Abstract
BK virus (BKV) is a ubiquitous human pathogen that establishes a lifelong persistent infection in kidney epithelial cells. BKV reactivation within these cells results in a lytic infection in immunocompromised patients. Little is known about the specific interactions of BKV and the host cell during persistence and reactivation. We performed global cellular gene expression analyses using microarrays to characterize the global effect of BKV on primary kidney epithelial cells during the viral life cycle. Our results demonstrate that BKV primarily activates genes involved in cell cycle regulation and apoptosis (58% and 44% of upregulated genes at 48 and 72 hours post-infection, respectively). Surprisingly, we observed that only four genes were downregulated during infection and that only two genes directly involved in the inflammatory response were differentially expressed. These results provide information about how BKV interacts with a cell type in which it both establishes persistence and undergoes lytic reactivation.
Keywords: BK virus, T antigen, microarray, kidney epithelial cell
Introduction
BK virus (BKV) is a member of the human polyomavirus family that was first isolated in 1971 from the urine of a renal transplant recipient and is ubiquitous in the human population (Gardner et al., 1971). Primary exposure to BKV likely results in an asymptomatic infection during early childhood, with antibodies against BKV detected in 50% of children by age 3 and over 90% by age 10 (Knowles, 2001).
It is widely believed that the kidney is the major reservoir for BKV: a variety of studies have demonstrated the presence of BKV DNA and proteins in normal and diseased human kidneys, and reactivation of BKV leads to diseases of the kidney and urinary tract (Bedi et al., 1995; Heritage et al., 1981; Shinohara et al., 1993). BKV lytic infection following reactivation can be subclinical but is often associated with hemorrhagic cystitis, an infection of the bladder characterized by inflammation and hematuria in bone marrow transplant patients, or polyomavirus nephropathy (PVN) in renal transplant patients, a destruction of kidney tissue frequently leading to deterioration and eventual loss of the graft (reviewed in Jiang et al., 2009a). Clinical BKV infection has become a greater problem in recent years due to the increasing prevalence of bone marrow and renal transplants, as well as the development of more effective immunosuppressive therapies used in these patients. The frequency of BKV viruria and viremia after renal transplantation can be as high as 80%, with 10% of patients progressing to PVN (Binet et al., 1999; Bressollette-Bodin et al., 2005; Egli et al., 2007; Hirsch et al., 2002). Due to the lack of effective antiviral drugs, the primary course of action for physicians is to decrease immunosuppression, thereby increasing the risk of rejection. As a result, up to 90% of patients that develop PVN lose allograft function, and overall, BKV reactivation may be responsible for up to 10% of all renal graft dysfunction (Egli et al., 2007; Nickeleit et al., 2000). BKV reactivation has also been detected in recipients of other solid organ transplants, including heart, lung, liver, and pancreas (Pavlakis et al., 2006; Vilchez and Kusne, 2006).
Transcription of the BKV genome occurs divergently from a single non-coding control region: the early proteins, large tumor antigen (TAg), small tumor antigen (tAg), and truncated tumor antigen (truncTAg) are coded for by one strand, while the late proteins, VP1, VP2, VP3, and the agnoprotein, are encoded by the opposite strand (reviewed in Imperiale and Major, 2007). Upon transport of the viral genome to the nucleus, the virus expresses the three T antigens. TAg and tAg induce the non-dividing epithelial cell to enter S phase, which facilitates replication of the genome. BKV TAg is a 695 amino acid protein that binds to the retinoblastoma family of tumor suppressor proteins: pRB, p107, and p130 (Harris et al., 1996). The interaction of TAg with the pRB family members is one of the critical early events that induce S phase progression. The binding of TAg to pRB causes displacement of E2F transcription factor family members, which subsequently activate transcription of genes involved in S phase progression and DNA synthesis (Bracken et al., 2004; DeCaprio et al., 1988; Harris et al., 1996). TAg has also been shown to prevent the cell from undergoing cell cycle arrest or apoptosis through interactions with the tumor suppressor protein p53 (Harris et al., 1998; Nakshatri et al., 1988). In normal cells, p53-mediated cell cycle arrest occurs through transcriptional activation of the genes encoding p21Waf1, MDM2, and cyclin G1, while p53-mediated apoptosis occurs through transcriptional activation of the Bcl-2 family, p53AIP1, and the Fas death receptor (Vousden, 2000).
In the absence of lytic replication, the expression of BKV early proteins can result in the oncogenic transformation of mammalian cells in culture (Harris et al., 1996; Major and Di Mayorca, 1973) and the development of tumors in transgenic mice (Dalrymple and Beemon, 1990; Small et al., 1986). When BKV is injected intracerebrally or intravenously into hamsters, rats, or newborn mice, species in which the virus cannot lytically replicate, 75% of the animals develop tumors containing viral sequences and proteins (Imperiale and Major, 2007; Uchida et al., 1976). In the presence of activated c-H-Ras or c-Myc, BKV transforms human embryonic kidney cells, demonstrating that under certain conditions the viral proteins may behave similarly in human and rodent cells (Corallini et al., 1991; Pater and Pater, 1986). Although a role for BKV in the formation of tumors in humans remains controversial, BKV sequences have been detected in a number of human tumors, including those of the urinary tract (Abend et al., 2009). The transforming potential of BKV in the absence of productive infection is directed primarily by changes in gene expression associated with cell cycle regulation by the tumor antigens.
Due to the increasing prevalence of BKV-associated diseases, we are interested in global changes in cellular gene expression that occur as a result of BKV lytic infection. In this report, we used array analyses to characterize the effect of BKV infection on primary human proximal tubule epithelial (HPTE) cells, which we have previously described as a model system for BKV lytic infection (Low et al., 2004). To include both the early and late stages of the viral life cycle in the analyses, we chose to examine changes in cellular gene expression induced by BKV lytic replication up to 72 hours post-infection (hpi). We used quantitative RT-PCR and immunoblotting to confirm the microarray results for several genes with significant changes in expression in the presence of BKV. As expected, a large number of genes involved in cell cycle regulation and proliferation were upregulated during infection. Surprisingly, there was no effect on the expression of pro-inflammatory genes and only four genes were significantly downregulated during BKV lytic infection. These findings are important for understanding the host-pathogen interactions that occur between BKV and human kidney epithelial cells, and more specifically, the cellular environment established and required for lytic infection by BKV.
Results
Microarray analysis of BKV-infected HPTE cells
Our previous studies examined the time course of BKV infection in HPTE cells: early viral gene expression is first detectable at 24 hpi, with levels increasing out to 72 hpi; synthesis of viral DNA and late gene expression begin concurrently at 36 hpi, with levels increasing through the late stages of infection; and viral progeny production begins at 48 hpi (Low et al., 2004). Therefore, we chose to examine the effects of BKV lytic infection on human kidney epithelial cells at 24, 48, and 72 hpi to encompass both early and late stages of the infectious cycle. HPTE cells were infected with purified BKV at an MOI of 5 IU/cell. Total cell RNA was extracted at 24, 48, and 72 hpi and subjected to microarray analysis using the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array, which measures the expression level of more than 47,000 transcripts and variants. Principal component analysis of the data from these three time points suggested significant differences between mock- and BKV-infected samples at 48 and 72 hpi, but few changes at 24 hpi. A two-fold cutoff was used to define genes as significantly affected in the presence of BKV. According to this definition, zero genes were significantly affected by BKV in the first microarray analysis at 24 hpi. A repeat analysis at 24 hpi produced a similar result: the expression of only 16 genes, mainly involved in cell cycle progression, was upregulated by slightly more than two-fold at 24 hpi (data not shown). However, a large number of genes were affected by BKV infection at both the 48 and 72 hour time points (Table 1, Supplemental Tables 1, 2).
Table 1. Genes in HPTE cells affected 48 and 72 hours post BKV infection.
| Gene Symbol | Change (n-fold) 48 hours | Change (n-fold) 72 hours | Full Gene Name |
|---|---|---|---|
| Cell Cycle/Proliferation | |||
| CCNE2 | 14.1 | 24.3 | cyclin E2 |
| MCM10 | 9.1 | 13.6 | minichromosome maintenance deficient 10 |
| CDC6 | 9.0 | 16.2 | cell division cycle 6 |
| ANLN | 8.3 | 15.8 | anillin, actin binding protein |
| CDC2 | 8.3 | 13.6 | cell division cycle 2 |
| E2F8 | 7.1 | 16.5 | E2F transcription factor 8 |
| CCNA2 | 7.1 | 11.4 | cyclin A2 |
| E2F7 | 5.3 | 10.6 | E2F transcription factor 7 |
| BUB1B | 5.0 | 9.0 | budding uninhibited by benzimidazoles 1 beta |
| BUB1 | 4.6 | 5.0 | budding uninhibited by benzimidazoles 1 |
| MKI67 | 4.2 | 7.9 | antigen identified by monoclonal antibody Ki-67 |
| PCNA | 3.3 | 4.7 | proliferating cell nuclear antigen |
| Apoptosis | |||
| BIRC5 | 4.7 | 8.9 | baculoviral IAP repeat-containing 5 (survivin) |
| GTSE1 | 3.5 | 7.8 | G-2 and S-phase expressed 1 |
| BCL2L12 | 2.1 | 3.6 | BCL2-like 12 (proline-rich) |
| Signal Transduction | |||
| SHCBP1 | 8.3 | 17.3 | SHC SH2-domain binding protein 1 |
| PBK | 6.6 | 13.4 | PDZ binding kinase |
| Chromatin Remodeling | |||
| HCAP-G | 7.7 | 13.3 | chromosome condensation protein G |
| BRRN1 | 5.0 | 9.6 | barren homolog, HCAP-H |
| HELLS | 4.9 | 8.1 | helicase, lymphoid-specific |
| TCF19 | 4.1 | 9.0 | transcription factor 19 (SC1) |
| HMGB2 | 3.9 | 4.7 | high-mobility group box 2 |
| DNMT1 | NC | 2.8 | DNA (cytosine-5-)-methyltransferase 1 |
| DNA Damage Repair | |||
| RAD51AP1 | 7.8 | 23.4 | RAD 51 associated protein 1 |
| BRIP1 | 6.1 | 10.5 | BRCA1 interacting protein C-terminal helicase 1 |
| BARD1 | 5.9 | 8.8 | BRCA1 associated RING domain 1 |
| RAD51 | 4.4 | 5.5 | RAD51 homolog (RecA homolog) |
| BRCA1 | 2.7 | 4.8 | breast cancer 1, early onset |
| Protein Ubiquitination | |||
| UBE2C | 6.5 | 8.0 | ubiquitin-conjugating enzyme E2C |
| FBXO5 | 5.9 | 9.9 | F-box protein 5, early mitotic inhibitor 1 |
| USP1 | 4.3 | 3.8 | ubiquitin specific peptidase 1 |
| RNA Processing | |||
| APOBEC3B | 3.1 | 6.9 | apolipoprotein B mRNA editing enzyme, 3B |
| RNASEH2A | 3.0 | 4.1 | ribonuclease H2, large subunit |
| Immune Response | |||
| PTX3 | NC | 3.6 | pentraxin-related gene, induced by IL-1 beta |
| MICB | NC | 2.6 | MHC class I polypeptide-related sequence B |
| Downregulated Genes | |||
| ADAMTS5 | -2.0 | -2.3 | a disintegrin and metalloproteinase-thrombospondin motif, 5 |
| COL3A1 | -1.8 | -3.1 | collagen, type III, alpha 1 |
| COL1A2 | -1.9 | -2.2 | collagen, type I, alpha 2 |
NC: no change in gene expression.
The largest category of genes upregulated in the presence of BKV contained genes involved in regulation of cell cycle, proliferation, and apoptosis. During BKV lytic infection, 114 out of 195 genes significantly upregulated at 48 hours and 165 out of 372 genes significantly upregulated at 72 hours belonged to this category (Supplemental Tables 1, 2). Among the genes involved in regulation of cell cycle and proliferation were multiple members of the cell division cycle (CDC), cyclin, and minichromosome maintenance deficient (MCM) families. Genes with altered expression also included those involved in signal transduction, regulation of transcription and chromatin remodeling, DNA damage repair mechanisms, protein ubiquitination, protein folding and synthesis, RNA processing, and cellular trafficking and structure, as well as a number of genes for which a function has not yet been defined.
As would be anticipated, many of the genes induced at 48 hpi were also significantly, and in many cases more dramatically, upregulated at 72 hpi (Table 1). For example, transcript levels for CDC6, an E2F target that is required for cellular DNA replication, were elevated by 9-fold at 48 hpi and more than 16-fold at 72 hpi. Transcript levels for CDC2, an essential component of the M-phase promoting factor, which regulates progression of the cell cycle, were upregulated by more than 8-fold at 48 hpi and 13-fold at 72 hpi. Cyclin E2 (CCNE2), an activator of cyclin-dependent kinase 2 that is also upregulated by human papillomavirus E6 and E7 oncoproteins (Hunter and Pines, 1994; Zariwala et al., 1998), was upregulated by over 14-fold at 48 hpi and 24-fold at 72 hpi. Table 1 presents a summary of the expression levels of select genes that were significantly affected by BKV infection and are later discussed.
Very few genes were downregulated in the presence of BKV at 48 or 72 hpi. The only gene significantly downregulated at both time points was that encoding a disintegrin-like and metalloprotease with thrombospondin type 1 motif, 5 (ADAMTS5), for which the major known function is cleavage of aggrecan, a component of cartilage (Malfait et al., 2002). In addition, the transcript levels of type I and type III collagen genes (COL1A2, COL3A1) and a gene of unknown function, C9orf71, were significantly decreased at 72 hpi.
We were intrigued by the apparent lack of virus-mediated regulation of genes involved in the immune response. To explore this observation further, we performed PCR array analysis targeted to common human cytokines at a very early stage of BKV infection to determine if there is any change in cellular gene expression as a result of viral binding and entry. HPTE cells were incubated with purified BKV at 4°C for 1 hour to allow for binding of virions and synchronization of viral entry. Cells were then incubated at 37°C to initiate BKV infection and total cell RNA was harvested at 4 hpi, when the virus is still trafficking through the cytoplasm (Jiang et al., 2009b) and would be likely to engage innate immune receptors. Changes in expression of various cytokines in response to the binding, entry, and trafficking of BKV were analyzed using the Common Human Cytokines quantitative RT-PCR array (SABiosciences). In agreement with the results from the microarray analyses at 24, 48, and 72 hpi, there were no significant changes in the transcript levels at 4 hpi of the 85 cytokine genes analyzed on the array (Supplemental Table 3). We also performed ribonuclease protection assays (RPAs) on RNA isolated from mock- and BKV-infected HPTE cells throughout the course of infection (from 12 to 96 hpi). Using probe sets that detect almost 50 chemokines and cytokines, including those reported to be produced by proximal tubule epithelial cells (Daha and van Kooten, 2000; Frank et al., 1993; Gerritsma et al., 1998), we observed no reproducible differences of more than two-fold for detectable genes (data not shown).
Quantitative RT-PCR confirmation of microarray results
To confirm the results of the microarray analysis, we performed quantitative RT-PCR on selected genes that were significantly changed during BKV infection. cDNA was synthesized from the RNA samples isolated at 48 and 72 hpi and the relative levels of CDC2, CDC6, CCNE2, COL1A2, and COL3A1 transcripts in BKV-infected or mock-infected samples were determined, normalizing to levels of β2-microglobulin transcripts, which remained unchanged during infection. Table 2 shows a comparison of the results from the quantitative RT-PCR assay with those of the microarray analysis. The quantitative RT-PCR assay confirmed that the steady-state levels of CDC2, CDC6, and CCNE2 transcripts were upregulated in BKV-infected HPTE cells at both 48 and 72 hpi, while the levels of COL1A2 and COL3A1 were downregulated at both time points. In addition, the relative change in transcript levels at each time point was very similar between the two assays, further supporting the results of the microarray analysis. The changes in levels of β-2-microglobulin transcripts were determined by averaging the fold difference between BKV-infected and mock-infected samples in all five assays at each time point.
Table 2. Comparison of Quantitative RT-PCR and Microarray Results.
| Gene Symbol | qRT-PCR Change* 48 hpi | Microarray Change 48 hpi | qRT-PCR Change 72 hpi | Microarray Change 72 hpi |
|---|---|---|---|---|
| CDC2 | 4.6 | 8.3 | 12.9 | 13.6 |
| CDC6 | 8.0 | 9.0 | 20.0 | 16.2 |
| CCNE2 | 12.3 | 14.1 | 31.2 | 24.3 |
| COL3A1 | -1.4 | -1.8 | -3.1 | -3.1 |
| COL1A2 | -1.9 | -1.9 | -4.0 | -2.2 |
| B2M | 1.3 | 1.0 | -1.3 | 1.0 |
Expressed as fold difference between BKV-infected and mock-infected cells, with relative levels of CDC2, CDC6, cyclin E2, collagen type I, alpha 2, and collagen type III, alpha 1 transcripts normalized to the levels of β-2-microglobulin transcripts in each sample.
hpi, hours post-infection
Confirmation of microarray results at the level of protein expression
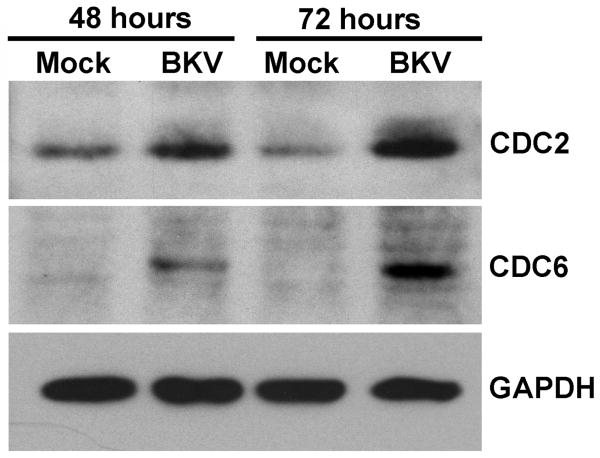
To determine whether the levels of CDC2 and CDC6 protein were changed in a manner consistent with the microarray and quantitative RT-PCR results, we performed Western blot analysis. HPTE cells were infected with purified BKV at an MOI of 5 IU/cell and total cellular protein was extracted at 48 and 72 hpi, and we examined the expression of CDC2, CDC6, or GAPDH as a loading control (Figure 1). At both 48 and 72 hpi, we observed increased expression of CDC2 and CDC6 in the BKV-infected sample as compared to the mock-infected sample, verifying the results of the RNA analyses.
Figure 1. Increased expression of CDC2 and CDC6 after BKV infection.
HPTE cells were infected with BKV at an MOI of 5 IU/cell and total cellular protein was extracted at 0 and 72 hpi. 25 μg of protein lysate was analyzed for the expression of CDC2, CDC6, or GAPDH by Western blotting.
Discussion
In this study, we were interested in determining how BKV affects the cellular environment in the context of a lytic infection. We expected that TAg interactions with pRB and p53, both of which control the transcription of various target genes, would change the transcriptional profile of the cell. In addition, most pathogens trigger an intracellular innate immune response by binding to pattern recognition receptors in the cell (Mogensen, 2009). Our studies show that infection of primary kidney epithelial cells with BKV upregulated the expression of a large number of cellular genes. Many of these genes can be categorized as cell cycle and proliferation factors and, more specifically, targets of p53 and E2F transcription factors. Genes involved in apoptosis, signal transduction, transcriptional regulation, DNA damage repair, protein ubiquitination and folding, RNA processing, and cellular trafficking and structure were also upregulated during infection. In Supplemental Tables 1 and 2, we have provided the comprehensive results of the microarray analysis organized into these functional categories. It should be noted that some genes could be placed in more than one category. Interestingly, while the expression of many genes was activated, the transcript levels of only four genes decreased during BKV infection. To confirm the results of the microarray analysis, the steady-state RNA levels of CDC2, CDC6, CCNE2, COL1A2, and COL3A1 were analyzed by quantitative RT-PCR. Furthermore, the changes in levels of protein expression of CDC2 and CDC6 were confirmed by Western blot analysis. The results of this study provide information about how BKV manipulates the host cell environment during a lytic infection.
We anticipated that the expression of genes that regulate cell cycle and proliferation would be elevated in the context of BKV lytic infection, based on our knowledge of the BKV genome and the activities of TAg. Like other DNA viruses, BKV does not encode enzymes for DNA synthesis and therefore must rely on the host cell replication machinery during productive infection. The interaction of TAg with pRB results in the activation of the E2F transcription factor family, which regulates the expression of many genes involved in cell cycle progression and DNA replication. The subsequent change in the transcriptional profile of the cell provides an optimal environment for viral replication. Others have reported the upregulation of a similar set of genes in the presence of JCV and SV40. Two independent microarray studies of JCV infection have shown elevated levels of cell cycle-associated genes that our results indicate are also affected in the presence of BKV, including members of the cyclin and CDC families (Radhakrishnan et al., 2003; Verma et al., 2006). Studies examining gene expression in SV40 TAg transformed cells or transgenic mouse models by microarray analysis have made similar observations (Guo et al., 2006; Larsson et al., 2004).
Another study described the microarray analysis of cellular transcriptional changes during BKV infection of HUV-EC-C cells, a spontaneously immortalized cell line derived from human umbilical vein endothelial cells (Grinde et al., 2007). There are similarities between the results of that study and ours, including the upregulation of genes involved in cell cycle regulation. However, in the BKV-infected HUV-EC-C cells, a large number of genes were also downregulated: 249 genes at 24 hpi and 104 genes at 40 hpi. The authors also noted that transcripts involved in viral defense were among those downregulated, suggesting that BKV was actively diminishing the cellular antiviral response. While this study is similar to ours in nature and intent, we believe that the primary kidney proximal tubule epithelial cells we used are more relevant for the examination of BKV lytic infection. The differences in cell type (epithelial versus endothelial) and the state of the cell (primary versus immortalized), and the subsequent differences in the stage of infection examined could account for the differences between the two analyses.
It has previously been shown that BKV TAg interaction with p53 can result in inhibition of p53-mediated apoptosis (Harris et al., 1998). In our study, increased levels of anti-apoptotic transcripts, such as BIRC5 (survivin), GTSE1, and BCL2L12, were observed at both 48 and 72 hpi of kidney epithelial cells. The upregulation of these anti-apoptotic genes may prolong the life of the host cell during lytic infection and maximize the production of viral progeny. Simultaneously, however, there were increased levels of pro-apoptotic transcripts, such as CASP8AP2, APAF1, and SIVA, in BKV-infected cells in our studies. Such an opposing effect may indicate a cellular response to the deregulation of the cell cycle resulting from the interaction of TAg and pRB family members.
It is interesting to note the elevated expression of genes involved in chromatin remodeling, structure, and assembly during infection. The BKV genome is associated with cellular histones H2A, H2B, H3, and H4 to form a cellular chromatin-like structure known as the minichromosome (Meneguzzi et al., 1978). Therefore, the induction of chromatin remodeling enzymes may be an important regulatory mechanism for viral DNA replication and transcription, as well as for cellular gene expression. In particular, levels of DNA methyltransferase 1 (DNMT1), which is mainly associated with transcriptional repression, were increased at 48 and 72 hpi in the microarray analysis, which correlates with our previous results demonstrating increased steady-state levels of DNMT1 in infected cells (McCabe et al., 2006). The activation of genes involved in chromatin remodeling and modification could contribute to BKV-associated disease in several ways. For example, expression of such proteins could result in the repression of cytokine production, allowing BKV to evade the immune system. This effect may be especially important in the context of infected kidney proximal tubule cells, which function in immune activation (Daha and van Kooten, 2000; Frank et al., 1993). Modification of viral chromatin may control the level of viral DNA replication or the state of infection under different conditions. Furthermore, in a nonpermissive cell, hypermethylation of tumor suppressor genes may promote BKV transformation (Herman, 1999).
Genes involved in DNA damage repair were also highly upregulated during infection, including multiple members of the RAD family of proteins (RAD51, RAD51AP1, RAD54B, RAD18), BRCA1, BRCA1-associated proteins (BRIP1, BARD1), and the histone variant H2AX. The elevated expression of these proteins may indicate the cellular response to TAg-mediated disruption of DNA repair mechanisms, which are documented for SV40 and JCV (Digweed et al., 2002; Reiss et al., 2006). It has recently been suggested that SV40 uses the DNA damage repair pathway to effect its own replication (Zhao et al., 2008), and BKV may use the same strategy. Interestingly, SV40 TAg binds the spindle checkpoint protein, Bub1, leading to a DNA damage response (Hein et al., 2009), and BKV activates expression of both the BUB1 gene and its homolog, BUB1B, to high levels.
Remarkably, only four genes were significantly downregulated over the course of BKV infection of proximal tubule cells. The metalloprotease ADAMTS5, which has a putative role in cartilage degradation in osteoarthritis, was the only gene downregulated by more than two-fold at both 48 and 72 hpi. Previous studies characterizing ADAMTS5 suggest that regulation typically occurs at the post-translational level (Vankemmelbeke et al., 2001; Yamanishi et al., 2002). Our results, however, suggest instead that BKV regulates ADAMTS5 at the level of transcription, though it is unclear what role this protein plays during infection. Two collagen transcripts, COL1A2 and COL3A1, were also expressed at lower levels in the presence of BKV. In agreement with our findings, a decrease in collagen expression has previously been observed in fibroblasts transformed with SV40 (Bankowski et al., 1978; Parker and Gevers, 1984; Parker et al., 1982). Furthermore, transcript levels of GSTA4, a glutathione S-transferase that detoxifies lipid peroxidation products, were decreased during both BKV infection of HPTE cells and SV40 infection of human fibroblasts and liver cells (Bravard et al., 1993; Sompayrac, 1997). The similarities between our results and those of previous reports in which microarray analysis was not employed support our overall findings, including the paucity of downregulated genes. Studies involving cellular changes induced by expression of human cytomegalovirus IE86 in human fibroblasts show the same pattern: upregulation of hundreds of genes, mainly those involved in cell cycle and proliferation, and downregulation of very few transcripts (Song and Stinski, 2002). It is possible that at later stages of infection, BKV may downregulate additional transcripts, perhaps through interactions with methyltransferases and other chromatin remodeling genes.
During BKV infection of the human host, the virus establishes a persistent infection primarily in kidney epithelial cells until the host environment changes in immunosuppressed patients to favor lytic infection. Considering that kidney proximal tubule cells play a role in immune monitoring and activation, we anticipated that genes involved in regulation of the immune response would be affected by BKV infection. Our analysis, however, revealed that BKV has no overall effect on genes in this category. Only two pro-inflammatory genes were significantly upregulated during infection: pentraxin 3 (PTX3), a cytokine-inducible protein implicated in innate immunity and inflammation (Bottazzi et al., 2006), and MICB, an MHC class I-related molecule that regulates the innate immune response by interacting with the NKG2D receptor on NK and cytotoxic T cells (Collins, 2004). The cytokine-targeted PCR array analysis performed at 4 hpi and the RPA analyses confirmed that BKV does not appear to induce a pro-inflammatory response. Using ELISA, we also observed that BKV infection of HPTE cells had no effect on levels of MCP-1, IL-6, and IL-8, cytokines known to be expressed by proximal tubule cells (data not shown; Frank et al., 1993; Gerritsma et al., 1998).
There are at least three possible explanations for the lack of regulation of genes involved in cytokine expression and immune activation during infection. First, BKV may have developed mechanisms that prevent immune activation during infection, which could include both modulation of cellular gene expression by viral proteins and, especially at very early times post-infection, an entry pathway that avoids the cellular innate immune surveillance mechanisms (Mogensen, 2009). We have recently demonstrated that BKV traffics through an endocytic compartment to the endoplasmic reticulum, where disassembly of the virus begins (Jiang et al., 2009b). It is possible that this route through the cytoplasm does not bring any viral components in contact with innate immune receptors. Second, BKV may actively inhibit pro-inflammatory signaling in the cell. Third, it is possible that HPTE cells are not able to elicit an antiviral immune response in the given experimental context.
In this study, we have characterized the effect of BKV on gene expression in primary human kidney cells at the early and late stages of infection. We observed the upregulation of a large number of genes, particularly those involved in cell cycle and proliferation, and speculate that this is a result of the stimulatory nature of BKV early proteins and subsequent activation of E2F. Very few transcripts were downregulated during infection and there was no activation of a pro-inflammatory response in the cells, suggesting that BKV may evade the immune response by avoiding the regulation of certain transcripts. This study provides an important collection of data detailing the host-pathogen interaction between BKV and kidney epithelial cells, the major site of lytic replication in patients with PVN.
Materials and Methods
Microarray analysis
Primary human proximal tubule epithelial (HPTE) cells were grown as previously described (Low et al., 2004). 1.0 × 106 HPTE cells plated in T-25 tissue culture flasks were infected at 70% confluence using purified BKV strain TU at an MOI of 5 infectious units per cell (IU/cell). Cells were incubated with the virus in 1 ml of growth media at 37°C for 1 hour, with rocking every 15 minutes. Inoculating medium was removed and the cells were washed three times with 1 ml PBS. 3 ml of growth media were then added and the cells were incubated at 37°C with 5% CO2 in a humidified incubator. Infections were performed in duplicate or triplicate and allowed to progress for 24, 48, or 72 hours. RNA was isolated using the Qiagen RNeasy Mini Kit according to the manufacturer's instructions. RNA samples were quantified using an Eppendorf spectrophotometer by measuring the absorbance at 260 nm. RNA purity and quality were determined using Agilent RNA 6000 Nano Eukaryote Total RNA LabChips analyzed on an Agilent 2100 BioAnalyzer. Biotin-labeled cDNA was produced from 10 μg of total cellular RNA and hybridized to the Affymetrix GeneChip Human Genome U133 Plus 2.0 Array according to the manufacturer's suggested protocol (http://gg.bu.edu/microarray/expression_manual.pdf). Sequence clusters were obtained from the Unigene database (Build 133, April 20, 2001). These procedures were performed at the University of Michigan Comprehensive Cancer Center DNA Microarray Core Facility. The arrays were scanned using an Affymetrix GeneChip Scanner to obtain .cel files. The files were then analyzed using dChip software (www.dchip.org) and expression profiles for each transcript were generated using a robust microarray average. The expression values were analyzed with a principal components analysis to determine if the replicated samples grouped together. Once appropriate grouping was determined, the data were filtered to remove from the analysis any genes with expression levels less than 26 relative intensity units, those genes expressed at levels similar to background. The remaining genes were analyzed using a T test of variance, and a two-fold cutoff was used to define significant changes.
Targeted PCR array analysis
HPTE cells were grown to 70% confluence in 12 well plates. To synchronize the infection due to the short duration of the infection, cells were prechilled at 4°C for 15 minutes and then incubated for 1 hour at 4°C with purified BKV TU at an MOI of 5 IU/cell. Inoculating medium was removed and cells were washed once with cold media. 1 ml of 37°C growth media was then added to initiate viral entry and the cells were incubated at 37°C with 5% CO2 in a humidified incubator for 4 hours, at which point total cell RNA was harvested as described above. RNA integrity was confirmed by electrophoresis on an agarose gel and 1 μg of RNA was converted to cDNA using the RT2 First Strand Kit (SABiosciences, cat# C-03), according to manufacturer's instructions. Diluted cDNA was added to the RT2 SYBR Green/ Fluorescein qPCR Master Mix (SABiosciences, cat# PA-011). Human Common Cytokines RT2 Profiler PCR Arrays (SABiosciences, cat# PAHS-021A) were loaded with 25 μl/well of cDNA-master mix and the following PCR program was used for amplification: 10 minutes at 95°C; 40 cycles of denaturation at 95°C for 15 seconds and annealing and extension at 60°C for 1 minute, followed by standard melt curve analysis. Results were analyzed using RT2 Profiler PCR Array Data Analysis Template v3.0, using levels of β-actin (ACTB) and gylceraldehyde-3-phosphate dehydrogenase (GAPDH) to normalize mock and infected sample pairs. PCR arrays were performed on mock- and BKV-infected total cell RNA from two independent experiments; the results are shown in separate columns in Supplemental Table 3.
Quantitative RT-PCR
RNA samples were treated with DNase I (Promega) to reduce contaminating DNA, and RNA integrity was confirmed by electrophoresis on an agarose gel. To generate cDNA, a reverse transcription reaction was performed using 500 ng RNA as template and the iScript cDNA Synthesis Kit (Bio-Rad), according to manufacturer's instructions. Primers and probes were designed to amplify 85- to 145-base pair fragments of CDC2, CDC6, cyclin E2 (CCNE2), collagen type I, alpha 2 (COL1A2), collagen type III, alpha 1 (COL3A1), and β-2-microglobulin (B2M) using Primer3 software (Table 3; Rozen and Skaletsky, 2000). Primers were synthesized by Invitrogen. Oligonucleotide probes, synthesized by Integrated DNA Technologies, Inc., were tagged with 6-fluorescein (FAM) as the reporter dye at the 5′ end and TAMRA-Sp as the quencher dye at the 3′ end. Quantitative PCR reactions were performed in a total volume of 25 μl using TaqMan Universal PCR 2× master mix (Applied Biosystems), 200 nM of probe, either 1200 nM (for amplification of the gene of interest) or 500 nM (for amplification of B2M) of each primer, and 2.5 μl of the cDNA template. Amplification was performed in 96-well PCR plates using the iCycler iQ5 Real Time Detection System (Bio-Rad). The following PCR program was used for amplification: 2 minutes at 50°C; 10 minutes at 95°C; 50 cycles of denaturation at 95°C for 15 seconds and annealing and extension at 58°C (or 60°C for COL3A1) for 1 minute. Results are presented as fold change in target gene transcript levels relative to levels in mock-infected samples, normalized to B2M using the 2-ΔΔC(t) method (Livak and Schmittgen, 2001).
Table 3. Primers and Probes Used for Quantitative RT-PCR (Taqman Assay).
| Target Gene | Primers/Probes | Sequence (5′ to 3′) | Amplicon Size (bp) |
|---|---|---|---|
| CDC2 | Forward Reverse Probe |
TGACATTTGGAGTATAGGCACCATA TCTGAAAATCCTGAAGAGTTGATCA TTGCTGAACTAGCAACTAAGAAACCACTTTT CCAT |
100 |
| CDC6 | Forward Reverse Probe |
CCATGATTGTGTTGGTATTG GCACCAAGTGAGAATTGCTT CAAATAGCGTGTACAATACATCCTGGCCTT |
110 |
| CCNE2 | Forward Reverse Probe |
TGTTTGTGAAACTGTTAAGGTCCTTT CCTTAAGAAAATAGAAGTTTAGGTCAAGTGT AAGCTTTATCACTTTGACACTGTCCTTATCTC ACAAT |
107 |
| COL1A2 | Forward Reverse Probe |
GCAGATTATTTGCCCAAATCT CTGTGGAACCATGGAAGAAG CAGATTCAGCATTTGTTCTTTGCCAGTC |
85 |
| COL3A1 | Forward Reverse Probe |
TAGCTACGGCAATCCTGAAC TACATTTCCACTGGCCTGAT CTTCCCAGAACATCACATATCACTGCAA |
145 |
| B2M | Forward Reverse Probe |
TGGAGGCTATCCAGCGTACT TCAATGTCGGATGGATGAAA TCCAGCAGAGAATGGAAAGTCAAATTTCCT |
115 |
Western blotting
Total cell protein was extracted using E1A lysis buffer (Harlow et al., 1986) containing 0.05 M NaF and Complete EDTA-free Proteinase Inhibitors (Roche). Protein concentration was determined using the Bio-Rad protein assay, and 25 μg of protein were electrophoresed on an 8% SDS/polyacrylamide gel. Proteins were transferred to nitrocellulose membrane as previously described (Low et al., 2006). Membranes were probed overnight at 4°C with the following primary antibodies in PBS-TM (PBS containing 0.1% Tween 20 and 5% non-fat dry milk): anti-Cdk1 (Sigma, catalog #C4973) to detect CDC2, DCS-180 (Sigma, catalog #C0224) to detect CDC6, and 6C5 (Abcam, catalog #Ab8245) to detect GAPDH. Primary antibodies were detected using horseradish peroxidase-conjugated secondary antibodies (Amersham) in PBS-TM and observed using the ECL Plus Western blotting detection system (Amersham).
Supplementary Material
Acknowledgments
We would like to thank the members of the Imperiale lab for assistance and Kathy Spindler and Mary O'Riordan for critical reading of the manuscript. In addition, we would like to thank the University of Michigan Comprehensive Cancer Center Affymetrix and Microarray Core Facility for assistance with the microarray analyses. This work was supported by a research grant to M.J.I. from the NIH (AI060584) and in part by The University of Michigan Comprehensive Cancer Center Core grant (CA46592). J.R.A. supported by the NIH National Research Service Award T32-GM07544 and the F.G. Novy Fellowship.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abend JR, Jiang M, Imperiale MJ. BK virus and human cancer: innocent until proven guilty. Semin Cancer Biol. 2009;19(4):252–60. doi: 10.1016/j.semcancer.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bankowski E, Rzeczycki W, Nowak HK, Jodczyk KJ. Decrease of collagen biosynthesis ability of rat kidney fibroblasts transformed with SV--40 virus. Mol Cell Biochem. 1978;20(2):77–83. doi: 10.1007/BF00241385. [DOI] [PubMed] [Google Scholar]
- Bedi A, Miller CB, Hanson JL, Goodman S, Ambinder RF, Charache P, Arthur RR, Jones RJ. Association of BK virus with failure of prophylaxis against hemorrhagic cystitis following bone marrow transplantation. J Clin Oncol. 1995;13:1103–1109. doi: 10.1200/JCO.1995.13.5.1103. [DOI] [PubMed] [Google Scholar]
- Binet I, Nickeleit V, Hirsch HH, Prince O, Dalquen P, Gudat F, Mihatsch MJ, Thiel G. Polyomavirus disease under new immunosuppressive drugs: a cause of renal graft dysfunction and graft loss. Transplantation. 1999;67(6):918–22. doi: 10.1097/00007890-199903270-00022. [DOI] [PubMed] [Google Scholar]
- Bottazzi B, Garlanda C, Salvatori G, Jeannin P, Manfredi A, Mantovani A. Pentraxins as a key component of innate immunity. Curr Opin Immunol. 2006;18(1):10–5. doi: 10.1016/j.coi.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Bracken AP, Ciro M, Cocito A, Helin K. E2F target genes: unraveling the biology. Trends Biochem Sci. 2004;29(8):409–17. doi: 10.1016/j.tibs.2004.06.006. [DOI] [PubMed] [Google Scholar]
- Bravard A, Hoffschir F, Ricoul M, Cassingena R, Estrade S, Luccioni C, Dutrillaux B. Alterations of the glutathione cycle enzymes during and after SV40-transformation of human fibroblasts. Carcinogenesis. 1993;14(1):21–4. doi: 10.1093/carcin/14.1.21. [DOI] [PubMed] [Google Scholar]
- Bressollette-Bodin C, Coste-Burel M, Hourmant M, Sebille V, Andre-Garnier E, Imbert-Marcille BM. A prospective longitudinal study of BK virus infection in 104 renal transplant recipients. Am J Transplant. 2005;5(8):1926–33. doi: 10.1111/j.1600-6143.2005.00934.x. [DOI] [PubMed] [Google Scholar]
- Collins RW. Human MHC class I chain related (MIC) genes: their biological function and relevance to disease and transplantation. Eur J Immunogenet. 2004;31(3):105–14. doi: 10.1111/j.1365-2370.2004.00457.x. [DOI] [PubMed] [Google Scholar]
- Corallini A, Gianni M, Mantovani C, Vandini A, Rimessi P, Negrini M, Giavazzi R, Bani M, Milanesi G, Dal Cin P, van den Berghe H, Barbanti-Brodano G. Transformation of human cells by recombinant DNA molecules containing BK virus early region and the human activated c-H-ras or c-myc oncogene. Cancer J. 1991;4:24–34. [Google Scholar]
- Daha MR, van Kooten C. Is the proximal tubular cell a proinflammatory cell? Nephrol Dial Transplant. 2000;15 6:41–3. doi: 10.1093/ndt/15.suppl_6.41. [DOI] [PubMed] [Google Scholar]
- Dalrymple SA, Beemon KL. BK virus T antigens induce kidney carcinomas and thymoproliferative disorders in transgenic mice. J Virol. 1990;64(3):1182–91. doi: 10.1128/jvi.64.3.1182-1191.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeCaprio JA, Ludlow JW, Figge J, Shew JY, Huang CM, Lee WH, Marsilio E, Paucha E, Livingston DM. SV40 large tumor antigen forms a specific complex with the product of the retinoblastoma susceptibility gene. Cell. 1988;54(2):275–83. doi: 10.1016/0092-8674(88)90559-4. [DOI] [PubMed] [Google Scholar]
- Digweed M, Demuth I, Rothe S, Scholz R, Jordan A, Grotzinger C, Schindler D, Grompe M, Sperling K. SV40 large T-antigen disturbs the formation of nuclear DNA-repair foci containing MRE11. Oncogene. 2002;21(32):4873–8. doi: 10.1038/sj.onc.1205616. [DOI] [PubMed] [Google Scholar]
- Egli A, Binggeli S, Bodaghi S, Dumoulin A, Funk GA, Khanna N, Leuenberger D, Gosert R, Hirsch HH. Cytomegalovirus and polyomavirus BK posttransplant. Nephrol Dial Transplant. 2007;22(Suppl 8):viii72–viii82. doi: 10.1093/ndt/gfm648. [DOI] [PubMed] [Google Scholar]
- Frank J, Engler-Blum G, Rodemann HP, Muller GA. Human renal tubular cells as a cytokine source: PDGF-B, GM-CSF and IL-6 mRNA expression in vitro. Exp Nephrol. 1993;1(1):26–35. [PubMed] [Google Scholar]
- Gardner SD, Field AM, Coleman DV, Hulme B. New human papovavirus (B.K.) isolated from urine after renal transplantation. Lancet. 1971;1:1253–1257. doi: 10.1016/s0140-6736(71)91776-4. [DOI] [PubMed] [Google Scholar]
- Gerritsma JS, van Kooten C, Gerritsen AF, van Es LA, Daha MR. Transforming growth factor-beta 1 regulates chemokine and complement production by human proximal tubular epithelial cells. Kidney Int. 1998;53(3):609–16. doi: 10.1046/j.1523-1755.1998.00799.x. [DOI] [PubMed] [Google Scholar]
- Grinde B, Gayorfar M, Rinaldo CH. Impact of a polyomavirus (BKV) infection on mRNA expression in human endothelial cells. Virus Res. 2007;123:86–94. doi: 10.1016/j.virusres.2006.08.005. [DOI] [PubMed] [Google Scholar]
- Guo C, Wu G, Chin JL, Bauman G, Moussa M, Wang F, Greenberg NM, Taylor SS, Xuan JW. Bub1 up-regulation and hyperphosphorylation promote malignant transformation in SV40 tag-induced transgenic mouse models. Mol Cancer Res. 2006;4(12):957–69. doi: 10.1158/1541-7786.MCR-06-0168. [DOI] [PubMed] [Google Scholar]
- Harlow E, Whyte P, Franza BR, Jr, Schley C. Association of adenovirus early-region 1A proteins with cellular polypeptides. Mol Cell Biol. 1986;6:1579–1589. doi: 10.1128/mcb.6.5.1579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris KF, Chang E, Christensen JB, Imperiale MJ. BK virus as a potential co-factor in human cancer. Dev Biol Stand. 1998;94:81–91. [PubMed] [Google Scholar]
- Harris KF, Christensen JB, Imperiale MJ. BK virus large T antigen: interactions with the retinoblastoma family of tumor suppressor proteins and effects on cellular growth control. J Virol. 1996;70:2378–2386. doi: 10.1128/jvi.70.4.2378-2386.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hein J, Boichuk S, Wu J, Cheng Y, Freire R, Jat PS, Roberts TM, Gjoerup OV. Simian virus 40 large T antigen disrupts genome integrity and activates a DNA damage response via Bub1 binding. J Virol. 2009;83(1):117–27. doi: 10.1128/JVI.01515-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heritage J, Chesters PM, McCance DJ. The persistence of papovavirus BK DNA sequences in normal human renal tissue. J Med Virol. 1981;8:143–150. doi: 10.1002/jmv.1890080208. [DOI] [PubMed] [Google Scholar]
- Herman JG. Hypermethylation of tumor suppressor genes in cancer. Semin Cancer Biol. 1999;9(5):359–67. doi: 10.1006/scbi.1999.0138. [DOI] [PubMed] [Google Scholar]
- Hirsch HH, Knowles W, Dickenmann M, Passweg J, Klimkait T, Mihatsch MJ, Steiger J. Prospective study of polyomavirus type BK replication and nephropathy in renal-transplant recipients. N Engl J Med. 2002;347(7):488–96. doi: 10.1056/NEJMoa020439. [DOI] [PubMed] [Google Scholar]
- Hunter T, Pines J. Cyclins and cancer. II: Cyclin D and CDK inhibitors come of age. Cell. 1994;79(4):573–82. doi: 10.1016/0092-8674(94)90543-6. [DOI] [PubMed] [Google Scholar]
- Imperiale MJ, Major EO. Polyomaviruses. In: Knipe DM, Howley PM, editors. Fields Virology. Fifth. Vol. 2. Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 2263–2298. 2 vols. [Google Scholar]
- Jiang M, Abend JR, Johnson SF, Imperiale MJ. The role of polyomaviruses in human disease. Virology. 2009a;384(2):266–73. doi: 10.1016/j.virol.2008.09.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang M, Abend JR, Tsai B, Imperiale MJ. Early events during BK virus entry and disassembly. J Virol. 2009b;83(3):1350–8. doi: 10.1128/JVI.02169-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knowles WA. The epidemiology of BK virus and the occurrence of antigenic and genomic subtypes. In: Khalili K, Stoner GL, editors. Human Polyomaviruses: Molecular and Clinical Perspectives. Wiley-Liss Inc.; New York: 2001. pp. 527–559. [Google Scholar]
- Larsson O, Scheele C, Liang Z, Moll J, Karlsson C, Wahlestedt C. Kinetics of senescence-associated changes of gene expression in an epithelial, temperature-sensitive SV40 large T antigen model. Cancer Res. 2004;64(2):482–9. doi: 10.1158/0008-5472.can-03-1872. [DOI] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Low J, Humes HD, Szczypka M, Imperiale M. BKV and SV40 infection of human kidney tubular epithelial cells in vitro. Virology. 2004;323(2):182–8. doi: 10.1016/j.virol.2004.03.027. [DOI] [PubMed] [Google Scholar]
- Low JA, Magnuson B, Tsai B, Imperiale MJ. Identification of gangliosides GD1b and GT1b as receptors for BKV virus. J Virol. 2006;80(3):1361–1366. doi: 10.1128/JVI.80.3.1361-1366.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Major EO, Di Mayorca G. Malignant transformation of BHK21 clone 13 cells by BK virus--a human papovavirus. Proc Natl Acad Sci U S A. 1973;70(11):3210–2. doi: 10.1073/pnas.70.11.3210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malfait AM, Liu RQ, Ijiri K, Komiya S, Tortorella MD. Inhibition of ADAM-TS4 and ADAM-TS5 prevents aggrecan degradation in osteoarthritic cartilage. J Biol Chem. 2002;277(25):22201–8. doi: 10.1074/jbc.M200431200. [DOI] [PubMed] [Google Scholar]
- McCabe MT, Low JA, Imperiale MJ, Day ML. Human polyomavirus BKV transcriptionally activates DNA methyltransferase 1 through the pRb/E2F pathway. Oncogene. 2006;25(19):2727–35. doi: 10.1038/sj.onc.1209266. [DOI] [PubMed] [Google Scholar]
- Meneguzzi G, Pignatti PF, Barbanti-Brodano G, Milanesi G. Minichromosome from BK virus as a template for transcription in vitro. Proc Natl Acad Sci U S A. 1978;75:1126–1130. doi: 10.1073/pnas.75.3.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22(2):240–73. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakshatri H, Pater MM, Pater A. Functional role of BK virus tumor antigens in transformation. J Virol. 1988;62(12):4613–21. doi: 10.1128/jvi.62.12.4613-4621.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickeleit V, Hirsch HH, Zeiler M, Gudat F, Prince O, Thiel G, Mihatsch MJ. BK-virus nephropathy in renal transplants-tubular necrosis, MHC-class II expression and rejection in a puzzling game. Nephrol Dial Transplant. 2000;15(3):324–32. doi: 10.1093/ndt/15.3.324. [DOI] [PubMed] [Google Scholar]
- Parker MI, Gevers W. Demethylation of the type I procollagen genes in transformed fibroblasts treated with 5-azacytidine. Biochem Biophys Res Commun. 1984;124(1):236–43. doi: 10.1016/0006-291x(84)90942-2. [DOI] [PubMed] [Google Scholar]
- Parker MI, Judge K, Gevers W. Loss of type I procollagen gene expression in SV40-transformed human fibroblasts is accompanied by hypermethylation of these genes. Nucleic Acids Res. 1982;10(19):5879–91. doi: 10.1093/nar/10.19.5879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pater A, Pater MM. Transformation of primary human embryonic kidney cells to anchorage independence by a combination of BK virus DNA and the Harvey-ras oncogene. J Virol. 1986;58(2):680–3. doi: 10.1128/jvi.58.2.680-683.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlakis M, Haririan A, Klassen DK. BK virus infection after non-renal transplantation. Adv Exp Med Biol. 2006;577:185–189. doi: 10.1007/0-387-32957-9_13. [DOI] [PubMed] [Google Scholar]
- Radhakrishnan S, Otte J, Enam S, Del Valle L, Khalili K, Gordon J. JC virus-induced changes in cellular gene expression in primary human astrocytes. J Virol. 2003;77(19):10638–10644. doi: 10.1128/JVI.77.19.10638-10644.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiss K, Khalili K, Giordano A, Trojanek J. JC virus large T-antigen and IGF-I signaling system merge to affect DNA repair and genomic integrity. J Cell Physiol. 2006;206(2):295–300. doi: 10.1002/jcp.20455. [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the WWW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Humana Press; Totowa: 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Shinohara T, Matsuda M, Cheng SH, Marshall J, Fujita M, Nagashima K. BK virus infection of the human urinary tract. J Med Virol. 1993;41:301–305. doi: 10.1002/jmv.1890410408. [DOI] [PubMed] [Google Scholar]
- Small JA, Khoury G, Jay G, Howley PM, Scangos GA. Early regions of JC virus and BK virus induce distinct and tissue-specific tumors in transgenic mice. Proc Natl Acad Sci U S A. 1986;83(21):8288–92. doi: 10.1073/pnas.83.21.8288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sompayrac L. SV40 and adenovirus may act as cocarcinogens by downregulating glutathione S-transferase expression. Virology. 1997;233(1):130–5. doi: 10.1006/viro.1997.8610. [DOI] [PubMed] [Google Scholar]
- Song YJ, Stinski MF. Effect of the human cytomegalovirus IE86 protein on expression of E2F-responsive genes: a DNA microarray analysis. Proc Natl Acad Sci U S A. 2002;99(5):2836–41. doi: 10.1073/pnas.052010099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uchida S, Watanabe S, Aizawa T, Kato K, Furuno A. Induction of papillary ependymomas and insulinomas in the Syrian golden hamster by BK virus, a human papovavirus. Gann. 1976;67(6):857–65. [PubMed] [Google Scholar]
- Vankemmelbeke MN, Holen I, Wilson AG, Ilic MZ, Handley CJ, Kelner GS, Clark M, Liu C, Maki RA, Burnett D, Buttle DJ. Expression and activity of ADAMTS-5 in synovium. Eur J Biochem. 2001;268(5):1259–68. doi: 10.1046/j.1432-1327.2001.01990.x. [DOI] [PubMed] [Google Scholar]
- Verma S, Ziegler K, Ananthula P, Co JK, Frisque RJ, Yanagihara R, Nerurkar VR. JC virus induces altered patterns of cellular gene expression: interferon-inducible genes as major transcriptional targets. Virology. 2006;345(2):457–467. doi: 10.1016/j.virol.2005.10.012. [DOI] [PubMed] [Google Scholar]
- Vilchez RA, Kusne S. Molecular and clinical perspectives of polyomaviruses: emerging evidence of importance in non-kidney transplant populations. Liver Transpl. 2006;12:1457–1463. doi: 10.1002/lt.20915. [DOI] [PubMed] [Google Scholar]
- Vousden KH. p53: death star. Cell. 2000;103(5):691–4. doi: 10.1016/s0092-8674(00)00171-9. [DOI] [PubMed] [Google Scholar]
- Yamanishi Y, Boyle DL, Clark M, Maki RA, Tortorella MD, Arner EC, Firestein GS. Expression and regulation of aggrecanase in arthritis: the role of TGF-beta. J Immunol. 2002;168(3):1405–12. doi: 10.4049/jimmunol.168.3.1405. [DOI] [PubMed] [Google Scholar]
- Zariwala M, Liu J, Xiong Y. Cyclin E2, a novel human G1 cyclin and activating partner of CDK2 and CDK3, is induced by viral oncoproteins. Oncogene. 1998;17(21):2787–98. doi: 10.1038/sj.onc.1202505. [DOI] [PubMed] [Google Scholar]
- Zhao X, Madden-Fuentes RJ, Lou BX, Pipas JM, Gerhardt J, Rigell CJ, Fanning E. Ataxia telangiectasia-mutated damage-signaling kinase- and proteasome-dependent destruction of Mre11-Rad50-Nbs1 subunits in Simian virus 40-infected primate cells. J Virol. 2008;82(11):5316–28. doi: 10.1128/JVI.02677-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.