Abstract
Chromosome microdeletions or duplications are detected in 10–20% of patients with mental impairment and normal karyotypes. A few cases have been reported of mental impairment with microdeletions comprising tumor suppressor genes. By array-CGH we detected 4 mentally impaired individuals carrying de novo microdeletions sharing an overlapping segment of ∼180 kb in 17p13.1. This segment encompasses 18 genes, including 3 involved in cancer, namely KCTD11/REN, DLG4/PSD95, and GPS2. Furthermore, in 2 of the patients, the deletions also included TP53, the most frequently inactivated gene in human cancers. The 3 tumor suppressor genes KCTD11, DLG4, and GPS2, in addition to the GABARAP gene, have a known or suspected function in neuronal development and are candidates for causing mental impairment in our patients. Among our 4 patients with deletions in 17p13.1, 3 were part of a Brazilian cohort of 300 mentally retarded individuals, suggesting that this segment may be particularly prone to rearrangements and appears to be an important cause (∼1%) of mental retardation. Further, the constitutive deletion of tumor suppressor genes in these patients, particularly TP53, probably confers a significantly increased lifetime risk for cancer and warrants careful oncological surveillance of these patients. Constitutional chromosome deletions containing tumor suppressor genes in patients with mental impairment or congenital abnormalities may represent an important mechanism linking abnormal phenotypes with increased risks of cancer.
Key Words: Array CGH, Mental retardation, Microdeletion, TP53, Tumor suppressor genes
Germline mutations in highly penetrant tumor suppressor genes are rare events that, when not lethal before adulthood, may be associated with hereditary cancers. This is the case for mutations involving the genes TP53, BRCA1, and BRCA2. Chromosome microdeletion is an alternative mechanism for the constitutional loss of function of tumor suppressor genes. This has been described in a few microdeletion syndromes associated with the occurrence of tumors, such as the WAGR syndrome (Wilms tumor, aniridia, genitourinary anomalies, and mental retardation syndrome, MIM 194072), caused by deletions in 11p13 encompassing the WT1 gene. Another example is the NF1 microdeletion syndrome, affecting 5–20% of patients with neurofibromatosis (MIM 162200).
Since the introduction of array-CGH in routine diagnosis, it has been shown that 10–20% of patients with idiopathic mental retardation and/or congenital abnormalities show submicroscopic chromosome imbalances [Shaw-Smith et al., 2004; Rosenberg et al., 2006]. Here we report 4 patients with syndromic mental impairment and chromosomal microdeletions in 17p13.1 of variable size encompassing several tumor suppressor genes, including TP53. Although acquired deletions of this chromosomal segment have been reported in tumors [De Smaele et al., 2004; Ferretti et al., 2005], constitutional deletions have not been described and their long term consequences are undetermined to date.
Patients and Methods
Patients
The patients presenting with mental retardation, additional clinical signs, and normal G-banded karyotypes were selected for chromosome submicroscopic investigation by array-CGH. The research protocol was approved by the ethics committees of the institutions. Informed consent for publishing results and photos were obtained from the patients’ legal guardians. Data on the patients were deposited in the DECIPHER database (Database of Chromosomal Imbalances and Phenotype in Humans using Ensembl Resources, https://decipher.sanger.ac.uk), and the corresponding DECIPHER number is given. Patient 1 was studied at the Wessex Regional Genetics Laboratory (WRGL), UK, and patients 2, 3, and 4 were part of a sample of 300 Brazilian patients with syndromic mental retardation and normal karyotypes investigated by a 1 Mb BAC array-CGH at the University of São Paulo.
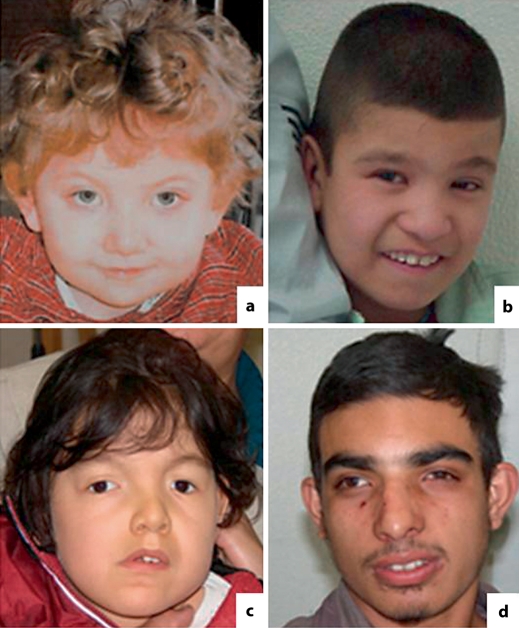
The clinical and familial data on the patients are described below. For patients 2 and 4, records of measurements at birth were not available. Figure 1 shows the facial features of the patients.
Fig. 1.
Facial features of patients carrying deletions in 17p13.1. a Patient 1, 3 years of age. b Patient 2, 8 years of age. c Patient 3, 6 years of age. d Patient 4, 18 years of age.
Patient 1 (DECIPHER 2346). A girl born with 3,650 g, following a pregnancy complicated by decreased fetal movements. Bilateral hip dysplasia was noted and required splinting, but surgery was not required. By 6 weeks of age unusual eye movements were present that subsequently improved, but a squint appeared to be secondary to an inability to focus; the retinal examination was normal. By the age of 8 months she had poor head control, and by 2 years of age she was sitting up without support. At 3 years of age she could not walk or speak; dysmorphology examination showed brachycephaly, mild ptosis, and a full nasal tip. A spinal tail was noted and she wore a brace for mild scoliosis. She had generally low tone and her reflexes were difficult to elicit. X rays of the spine revealed 4 extra distal sacral vertebrae. Brain MRI detected white matter changes (bilateral but not symmetrical patchy areas of signal, most prominent in the peritrigonal regions). At 5 years her height was 100 cm, weight 14.5 kg, and head circumference 49 cm, which are all between the 3rd and 9th centiles.
Patient 2(DECIPHER 2173). A boy, the second child of a healthy non-consanguineous couple; his elder and younger sisters were clinically normal. He was born at term after an uneventful pregnancy, and was cyanotic and hypotonic. His cry was weak and he could not be breast-fed. Milestones were delayed: he held up his head at about 5 months of age and walked at 3 6/12 years. When examined at 8 8/12 years, mental deficiency appeared to be profound; he was hyperactive and interactions were practically absent. He appeared to listen to music and to be happy when touched. Speech was absent. His head circumference was 50 cm (2nd centile), height 126 cm (25th centile), and weight 25.5 kg (50th centile). On examination, he had turricephaly with narrow forehead, a low frontal hairline, epicanthic folds, telechanthus, narrow palpebral fissures, a bulbous nose, narrow high palate, posteriorly rotated dysplastic ears, and long slender fingers.
Patient 3 (DECIPHER 2009). A boy, the second child of a healthy non-consanguineous couple; his older sister was clinically normal. Pregnancy was complicated by oligohydramnios. Delivery was normal, at term, and cyanosis, jaundice, and hypotonia were recorded. His length (50 cm) and weight (3,180 g) were in the 50th centile for Brazilian newborns. His psychoneuromotor development was delayed; he held up his head at 11 months of age, and when examined at 1 11/12 years, he could not sit up without support or change position when lying on bed. Speech was absent, and contact was poor. He had microcephaly (47 cm, 2nd centile), plagiocephaly, facial asymmetry (left side > right side), downward slanting palpebral fissures, epicanthic folds, telecanthus, low nasal bridge, anteverted nostrils, microretrognathia, high-arched palate, gingival hypertrophy, umbilical hernia, lumbar/sacral pilonidal dimple, hydrocele, and genital hypoplasia. His eye examination was normal. MRI revealed slight diffuse cerebral atrophy, signs of delayed myelination, and persistence of the cavum septum pellucidum and vergae; spina bifida was present at L5-S1. His parents reported that his neuropsychomotor development at 6 10/12 years had not progressed. His height was 118 cm (25th centile) and weight 18 kg (5th centile).
Patient 4 (DECIPHER 2203). A boy, the only child of a non-consanguineous couple. He was born at term after an uneventful pregnancy by normal delivery. His early milestones were only mildly delayed; he sat up without support at 1 year of age, walked at the age of 1 1/2 years, and said his first words when he was 1 year old. However, by the age of 5 years, learning difficulties were noted, and he was assessed as requiring special educational needs. He was seen at age 18 1/2 years, when he presented with severe mental deficiency. He needed support for bathing, dressing, and using the toilet. His understanding of orders was highly impaired. He articulated words poorly, and talked almost uninterruptedly about unintelligible subjects. His height was 165 cm (10th–25th centile), weight 46 kg (2.5th–10th centile), and head circumference 54 cm (2nd–10th centile). Physical examination showed a long face, narrow forehead, synophrys, divergent strabismus on the right, prominent dysplastic ears, high nasal bridge, short philtrum, protruding lower lip, asymmetrical smile, short and broad neck, shortened intermammilar distance, long and tapering fingers with interdigital webbing and hypoplastic dermal ridges, bilateral hallux valgus, an enlarged interval between the 1st and 2nd toe on the left, and limitation of joint extension. He had generalized muscle atrophy and weakness, with difficulty going up staircases and a positive Gower sign. Visual evaluation revealed myopia. His mother had moderate mental deficiency herself and had some of the same clinical features observed in her son, namely narrow forehead, synophrys, high nasal bridge, short philtrum, asymmetrical smile, and short and broad neck.
Array-CGH
The deletions in the Brazilian patients (patients 2, 3, and 4) were detected by 1 Mb BAC arrays as previously described [Krepischi-Santos et al., 2006] and were further mapped using oligonucleotide arrays (Human Genome Microarray CGH 244A, Agilent Technologies). The European patient (patient 1) was directly investigated using a customized constitutional version of Agilent Human Genome Microarray CGH 44 K (http://www.ngrl.org.uk/Wessex/arraycgh.htm). Samples were labeled with Cy3 and Cy5-dCTPs by random priming, and purification, hybridization, and washing were carried out as recommended by the manufacturer. Scanned images of the arrays were processed with Agilent Feature Extraction v9.5.1 and analyzed using CGH Analytics software 3.4.40 (both from Agilent Technologies) with the statistical algorithm ADM-2, and sensitivity threshold 6.0. At least 3 consecutive oligonucleotides had to have aberrant log2 ratios to be called by the software. The average resolution was ∼40 kb and ∼8 kb for the 44K- and 244K-oligonucleotide platforms, respectively.
The 4 deletions in 17p13.1 detected by array-CGH were confirmed either by MLPA (patient 1) or FISH with clones that map to the imbalanced regions (patients 2–4).
Results
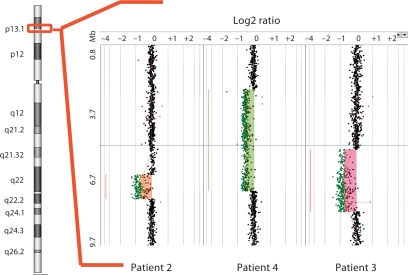
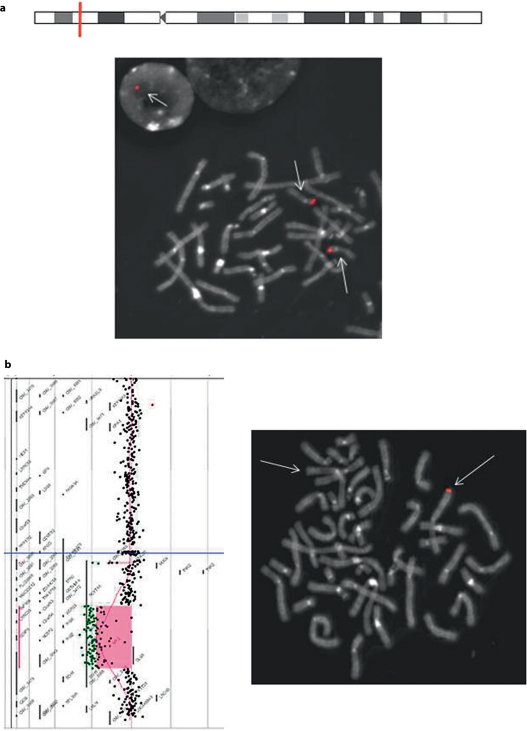
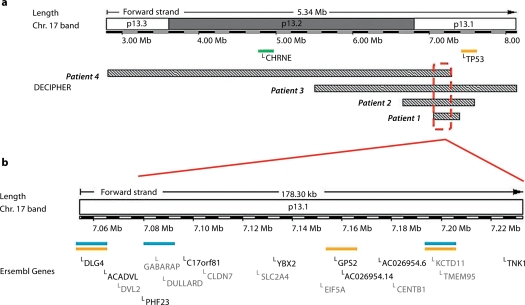
Deletions in chromosome 17p13.1 were identified by array-CGH in four patients (table 1; fig. 2). All the deletions were shown to be de novo (parents of patient 1 were investigated by MLPA using primers specific to TNK1, and those of patients 2, 3, and 4, were analyzed by FISH). Although the array profile of patient 4 clearly revealed the deletion in 17p13.1, its log2 ratio value was higher than the values obtained for the deletions in the two other Brazilian patients investigated in simultaneous hybridization experiments (fig. 2). In fact, this deletion was present in a mosaic state in about 50% of blood cells, as revealed by FISH (fig. 3a). The deletions mapped to chromosome 17p13.2→p13.1 covering a 5.3 Mb region, as estimated by array-CGH (fig. 4a). The breakpoints were unique for each of the 4 deletions. However, there was an overlapping segment of approximately 180 kb in 17p13.1, encompassing 18 genes (fig. 4b).
Table 1.
Genomic mapping of 17p13 deletions and additional chromosomal alterations
| Patient |
Genomic position of the 17p13 deletion (kb) |
Size of the 17p13 deletion (kb) |
Additional cytogenetic findings |
|---|---|---|---|
| 1 | 7054.4–7341.0 | 287 | – |
| 2 | 6507.6–7567.6 | 1060 | Mother with deletion in 17p12 (14014.3–15479.4 kb) |
| 3 | 5405.1–8133.5 | 2728 | Maternally inherited duplication in 20p12.1 (8670.1–9349.9 kb) |
| 4 | 2796.6–7232.7 (50% mosaic) | 4436 | Maternally inherited deletion in 3q29 (19760.4–19859.2 kb) |
Fig. 2.
Array-CGH profile of the most distal 10 Mb of 17p13 in the Brazilian patients. The red box on the idiogram shows the segment containing the deletions. For patient 4, who has the deletion in ∼50% of the cells, the log2 ratio value is intermediate between normal copy number and non-mosaic deletions.
Fig. 3.
Chromosomal alterations detected in patient 4. a Mosaic deletion at 17p13.1, as revealed by FISH (∼50% of metaphases of cultured lymphocytes): metaphase with the fluorescent signal of BAC RP11-144K9 on both chromosomes 17 and a nucleus with a single signal (arrows). b Deletion at 3q29 present in the patient and his mother. Left side: array-CGH profile in the patient; right side: FISH on a metaphase from the patient's mother, showing the red fluorescent signal of BAC GS-56-H22 on only one chromosome 3 (arrows).
Fig. 4.
Mapping of deletions in 17p13. a Schematic representation of the deletions in the 4 patients depicting the minimum overlapping region in 17p13.1. The green bar represents a gene relevant for the phenotype of patient 4. b Enlargement of this region, showing the 18 genes therein (Ensembl Genome Browser; August 2008). The blue bars represent genes that have a role in neuronal development/function, while orange bars indicate genes with known or suspected tumor suppression activity.
Patient 4 carried an additional deletion in chromosome 3q29, which was maternally inherited (fig. 3b). The mother of patient 2 was found to carry the recurrent deletion of PMP22 in 17p12, lying approximately 6.4 Mb proximal to the 17p deletion in her son.
Table 1 summarizes the genomic positions of the detected chromosomal alterations.
Discussion
We described 4 patients with differently sized deletions in 17p13.1. Copy number variations of segments within this region have neither been detected among our in-house controls nor described in the Database of Genomic Variants (DGV http://projects.tcag.ca/variation/). The phenotypes of the patients were variable, which might be partially explained by the different genes involved in the specific deletions. In spite of this, all patients presented with severe to profound mental retardation and absent or very poor speech. A small head and poor growth were also common features.
Eighteen genes map to the deleted segment common to all 4 patients (fig. 4), and 4 of them appear as candidates for causing mental impairment, namely KCTD11/REN (potassium channel tetramerization domain-containing protein 11, MIM 609848), DLG4/PSD95 (postsynaptic density protein 95, MIM 602887), GPS2 (G protein pathway suppressor 2), and GABARAP (GABA-A receptor-associated protein). KCTD11 is a developmentally regulated gene that promotes neuronal differentiation [Gallo et al., 2002], DLG4is a major protein found in virtually all mature excitatory glutamatergic synapses in the brain [Beique and Andrade, 2003], GPS2 was shown in mice to modulate transactivation of genes involved in brain morphogenesis [Zhang et al., 2008], and GABARAP mediates inhibitory neurotransmission [Weiergraber et al., 2008]. These neurodevelopmental genes may well play a role in the mental impairment of our patients.
A further 17p gene known to be disease-associated is deleted in patient 4, the cholinergic receptor, nicotinic, epsilon polypeptide gene (CHRNE; MIM 100725). It is one of the genes responsible for postsynaptic congenital myasthenic syndrome. The generalized muscle atrophy and weakness of patient 4 may be related to this; however, the phenotype-genotype correlation in the patient is quite complex, both because his 17p deletion is in mosaic form and is combined with a 3q29 deletion [Ballif et al., 2008].
The 17p13.1 deletions have a further challenging aspect for genetic counseling, as several of the deleted genes are known or suspected to have a role in tumorigenesis. The deletion of this chromosomal segment is the most frequent genetic lesion in medulloblastomas, and KCTD11 was found to be either deleted or down-regulated in the majority of these tumors [Di Marcotullio et al., 2006]. KCTD11 is known to induce apoptosis and growth arrest and is considered a tumor suppressor gene [Beique and Andrade, 2003]. DLG4, although less well characterized, is likely a tumor suppressor gene implicated in the development of HPV-associated cancers [Handa et al., 2007]. Furthermore and possibly with higher clinical impact, two of the patients (patients 2 and 3) have a deletion of the TP53 tumor suppressor gene. Germline mutations in TP53 are associated with the Li-Fraumeni syndrome (LFS, MIM 151623), a complex autosomal dominant syndrome characterized by familial predisposition to multiple early-onset cancers [Malkin, 2001; Nichols et al., 2001]. In LFS, penetrance is about 50 and 90% by the ages of 30 and 70 years, respectively, and probably higher in cases of full loss of TP53 [Olivier et al., 2003]. A recent publication reports the genome-wide profile of germline copy number variations (CNVs) in LFS families [Shlien et al., 2008]. This study showed that the number of CNVs was strikingly enriched in patients with TP53 mutations. In this cohort, however, CNVs are likely a consequence of impairment of apoptosis caused by TP53 loss-of-function mutations. As a complicating factor, the GPS2 gene, also deleted in all our patients, modulates TP53 activity and contributes to apoptosis [Peng et al., 2001].
Constitutional chromosome deletions containing tumor suppressor genes in patients with mental impairment or congenital abnormalities may represent an important mechanism linking abnormal phenotypes with increased risks of cancer. In fact, several works show an association between childhood neoplasia and congenital abnormalities [Mehes et al., 1987; Altmann et al., 1998; Agha et al., 2005]. A recent publication [Bjorge et al., 2008] comprising over 5 million children and their families from Sweden and Norway showed that children with birth defects have a 1.6–1.7-fold higher risk of developing cancer in childhood and early adulthood than the control population, and this risk seems to be over 5 times higher when the patient presents multiple congenital abnormalities.
Considering the relatively young age of our 4 patients, the fact that no tumor has so far been reported is within expectation. Because no cases of constitutional deletion of KCTD11 or DLG4 have been reported, no empirical risk estimates of cancer can be inferred for these conditions. However, for what we know of the deleted genes, it is likely that our patients have a significantly increased risk for cancer. The identification of potential tumor suppressor genes within microdeletions brings an unusual aspect in genetic counseling for mental impairment. We provided the families of our patients with information concerning cancer risks as well as referral to oncological services for proper follow-up. Besides the clinical implications, follow-up of these patients will contribute to better cancer risk estimates.
We detected deletions on 17p13.1 in 3 out of 300 (1%) mentally impaired Brazilian patients, suggesting that such deletions represent a major cause of mental retardation in addition to likely conferring a significant risk to cancer. Long-term oncological surveillance is necessary.
Acknowledgements
The authors wish to thank the patients and their families for cooperation. We are indebted to Ms. Silvia Sousa Costa and Ms. Maria Raimunda Pinheiro for technical support. We also would like to acknowledge the DECIPHER database for bringing together similar patients from different groups.
References
- Agha MM, Williams JI, Marrett L, To T, Zipursky A, Dodds L. Congenital abnormalities and childhood cancer. Cancer. 2005;103:1939–1948. doi: 10.1002/cncr.20985. [DOI] [PubMed] [Google Scholar]
- Altmann AE, Halliday JL, Giles GG. Associations between congenital malformations and childhood cancer. A register-based case-control study. Br J Cancer. 1998;78:1244–1249. doi: 10.1038/bjc.1998.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ballif BC, Theisen A, Coppinger J, Gowans GC, Hersh JH, et al. Expanding the clinical phenotype of the 3q29 microdeletion syndrome and characterization of the reciprocal microduplication. Mol Cytogenet. 2008;1:8. doi: 10.1186/1755-8166-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beique JC, Andrade R. PSD-95 regulates synaptic transmission and plasticity in rat cerebral cortex. J Physiol. 2003;546:859–867. doi: 10.1113/jphysiol.2002.031369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bjorge T, Cnattingius S, Lie RT, Tretli S, Engeland A. Cancer risk in children with birth defects and in their families: a population based cohort study of 5.2 million children from Norway and Sweden. Cancer Epidemiol Biomarkers Prev. 2008;17:500–506. doi: 10.1158/1055-9965.EPI-07-2630. [DOI] [PubMed] [Google Scholar]
- De Smaele E, Di Marcotullio L, Ferretti E, Screpanti I, Alesse E, Gulino A. Chromosome 17p deletion in human medulloblastoma: a missing checkpoint in the Hedgehog pathway. Cell Cycle. 2004;3:1263–1266. doi: 10.4161/cc.3.10.1200. [DOI] [PubMed] [Google Scholar]
- Di Marcotullio L, Ferretti E, De SE, Screpanti I, Gulino A. Suppressors of hedgehog signaling: linking aberrant development of neural progenitors and tumorigenesis. Mol Neurobiol. 2006;34:193–204. doi: 10.1385/MN:34:3:193. [DOI] [PubMed] [Google Scholar]
- Ferretti E, De Smaele E, Di Marcotullio L, Screpanti I, Gulino A. Hedgehog checkpoints in medulloblastoma: the chromosome 17p deletion paradigm. Trends Mol Med. 2005;11:537–545. doi: 10.1016/j.molmed.2005.10.005. [DOI] [PubMed] [Google Scholar]
- Gallo R, Zazzeroni F, Alesse E, Mincione C, Borello U, et al. REN: a novel, developmentally regulated gene that promotes neural cell differentiation. J Cell Biol. 2002;158:731–740. doi: 10.1083/jcb.200202024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Handa K, Yugawa T, Narisawa-Saito M, Ohno S, Fujita M, Kiyono T. E6AP-dependent degradation of DLG4/PSD95 by high-risk human papillomavirus type 18 E6 protein. J Virol. 2007;81:1379–1389. doi: 10.1128/JVI.01712-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krepischi-Santos AC, Vianna-Morgante AM, Jehee FS, Passos-Bueno MR, Knijnenburg J, et al. Whole-genome array-CGH screening in undiagnosed syndromic patients: old syndromes revisited and new alterations. Cytogenet Genome Res. 2006;115:254–261. doi: 10.1159/000095922. [DOI] [PubMed] [Google Scholar]
- Malkin D. The role of p53 in human cancer. J Neurooncol. 2001;51:231–243. doi: 10.1023/a:1010636426860. [DOI] [PubMed] [Google Scholar]
- Mehes K, Szule E, Torzsok F, Meggyessy V. Supernumerary nipples and urologic malignancies. Cancer Genet Cytogenet. 1987;24:185–188. doi: 10.1016/0165-4608(87)90097-5. [DOI] [PubMed] [Google Scholar]
- Nichols KE, Malkin D, Garber JE, Fraumeni JF, Jr, Li FP. Germ-line p53 mutations predispose to a wide spectrum of early-onset cancers. Cancer Epidemiol Biomarkers Prev. 2001;10:83–87. [PubMed] [Google Scholar]
- Olivier M, Goldgar DE, Sodha N, Ohgaki H, Kleihues P, et al. Li-Fraumeni and related syndromes: correlation between tumor type, family structure, and TP53 genotype. Cancer Res. 2003;63:6643–6650. [PubMed] [Google Scholar]
- Peng YC, Kuo F, Breiding DE, Wang YF, Mansur CP, Androphy EJ. AMF1 (GPS2) modulates p53 transactivation. Mol Cell Biol. 2001;21:5913–5924. doi: 10.1128/MCB.21.17.5913-5924.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenberg C, Knijnenburg J, Bakker E, Vianna-Morgante AM, Sloos W, et al. Array-CGH detection of micro rearrangements in mentally retarded individuals: clinical significance of imbalances present both in affected children and normal parents. J Med Genet. 2006;43:180–186. doi: 10.1136/jmg.2005.032268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw-Smith C, Redon R, Rickman L, Rio M, Willatt L, et al. Microarray based comparative genomic hybridisation (array-CGH) detects submicroscopic chromosomal deletions and duplications in patients with learning disability/mental retardation and dysmorphic features. J Med Genet. 2004;41:241–248. doi: 10.1136/jmg.2003.017731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shlien A, Tabori U, Marshall CR, Pienkowska M, Feuk L, et al. Excessive genomic DNA copy number variation in the Li-Fraumeni cancer predisposition syndrome. Proc Natl Acad Sci USA. 2008;105:11264–11269. doi: 10.1073/pnas.0802970105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiergraber OH, Stangler T, Thielmann Y, Mohrluder J, Wiesehan K, Willbold D. Ligand binding mode of GABAA receptor-associated protein. J Mol Biol. 2008;381:1320–1331. doi: 10.1016/j.jmb.2008.06.086. [DOI] [PubMed] [Google Scholar]
- Zhang D, Harry GJ, Blackshear PJ, Zeldin DC. G-protein pathway suppressor 2 (GPS2) interacts with the regulatory factor X4 variant 3 (RFX4_v3) and functions as a transcriptional co-activator. J Biol Chem. 2008;283:8580–8590. doi: 10.1074/jbc.M708209200. [DOI] [PMC free article] [PubMed] [Google Scholar]