Abstract
Fusobacterium nucleatum is a Gram-negative anaerobic rod found in dental plaque biofilms, and is an opportunistic pathogen implicated in periodontitis as well as a wide range of systemic abscesses and infections. Genomic analyses of F. nucleatum indicate considerable genetic diversity and a prominent role for horizontal gene transfer in the evolution of the species. Several plasmids isolated from F. nucleatum, including pFN1, harbor relaxase gene homologues that may function in plasmid mobilization. In this investigation we examined the RP4-mediated mobilization properties of pFN1 and the prevalence of pFN1-related sequences in a panel of F. nucleatum clinical isolates. The fusobacterial plasmid pFN1 was mobilized by RP4 at a high frequency. Deletion analyses were used to delineate the core mobilon of pFN1, which consisted of the relaxase gene (rlx), an upstream open reading frame ORF4 and a region of DNA upstream of ORF4 with potential nic sites. To examine the prevalence of pFN1 in a panel of clinical isolates, total DNA isolated from the strains was hybridized with pFN1 replication (repA) and rlx gene probes. DNA from strains harboring plasmids known to be homologous to pFN1 hybridized with both the repA and rlx probes. Five additional strains were rlx-positive but repA-negative, indicating a greater prevalence of rlx-related genes in comparison with repA-related genes. Plasmid or plasmid-related sequences were identified in 11.5% of the strains examined. These findings demonstrate mobilization properties of a fusobacterial plasmid that may be important in horizontal gene transfer.
Keywords: Fusobacterium nucleatum, mobilization, conjugation, plasmid, pFN1
1. INTRODUCTION
Fusobacterium nucleatum is a Gram-negative anaerobic rod that plays a significant role in the ecology of dental plaque biofilms and human infectious diseases (Bearfield et al., 2002; Brook and Frazier, 1998; Fouad et al., 2002; Kolenbrander and London, 1993; Koornstra et al., 1998; Lark et al., 2001; Mikamo et al., 1998; Pickering et al., 2000; Sakamoto et al., 2006). Studies indicate considerable phenotypic variability between F. nucleatum strains (Bolstad et al., 1996), and biochemical analyses have suggested that F. nucleatum represents a “species complex” (Morris et al., 1996). Comparative genomic analyses support this assertion. For example, 25% of the genes encoded by F. nucleatum ATCC 10953 ssp. polymorphum are not found in either of the sequenced genomes of F. nucleatum ssp. nucleatum and vincentii (Karpathy et al., 2007). In addition, 21% of these unique ORFs mapped to clusters of five or more genes, suggesting that the may have been introduced into the genome by horizontal gene transfer.
One mechanism contributing to horizontal gene transfer is conjugative mobilization of DNA within and between bacterial species, which may occur in association with plasmids. The first report of plasmids from F. nucleatum indicated that plasmids or plasmid-related sequences were found in 18% of the oral isolates examined (McKay et al., 1995). Plasmids isolated from F. nucleatum are reported to range from 5 to 15 kb (Bachrach et al., 2004; Haake et al., 2000; McKay et al., 1995), which is insufficient to encode all the genes needed for conjugation of self-transmissible plasmids. However, sequence analyses of several F. nucleatum plasmids revealed homologues of plasmid relaxases suggesting that the plasmids may be mobilizable with conjugative functions provided in trans (Haake et al., 2000; Karpathy et al., 2007). The initial phase in conjugative dissemination of plasmid DNA involves DNA processing reactions resulting in the formation of the relaxosome, a DNA-protein intermediate consisting of a single-strand of the plasmid DNA complexed with proteins that include the plasmid-encoded relaxase. The relaxase is a key component of the relaxosome as it is responsible for the single-stranded nicking of the plasmid by a reversible transesterification reaction. The relaxase is covalently bound to the single-stranded DNA at the 5’ end, and after transfer of the relaxosome to a recipient cell it mediates the recircularization of the transferred DNA (Byrd and Matson, 1997).
The 5.9 kb fusobacterial plasmid pFN1 was identified in a transtracheal isolate of F. nucleatum, and the replicon of this plasmid proved useful in the development of gene transfer systems for the species (Haake et al., 2000; Kinder Haake et al., 2006). The pFN1 plasmid also encodes a putative relaxase. The predicted protein sequence of the pFN1 relaxase gene indicates the presence of the amino acid motifs characteristic of the MOBP family of relaxases, including the signature 3H motif and the active site tyrosine residue (Francia et al., 2004; Haake et al., 2000; Ilyina and Koonin, 1992; Karpathy et al., 2007). The prototype for the MOBP relaxases is the TraI protein encoded on the conjugative broad-host range plasmid RP4. We hypothesized that pFN1 would be mobilized by the RP4 conjugative system in trans. The present investigation was undertaken to examine the RP4-mediated mobilization properties of pFN1, and the prevalence of plasmids and pFN1 plasmid-related sequences in a panel of F. nucleatum strains isolated from oral and non-oral infections.
2. MATERIAL AND METHODS
2.1. Bacterial strains, media, growth conditions, and DNA isolation
Escherichia coli strain DH5α (Invitrogen, Carlsbad, CA) was used as the host strain for all plasmid constructions and manipulations. Mobilization studies were conducted with filter matings using E. coli strain S17-1 (Simon et al., 1983) as a donor strain and E. coli strain EM24NR (Leung et al., 2002) as the recipient strain. Luria-Bertani (LB) medium (BD, Sparks, MD) was used for all liquid cultures, which were shaken aerobically at 37°C. Luria-Bertani agar plates were used for solid media requirements and the plates were incubated at 37°C under aerobic conditions. Trimethoprim (trim; 10 µg/ml Sigma Aldrich, St. Louis, MO) was used to select for S17-1 and erythromycin (erm; 300µg/ml; Fisher Scientific, Tustin, CA) was used to select for all plasmids. Antibiotics used in the selection of EM24NR were streptomycin (str; 10 µg/ml, Sigma-Aldrich), naladixic acid (nal; 50 µg/ml, Acrōs Organics, Geel, Belgium), and rifampicin (rif; 20 µg/ml, Fisher Scientific). Antibiotics in liquid media were used at half their stated concentrations. Plasmid DNA isolated from E. coli was prepared using QIAGEN Plasmid Maxi, Midi, or Mini-prep Kits (QIAGEN, Valencia, CA). F. nucleatum strains were identified according to the Wadsworth Anaerobe Manual (Jousimmies-Somer et al., 2002) and were maintained anaerobically as previously described (Kinder Haake et al., 2006). Plasmid DNA isolated from F. nucleatum strains was prepared using the StrataPrep® EF Plasmid Midiprep Kit (Stratagene, La Jolla, CA), and total cell DNA was prepared as previously described (Haake and Wang, 1997).
2.2. Plasmid constructs
The plasmids used in this study and their relevant properties are listed in Table 1. PCR was performed using KOD XL DNA Polymerase according to conditions specified by the manufacturer (Novagen, San Diego, CA). Restriction endonucleases and DNA modifying enzymes were purchased from New England Biolabs (Beverly, MA) and Fermentas (Glen Burnie, MD). Gel-purification of all DNA fragments was done using either the S.N.A.P. UV-Free Gel Purification Kit (Invitrogen, Carlsbad, CA), the QIAquick Gel Extraction Kit (QIAGEN), or the GENECLEAN III Kit (MP Biomedicals, Solon, OH) according to the manufacturers’ protocols.
Table 1.
Bacterial Strains, Plasmids, and Oligonucleotide Primers
| Strain, plasmid, or primer |
Relevant Characteristics | Source or Reference |
|---|---|---|
| E. coli Strains | ||
| DH5α | RecA-; used for cloning | Invitrogen |
| S17-1 | RecA-,Trimr, used as mating donor; IncP RP4 inserted into chromosome | (Simon et al., 1983) |
| EM24NR | RecA-, Stpr, Nalr, Rifr | (Leung et al., 2002) |
| Plasmids | ||
| pCR-2.1 TOPO | Ampr, Kanr; 3.9 kb plasmid used for cloning PCR products | Invitrogen |
| pBluescript SK(-) | Ampr; 3.0-kb E. coli cloning vector | Stratagene Cloning Systems |
| pACYC184 | 4.2 kb low copy number plasmid used as a source of the p15A oriV | (Chang and Cohen, 1978) |
| pVA2198 | Ermr, Mob+; 9.2 kb Bacteriodes shuttle vector, source of ermF- ermB cassette | (Fletcher et al., 1995) |
| pHS19 | Ermr, Amps; 4.1 kb plasmid constructed by cloning ermF-ermB into pBluescriptΔbla backbone | (Haake et al., 2000) |
| pFN1 | 5.9 kb plasmid isolated from F. nucleatum WAL12230 | (Haake et al., 2000) |
| pHS17 | Ermr, Mob+; 10.0 kb shuttle plasmid constructed by cloning pFN1 into pHS19 | (Haake et al., 2000) |
| pHS23 | Ermr, Mob-; 8.0 kb shuttle plasmid derived from pHS17 by deletion of 2.0 kb rlx-ORF2-ORF3 pFN1 fragment | (Kinder Haake et al., 2006) |
| pHS25 | Ermr, Mob+; 8.6 kb shuttle plasmid derived from pHS17 by deletion of 1.4 kb ORF6-oriVFn-repAFn pFN1 fragment | (Kinder Haake et al., 2006) |
| pTL1 | Ermr, Mob-; 3.3 kb plasmid constructed by cloning the pACYC184 p15A oriV into pHS19 ΔColE1oriV | This study |
| pBC7 | Ermr, 6.2 kb plasmid constructed by cloning 2.2 kb pFN1 PCR-amplified ORF7, ORF4 and rlx into pHS19. | This study |
| pBC17 | Ermr, Mob+; 9.1 kb plasmid constructed by cloning the 5.9 kb pFN1 fragment of pHS17 into pTL1 | This study |
| pBC23 | Ermr, Mob-; 7.2 kb plasmid constructed by cloning the 4.0 kb pHS23 pFN1 fragment into pTL1 | This study |
| pBC25 | Ermr, Mob+; 7.8 kb plasmid constructed by cloning the 4.6 kb pHS25 pFN1 fragment into pTL1 | This study |
| pBC27 | Ermr, Mob+; 5.5 kb plasmid constructed by cloning a 2.2 kb ORF7-ORF4-rlx pFN1 fragment into pTL1 | This study |
| pBC31 | Ermr, Mob-; 4.6 kb plasmid constructed by cloning a 1.4 kb ORF7-ORF4 pFN1 fragment into pTL1 | This study |
| pBC32 | Ermr, Mob+; 7.1 kb plasmid construction from pBC25 by deletion of pFN1 ORF3 fragment | This study |
| pBC33 | Ermr, Mob+; 6.6 kb plasmid constructed by cloning PCR-amplified pFN1 ORF3 into pBC27 | This study |
| pFN2 | 7.2 kb native plasmid from F. nucleatum WAL10113 | (Haake et al., 2000) (Haake et al., 2000) |
| pFN3 | 11.9 kb plasmid from F. nucleatum ATCC10953 | Karpathy et al., 2007) |
| pFN4 | 9.6 kb plasmid from F. nucleatum RMA11532 | This study |
| pFN5 | 5.8 kb plasmid from F. nucleatum RMA8409 | This study |
| pFN6 | 4.0 kb plasmid from F. nucleatum RMA5369 | This study |
|
Oligonucleotide Primersa |
||
| BC-390 | 5'- TAGCTTCCATACATATCACACTCC -3'; pBC7 upper primer | Invitrogen |
| BC-391 | 5'- TTCTCAAATATTTCTGCTCTACTC -3'; pBC7 lower primer | Invitrogen |
| BC-476 | 5'- TACTGACAGCTTCGGGGATCCACT -3'; pBC31 upper primer | Invitrogen |
| BC-477 | 5'- TTGCCAGCTCCATTTTATAACTTG -3'; pBC31 lower primer | Invitrogen |
| BC-648 | 5'- aaaaCTGCAGAAGTTTTGGTACAGTGAAGA -3'; pBC33 upper primer | Invitrogen |
| BC-649 | 5'-aaggaaaaaaGCGGCCGCTGAAACATAGCAACAAAGTA-3'; pBC33 lower primer | Invitrogen |
Underlined sequences indicate PstI or NotI restriction sites; nucleotides shown in lowercase added 5' to these sites to ensure complete cleavage of PCR products
For the mobilization studies we used a backbone plasmid, pTL1, comparable to plasmids previously shown to not be mobilized by the RP4 conjugative system (Leung et al., 2002). pTL1 was generated by cloning the 0.8 kb EcoRV/SacII fragment from pACYC184 encoding the p15A oriV (Chang and Cohen, 1978) into the 2.4 kb SacII/FspI fragment of pHS19 that encodes the ermF-ermB cassette (previously ermF-ermAM) (Haake et al., 2000; Roberts et al., 1999). The ermF-ermB cassette of pHS19 was originally isolated from pVA2198 as an EcoRI-BamHI fragment (Fletcher et al., 1995; Haake et al., 2000) that was cloned into a similarly digested pBluescript backbone, followed by deletion of the pBluescript bla gene with BspHI digestion. The pFN1-based plasmids generated in this investigation were constructed in the pTL1 backbone as follows (Table 1, Figure 1). pBC17 and pBC23 were constructed by ligating the 5.9 kb and 4.0 kb AvaI/ SacI pFN1 fragments of pHS17 and pHS23 (Kinder Haake et al., 2006), respectively, into the 3.2 kb AvaI/ SacI pTL1 backbone. pBC25 was constructed by ligating the 4.6 kb NotI/BamHI fragment of pHS25 into the similarly digested 3.3 kb pTL1 backbone. An intermediary plasmid, pBC7, was used in the construction of pBC27 and pBC31. The pFN1 ORF7, ORF4, and relaxase region amplified from pHS25 was cloned in pCR2.1-TOPO (Invitrogen, Carlsbad, CA), then the 2.2 kb NotI/BamHI fragment was isolated and cloned into the similarly digested pHS19 backbone to generate pBC7. pBC27 was constructed by isolating the 2.2 kb pFN1 NotI/BamHI fragment of pBC7 and cloning it in the similarly digested pTL1 backbone. pBC31 was generated by amplifying the 1.4 kb pFN1 ORF7 and ORF4 regions from pBC7, cloning it into pCR2.1-TOPO, subsequently isolating the 1.4 kb NotI/BamHI fragment and cloning it in the similarly digested pTL1 backbone. The 0.7 kb deletion of ORF3 from pBC25 via a BplI/ NotI sequential digest formed pBC32. Finally, the ORF3 region of pFN1 was amplified with flanking NotI and PstI sites, digested and cloned into pBC27 to form pBC33. All PCR-amplified DNA was confirmed by sequence analysis (UCLA Genotyping and Sequencing CORE, Los Angeles, CA; Agencourt Bioscience Corporation, Beverly, MA) and the final constructs were all identically oriented in the pTL1 backbone. Sequence analyses were conducted with Lasergene v.7 software (DNASTAR, Inc., Madison, WI).
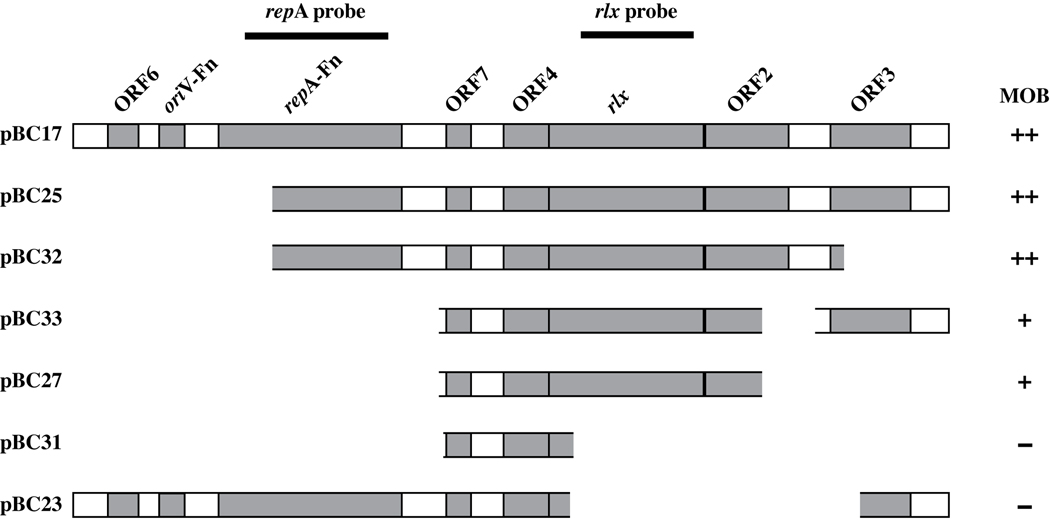
Figure 1. Schematic Illustration of pFN1 Components Associated with Plasmid Mobilization.
The diagram illustrates the portions of pFN1 cloned into the backbone vector pTL1 for each of the plasmid constructs evaluated, and indicates the DNA fragments used for the repA and rlx probes in hybridization studies (see Figure 3). MOB, mobilization above background levels by the E. coli S17-1 host strain possessing the chromosomally-integrated RP4 plasmid with transfer frequencies as follows: ++: > 10−2, +: > 10−6 but less than 10−2; − < 10−6. The backbone vector pTL1 was either not mobilized or mobilized with a transfer frequency < 10−6. This level is used to define the background level in interpreting the contribution of pFN1 to mobilization properties. See Materials and Methods and Table 2 for additional details on plasmid construction and transfer frequency, respectively.
2.3. Mobilization Studies
The mobilization properties of the pFN1-based plasmids were determined in an adaptation of a previously described spot filter mating procedure using the E. coli donor strain, S17-1 (Simon et al., 1983), and the E. coli recipient strain EM24NR (Leung et al., 2002). The plasmid pVA2198, which encodes the E. coli mobilization region from the RK2 broad host range plasmid (C.J. Smith, personnel communication), was used as a positive control. The backbone plasmid pTL1 was used as a negative control. Donor cultures of S17-1 harboring the plasmid to be tested and recipient cultures were grown overnight with the relevant antibiotic selection. The cultures were diluted 1:10, grown to mid-logarithmic phase, then standardized by volume to an OD600 = 0.600. Donor and recipient cells were combined in equal parts of 1 ml each, washed once with sterile PBS to remove residual antibiotics, and resuspended in 2 ml of sterile PBS. Aliquots (10 µl) were removed to determine the number of input donor and recipient colony-forming units (CFU’s) by plating with selection (donor, trim and erm; recipient, str and nal). The remaining mating mixture was centrifuged, resuspended in 50 µl of sterile PBS, spotted onto sterile nitrocellulose filters (0.45 µm, Millipore, Billerica, MA), incubated overnight on LB agar without selection at 37°C, then removed and the cells resuspended in sterile PBS. The number of output donor and recipient CFU’s was determined after plating on selective media as above. The number of transconjugant CFU’s was determined after plating on LB agar with antibiotics specific for the detection of transconjugant cells (stp, nal, and erm). The transfer frequency was determined by dividing the number of transconjugant CFU’s by the number of output recipient CFU’s. Control matings to rule out the possibility of natural transformation were done with the addition of DNase to the mating mixture (50 mg/ml; Roche Applied Science, Indianapolis, IN) as indicated. At minimum two independent filter matings were performed for each of the test plasmids.
2.4. Southern dot blots, DIG-probe labeling and hybridization
One µg of total cellular DNA from each F. nucleatum strain analyzed was alkali-denatured with a solution of 0.4 M NaOH/ 10 mM EDTA, heated for 10 minutes at 100°C, and applied to a positively charged nylon membrane (Roche Applied Science, Indianapolis, IN) housed in a dot blot apparatus (Bio-Rad, Bio-Dot Microfiltration Apparatus). Each well was rinsed with 0.4 M NaOH before the membrane was washed briefly in 2X SSC and then neutralization buffer (1M NaCl, 0.5M Tris-Cl pH7.2). Membranes were sandwiched between two pieces of Whatman paper that had been soaked in 10X SSC and were then UV cross-linked. The repA probe consisted of a 1.1 kb AseI fragment isolated from pHS23 and the rlx probe consisted of a 843 bp HincII-SwaI fragment isolated from pHS25 (Table 1, Figure 1). DIG labeling of DNA fragments to be used as probes and the hybridization of these probes was performed according to the DIG High Prime DNA Labeling and Detection Starter Kit I protocol (Roche), except that chemiluminescent detection was used. Chemiluminescent detection was performed using a 1:100 dilution of the disodium 3-(4-methoxyspiro {1,2-dioxetane-3,2′-(5'-chloro)tricyclo [3.3.1.13,7]decan}-4-yl)phenyl phosphate (CSPD; Roche) stock solution, exposing the membrane to X-RAY film from 2 minutes to several hours, followed by development of the film using standard techniques. Scanned images of developed films were subject to densitometric analysis (Quantity One, Bio-Rad Laboratories) and positive hybridization was determined based on spot intensity 2-fold or greater as compared to background on duplicate or triplicate independent blots.
3. RESULTS
3.1. Mobilization properties of pFN1
E. coli S17-1 harbors the RP4 plasmid in its chromosome, providing conjugative functions in trans, and was used as the donor strain to examine the mobilization properties of pFN1. A filter mating procedure was used to mobilize plasmids from E. coli S17-1 to the recipient strain E. coli EM24NR. In all experiments, representative transconjugants were subcultured on selective media and the plasmid content confirmed by purification and analysis by restriction endonuclease mapping (data not shown). The mobilizable plasmid pVA2198 (Fletcher et al., 1995), used as a positive control, had an average transfer frequency of 2.56 × 10−1 (Table 2). In our initial studies we used pHS19, a high copy number plasmid with a ColE1 oriV that lacks a mobilization region, as a negative control. However, we isolated a background level of transconjugants (data not shown). Because previous reports indicated no transconjugants isolated with a low copy number plasmid with a p15a oriV using the same donor and recipient strains (Leung et al., 2002), we switched to pTL1 which also possesses the p15a oriV and lacks a mobilization gene. Filter matings with the pTL1 backbone plasmid, resulted in either no transconjugants or a background level of <10−6 with the isolation of a single transconjugant colony. Thus, transfer levels of 10−6 or less were considered to represent background levels. To assess the mobilization properties of pFN1, we initially cloned the entire pFN1 plasmid into pTL1, resulting in the chimeric plasmid pBC17. Using the filter mating procedure, pBC17 was transferred from the E. coli S17-1 donor strain to E. coli EM24NR recipient with an average transfer frequency of 1.89 × 10−2 (Table 2).
Table 2.
Mobilization of pFN1-based Constructs by the Chromosomally-integrated Conjugative Plasmid RP4
| Donora | Average Transfer Frequencyb |
|---|---|
| pVA2198* | 2.56 × 10−1 |
| pTL1* | NTc or <10−6 |
| pBC17 | 1.89 × 10−2 |
| pBC25 | 5.68 × 10−2 |
| pBC32 | 1.94 × 10−1 |
| pBC33 | 8.98 × 10−4 |
| pBC27* | 2.78 × 10−3 |
| pBC31* | <10−6 |
| pBC23 | NT |
All donor plasmids carrying the erythromycin resistance cassette were present in the E. coli host strain S17-1. The recipient for filter matings was EM24NR (strr, nalr, erms).
Transfer frequency is the number of transconjugants (ermr, strr, nalr) divided by the number of output recipients (strr, nalr). Matings were performed in 2 to 6 independent experiments with similar results. < indicates that one trial resulted in transfer while the remaining trails resulted in no transfer (NT). See methods and materials for additional details.
Not transferable.
These plasmids were tested in the presence of DNase, added during incubation of the mating mixture on the nitrocellulose filter, with similar results to incubations without DNase.
To delineate the pFN1 genes contributing to mobilization, a series of pFN1-based constructs were derived from pBC17 (Figure 1) and tested by filter mating (Table 2). Deletion of the pFN1 oriV and 5’ region of the repA homologue had no effect on mobilization properties as demonstrated by pBC25. Deletion of ORF3 (pBC32) resulted in an increase in transfer frequency, suggesting that this region of DNA may be inhibitory to mobilization. Deletion of the 3’ region ORF2 alone (pBC33) or in combination with ORF3 (pBC27), resulted in diminished mobilization with average transfer frequencies of 8.98 × 10−4 and 2.78 × 10−3, respectively. These data suggest that the 3’ end of ORF2 is not essential for, but contributes to, efficient mobilization. Constructs with deletions in the relaxase gene (pBC23 and pBC31) were non-mobilizable or at background levels, indicating a central role for this gene in plasmid transfer. Plasmid mobilization was unaffected by the presence of DNaseI, supporting the assertion that the mechanism of transfer was conjugative.
3.2. Sequence Analysis of the pFN1 Mobilon
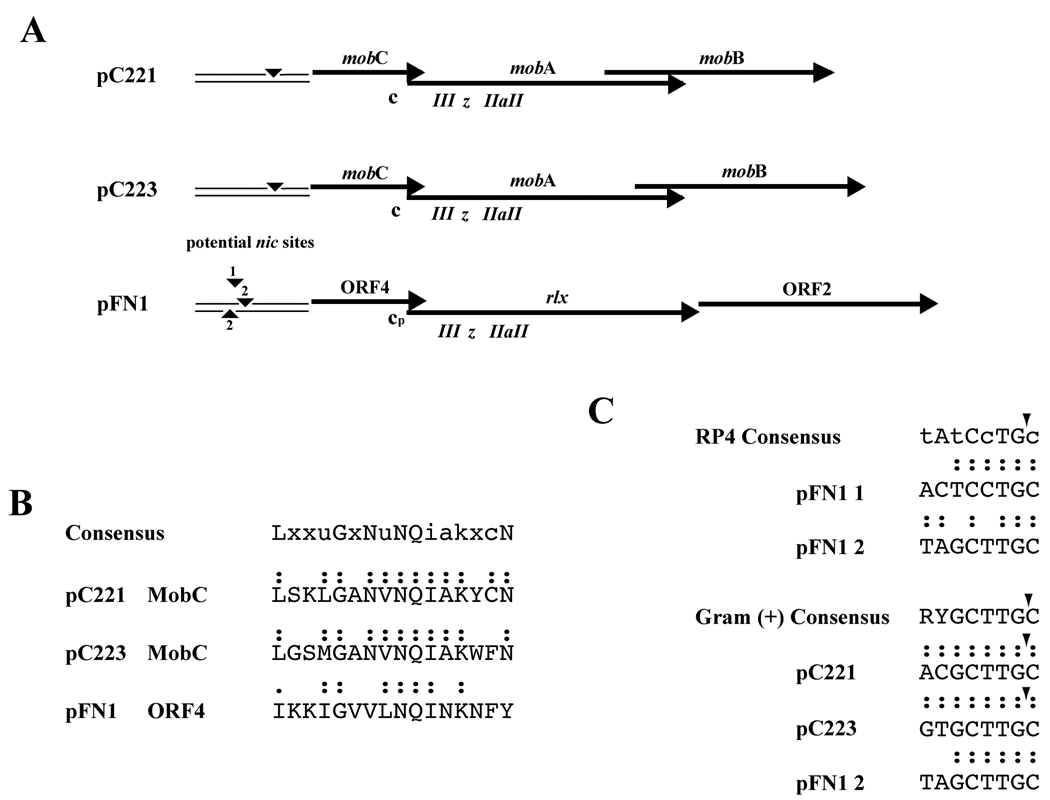
The deletion analyses indicated a core mobilon of pFN1 that included the relaxase, and DNA directly upstream containing ORF4 and ORF7. The organization of the pFN1 mobilon appeared similar to that of the staphylococcal plasmids pC221 and pC223 (Figure 2, panel A). The relaxase proteins of pC221 and pC223 are termed MobA and are 30 and 23% identical, respectively, to the pFN1 relaxase, in addition to homology within defined relaxase functional motifs (Haake et al., 2000). The staphylococcal plasmid MobC protein sequences demonstrate 20 to 21% identity with the pFN1 ORF4 and a possible MobC motif was identified in the pFN1 ORF4 (Figure 2, panel B). The staphylococcal MobB proteins demonstrated 13 to 14% identity with the pFN1 ORF2. No homology to the pFN1 ORF7 was identified in the staphylococcal plasmids, but several potential nic sites (designated 1 and 2, with sequence 2 occurring on both strands; Figure 2, panel A and C) were identified in the region upstream of ORF4 based on homology to previously characterized RP4 and staphylococcal MOBp oriT nic sites. Thus, the sequence analysis provides predictions consistent with the mobilon defined for the staphylococcal plasmids, including that ORF4 of pFN1 is a mobC homologue.
Figure 2. Comparison of pFN1 Mobilon with Staphylococcal Plasmid Mobilons.
(A) Alignment of pFN1 genes and elements predicted to be involved in plasmid mobilization with those of the mobilizable S. aureus plasmid pC221 and pC223. Elements indicated include nic sites (▾) and potential pFN1 nic sites 1 and 2; c, MobC motif; cp, putative pFN1 MobC motif; relaxase motifs, III, z, IIa and II. (B) Alignment of MobC protein motif of pC221 and pC223 (Varsaki et al., 2009) with predicted MobC motif of pFN1 ORF2. Colon (:) indicates identity with consensus; period (.) indicates conservative substitution. (C) Alignment of potential pFN1 nic sites with previously defined consensus sequences for MOBp family nic sites. The RP4 consensus sequence, representative of a Gram-negative nic site, indicates conserved nucleotides in capital letters and semi-conserved nucleotides in lower case letters (Lawley et al., 2004). The Gram-positive consensus is based on an analysis of staphylococcal plasmids (Smith and Thomas, 2004) and for pC221 and pC223 the nic site corresponding to that of the consensus has been demonstrated. The positions of confirmed nic sites (▾) are indicated. Colon (:) indicates identity with consensus.
3.3. Prevalence of Plasmids and Plasmid-Related Sequences
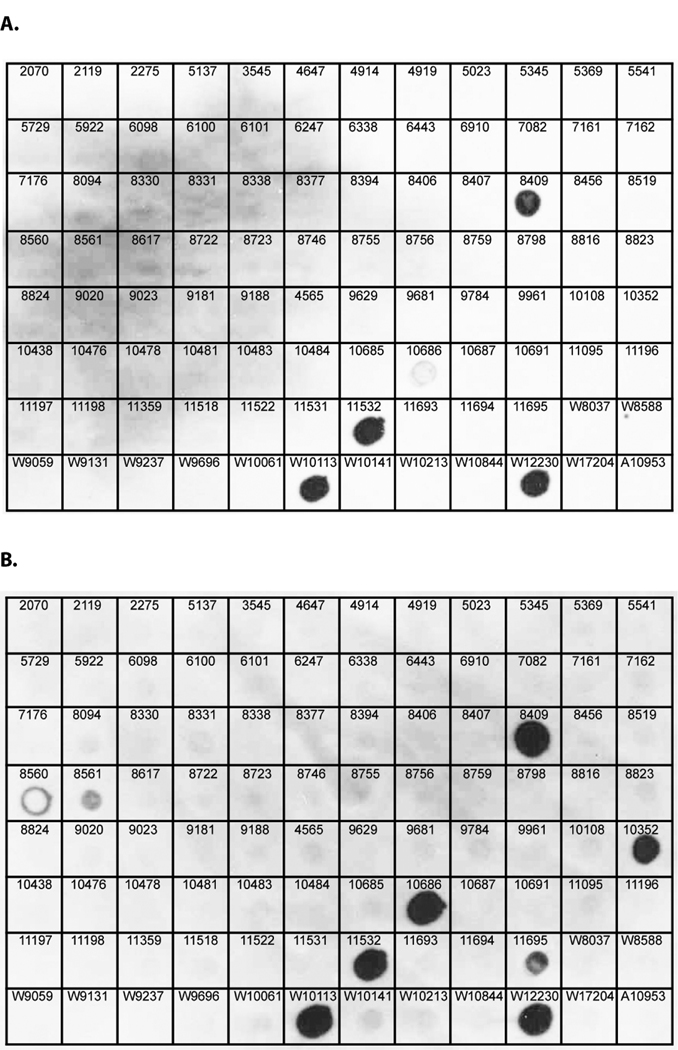
We screened 94 clinical and 4 laboratory isolates of F. nucleatum for plasmid content by visualization on ethidium bromide-stained agarose gels of chromosomal DNA and plasmid preparations. Plasmids were identified and isolated from 6 of the strains of the strains examined, and were designated pFN1 to pFN6 (Table 3). To examine the prevalence of pFN1-related gene sequences, we used a dot blot technique to probe preparations of total DNA from clinical isolates of F. nucleatum using probes for the pFN1 repA and rlx genes (Fig. 1 and Fig. 3, Table 3). Both the repA and rlx probes hybridized to DNA preparations from strains harboring pFN1, pFN2, pFN4 and pFN5, but consistent with our previous Southern analyses (Haake et al., 2000), did not hybridized to DNA from strains harboring pFN3 or pFN6. Additionally, the rlx probe hybridized to DNA from five strains from which plasmids were not isolated. The dot blot analyses indicate a prevalence of pFN1 repA- and rlx-related DNA in 4.2% and 9.4% of the strains, respectively. Overall, plasmid or plasmid-related sequences were identified in 11.5% of the strains examined.
Table 3.
Distribution of pFN1 repA- and rlx-related DNA in Strains of F. nucleatum
| Strain | Source of strain | Plasmid | Plasmid Size (kb) |
repA Homology |
Rlx Homology |
|---|---|---|---|---|---|
| WAL 12230 | Transtracheal isolate | pFN1 | 5.9 kb | + | + |
| WAL 10113 | Periodontal isolate | pFN2 | 7.2 kb | + | + |
| ATCC 10953 | Inflammed gingiva | pFN3 | 11.9 kb | − | − |
| RMA 11532 | Peritoneal fluid | pFN4 | 9.6 kb | + | + |
| RMA 8409 | Inguinal abscess | pFN5 | 5.8 kb | + | + |
| RMA 5369 | Facial abscess | pFN6 | 4.0 kb | − | − |
| RMA 8560 | Blood | ND* | ------ | − | + |
| RMA 8561 | Blood | ND | ------ | − | + |
| RMA 10352 | Peritoneal fluid | ND | ------ | − | + |
| RMA 10686 | Abdomenal fluid | ND | ------ | − | + |
| RMA 11695 | Respiratory-bronchial wash | ND | ------ | − | + |
ND: Plasmid not detected in this strain.
Figure 3. Dot Blot Hybridization of pFN1 repA and rlx probes to total DNA from isolates of F. nucleatum.
Total cellular DNA (1 µg) from clinical isolates of F. nucleatum were spotted on nylon membranes and hybridized to DIG-labelled pFN1 repA (panel A) or pFN1 rlx probes. Strain designations indicated on the figure are as follows: numbers only indicate RMA strains, W indicates WAL strains. The clinical isolate WAL12230, known to harbor pFN1, was included as a positive control. The laboratory strain ATCC 10953 (A10953), known to harbor pFN3, was included as a negative control based on prior hybridization studies (Haake et al., 2000). The repA and rlx probes hybridized to DNA from the positive control strain as well as strains known to harbor plasmids pFN2 (WAL10113), pFN4 (RMA11532) and pFN5 (RMA8409). The rlx probe hybridized to DNA from 5 additional strains from which plasmids have not been identified, including RMA8560, RMA8561, RMA10352, RMA10686 and RMA11695.
4. DISCUSSION
The studies described here demonstrate for the first time that the fusobacterial plasmid pFN1 encodes genes enabling efficient mobilization between strains of E. coli by the broad-host range plasmid RP4. The pFN1 mobilon is related by sequence to the mobilons of the staphylococcal plasmids pC221 and pC223. While the amino acid sequence identity between the staphylococcal and fusobacterial homologues was limited, relevant functional motifs were identified in both the pFN1 rlx and ORF4 predicted protein sequences. The staphylococcal plasmid MobC proteins are members of a family of nicking accessory proteins associated with MOBp relaxases (Varsaki et al., 2009). The mobC genes, found directly upstream of the mobA gene, encode the accessory protein required for DNA processing reactions to form the relaxosome (Smith and Thomas, 2004). MobA is required for DNA nicking, but unable to bind the oriT itself. Rather, the MobC protein binds specific sites within the oriT region, and MobA binds to the MobC-oriT complex (Caryl et al., 2004; Caryl and Thomas, 2006). The functional role of the staphylococcal MobC protein has not been clearly defined, but is hypothesized to involve altering the MobA binding site to facilitate recruitment of the MobA protein or positioning of the MobA protein in proximity to the nic site (Caryl and Thomas, 2006). The mobB gene of pC221 is not required for relaxation and its role in mobilization remains unclear (Projan and Archer, 1989).
Our studies on pFN1 delineated a core mobilon comparable to those of the staphylococcal plasmids, consisting in pFN1 of the relaxase gene homologue (rlx) as well as the upstream DNA predicted to encode a mobC homolog (ORF4) and possibly the oriT. Further analysis is required to define the oriT as well as determine the role of the pFN1 MobC homologue and if it is essential for mobilization. The plasmid pPA52 (McKay et al., 1995) isolated from a fusobacterial strain is nearly identical to pFN1 (98.6% sequence identity), but the major differences occur within the mobilization region. There is a 422 bp insertion in pPA52 between the rlx homolog and ORF2, which results in an extra ORF that is not found in pFN1. There is also a 31 bp deletion in the predicted mobC homolog of pPA52, which eliminates a direct repeat and introduces a stop codon at the beginning of the ORF. The disruption of the pPA52 mobC homolog suggests that this plasmid may not be mobilizable, and analysis of its mobilization properties could provide additional insight into the function of this gene.
We undertook our survey of F. nucleatum strains initially to identify plasmids for the development of vectors to use in molecular analyses (Haake et al., 2000; Kinder Haake et al., 2006), and plasmids or plasmid-related sequences were identified in 11.5% of the strains examined. Previously published studies on the detection of F. nucleatum plasmids reported higher prevalences, at 18% (McKay et al., 1995) or 27% (Paula et al., 2003) of the strains examined. A number of variables may account for these differences, including the inherently distinctive microbial populations isolated in different geographic regions. In addition the previous studies focused on oral isolates, whereas our investigation focused on strains that were isolated from sites of infection. Our panel of strains included oral isolates, but approximately 82% of the strains were non-oral and included isolates from sinus, respiratory, abdominal, genitourinary tract, soft tissue, blood and brain infections. One bacterial strain in common to all these studies was F. nucleatum ATCC 10953, and we were able to detect a plasmid from this strain (Haake et al., 2000; Karpathy et al., 2007) whereas the previously published studies did not (McKay et al., 1995; Paula et al., 2003). We repeated our attempts to isolate plasmid DNA from the strains identified solely by hybridization with the pFN1-rlx probe, but without success. However, we cannot rule out the possibility that the rlx-positive but repA-negative strains may harbor as yet undetected plasmids with pFN1-related rlx genes but divergent replication regions. Another consideration is the use of the hybridization approach (McKay et al., 1995), which does not discriminate between DNA localized to the chromosome or to an extrachromosomal element such as a plasmid. Our finding that pFN1-like relaxases are more prevalent in F. nucleatum than detectable plasmids may in some instances reflect integration of plasmid elements into the bacterial chromosome. Additional hybridization studies using sequences from other fusobacterial plasmids such as pFN3 (Haake et al., 2000; Karpathy et al., 2007) and pKH9 (Bachrach et al., 2004) may indicate more extensive prevalence of plasmid-related sequences in this species.
The plasmids thus far identified in F. nucleatum are small in size and cryptic, and do not appear to be essential for viability because the majority of strains lack identifiable plasmids. Conjugative mobilization may have contributed to the distribution of plasmids in F. nucleatum. Conjugal transfer of a TetM-like determinant between strains of F. nucleatum was previously demonstrated (Roberts and Lansciardi, 1990). In addition, components of a Type 4 secretion system, which mediates conjugative transfer of mobilizable plasmid DNA, can be found in the genome sequence of F. nucleatum ssp. polymorphum ATCC 10953. These include genes for the inner membrane and periplasmic components that are encoded on a large conjugal plasmid integrated into the chromosome (FNP_1868-1871, 1873, 1875), as well as genes which may encode Type IV pili (FNP_2389-2399, 1034) (Desvaux et al., 2005; Karpathy et al., 2007). Preliminary attempts in our laboratory to mobilize pFN1-based plasmids from ATCC 10953 have not been successful, but further investigation is in progress. In addition, the relationship of pFN1 with staphylococcal plasmids may reflect a heterologous origin. Although we are unaware of any reports of conjugal transfer of DNA from staphylococci to fusobacteria, conjugal transfer of a TetM-like determinant from Enterococcus faecalis to F. nucleatum was previously demonstrated (Roberts and Lansciardi, 1990). There is additional evidence from the F. nucleatum ATCC 25586 genome sequence of plasmid-related genes integrated into the chromosome. These include multiple ORFs related to plasmid addiction system proteins (data not shown), as well as a possible plasmid-related replication protein (FN0633) based on the presence of a plasmid replication protein conserved domain (COG5527) and weak similarity to a predicted replication protein from the Pasturella multocida plasmid pJR2 (Wu et al., 2003). Overall, these data are consistent with the hypothesis that plasmids have played a role in horizontal gene transfer in F. nucleatum. Further studies to define the origins of pFN1 and related plasmids may shed light on the issue of horizontal gene transfer in the oral environment.
5. CONCLUSIONS
This work demonstrates, for the first time, the mobilization properties encoded on a fusobacterial plasmid, pFN1. The prevalence of pFN1 replication- and relaxase-related sequences in clinical isolates of F. nucleatum, in addition to findings from prior genome analyses, suggests a role for plasmid mobilization in DNA transfer in F. nucleatum.
6. ACKNOWLEDGEMENTS
We thank Dr. David Haake for his review of this manuscript. We additional acknowledge Dr. Sharon Hunt Gerardo and Ms. Anna Kreymer for suggestions and technical assistance, respectively. This investigation was supported by NIH/NIDCR PHS Grant No. DE015348 (S.K.H.).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bachrach G, et al. Characterization of the novel Fusobacterium nucleatum plasmid pKH9 and evidence of an addiction system. Appl Environ Microbiol. 2004;70:6957–6962. doi: 10.1128/AEM.70.12.6957-6962.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bearfield C, et al. Possible association between amniotic fluid micro-organism infection and microflora in the mouth. BJOG. 2002;109:527–533. doi: 10.1111/j.1471-0528.2002.01349.x. [DOI] [PubMed] [Google Scholar]
- Bolstad AI, et al. Taxonomy, biology, and periodontal aspects of Fusobacterium nucleatum. Clin Microbiol Rev. 1996;9:55–71. doi: 10.1128/cmr.9.1.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brook I, Frazier EH. Microbiology of liver and spleen abscesses. J Med Microbiol. 1998;47:1075–1080. doi: 10.1099/00222615-47-12-1075. [DOI] [PubMed] [Google Scholar]
- Byrd DR, Matson SW. Nicking by transesterification: the reaction catalysed by a relaxase. Mol Microbiol. 1997;25:1011–1022. doi: 10.1046/j.1365-2958.1997.5241885.x. [DOI] [PubMed] [Google Scholar]
- Caryl JA, et al. Reconstitution of a staphylococcal plasmid-protein relaxation complex in vitro. J Bacteriol. 2004;186:3374–3383. doi: 10.1128/JB.186.11.3374-3383.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caryl JA, Thomas CD. Investigating the basis of substrate recognition in the pC221 relaxosome. Mol Microbiol. 2006;60:1302–1318. doi: 10.1111/j.1365-2958.2006.05188.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang AC, Cohen SN. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvaux M, et al. Protein secretion systems in Fusobacterium nucleatum: genomic identification of Type 4 piliation and complete Type V pathways brings new insight into mechanisms of pathogenesis. Biochim Biophys Acta. 2005;1713:92–112. doi: 10.1016/j.bbamem.2005.05.002. [DOI] [PubMed] [Google Scholar]
- Fletcher HM, et al. Virulence of a Porphyromonas gingivalis W83 mutant defective in the prtH gene. Infect Immun. 1995;63:1521–1528. doi: 10.1128/iai.63.4.1521-1528.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fouad AF, et al. PCR-based identification of bacteria associated with endodontic infections. J Clin Microbiol. 2002;40:3223–3231. doi: 10.1128/JCM.40.9.3223-3231.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Francia MV, et al. A classification scheme for mobilization regions of bacterial plasmids. FEMS Microbiol Rev. 2004;28:79–100. doi: 10.1016/j.femsre.2003.09.001. [DOI] [PubMed] [Google Scholar]
- Haake SK, Wang X. Cloning and expression of FomA, the major outer-membrane protein gene from Fusobacterium nucleatum T18. Arch Oral Biol. 1997;42:19–24. doi: 10.1016/s0003-9969(96)00105-7. [DOI] [PubMed] [Google Scholar]
- Haake SK, et al. Native plasmids of Fusobacterium nucleatum: characterization and use in development of genetic systems. J Bacteriol. 2000;182:1176–1180. doi: 10.1128/jb.182.4.1176-1180.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ilyina TV, Koonin EV. Conserved sequence motifs in the initiator proteins for rolling circle DNA replication encoded by diverse replicons from eubacteria, eucaryotes and archaebacteria. Nucleic Acids Res. 1992;20:3279–3285. doi: 10.1093/nar/20.13.3279. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jousimmies-Somer HR, et al. Wadsworth-KTL anaerobic bacteriology manual. 6th Edition. Belmont, CA: Star Publishing Co.; 2002. [Google Scholar]
- Karpathy SE, et al. Genome sequence of Fusobacterium nucleatum subspecies polymorphum - a genetically tractable fusobacterium. PLoS ONE. 2007;2:e659. doi: 10.1371/journal.pone.0000659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kinder Haake S, et al. Efficient gene transfer and targeted mutagenesis in Fusobacterium nucleatum. Plasmid. 2006;55:27–38. doi: 10.1016/j.plasmid.2005.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolenbrander PE, London J. Adhere today, here tomorrow: oral bacterial adherence. J Bacteriol. 1993;175:3247–3252. doi: 10.1128/jb.175.11.3247-3252.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornstra JJ, et al. Septic arthritis due to Fusobacterium nucleatum. Br J Rheumatol. 1998;37:1249. doi: 10.1093/rheumatology/37.11.1249. [DOI] [PubMed] [Google Scholar]
- Lark RL, et al. Risk factors for anaerobic bloodstream infections in bone marrow transplant recipients. Clin Infect Dis. 2001;33:338–343. doi: 10.1086/322595. [DOI] [PubMed] [Google Scholar]
- Lawley T, et al. Bacterial conjugation in Gram-negative bacteria. In: Funnell BE, Phillips GJ, editors. Plasmid Biology. Washington, DC: ASM Press; 2004. [Google Scholar]
- Leung KP, et al. Prevotella intermedia native plasmid can be mobilized by an Escherichia coli conjugal IncP plasmid. Plasmid. 2002;48:64–72. doi: 10.1016/s0147-619x(02)00011-2. [DOI] [PubMed] [Google Scholar]
- McKay TL, et al. Mobile genetic elements of Fusobacterium nucleatum. Plasmid. 1995;33:15–25. doi: 10.1006/plas.1995.1003. [DOI] [PubMed] [Google Scholar]
- Mikamo H, et al. Preterm labor and bacterial intraamniotic infection: arachidonic acid liberation by phospholipase A2 of Fusobacterium nucleatum. Am J Obstet Gynecol. 1998;179:1579–1582. doi: 10.1016/s0002-9378(98)70028-6. [DOI] [PubMed] [Google Scholar]
- Morris ML, et al. The use of allozyme electrophoresis to assess genetic heterogeneity among previously subspeciated isolates of Fusobacterium nucleatum. Oral Microbiol Immunol. 1996;11:15–21. doi: 10.1111/j.1399-302x.1996.tb00331.x. [DOI] [PubMed] [Google Scholar]
- Paula MO, et al. Plasmid profile in oral Fusobacterium nucleatum from humans and Cebus apella monkeys. Rev Inst Med Trop Sao Paulo. 2003;45:5–9. doi: 10.1590/s0036-46652003000100002. [DOI] [PubMed] [Google Scholar]
- Pickering MC, et al. Bilateral gluteal abscesses as a unique manifestation of fusobacterium septicaemia. Rheumatology (Oxford) 2000;39:224–225. doi: 10.1093/rheumatology/39.2.224. [DOI] [PubMed] [Google Scholar]
- Projan SJ, Archer GL. Mobilization of the relaxable Staphylococcus aureus plasmid pC221 by the conjugative plasmid pGO1 involves three pC221 loci. J Bacteriol. 1989;171:1841–1845. doi: 10.1128/jb.171.4.1841-1845.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC, Lansciardi J. Transferable Tet M in Fusobacterium nucleatum. Antimicrob Agents Chemother. 1990;34:1836–1838. doi: 10.1128/aac.34.9.1836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts MC, et al. Nomenclature for macrolide and macrolide-lincosamide-streptogramin B resistance determinants. Antimicrob Agents Chemother. 1999;43:2823–2830. doi: 10.1128/aac.43.12.2823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto M, et al. Molecular analysis of bacteria in asymptomatic and symptomatic endodontic infections. Oral Microbiol Immunol. 2006;21:112–122. doi: 10.1111/j.1399-302X.2006.00270.x. [DOI] [PubMed] [Google Scholar]
- Simon R, et al. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Biotechnology. 1983 November;:784–791. [Google Scholar]
- Smith MC, Thomas CD. An accessory protein is required for relaxosome formation by small staphylococcal plasmids. J Bacteriol. 2004;186:3363–3373. doi: 10.1128/JB.186.11.3363-3373.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varsaki A, et al. Analysis of ColE1 MbeC unveils an extended ribbon-helix-helix family of nicking accessory proteins. J Bacteriol. 2009;191:1446–1455. doi: 10.1128/JB.01342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JR, et al. Molecular characterization of plasmids with antimicrobial resistant genes in avian isolates of Pasteurella multocida. Avian Dis. 2003;47:1384–1392. doi: 10.1637/z7035. [DOI] [PubMed] [Google Scholar]