGCCAGGCTTGAAC). The RTPCR products were cloned and sequenced and the sequence was deposited into GenBank (accession FJ155068). The translated sequence was used to identify a unique peptide, SKPTPKKQLKFGRSPGQTAE, which was subsequently used to generate a polyclonal antibody (Biosource, now Invitrogen).
Immunolocalization
Immunolocalization on young anther tissue was performed essentially as described [Shi and Dawe, 2006], except that dissected, unfixed anthers were treated with 2% cellulase R10, 1% pectolyase Y23 (Karlan) in 1× PHEM buffer for 15–30 min to degrade cell walls prior to fixation.
Measurements and Statistical Analysis of Kinetochores
Fluorescent signal intensities were measured using SlideBook 4.0 software (Intelligent Imaging Innovations, California), essentially as described previously [Du and Dawe, 2007]. In brief, a projection image of relevant z-stacks from a raw 3D-image was created for each cell preparation. Total CY3 intensity values were measured for non-overlapping kinetochores using a brush tool to paint each signal. Kinetochore intensities were each divided by the area and then averaged to calculate densities. When the signal was split (in anaphase I lagging chromosomes), the values were averaged and taken as one data point. Excel software was used to calculate the variance of the neocentromere and averaged oat kinetochore intensities among cells. An F-test of equality of the kinetochore variances was conducted as follows, where A is the neocentromere value, and B is the average oat value: H0 : δ2A/δ2B = 1 versus HA : δ2A/δ2B > 1; F = s2A/s2B = 4.5019, d.f. = (6,6); F0.05(6,6) = 4.2839 [F-table: Bhattacharyya and Johnson, 1977]; 4.5019 > 4.2839, therefore H0 is rejected at α = 0.05.
Results
A Neocentromere Event on Maize Chromosome Arm 3S
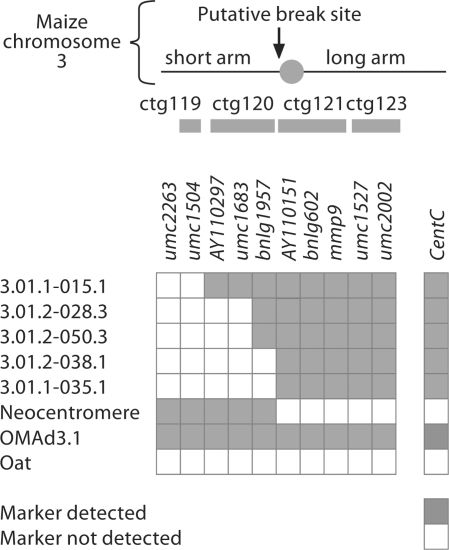
GISH analysis of progeny resulting from self mating a monosomic oat-maize chromosome 3 addition line detected a small chromosome comprised solely of maize DNA [Muehlbauer et al., 1999]. The lack of a primary constriction indicates it formed by a breakage event within or near the centromere [fig. 5D from Muehlbauer et al., 1999]. We became interested in this line, BC1F2–4/9, when a Southern blot of a sibling line suggested that it had lost its entire native centromere (summarized in fig. 1). Further mapping with SSR markers ordered along the length of maize chromosome 3 suggests that only DNA from the short arm (ch3S) remains. We hypothesize that the full maize chromosome underwent a breakage event on the short arm of ch3, distal to the centromere but proximal to marker bnlg1957, placing it between bnlg1957 and AY110151 (between contig 120 and 121, IBM2 map) (fig. 1). A new centromere stabilizing the acentric fragment is the most plausible explanation for the origin of the truncated chromosome. We wondered if newly formed centromeres have any unusual properties compared to those long established.
Fig. 1.
Neocentromere material does not contain maize centromere DNA or SSR marker DNA from the maize chromosome 3 long arm. A series of SSR markers spanning maize chromosome 3 were used to probe a panel of chromosome 3 truncation lines, BC1F2–4/9 ‘neocentromere’ progeny, a full maize chromosome 3 disomic addition line (OMAd3.1), and the oat parent of the addition cross (Sun II). The grey bars represent BAC contigs (ctg) that correlate the SSR marker position to the IBM2 physical map. The putative ch3 breakage site maps to the gap between ctg120 and ctg121 on the short arm. Each line was also Southern blotted and probed with the major maize centromere DNA element, CentC, demonstrating the loss of the original centromere in neocentromere lines.
Stability of the Neocentromere
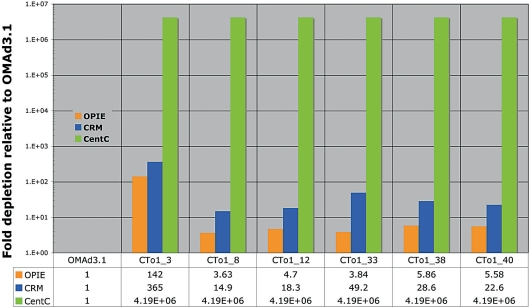
We were especially intrigued by Muehlbauer and colleagues’ [1999] evidence suggesting that the truncated fragment was completely absent in older tissue, such as tillers. Somatic loss could result from defects in the neocentromere. As a test we screened young leaves of T2 progeny selfed from the plant containing the original neocentromere event, and compared them to a full disomic ch3 addition line, OMAd3.1. A primer pair that amplifies maize, but not oat, members of the abundant OPIE family of retroelements identified 6 of 40 positive progeny. Quantitative PCR analysis confirmed that the truncated fragment had lost its native centromere. Maize centromere DNA is comprised of a tandem repeated satellite named CentC, interspersed with a centromere-specific retroelement family named CRM [Zhong et al., 2002a; Jin et al., 2004]. CentC did not amplify in any line and CRM retroelements amplified at very low levels (similar to the oat parental background line Sun II, not shown). In contrast both elements robustly amplified from a full ch3 addition line (fig. 2). OPIE elements were similarly amplified in 5 of 6 neocentromere progeny (fig. 2) as well as in another leaf from the same tiller (not shown). Consistent OPIE amplification suggested that the original neocentromere reported by Muehlbauer and colleagues had stabilized, and is now transmitting at a much more stable frequency. The sole exception was line CTo1_3, which showed dramatically reduced levels of OPIE and CRM elements (fig. 2). It appeared that the neocentomere in CTo1_3 had spontaneously destabilized.
Fig. 2.
Maize neocentromere lines lack centromere DNA but are stably inherited. Quantitative PCR analyses of leaf tissue from 6 neocentromere siblings (CTo1_3,8,12,33, 38 and 40) show a sharp decrease in OPIE and CRM retroelements, and did not detect the CentC centromere repeat (Ct = 35 was used arbitrarily for illustration, see Materials and Methods). The full disomic addition line, OMAd3.1 is used as a control. Quantitative measurements presented below the graph are expressed as depletions relative to OMAd3.1 after normalization to the 5S rDNA content of each sample. The maize chromosome of Cto1_3 seems to be inherited at lower frequency than the other neocentromere chromosomes, all of which share the same parent.
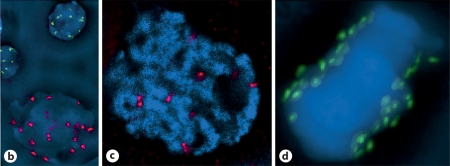
To directly analyze the stable and putative unstable forms of the neocentromere chromosome, we performed FISH on root tip spreads of the 5 stable lines, CTo1_3, and the OMAd3.1 control. Neither CentC nor CRM elements were detected in any neocentromere line, stable or putative unstable, as opposed to the clear signals detected in OMAd3.1 (fig. 3a, b; CRM not shown). The neocentromere chromosome is consistently seen in all 5 stable lines by its reduced size relative to oat chromosomes and its OPIE signal (fig. 3a). Because the OPIE signal appears symmetrical, we wondered if the ch3 short arm was duplicated around the center, despite the lack of a primary constriction typical of isochromosomes. In a second assay, we hybridized the chromosomes with a FISH probe for CentA. CentA is a family of retrotransposons that are closely related to the centromeric CRM elements, and generally localize in the centromere/pericentromere regions of grasses, including oat, but also speckle chromosome arms in maize (fig. 3c). The speckled, unsymmetrical CentA signal obtained from stable neocentromere lines confirms its origin as a simple truncation of maize chromosome 3 (fig. 3d). Upon its detection, the putative unstable chromosome of CTo1_3 was seen as 2 tiny symmetrical arms flanking a primary constriction (fig. 3e). Using OPIE signals of interphase cells, we calculated somatic transmission rates of nearly 100% in stable lines CTo1_12 and CTo1_33, as opposed to 3% in CTo1_3 (Table 1). Apparently a secondary breakage event occurred in Cto1_3 that further truncated the chromosome and either destabilized the original neocentromere or formed a new and unstable centromere. The dramatic increase in stability of the neocentromere from the T0 (original Muehlbauer line) to the T2 generation, combined with the rapid loss of stability in Cto1_3, raise the possibility that some neocentromere events may be initially unstable and subject to a period of flux, during which time they may be lost.
Fig. 3.
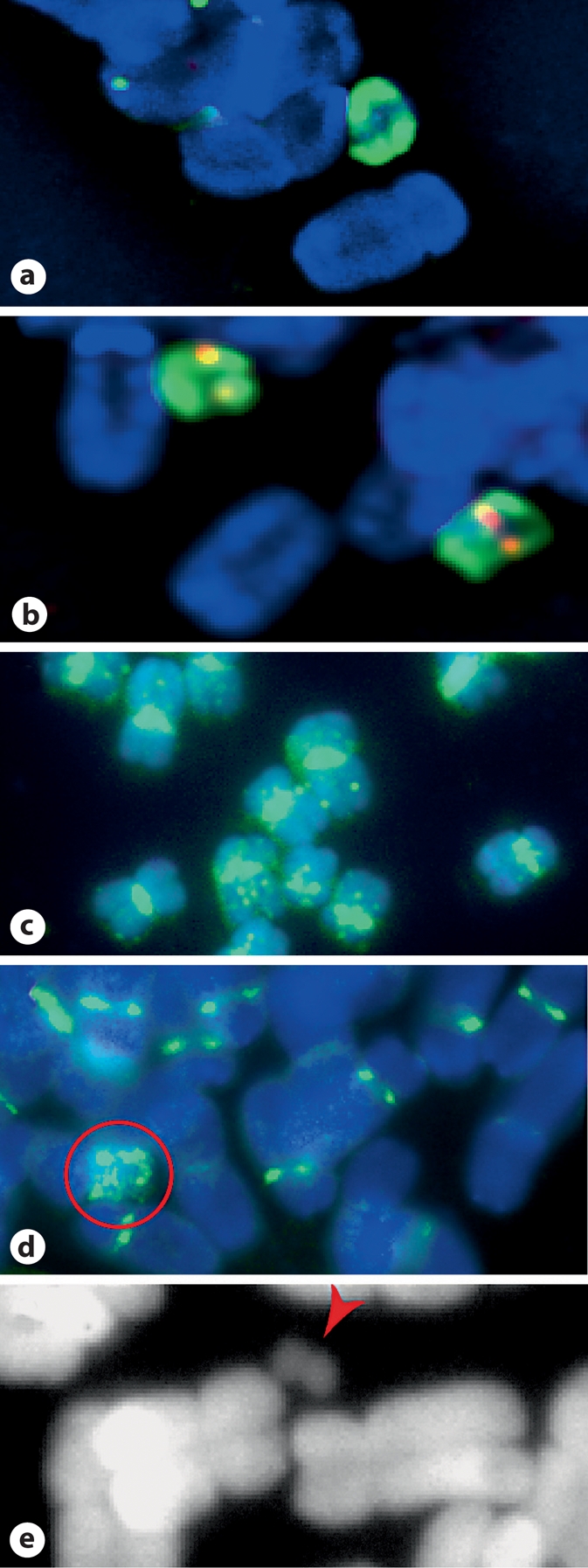
Neocentromere-containing chromosomes are distinguished from oat chromosomes. a DAPI (blue) staining and FISH using an OPIE probe (green) identifies stable neocentromere chromosomes in a metaphase spread, whereas a CentC FISH probe (red) does not stain. b Full addition line chromosomes are larger than their neocentromere counterparts and their native centromeres are identified by CentC staining. c FISH with a CentA probe (green) stains centromeres, pericentromeres, and speckles chromosome arms of maize B73 chromosomes. d The same CentA probe stains oat centromeres/pericentromeres, and stains the maize ch3S neocentromere chromosome asymmetrically (circled in red). e The unstable neocentromere chromosome CTo1_3 (red arrowhead) is a fraction of the stable size (a), and is symmetrical around a primary constriction.
Table 1.
Stability of neocentromere lines
| Plant line |
Number of cells with OPIE signal |
Number of cells without OPIE signal |
Percent transmission |
|---|---|---|---|
| OMAd3.1 – control | 331 | 0 | 100 |
| CTo1_3 – unstable | 4 | 131 | 3.0 |
| CTo1_12 – stable | 408 | 2 | 99.5 |
| CTo1_33 – stable | 318 | 0 | 100 |
Position of the Neocentromere
To investigate the functional aspects of neocentromeres, we developed polyclonal antibodies against a poorly conserved region of the oat CENH3 protein (here designated Avena sativa CENH3, or oat CENH3; fig. 4a). Oat CENH3 antibodies stain oat, but not maize cells (fig. 4b). Oat CENH3 antibodies also show the constitutive localization pattern that is typical for CENH3 [Talbert et al., 2002; Zhong et al., 2002b], clearly labeling the primary constrictions of all chromosomes (fig. 4c, d).
Fig. 4.
Identification of the oat CENH3 protein. a A CLUSTAL [Larkin et al., 2007] alignment using the N-termini of the rice, maize, and oat forms of CENH3 proteins. b The underlined sequence was used to generate a peptide antibody that detects oat but not maize CENH3 in mixed cell immunolocalization-FISH experiments. Oat cells are larger and stain for CENH3 (red), whereas maize cells are smaller and stain for CentC (green) but not oat CENH3. c Oat pachytene and d metaphase I immunolocalizations using the oat CENH3 antibody (red or green), stain oat centromeres.
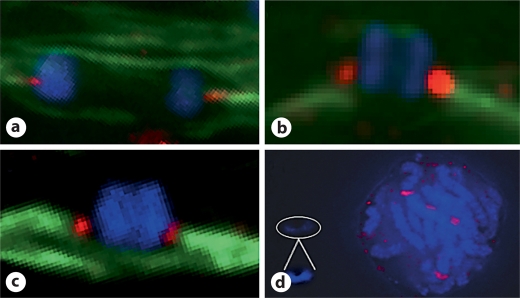
In Drosophila and barley the neocentromere formed close to the site of the original centromere. Based on our SSR work, we assumed the oat CENH3 signal would be on one end of the maize fragment. However, our cytological analysis of stable lines CTo1_12, CTo1_33, and CTo1_40, revealed a submetacentric location for oat CENH3 (fig. 5a–c). Thus the maize neocentromere appears to have formed de novo at a site unlinked to the original (lost) centromere.
Fig. 5.
The maize neocentromere formed submetacentrically. a, b, c CENH3 immunolocalization on meiocytes of stable lines reveal that the neocentromere is located at a submetacentric position on the chromosome arm. CENH3 is shown in red and tubulin is shown in green. d No CENH3 was detected on the unstable CTo1_3 neocentromere from a pachytene cell, despite clear staining on corresponding oat chromosomes (signal intensity of the maize chromosome has been enhanced for viewing – inset).
We found only one example of the highly unstable CTo1_3 isochromosome in a meiotic preparation. Despite strong staining of the oat kinetochores, oat CENH3 staining of the isochromosome was below the detectable limit (fig. 5d). Although plant spindles are capable of exerting polar forces on acentric fragments [Khodjakov et al., 1996], it is doubtful whether such a mechanism is capable of sustained transmission. We think it more likely that the unstable kinetochore has been reduced to a barely functional size.
Kinetochore Size within Stable Neocentromere Lines Varies
Classical plant cytogenetic research recognized a positive correlation between centromere size and somatic stability, but could only speculate as to why [Rhoades, 1940; Steinitz-Sears, 1966]. A potential mechanism for small centromere instability is suggested by later work that supports a minimum size requirement for kinetochore and CENH3 domains of stable chromosomes [Cherry et al., 1989; McEwen et al., 1998; Okamoto et al., 2007]. We could not analyze the kinetochore size of the original unstable neocentromere, but reasoned that analyzing the kinetochores of stable progeny would help us understand post-formation events.
The fluorescence intensity of immunodetected CENH3 is a good estimate of kinetochore size because CENH3 chromatin is the structural basis of kinetochores. The primary function of kinetochores is to bind microtubules, and microtubule binding capacity is dependent in large part on the structural densities of kinetochores [McEwen et al., 1998; Mitchison and Kirschner, 1985]. We compared CENH3 intensities of neocentromeres from stable lines and their oat counterparts to approximate their functional sizes (see Materials and Methods). The CENH3 staining intensity of most neocentromeres was 2 or more standard deviations smaller than the corresponding mean oat centromere value from the same cells, but some neocentromeres were much smaller than others (expressed as % of mean oat; table 2; fig. 6). The apparent size variation among neocentromeres is striking (table 2; fig. 6), considering that previously measured human neocentromeres were much less variable [Irvine et al., 2004]. We used an F-test to compare the CENH3 staining of neocentromeres to the mean staining of oat centromeres, with a null hypothesis that these values were equal. The variation of maize neocentromere size is greater than that of native oat centromeres at a 95% significance level (see Materials and Methods). Considering the apparent size plasticity of neocentromeres, we speculate that a sub-minimal kinetochore caused instability of the maize chromosome in the T0 generation, and that an increase in kinetochore size is responsible for its stability in the T1 and T2 generations.
Table 2.
Relative neocentromere intensities
| Cell |
Percent of mean oat intensity |
Number of standard deviations from mean oat |
|---|---|---|
| 1 | 75.2 | 3.1 |
| 2 | 59.6 | 3.2 |
| 3 | 87.0 | 2.0 |
| 4 | 32.5 | 2.8 |
| 5 | 54.3 | 6.8 |
| 6 | 79.4 | 3.4 |
| 7 | 80.7 | 2.3 |
| 8 | 90.3 | 1.1 |
The number of standard deviations (σ) of each neocentromere kinetochore from the mean oat intensity was calculated from the following formula: x = (mean oat intensity − neocentromere intensity)/σoat.
Fig. 6.
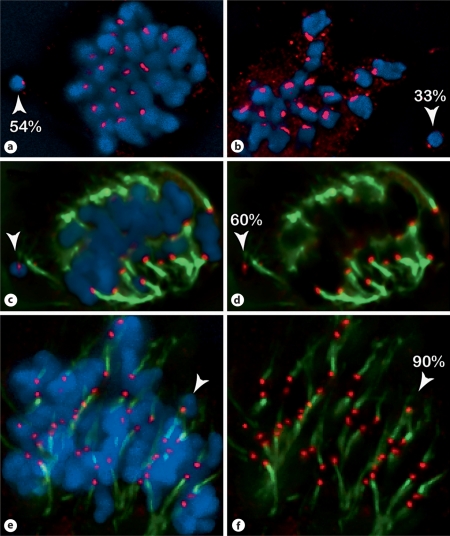
CENH3 staining of maize neocentromeres is more variable than staining of established oat centromeres. Cell spreads were immunostained with oat CENH3 (red), tubulin (green, c–f only), and counterstained with DAPI (blue). The fluorescence intensity was measured to estimate kinetochore size. Numbers indicate the relative intensity of the CENH3 of neocentromeres (arrowheads) relative to the mean intensity of oat kinetochores. a, b Two spreads from the same slide illustrating variation in CENH3 intensity. c, d and e, f Two similarly staged cells from the same neocentromere line illustrating a correlation between kinetochore size and attachment to the spindle. The smaller neocentromere (60% of mean oat; c, d) appears to be detached from the spindle, whereas a proportionally much larger neocentromere (90% of mean oat; e, f) is attached to a kinetochore fiber.
In anther tissue the neocentromere chromosome is often marginalized from the main nuclear body as the spindle forms in early prophase. We noticed that neocentromeres attached to the spindle were larger than neocentromeres isolated from the spindle (fig. 6). Though it seems likely that larger neocentromeres are simply better at binding microtubules, there is the additional possibility that microtubule attachments somehow enhance kinetochore size. Regardless, this observation supports the suggestion that neocentromere stability depends on its ability to interact with the spindle.
Discussion
The stability of centromeres provides the foundation for chromosomal, and hence genetic, inheritance. Yet because centromeres have an epigenetic basis, they are capable of rapid adaptation to genomic instabilities such as species hybridization and chromosome breakage. While neocentromere adaptation may be detrimental in some cases, cancers for example, they may also allow centromeres to affect evolutionary change. The current paradigm of centromere formation involves 1) establishing a critical mass of CENH3 chromatin sufficient to form a functional centromere/kinetochore, and 2) reinforcement of the centromere position over evolutionary timescales by the accumulation of specific repeats within the centromere and the expansion of flanking heterochromatin domains by genomic rearrangements and transposons [Nagaki et al., 2004; Topp and Dawe, 2006; Marshall et al., 2008]. It has been shown that the establishment phase may be accomplished by overwhelming a locus with CENH3 proteins [Heun et al., 2006]. Our results suggest that CENH3 accumulates over time and that the process is very dynamic, at least in the first two generations.
The idea of a flux period might have been expected given what is known about CENH3 incorporation into chromatin. The CENH3 nucleosome density of centromeres is diluted during chromosome replication because CENH3 deposition is not coupled to the replication machinery like canonical histones [Shelby et al., 2000; Sullivan and Karpen, 2001; Lermontova et al., 2007]. Various scenarios are proposed for centromere replenishment [Sullivan et al., 2001; Smith, 2002; Mellone and Allshire, 2003], but the weight of evidence points to a mechanism in which general chromatin factors deposit CENH3 histones at accessible locations [Henikoff and Ahmad, 2005; Furuyama et al., 2006]. Established centromeres are a highly favorable environment for new CENH3 incorporation, reinforcing centromere position. Cells have robust mechanisms for removing CENH3 nucleosomes as well, as evidenced by the rapid inactivation of entire centromeres in plants and animals [Amor et al., 2004; Higgins et al., 2005; Han et al., 2006], and CENH3 proteolytic degradation in yeast and flies [Collins et al., 2004; Moreno-Moreno et al., 2006].
One interpretation of the stability of established centromeres is that they are protected from CENH3 removal by some inherent property. A newly formed centromere would therefore be especially susceptible to loss until it gained enough centromere ‘identity’ to resist removal forces. An obvious feature of established centromeres is flanking heterochromatin, and several lines of evidence have led researchers to suggest that heterochromatin is integral to both centromere stability and de novo formation [Henikoff et al., 2000; Maggert and Karpen, 2000; Nakashima et al., 2005; Okada et al., 2007; Ishii et al., 2008; Nakano et al., 2008]. While general heterochromatin is unlikely to be a centromere identity factor per se, it may help to protect fledgling neocentromeres from removal.
The interplay between centromere identity factors and CENH3 removal forces may help explain the mosaic inheritance patterns reported for some human neocentromeres [Amor and Choo, 2002], as well as the rapid stabilization and variance in CENH3 domain size of the maize neocentromere reported here. For example, neocentromeres that were apparently cis-templated from nearby centromeres are stable, perhaps due to their formation near pericentric heterochromatin [Maggert and Karpen, 2001; Nasuda et al., 2005]. We might expect de novo neocentromeres to have less centromere identity and less protection from CENH3 removal. Perhaps the initial instability of the maize neocentromere resulted from its local susceptibility to CENH3 removal, which would reduce the kinetochore to a sub-minimal size.
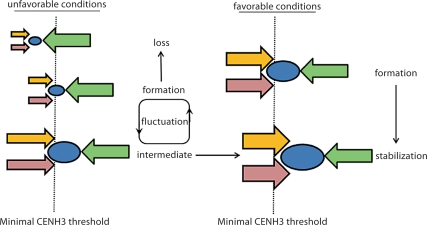
Based on our observations and the current understanding of CENH3 dynamics, we speculate on the processes that may occur during new centromere establishment (fig. 7). New centromeres undergo an intermediate period of CENH3 fluctuation, where reinforcement factors such as CENH3 from the last cell cycle and flanking heterochromatin counteract CENH3 removal. The period and outcome of the establishment process depends on the relative strengths of these forces and the size of the minimal CENH3 domain. If the neocentromere formed in a favorable environment, the initial reinforcement factors are strong and will rapidly push CENH3 density past the threshold. If the neocentromere formed in a less than favorable environment, the initial reinforcement factors are weak and the neocentromere will be unstable and may be lost. Alternatively, an unstable neocentromere may gain enough centromere identity through repeated cell divisions to reinforce itself [Mellone and Allshire, 2003]. Under this view, the location and initial events of neocentromere formation are critical to its ultimate stability.
Fig. 7.
A speculative model of neocentromere establishment. The conditions under which a centromere forms may contribute to its stability. Upon formation, a neocentromere (blue oval) is subject to removal by endogenous forces that normally suppress ectopic centromeres (green arrows). Under certain conditions, reinforcement factors (red and yellow arrows) counteract CENH3 removal and stabilize the neocentromere. If the reinforcement factors are relatively strong (size reflects its relative strength; right panel), establishment is favored and the centromere stabilizes rapidly. If reinforcement factors are weak to begin with (left panel), the centromere may be unstable and fall below the minimal threshold (dotted line). Alternatively, the reinforcement factors may gain strength over time, eventually resulting in a stable centromere.
Acknowledgements
This work was supported by a grant from the National Science Foundation (0421671) to R.K.D., and by a University of Georgia Graduate School Dissertation Completion Award to C.N.T. We sincerely thank Beau Brouillette and Eleanor Kuntz for helpful statistics discussions.
References
- Alonso A, Mahmood R, Li S, Cheung F, Yoda K, Warburton PE. Genomic microarray analysis reveals distinct locations for the CENP-A binding domains in three human chromosome 13q32. Hum Mol Genet. 2003;12:2711–2721. doi: 10.1093/hmg/ddg282. [DOI] [PubMed] [Google Scholar]
- Alonso A, Fritz B, Hasson D, Abrusan G, Cheung F, et al. Co-localization of CENP-C and CENP-H to discontinuous domains of CENP-A chromatin at human neocentromeres. Genome Biol. 8;2007:R148. doi: 10.1186/gb-2007-8-7-r148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Choo KH. Neocentromeres: Role in human disease, evolution, and centromere study. Am J Hum Genet. 2002;71:695–714. doi: 10.1086/342730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor DJ, Bentley K, Ryan J, Perry J, Wong L, et al. Human centromere repositioning ‘In progress’. Proc Natl Acad Sci USA. 2004;101:6542–6547. doi: 10.1073/pnas.0308637101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhattacharyya GK, Johnson RA. Statistical Concepts and Methods. New York: John Wiley & Sons; 1977. p. 604. [Google Scholar]
- Cherry LM, Faulkner AJ, Grossberg LA, Balczon R. Kinetochore size variation in mammalian chromosomes – an image-analysis study with evolutionary implications. J Cell Sci. 1989;92:281–289. doi: 10.1242/jcs.92.2.281. [DOI] [PubMed] [Google Scholar]
- Collins KA, Furuyama S, Biggins S. Proteolysis contributes to the exclusive centromere localization of the yeast Cse4/CENP-A histone H3 variant. Curr Biol. 2004;14:1968–1972. doi: 10.1016/j.cub.2004.10.024. [DOI] [PubMed] [Google Scholar]
- Dawe RK, Henikoff S. Centromeres put epigenetics in the driver's seat. Trends Biochem Sci. 2006;31:662–669. doi: 10.1016/j.tibs.2006.10.004. [DOI] [PubMed] [Google Scholar]
- Dawe RK, Hiatt EN. Plant neocentromeres: Fast, focused, and driven. Chromosome Res. 2004;12:655–669. doi: 10.1023/B:CHRO.0000036607.74671.db. [DOI] [PubMed] [Google Scholar]
- Du Y, Dawe RK. Maize NDC80 is a constitutive feature of the central kinetochore. Chromosome Res. 2007;15:767–775. doi: 10.1007/s10577-007-1160-z. [DOI] [PubMed] [Google Scholar]
- Furuyama T, Dalal Y, Henikoff S. Chaperone-mediated assembly of centromeric chromatin in vitro. Proc Natl Acad Sci USA. 2006;103:6172–6177. doi: 10.1073/pnas.0601686103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han FP, Lamb JC, Birchler JA. High frequency of centromere inactivation resulting in stable dicentric chromosomes of maize. Proc Natl Acad Sci USA. 2006;103:3238–3243. doi: 10.1073/pnas.0509650103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K. Assembly of variant histones into chromatin. Annu Rev Cell Dev Biol. 2005;21:133–153. doi: 10.1146/annurev.cellbio.21.012704.133518. [DOI] [PubMed] [Google Scholar]
- Henikoff S, Ahmad K, Platero JS, Steensel BV. Heterochromatic deposition of centromeric histone H3-like proteins. Proc Natl Acad Sci USA. 2000;97:716–721. doi: 10.1073/pnas.97.2.716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heun P, Erhardt S, Blower MD, Weiss S, Skora AD, Karpen GH. Mislocalization of the Drosophila centromere-specific histone CID promotes formation of functional ectopic kinetochores. Dev Cell. 2006;10:303–315. doi: 10.1016/j.devcel.2006.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins AW, Gustashaw KM, Willard HF. Engineered human dicentric chromosomes show centromere plasticity. Chromosome Res. 2005;13:745–762. doi: 10.1007/s10577-005-1009-2. [DOI] [PubMed] [Google Scholar]
- Irvine DV, Amor DJ, Perry J, Sirvent N, Pedeutour F, et al. Chromosome size and origin as determinants of the level of CENP-A incorporation into human centromeres. Chromosome Res. 2004;12:805–815. doi: 10.1007/s10577-005-5377-4. [DOI] [PubMed] [Google Scholar]
- Ishii K, Ogiyama Y, Chikashige Y, Soejima S, Masuda F, et al. Heterochromatin integrity affects chromosome reorganization after centromere dysfunction. Science. 2008;321:1088–1091. doi: 10.1126/science.1158699. [DOI] [PubMed] [Google Scholar]
- Jiang J, Birchler JA, Parrott WA, Dawe RK. A molecular view of plant centromeres. Trends Plant Sci. 2003;8:570–575. doi: 10.1016/j.tplants.2003.10.011. [DOI] [PubMed] [Google Scholar]
- Jin W, Melo JR, Nagaki K, Talbert PB, Henikoff S, et al. Maize centromeres: Organization and functional adaptation in the genetic background of oat. Plant Cell. 2004;16:571–581. doi: 10.1105/tpc.018937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khodjakov A, Cole RW, Bajer AS, Rieder CL. The force for poleward chromosome motion in haemanthus cells acts along the length of the chromosome during metaphase but only at the kinetochore during anaphase. J Cell Biol. 1996;132:1093–1104. doi: 10.1083/jcb.132.6.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JC, Meyer JM, Corcoran B, Kato A, Han FP, Birchler JA. Distinct chromosomal distributions of highly repetitive sequences in maize. Chromosome Res. 2007;15:33–49. doi: 10.1007/s10577-006-1102-1. [DOI] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lermontova I, Fuchs J, Schubert V, Schubert I. Loading time of the centromeric histone H3 variant differs between plants and animals. Chromosoma. 2007;116:507–510. doi: 10.1007/s00412-007-0122-8. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta c(t)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Maggert KA, Karpen GH. Acquisition and metastability of centromere identity and function: Sequence analysis of a human neocentromere. Genome Res. 2000;10:725–728. doi: 10.1101/gr.10.6.725. [DOI] [PubMed] [Google Scholar]
- Maggert K, Karpen G. The activation of a neocentromere in Drosophila requires proximity to an endogenous centromere. Genetics. 2001;158:1615–1628. doi: 10.1093/genetics/158.4.1615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik HS, Henikoff S. Adaptive evolution of CID, a centromere-specific histone in Drosophila. Genetics. 2001;157:1293–1298. doi: 10.1093/genetics/157.3.1293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marshall OJ, Chueh AC, Wong LH, Choo KHA. Neocentromeres: New insights into centromere structure, disease development, and karyotype evolution. Am J Hum Genet. 2008;82:261–282. doi: 10.1016/j.ajhg.2007.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McEwen BF, Ding Y, Heagle AB. Relevance of kinetochore size and microtubule-binding capacity for stable chromosome attachment during mitosis in PtK1 cells. Chromosome Res. 1998;6:123–132. doi: 10.1023/a:1009239013215. [DOI] [PubMed] [Google Scholar]
- Mellone BG, Allshire RC. Stretching it: Putting the CEN(P-A) in centromere. Curr Opin Genet Dev. 2003;13:191–198. doi: 10.1016/s0959-437x(03)00019-4. [DOI] [PubMed] [Google Scholar]
- Mitchison TJ, Kirschner MW. Properties of the kinetochore in vitro. II. Microtubule capture and ATP-dependent translocation. J Cell Biol. 1985;101:766–777. doi: 10.1083/jcb.101.3.766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moreno-Moreno O, Torras-Llort M, Azorin F. Proteolysis restricts localization of CID, the centromere-specific histone H3 variant of Drosophila, to centromeres. Nucleic Acids Res. 2006;34:6247–6255. doi: 10.1093/nar/gkl902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mroczek RJ, Dawe RK. Distribution of retroelements in centromeres and neocentromeres of maize. Genetics. 2003;165:809–819. doi: 10.1093/genetics/165.2.809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muehlbauer GJ, Fowler JE, Girard L, Tyers R, Harper L, Freeling M. Ectopic expression of the maize homeobox gene liguleless3 alters cell fates in the leaf. Plant Physiol. 1999;119:651–662. doi: 10.1104/pp.119.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagaki K, Cheng Z, Ouyang S, Talbert PB, Kim M, et al. Sequencing of a rice centromere reveals active genes. Nat Genet. 2004;36:138–145. doi: 10.1038/ng1289. [DOI] [PubMed] [Google Scholar]
- Nakano M, Cardinale S, Noskov VN, Gassmann R, Vagnarelli P, et al. Inactivation of a human kinetochore by specific targeting of chromatin modifiers. Dev Cell. 2008;14:507–522. doi: 10.1016/j.devcel.2008.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakashima H, Nakano M, Ohnishi R, Hiraoka Y, Kaneda Y, et al. Assembly of additional heterochromatin distinct from centromere-kinetochore chromatin is required for de novo formation of human artificial chromosome. J Cell Sci. 2005;118:5885–5898. doi: 10.1242/jcs.02702. [DOI] [PubMed] [Google Scholar]
- Nasuda S, Hudakova S, Schubert I, Houben A, Endo TR. Stable barley chromosomes without centromeric repeats. Proc Natl Acad Sci USA. 2005;102:9842–9847. doi: 10.1073/pnas.0504235102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill RJ, Eldridge MDB, Metcalfe CJ. Centromere dynamics and chromosome evolution in marsupials. J Hered. 2004;95:375–381. doi: 10.1093/jhered/esh063. [DOI] [PubMed] [Google Scholar]
- Okada T, Ohzeki JI, Nakano M, Yoda K, Brinkley WR, et al. CENP-B controls centromere formation depending on the chromatin context. Cell. 2007;131:1287–1300. doi: 10.1016/j.cell.2007.10.045. [DOI] [PubMed] [Google Scholar]
- Okamoto Y, Nakano M, Ohzeki JI, Larionov V, Masumoto H. A minimal CENP-A core is required for nucleation and maintenance of a functional human centromere. EMBO J. 2007;26:1279–1291. doi: 10.1038/sj.emboj.7601584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rhoades M. Studies of a telocentric chromosome in maize with reference to the stability of its centromere. Genetics. 1940;25:483–520. doi: 10.1093/genetics/25.5.483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H. Primer3 on the www for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- Saffery R, Irvine DV, Griffiths B, Kalitsis P, Wordeman L, Choo KH. Human centromeres and neocentromeres show identical distribution patterns of >20 functionally important kinetochore-associated proteins. Hum Mol Genet. 2000;9:175–185. doi: 10.1093/hmg/9.2.175. [DOI] [PubMed] [Google Scholar]
- Shelby RD, Monier K, Sullivan KF. Chromatin assembly at kinetochores is uncoupled from DNA replication. J Cell Biol. 2000;151:1113–1118. doi: 10.1083/jcb.151.5.1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Dawe K. Partitioning of the maize epigenome by the number of methyl groups on histone H3 lysines 9 and 27. Genetics. 2006;173:1571–1583. doi: 10.1534/genetics.106.056853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith M. Centromeres and variant histones: What, where, when and why? Curr Opin Cell Biol. 2002;14:279–285. doi: 10.1016/s0955-0674(02)00331-9. [DOI] [PubMed] [Google Scholar]
- Steinitz-Sears LM. Somatic instability of telocentric chromosomes in wheat and nature of centromere. Genetics. 1966;54:241–248. doi: 10.1093/genetics/54.1.241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B, Karpen G. Centromere identity in Drosophila is not determined in vivo by replication timing. J Cell Biol. 2001;154:683–690. doi: 10.1083/jcb.200103001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan B, Blower M, Karpen G. Determining centromere identity: Cyclical stories and forking paths. Nat Rev Genet. 2001;2:584–596. doi: 10.1038/35084512. [DOI] [PubMed] [Google Scholar]
- Talbert P, Masuelli R, Tyagi A, Comai L, Henikoff S. Centromeric localization and adaptive evolution of an Arabidopsis histone H3 variant. Plant Cell. 2002;14:1053–1066. doi: 10.1105/tpc.010425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Topp CN, Dawe RK. Reinterpreting pericentromeric heterochromatin. Curr Opin Plant Biol. 2006;9:647–653. doi: 10.1016/j.pbi.2006.09.008. [DOI] [PubMed] [Google Scholar]
- Van Hooser AA, Ouspenski II, Gregson HC, Starr DA, Yen TJ, et al. Specification of kinetochore-forming chromatin by the histone H3 variant CENP-A. J Cell Sci. 2001;114:3529–3542. doi: 10.1242/jcs.114.19.3529. [DOI] [PubMed] [Google Scholar]
- Ventura M, Archidiacono N, Rocchi M. Centromere emergence in evolution. Genome Res. 2001;11:595–599. doi: 10.1101/gr.152101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warburton PE, Cooke CA, Bourassa S, Vafa O, Sullivan BA, et al. Immunolocalization of CENP-A suggests a distinct nucleosome structure at the inner kinetochore plate of active centromeres. Curr Biol. 1997;7:901–904. doi: 10.1016/s0960-9822(06)00382-4. [DOI] [PubMed] [Google Scholar]
- Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002a;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhong CX, Marshall JB, Topp C, Mroczek R, Kato A, et al. Centromeric retroelements and satellites interact with maize kinetochore protein CENH3. Plant Cell. 2002b;14:2825–2836. doi: 10.1105/tpc.006106. [DOI] [PMC free article] [PubMed] [Google Scholar]