Abstract
The characterization of the subfamily of steroid hormone receptors has enhanced our understanding of how a set of hormonally derived lipophilic ligands controls cellular and molecular functions to influence development and help achieve homeostasis. The glucocorticopid receptor (GR), the first member of this subfamily, is a ubiquitously expressed intracellular protein, which functions as a ligand-dependent transcription factor that regulates the expression of glucocorticoid-responsive genes. The effector domains of the GR mediate transcriptional activation by recruiting coregulatory multi-subunit complexes that remodel chromatin, target initiation sites, and stabilize the RNA polymerase II machinery for repeated rounds of transcription of target genes. This review summarizes the basic aspects of the structure and of the human (h) GR, and the molecular basis of its biologic function.
Keywords: Human Glucocorticoid Receptor (hGR), Generalized Glucocorticoid Resistance, Mutations in the hGR Gene, Polymorphisms of the hGR Gene
Introduction
In humans, glucocorticoids regulate a broad spectrum of physiologic functions essential for life and play an important role in the maintenance of basal and stress-related homeostasis (1–3). Approximately 20% of the genes expressed in human leukocytes are regulated positively or negatively by glucocorticoids (4). Glucocorticoids are involved in almost every cellular, molecular and physiologic network of the organism and play a pivotal role in critical biologic processes, such as growth, reproduction, intermediary metabolism, immune and inflammatory reactions, as well as central nervous system and cardiovascular functions (1, 4). Physiologic amounts of glucocorticoids are also essential for normal renal tubular function and thus for water and electrolyte homeostasis. Furthermore, glucocorticoids represent one of the most widely used therapeutic compounds often employed in the treatment of inflammatory, autoimmune and lymphoproliferative disorders (1).
At the cellular level, the action of glucocorticoids is mediated by an intracellular protein, the glucocorticoid receptor (GR) (3, 5, 6). The human (h) GR belongs to the steroid/thyroid/retinoic acid nuclear receptor superfamily of transcription factor proteins and functions as a ligand-dependent transcription factor that regulates the expression of glucocorticoid-responsive genes positively or negatively. This review summarizes the basic aspects of structure of the hGR, its genomic actions and the molecular basis of its biologic function.
Structure of the Glucocorticoid Receptor Gene and Protein
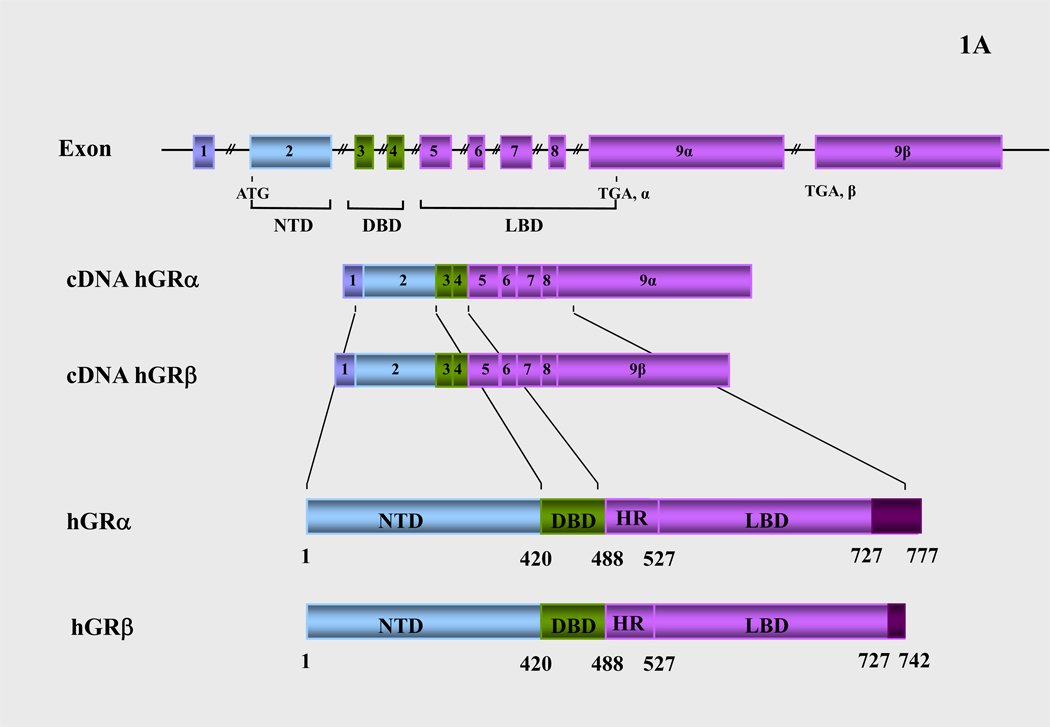
The human glucocorticoid receptor (hGR) cDNA was isolated by expression cloning in 1985 (13). The hGR gene consists of 9 exons and is located on chromosome 5. Alternative splicing of the hGR gene in exon 9 generates two highly homologous receptor isoforms, termed α and β. These are identical through amino acid 727, but then diverge, with hGRα having an additional 50 amino acids and hGRβ having an additional, nonhomologous 15 amino acids (Figure 1A). The molecular weights of these receptor isoforms are 97 and 94 kilo-Dalton, respectively. The hGRα resides primarily in the cytoplasm of cells and represents the classic glucocorticoid receptor that functions as a ligand-dependent transcription factor. The hGRβ, on the other hand, does not bind glucocorticoid agonists, may or may not bind the synthetic glucocorticoid antagonist RU486, has intrinsic, hGRα-independent, gene-specific transcriptional activity, and exerts a dominant negative effect upon the transcriptional activity of hGRα (3, 5, 6, 14, 15).
Figure 1.
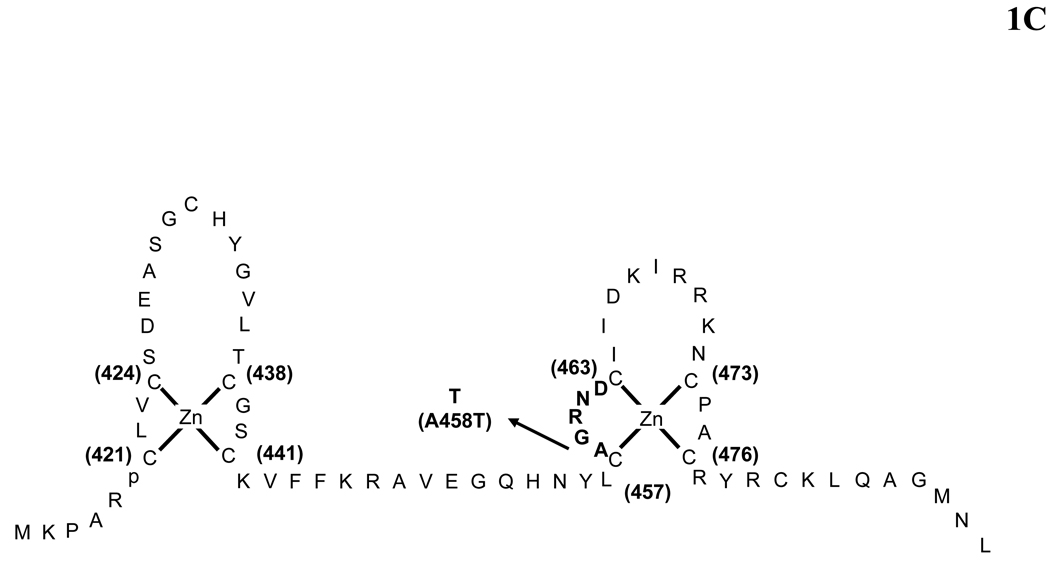
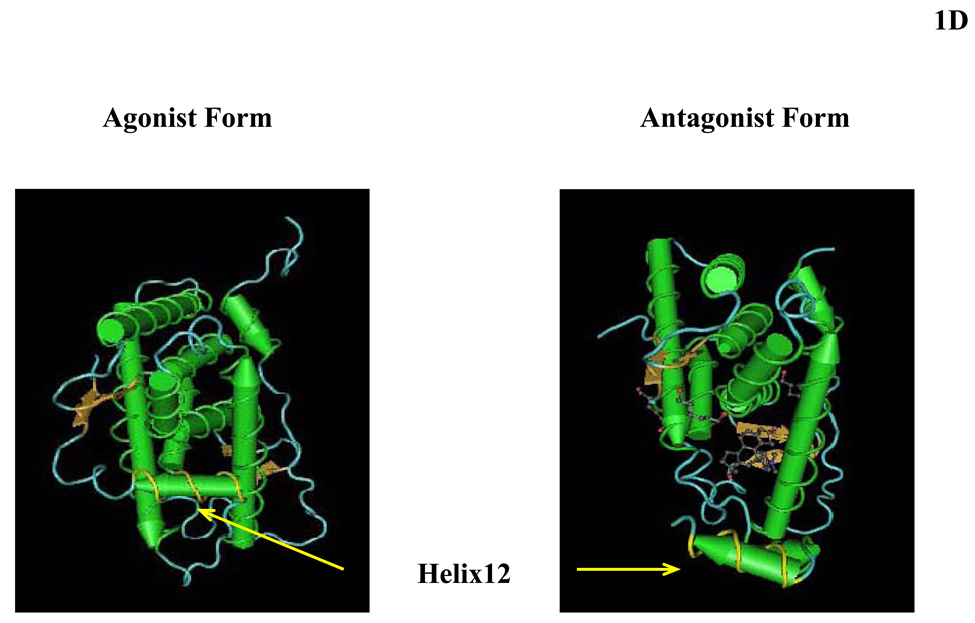
(A) Schematic representation of the structure of the human glucocorticoid receptor (hGR) gene. Alternative splicing of the primary transcript gives rise to the two mRNA and protein isoforms, hGRα and hGRβ (B) Functional domains of the hGRα. The functional domains and subdomains are indicated beneath the linearized protein structures. AF, activation function; DBD, DNA-binding domain; LBD, ligand-binding domain; NLS, nuclear localization signal. (C) Enlargement of part of the DBD showing the amino acid sequence (single letter codes) of the two zinc fingers and the dimerization loop (in bold). The A to T mutation at position 458 that could produce a dimerization defective receptor is shown. (D) Crystal structure of the ligand-binding domain (LBD) of the human glucocorticoid receptor-α (hGRα). Stereotactic conformation of the agonist (left) and antagonist (right) form of the LBD of hGRα. The yellow arrows indicate the position of Helix 12, which is critical for the formation of AF-2 surface that allows interaction with activators (Adapted from References 78–80).
The human GR is a modular protein composed of distinct regions illustrated in Figure 1B: the amino-terminal A/B region, also called immunogenic or N-terminal domain, and the C, D and E regions, which correspond to the DNA-binding domain, the hinge region and the ligand-binding domain, respectively.
The N-terminal domain (NTD) of the hGRα contains a major transactivation domain, termed activation function (AF)-1, which is located between amino acids 77 and 262 of the hGRα and is ligand-independent. The AF-1 plays an important role in the interaction of the receptor with molecules necessary for the initiation of transcription, such as coactivators, chromatin modulators and basal transcription factors, including RNA polymerase II, TATA-binding protein (TBP) and a host of TBP-associated proteins (TAFIIs) (3, 5, 6).
The DNA-binding domain (DBD) of the hGRα corresponds to amino acids 420–480 and contains two zinc finger motifs through which the hGRα binds to specific DNA sequences, the glucocorticoid-response elements (GREs) in the promoter region(s) of target genes (5, 6). The DBD is the most highly conserved domain throughout the steroid receptor family (Figure 2A). The two zinc finger motifs are able to tetrahedrally coordinate a zinc atom and are held by four cysteine (Cys) residues (Figure 1C). Only very few amino acids, termed the proximal (P)-box, within the first zinc finger are responsible for specific recognition of the cognate GREs. Another set of amino acids, called the distal (D)-box within the second zinc finger, forms the weak dimerization interface of the DBD. The DBD of the hGR also contains sequences important for receptor dimerization and nuclear translocation (5, 6).
Figure 2.
(A) Nucleocytoplasmic shuttling of the glucocorticoid receptor. Upon binding to the ligand, the activated hGRα dissociates from HSPs and translocates into the nucleus, where it homodimerizes and binds to GREs in the promoter region of target genes. (B) Schematic representation of the interaction of AF-1 and AF-2 of hGRα with coactivators. AF: activation function; DRIP/TRAP: vitamin D receptor-interacting protein/thyroid hormone receptor-associated protein; GR: glucocorticoid receptor; GREs: glucocorticoid response elements; HSP: heat shock protein; SWI/SNF: switching/sucrose non-fermenting; TF: transcription factor; TFRE: transcription factor-response element (Adapted from References 78–80).
The hinge region or region D is a flexible region between the DNA- and ligand-binding domains. Its amino terminus is an integral part of the DBD and is involved in its dimerization. The hinge region confers structural flexibility in the receptor dimmers, thereby allowing a single receptor dimmer to interact with multiple GREs (5, 6).
The ligand-binding domain (LBD) of the hGRα corresponds to amino acids 481–777, binds to glucocorticoids and plays a critical role in the ligand-induced activation of hGRα. The LBD also contains a second transactivation domain, termed AF-2, which is ligand-dependent, as well as sequences important for receptor dimerization, nuclear translocation, binding to the heat shock proteins and interaction with coactivators (5, 6).
Crystal Structure of the Ligand-binding Domain of the hGR
The crystal structure of the GRα LBD contains 12 α-helices and 4 small β-strands that fold into a three-layer helical domain (16). Helices 1 and 3 form one side of a helical sandwich, while helices 7 and 10 form the other side. The middle layer of helices (helices 4, 5, 8, and 9) are present in the top half but not in the bottom half of the protein. This arrangement of helices creates a cavity in the bottom half of the LBD, where the agonist molecule is bound (16, 17). Ligand binding of the LBD of hGRα induces conformational changes leading to a more compact structure characterized by an increased stability towards proteinases and a higher electrophoretic mobility under non-denaturing conditions. Binding of an agonist compound is required for the induction of the AF-2, while the integrity of the conserved amphipathic α-helix, which constitutes the core of the AF-2 activating domain (AD) (contained in Helix 12), is essential for AF-2-dependent transactivation. Helix (H) 12 plays a critical role in the formation of both the ligand-binding pocket and the AF-2 surface that facilitates interaction with coactivators. Upon ligand-binding, the receptor undergoes major conformational changes, which alter the position of H11 and H12 and generate an interaction surface that allows coactivators to bind to AF-2 through their LXXLL motifs (17). In the agonist-bound conformational state of hGR, H12 adopts a position over the ligand-binding pocket, which allows coactivators to interact within the coactivator cavity, thus forming a transcriptionally active receptor. In contrast, binding of RU486 induces structural changes leading to loss of the helical structure in the C-terminal portion of H11 and movement of H12 over the ligand-binding pocket, a position that prevents coactivator binding and enables corepressor binding (18) (Figure 2D).
Following helix 12, there is an extended strand that forms a conserved β sheet with a β strand between helices 8 and 9. This C-terminal β strand appears to play an important role in receptor activation by stabilizing helix 12 in the active conformation (16). Deletion of the last few residues that form the β strand lead to alterations in hormone binding specificity and significant reduction in the receptor-mediated transactivation of target genes (19), suggesting that the C-terminal region of hGRα, downstream of helix 12, is essential for ligand-binding specificity and agonist potential, although it does not appear to confer differential hormone-binding capacities to the receptor (18, 19).
Transcriptional and Translational Regulation of hGR Isoforms
As described above, the hGR gene expresses two mRNAs through alternative use of exons 9α and 9β, and produces two splice variants. The hGRα mRNA further expresses multiple isoforms by using at least 8 alternative translation initiation sites (20). Since hGRβ shares a common mRNA domain that contains the same translation initiation sites with the hGRα (13), the hGRβ variant mRNA might also be translated through the same initiation sites to a similar host of β isoforms. All these hGRα isoforms have different transcriptional activity in response to dexamethasone, are differentially distributed in the cytoplasm and/or the nucleus in the absence of ligand, translocate into the nucleus in response to the ligand, and display distinct transactivation or transrepression patterns on global gene expression examined by cDNA microarray analyses (20). Therefore, these N-terminal hGRα isoforms may differentially transduce the glucocorticoid signal to target tissues depending on their selective relative expression and inherent activities. Furthermore, it is likely that similar differential cell-specific production and functional differences might also be present between the putative hGRβ translational isoforms.
The hGR gene has at least three different promoters, A, B and C. Promoter A can be used with three untranslated exons, 1A1, 1A2 and 1A3, that contain unique promoter fragments (21). Therefore, the hGR gene can produce five different transcripts from different promoters that encode the same hGR proteins. Through differential use of these promoters, the levels of hGR proteins can vary considerably among tissues. Splice and translational hGR isoforms expressed from different promoters appear to form up to 256 different combinations of homo- and hetero-dimers with varying transcriptional activities. The marked complexicity in the transcription/translation of the hGR gene enables target tissues to differentially respond to circulating glucocorticoid concentrations and accounts for the highly stochastic nature of the glucocorticoid signaling pathway (22).
Genomic Actions of the hGR
Nucleocytoplasmic Shuttling of hGRα
In the absence of ligand, hGRα resides mostly in the cytoplasm of cells as part of a hetero-oligomeric complex, which contains chaperone heat shock proteins (HSPs) 90, 70 and 50, immunophilins, as well as other proteins (23). Immunophilin FKBP51 (FK506 binding protein of molecular weight 51) and the related protein FKBP52 are thought to be involved in the regulation of GR signaling (Davies et al., 2002). FKBP51 acts as a competitive inhibitor to its genetic relative FKBP52 (gene ID FKBP4) (Davies et al., 2002) and may account for the generalized glucocorticoid resisistance observed in species of New World primates (Brown et al., 1970) Expression of FKBP51 is 13-fold higher in these primates, and FKBP52 expression less than one-half compared to humans; effectively rendering the monkeys resistant to the effects of cortisol mediated by GR (Denny et al., 2000).
HSP90 regulates ligand binding, as well as cytoplasmic retention of hGRα by exposing the ligand-binding site and masking the two nuclear localization sequences (NLS), NL1 and NL2, which are located adjacent to the DBD and in the LBD of the receptor, respectively. Upon ligand-induced activation, the receptor undergoes a conformational change that results in dissociation from this multiprotein complex and translocation into the nucleus (23, 24). Within the nucleus, the receptor binds as a homodimer to GREs in the promoter regions of target genes, and regulates their expression positively or negatively depending on GRE sequence and promoter context (25, 26).
Alternatively, the ligand-activated hGRα can modulate gene expression independently of binding to GREs, by interacting possibly as a monomer with other transcription factors, such as activator protein-1 (AP-1), nuclear factor-κB (NF-κB), p53 and signal transducers and activators of transcription (STATs) (27–29). This GR nuclear action is independent of DNA-binding and involves modulation of transcriptional activity through direct protein-protein interaction with inducible transcription factors [Gottlicher M et al, 1998]. An important example of DNA-independent actions of steroids is transrepression of the proinflammatory transcription factors AP-1 and NF-κB. GR weakly interacts with and inhibits AP-1-dependent transcription without altering its DNA binding [Jonat C et al. 1990; Schule R et al. 1990; Kong H et al. 1992]. Importantly, DNA-binding inactive mutants of GR are fully capable of AP-1 transrepression [Heck S et al. 1994]. Interaction between activated GR and NF-κB is mediated by the second zinc-finger in the DBD of GR, and transrepression does not require DNA binding of GR. Instead, GR interferes with the transactivation potential of the p65/RelA subunit of NF-κB ([McKay et al. 1999] and references herein). In addition to transrepression, protein-protein interaction can also lead to synergistic induction of promoter activity, as shown for interaction of GR with Stat-5 on the β-casein promoter [Stocklin E et al. 1996].
Following transcriptional activation or inhibition of glucocorticoid-responsive genes, hGRα dissociates from the ligand and has a lower affinity for binding to GREs. The unliganded hGRα remains within the nucleus for a considerable length of time and is then exported to the cytoplasm; both within the nucleus and within the cytoplasm the hGR may be recycled and/or degraded in the proteasome (30). The nuclear export of hGR occurs slowly and is opposed actively by a nuclear retention signal (NRS) in the hinge region of the receptor, which overlaps closely with the NL1 (31) (Figure 2A).
Mechanisms of Transcriptional Activation by hGRα
The hGRα exerts its classic transcriptional activity on its responsive promoters following binding to GREs (25). Active endogenous GREs are present in the promoter region(s) of glucocorticoid-responsive genes. The optimal recognition site is an inverted hexameric palindrome separated by 3 base pairs, PuGNACAnnnTGTNCPy (Pu denotes purine and Py denotes pyramidine), with each hGR molecule binding to one of the palindromes (32). The GRE-bound hGRα stimulates the transcription of target genes by facilitating the formation of the transcription initiation complex, including the RNA polymerase II and its ancillary components via its AF-1 and AF-2 domains (33) (Figure 2B).
To initiate transcription, hGRα uses its transcriptional activation domains, AF-1 and AF-2, as surfaces to interact with nuclear receptor coactivators and chromatin-remodeling complexes. Several coactivators form a bridge between the DNA-bound hGRα and the transcription initiation complex, and facilitate the transmission of the glucocorticoid signal to the RNA polymerase II (34–37). These include: (1) The p300 and the homologous cAMP-responsive element-binding protein (CREB)-binding protein (CBP), which also serve as macromolecular docking “platforms” for transcription factors from several signal transduction cascades, including nuclear receptors, CREB, AP-1, NF-κB, p53, Ras-dependent growth factor, and STATs. In view of their central position in many signal transduction cascades, the p300/CBP coactivators are also called cointegrators; (2) The p300/CBP-associated factor (p/CAF), which interacts with p300/CBP, but is also a broad transcription coactivator; and (3) The p160 family of coactivators, which preferentially interact with the steroid hormone receptors (34–37). These include the steroid receptor coactivator-1 (SRC-1), SRC-2 and SRC-3 (34–37).
The p160 coactivators are the first to be attracted to the DNA-bound hGRα and help accumulate p300/CBP and p/CAF proteins to the promoter region. The p160 coactivators, interact directly with both the AF-1 of hGRα through their carboxyl-terminal domain and the AF-2 of hGRα through multiple amphipathic LXXLL signature motifs located in their nuclear receptor-binding (NRB) domain (38). These coactivators also have intrinsic histone acetyltransferase (HAT) activity, which promotes chromatin decondensation, and allows the transcription initiation complex of the RNA-polymerase II and its ancillary components to initiate and promote transcription (Figure 2B). HAT activity also modulates the binding of transcription factors to specific elements on their responsive promoters, as well as the dissociation of coactivators from nuclear receptors or other transcription factors (34–37).
The hGRα also interact with several other distinct chromatin modulators through its transactivation domains, such as the mating-type switching/sucrose non-fermenting (SWI/SNF) complex and components of the vitamin D receptor-interacting protein/thyroid hormone receptor-associated protein (DRIP/TRAP) complex (38). The SWI/SNF complex is an ATP-dependent chromatin remodeling factor with a multi-subunit structure. The DRIP/TRAP complex is also a multiprotein conglomerate, which consists of over 10 different proteins, including DRIP205/TRAP220/PBP and components of SMCC. The DRIP/TRAP complex may modulate transcription through interaction and modification of general transcription factors, such as TFIIH or the C-terminal tail of the RNA polymerase II. DRIP205/TRAP220 contains two LXXLL motifs through which it binds to the ligand-activated AF-2 directly (34–37).
Interaction of hGRα with Other Transcription Factors
The hGRα may affect signal transduction cascades through protein-protein interactions with specific transcription factors by influencing their ability to stimulate or inhibit the transcription rates of respective target genes. This activity may be more important than the GRE-mediated one, given that mice harboring a mutant GR, which is active in terms of protein-protein interactions but inactive in terms of transactivation via DNA, survive and procreate, in contrast to mice with a deletion of the entire GR gene that die immediately after birth from severe respiratory distress syndrome (39, 40).
The protein-protein interactions of GR with other transcription factors may take place on the promoters that do not contain GREs (tethering mechanism), as well as on the promoters that have both GRE(s) and responsive element(s) of transcription factors that interact with GR (“composite promoters”). Suppression of transactivation of other transcription factors through protein-protein interactions may be particularly important in suppression of immune function and inflammation by glucocorticoids (39, 41). Most of the effects of glucocorticoids on the immune system may be mediated by the interaction between GR and NF-κB, AP-1 and STATs (42–44).
Post-translational Modifications of GR
Although the transcriptional activity of GR is primarily governed by ligand binding, accumulating evidence suggests that post-translational modifications (PTMs) play an important additional role. The exact modification sites and the modifying enzymes involved have been identified in many cases. These covalent changes may affect receptor stability, subcellular localization, as well as the interaction between GR and other proteins.
Phosphorylation of the GR
The human GR has several phosphorylation sites, including S113, S141, S203, S211, S226 and S404, most of which are all located in the AF-1 domain of its NTD (45). Phosphorylation of hGRα typically occurs after binding to the ligand, and may determine turnover, subcellurar trafficking, target promoter specificity, cofactor interaction, strength and duration of receptor signaling, and receptor stability. In addition, it modifies protein-protein interactions, which can stabilize the hypophosphorylated form of the receptor in the absence of ligand, as well as facilitate transcriptional activation by the hyperphosphorylation of GR via cofactor recruitment upon ligand binding. Finally, GR phosphorylation may also participate in the nongenomic activation of cytoplasmic signaling pathways evoked by GR. Therefore, phosphorylation of the GR is a versatile mechanism for modulating and integrating multiple receptor functions (45, 46). There are several kinases that phosphorylate the hGRα in vitro and in vivo. These include: i) the yeast cyclin-dependent kinase p34CDC28, which is the product of the CDC28 gene and is a 34-kDa protein kinase homologous to p34CDC2, the catalytic subunit of the mammalian mitosis-promoting factor (45, 47); ii), the p38 mitogen-activated protein kinase (MAPK) (48); iii) the CNS-specific cyclin-dependent kinase 5 (CDK5) (49); iv) the glycogen synthase kinase 3β (GSK-3β) (50, 51); and v) the c-Jun N-terminal kinase (JNK) (52, 53), and either increase or decrease the transcriptional activity of hGRα. On the other hand, phosphatases, such as PP1, PP2a and PP5, may reverse the process of phosphorylation.
Ubiquitination of GR and Components of the Transcriptional Initiation Complex
The ubiquitin/proteasome pathway plays an important role in the transcriptional regulation. Following ligand binding, the GR is destabilised and directed towards the proteasome pathway for degradation. Several transcription factors (such as p53, cJun, cMyc and E2F-1) and transcriptional intermediate molecules, such as nuclear receptor coactivators, chromatin remodeling factors and some chromatin components, are also ubiquitinated and degraded by the proteasome (54, 55). Furthermore, the proteasome interacts with the C-terminal tail of the RNA polymerase II and is directly associated with the promoter regions of several genes, influencing their transcriptional activities (56). Thus, ubiquitination and proteasomal degradation regulate the transcriptional activity of GR, by promoting the rapid turnover of the receptor, which leads to down-regulation and a decrease in the transcriptional activity of GR. Ubiquitination also regulates the motility of GR inside the nucleus of living cells (57, 58). Inhibition of the proteasomal activity reduces the motility of the wild-type GRα, as well as the natural mutants GRαD641V, GRαV729I, GRαI747M, and GRαL773P (58).
Acetylation of GR
Acetylation of the GR occurs after ligand-binding and prior to nuclear translocation (59). The acetylated GR is deacetylated by histone deacetylase 2 (HDAC2) and this deacetylation is necessary for the GR to be able to inhibit NF-κB activation of inflammatory genes (59). The site of acetylation of the GR is the lysine rich ‘hinge’ region 492–495 (sequence KKTK), which is analogous to the acetylation sites identified in other nuclear hormone receptors. Site-directed mutagenesis of the lysine residues K494 and K495 prevents hGR acetylation and reduces the activation of the SLPI gene by glucocorticoids, whereas repression of NF-κB is unaffected (59). Recent studies have demonstrated that the circadian rhythm-generating transcription factors CLOCK and BMAL1 repress GR-induced transcriptional activity by acetylating several lysine residues located as a cluster in the ‘hinge’ region of the receptor (60). This post-translational modification attenuates the binding of GR to GREs and its ability to influence glucocorticoid-responsive gene expression. These findings indicate that CLOCK/BMAL1 may function as a circadian negative regulator of glucocorticoid action in target tissues (60).
Sumoylation of hGR
Sumoylation of the GR is not necessarily ligand-dependent and may affect both protein stability and transcriptional activation (61). The SUMO conjugase Ubc9 interacts with the GR (62, 63). Overexpression of SUMO-1 destabilises the GR and this can be prevented by a proteasome inhibitor (61). Three sumoylation sites have been identified in the GR (K277, K293 and K703), and two of them are located in synergy control motifs (64, 65). Mutations of the sumoylation sites present in synergy control motifs increase GR function on minimal promoters but not on the MMTV promoter (64). These results indicate that the effects of GR sumoylation are highly dependent on the promoter and possibly the cell context.
Tissue Sensitivity to Glucocorticoids
Pathologic Natural hGR Mutations
Alterations in any of the molecular mechanisms of hGRα action described above may lead to alterations in tissue sensitivity to glucocorticoids, which may take the form of resistance or hypersensitivity and may be associated with significant morbidity (66–69). One such condition that we have extensively investigated over the years is the primary Generalized Glucocorticoid Resistance (70–76).
Primary Generalized Glucocorticoid Resistance is a rare, familial or sporadic genetic condition characterized by generalized, partial, target-tissue insensitivity to glucocorticoids (70–76). This leads to activation of the hypothalamic-pituitary-adrenal (HPA) axis and compensatory elevations in circulating cortisol and adrenocorticotropic hormone (ACTH) concentrations, which maintain circadian rhythmicity and appropriate responsiveness to stressors. The excess ACTH secretion results in adrenal hyperplasia, and increased production of adrenal steroids with mineralocorticoid [cortisol, deoxycorticosterone (DOC) and corticosterone] and/or androgenic activity [androstenedione, dehydroepiandrosterone (DHEA) and DHEA-sulfate (DHEAS)], and the corresponding clinical manifestations (70–76) (Figure 3A) (Table 1). The clinical spectrum of Generalized Glucocorticoid Resistance is broad, ranging from most severe to mild forms, while a number of patients may be asymptomatic, displaying biochemical alterations only (70–76) (Table 1).
Figure 3.
(A) Location of the known mutations of the glucocorticoid receptor (hGR) gene. AF: activation function; DBD: DNA-binding domain; GR: glucocorticoid receptor; GREs: glucocorticoid response elements; HSP: heat shock protein; NTD: amino terminal domain; LBD: ligand-binding domain. (B) Alterations in the hypothalamic-pituitary-adrenal (HPA) axis in generalized glucocorticoid resistance and hypersensitivity. AVP: arginine vasopressin; DOC: deoxycorticosterone. (C) Location of the known mutations of hGRα in the agonist (upper panel) and antagonist (lower panel) form of the LBD of hGRα. Helices are indicated in red and are underlined, while β-sheets are indicated in green. Two mutations (I559 and V571A) are located within H5, while four mutations (V729I, F737L, I747M and L773P) are located within or close to helices 11 and 12. The ligand-binding pocket is formed by helices 3, 5, 11 and 12. Upon ligand-binding, the receptor undergoes major conformational changes, which alter the position of H11 and H12 and generate an interaction surface that allows coactivators to bind to the LBD through their LXXLL motifs. Helix 12 plays a critical role in the formation of both the ligand-binding pocket and the AF-2 surface that facilitates interaction with coactivators. The fact that most hGRα mutations are clustered around H5, H11 and H12 indicates that these helices play an important role in glucocorticoid signal transduction. (D) Schematic representation of the hGR gene polymorphisms located in the amino-terminal domain of the receptor and a summary of their clinical associations (Adapted from References 68, 78–80).
TABLE 1.
Clinical Manifestations and Diagnostic Evaluation of Generalized Glucocorticoid Resistance
| Clinical Presentation | |
|---|---|
| Apparently normal glucocorticoid function | |
| Asymptomatic | |
| Chronic fatigue (glucocorticoid deficiency?) | |
| Mineralocorticoid excess | |
| Hypertension | |
| Hypokalemic alkalosis | |
| Androgen excess | |
| Children: Ambiguous genitalia at birth, premature adrenarche, precocious puberty | |
| Females: Acne, hirsutism, male-pattern hair loss, menstrual irregularities, oligoanovulation, infertility | |
| Males: Acne, hirsutism, oligospermia, adrenal rests in the testes, infertility | |
| Increased HPA axis activity (CRH/ACTH hypersecretion) | |
| Anxiety | |
| Adrenal rests | |
| Diagnostic Evaluation | |
| Absence of clinical features of Cushing syndrome | |
| Normal or elevated plasma ACTH concentrations | |
| Elevated plasma cortisol concentrations | |
| Increased 24-hour urinary free cortisol excretion | |
| Normal circadian and stress-induced pattern of cortisol and ACTH secretion | |
| Resistance of the HPA axis to dexamethasone suppression | |
| Thymidine incorporation assays: Increased resistance to dexamethasone-induced suppression of phytohemaglutinin-stimulated thymidine incorporation compared to control subjects | |
| Dexamethasone-binding assays: Decreased affinity of the glucocorticoid receptor for the ligand compared to control subjects | |
| Molecular studies: Mutations/deletions of the glucocorticoid receptor | |
Adapted from Reference 65
The molecular basis of primary Generalized Glucocorticoid Resistance has been ascribed to mutations in the hGR gene, which impair the molecular mechanisms of hGR action, thereby altering tissue sensitivity to glucocorticoids. Inactivating mutations within the ligand-binding domain or the DNA-binding domain of the receptor, and a 4-base pair deletion at the 3’-boundary of exon 6 of the gene, have been described in five kindreds and five sporadic cases (70–88). The molecular defects elucidated in the reported cases are summarized in Table 2, while the corresponding mutations in the hGR gene are shown in Table 2.
TABLE 2.
Mutations of the Human Glucocorticoid Receptor Gene Causing Generalized Glucocorticoid Resistance
| Mutation Position |
|||
|---|---|---|---|
| Author (Reference) | cDNA | Amino acid | Molecular Mechanisms |
| Chrousos et al. (64) | 1922 (A→T) | 641 (D→V) | Transactivation ↓ |
| Hurley et al. (71) | Affinity for ligand ↓ (x 3) | ||
| Nuclear translocation: 22 min | |||
| Abnormal interaction with GRIP1 | |||
| Karl et al. (72) | 4 bp deletion in exon-intron 6 | hGRαnumber: 50% of control | |
| Inactivation of the affected allele | |||
| Malchoff et al. (73) | 2185 (G→A) | 729 (V→I) | Transactivation ↓ |
| Affinity for ligand ↓ (x 2) | |||
| Nuclear translocation: 120 min | |||
| Abnormal interaction with GRIP1 | |||
| Karl et al. (70) | 1676 (T→A) | 559 (I→N) | Transactivation ↓ |
| Decrease in hGR binding sites | |||
| Transdominance (+) | |||
| Nuclear translocation: 180 | |||
| Abnormal interaction with GRIP1 | |||
| Ruiz et al. (75) | 1430 (G→A) | 477 (R→H) | Transactivation ↓ |
| Charmandari et al. (80) | No DNA binding | ||
| Nuclear translocation: 20 min | |||
| Ruiz et al. (75) | 2035 (G→A) | 679 (G→S) | Transactivation ↓ |
| Charmandari et al. (80) | Affinity for ligand ↓(x 2) | ||
| Nuclear translocation: 30 min | |||
| Abnormal interaction with GRIP1 | |||
| Mendonca et al. (76) | 1712 (T→C) | 571 (V→A) | Transactivation ↓ |
| Affinity for ligand ↓ (x 6) | |||
| Nuclear translocation: 25 min | |||
| Abnormal interaction with GRIP1 | |||
| Vottero et al. (77) | 2241 (T→G) | 747 (I→M) | Transactivation ↓ |
| Transdominance (+) | |||
| Affinity for ligand ↓ (x 2) | |||
| Nuclear translocation ↓ | |||
| Abnormal interaction with GRIP1 | |||
| Charmandari et al. (79) | 2318 (T→C) | 773 (L→P) | Transactivation ↓ |
| Transdominance (+) | |||
| Affinity for ligand ↓ (x 2.6) | |||
| Nuclear translocation: 30 min | |||
| Abnormal interaction with GRIP1 | |||
| Charmandari et al. (81) | 2209 (T→C) | 737 (F→L) | Transactivation ↓ |
| Transdominance/time-dependent (+) | |||
| Affinity for ligand ↓ (x 1.5) | |||
| Nuclear translocation: 180 min | |||
Adapted from Reference 78
We have identified most hGR mutations associated with primary Generalized Glucocorticoid Resistance and have systematically investigated the molecular mechanisms through which these various natural hGR mutants affect glucocorticoid signal transduction in almost all reported cases of Generalized Glucocorticoid Resistance. We studied: i) the transcriptional activity of the mutant receptors; ii) the ability of the mutant receptors to exert a dominant negative effect upon the wild-type receptor; iii) the affinity of the mutant receptors for the ligand; iv) the subcellular localization of the mutant receptors and their nuclear translocation following exposure to the ligand; v) the ability of the mutant receptors to bind to GREs; and vi) the interaction of the mutant receptors with the GRIP1 coactivator, which belongs to the p160 family of nuclear receptor coactivators and plays an important role in the hGRα-mediated transactivation of glucocorticoid-responsive genes (70–88).
Transactivation studies showed that all mutant receptors demonstrated a variable reduction in their ability to transactivate the glucocorticoid-responsive MMTV promoter in response to dexamethasone compared to the wild-type receptor, with the most severe impairment observed in the cases of R477H, I559N, V571A and D641V mutations (77–88). The mutant receptors hGRαI559N, hGRαF737L, hGRαI747M and hGRαL773P further exerted a dominant negative effect upon the wild-type receptor, which in association with their impaired ability to transactivate glucocorticoid-responsive genes might have contributed to manifestation of the disease at the heterozygote state (77, 81, 83, 86, 88).
Dexamethasone-binding studies showed a variable reduction in the affinity of the mutant receptors for the ligand, with the most severe reduction observed in the cases of I559N, I747M and V571A mutations (77–88). The only mutant receptor that demonstrated normal affinity for the ligand was the hGRαR477H, in which the mutation is located in the DBD of the receptor (87). The decreased affinity of the mutant receptors for the ligand most likely reflects the location of the mutations in the LBD of hGRα.
We studied the subcellular localization and nuclear translocation of the wild-type and mutant receptors. In the absence of dexamethasone, hGRα was primarily localized in the cytoplasm of cells. Addition of dexamethasone resulted in translocation of the wild-type receptor into the nucleus within 12 min. The pathologic mutant receptors were also observed predominantly in the cytoplasm of cells in the absence of ligand, except for the mutant receptors hGRαV729I and hGRαF737L, which were localized both in the cytoplasm and in the nucleus of cells. Exposure to the same concentration of dexamethasone induced a slow translocation of the mutant receptors into the nucleus, which ranged from 20 min (R477H) to 180 min (I559N and F737L) (77–88). These findings indicate that all hGR mutations affect the nucleocytoplasmic shuttling of hGRα, probably through impairment of the NL1 and/or NL2 function (89–92).
DNA-binding studies showed that the wild-type and all mutant receptors in which the mutations were located in the LBD of hGRα preserved their ability to bind to DNA (77–88). The only mutant receptor that failed to bind to DNA was the hGRαR477H, in which the mutation is located at the C-terminal zinc finger of the DBD of the receptor, very close to the D-box or DBD dimerization interface (87).
To determine whether the mutant receptors displayed an abnormal interaction with the p160 coactivators, we investigated the interaction between the mutant receptors and the GRIP1 coactivator. GRIP1 contains two sites that bind to hGR: one site, the NRB site, interacts with the AF-2 of hGRα in a ligand-dependent fashion, while the other site interacts with the AF-1 of hGRα in a ligand-independent fashion (93–95). The wild-type and most mutant receptors interacted with the GRIP1 coactivator in vitro only through their AF-1 domain. Exceptions were the mutant receptors hGRαR477H, which interacted with GRIP1 through both its AF-1 and AF-2, and hGRαI559N, which did not interact with GRIP1 (77–88). These findings indicate that the hGR mutant receptors may form a defective complex with GRIP1, which is partially or completely ineffective.
Finally, we examined the association between the location of the known mutations in the crystal structure of the LBD of hGR and the molecular mechanisms through which these mutations impaired glucocorticoid signal transduction (Figure 3A). All mutations within the LBD of the receptor were shown to affect the affinity of the receptor for the ligand; however, this effect was more pronounced in the cases of I559N and V571A mutations located in H5 of the receptor. Nuclear translocation was more delayed in the cases of I559N, V729I and F737L mutations, implicating mostly H5, H10 and H11. All mutations within the LBD of the receptor affected the in vitro interaction of the receptor with the GRIP1 coactivator but preserved their ability to bind to DNA. The one mutation R477H identified in the DBD of the receptor impaired primarily the ability of the receptor to bind to GREs. The fact that most mutant receptors interacted with the GRIP1 coactivator in vitro only through their AF-1 domain highlights the importance of H10, H11 and H12 of the LBD of the receptor in facilitating the formation of the AF-2 surface that interacts with coactivators (75).
hGR Polymorphisms
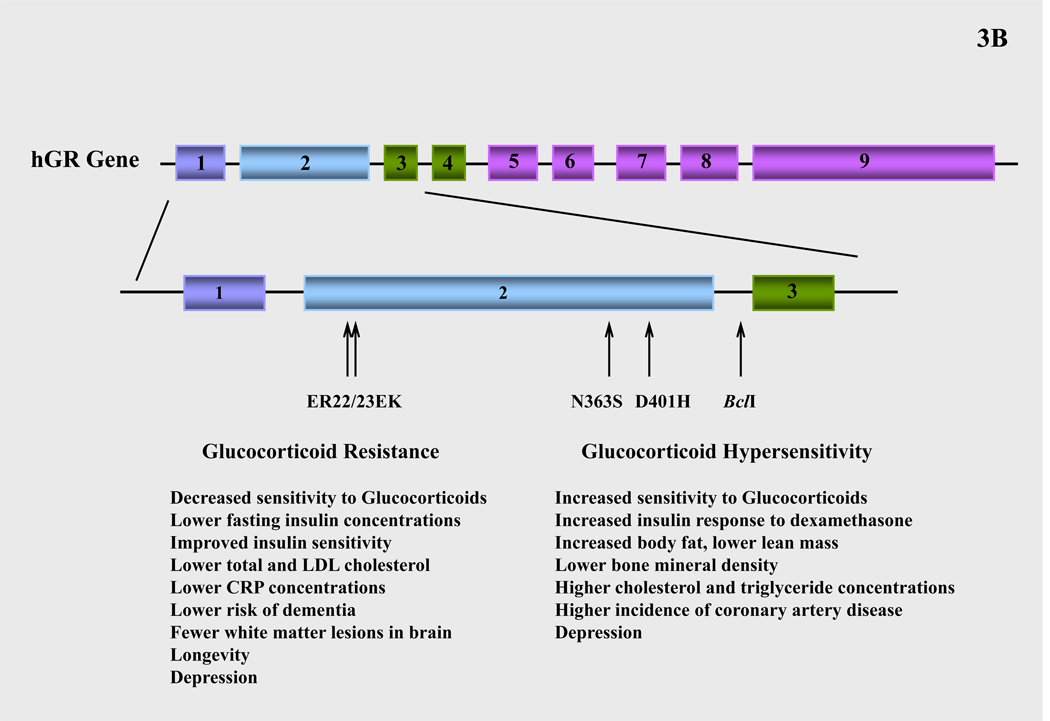
Inter-individual variations in tissue sensitivity to glucocorticoids have been described within the normal population and have been partly attributed to polymorphisms in the hGR gene. Several polymorphisms of the hGR gene have been reported (96–98). The N363S polymorphism is associated with higher sensitivity to glucocorticoids in vivo, increased insulin response to exogenous dexamethasone administration (98–100), a higher BMI (98, 100–103), higher waist-to-hip ratio (104), and a tendency toward lower bone mineral density in trabecular bone (100, 101). The N363S variant is also associated with elevated cholesterol and triglyceride concentrations and higher incidence of coronary artery disease independent of weight (101, 105) (Figure 3B).
A frequent BclI restriction fragment length polymorphism is also associated with increased sensitivity to glucocorticoids, hypertension, visceral adipocity (98, 106–108) and increased insulin concentrations in obese women (109). The exact mutation of this polymorphic site was identified as a C → G substitution in intron 2. In the elderly, the G allele of the BclI polymorphism is associated with a lower BMI and a tendency towards lower lean body mass, which is likely to arise as a result of the increased sensitivity to glucocorticoids (110) (Figure 3B).
A third polymorphism consists of two linked, single-nucleotide mutations in codons 22 and 23 in exon 2 of the hGR gene (98, 99, 111). The first mutation in codon 22 does not result in an amino acid change (GAG to GAA, both coding for glutamic acid (E)), but the mutation in codon 23 (AGG to AAG) results in arginine (R) to lysine (K) substitution (98, 99, 111). This polymorphism reduces sensitivity to glucocorticoids and results in a phenotype that is characterized by a more favorable metabolic profile, leading to longevity. Carriers of the ER22/23EK polymorphism display relative glucocorticoid resistance, lower fasting insulin concentrations and improved insulin sensitivity, lower total and LDL cholesterol concentrations and lower CRP concentrations (98, 112–114). The latter suggests that carriers of the ER22/23EK polymorphism have a lower tendency to develop impaired glucose tolerance, type 2 diabetes or cardiovascular disease. In line with this favorable metabolic profile, the ER22/23EK polymorphism is significantly higher in the oldest half of the population and is associated with increased survival (91). At young age, a sexually dimorphic pattern in body composition has been documented. Male ER22/23EK carriers are taller, have more muscle mass, and are stronger than noncarriers. Female ER22/23EK carriers have smaller waist and hip circumferences and lower body weight. Finally, at older age, carriers of the ER22/23EK polymorphism have lower risk of dementia and fewer white matter lesions in the brain compared to noncarriers (98). The molecular mechanisms through which the ER22/23EK polymorphism produces the above effects are likely to involve a higher expression of the hGRα-A (94 kDa) isoform at the expense of the hGRα-B (91 kDa) isoform. Given that the latter isoform has greater transactivational activity, the shift in hGRα-A to hGRα-B expression ratio leads to an overall decrease in transcriptional activity (115) (Figure 3B).
Finally, the TthIIII variant is a restriction site length polymorphism in the promoter region of the hGR gene, which is not functional by itself (98, 116, 117). However, the ER22/23EK variant was found to be invariably linked to the TthIIII polymorphism. Therefore, associations with glucocorticoid resistance and healthier metabolic profile observed in the TthIIII carriers are likely to arise as a result of the ER22/23EK polymorphism.
We have recently identified a novel, heterozygous hGR mutation (D401H) in a patient with symptomatology suggestive of glucocorticoid hypersensitivity (118). Similarly to most other polymorphic variants, the D401H mutation is located in the NTD of the receptor, in close proximity to the major transactivation domain AF-1. Functional studies demonstrated that natural mutant receptor hGRαD401H enhanced the transcriptional activity of hGRα at low expression levels in a dose-dependent manner (118). Therefore, this point mutation may predispose carriers to an adverse metabolic profile and an increased risk for metabolic syndrome-related atherosclerotic cardiovascular disease (Figure 3B).
Finally, a single nucleotide polymorphism that replaces A with G at the nucleoside 3669 (A3669G) located in the 3’ end of exon 9β has also been described (119). This polymorphism does not change the amino acid sequence but increases the stability of hGRβ mRNA and hGRβ protein expression, leading to greater inhibition of hGRα-induced transcriptional activity and glucocorticoid resistance. The presence of the A3669G allele is associated with reduced central obesity and a more favorable lipid profile in affected subjects (119).
In clinical practice, glucocorticoids are used widely to treat a number of pathologic conditions, as well as for replacement therapy purposes. The effects of glucocorticoid treatment may vary considerably between patients and may be partly attributed to polymorphisms in the hGR gene. Therefore, when the presence of these hGR gene variants is known in a patient, the dose of glucocorticoids should be adjusted accordingly to ensure optimal therapy and minimal adverse effects (96, 98).
Conclusions
The glucocorticoid receptor is a ubiquitously expressed intracellular, ligand-dependent transcription factor, which mediates the action of glucocorticoids and influences physiologic functions essential for life. The stochastic nature of glucocorticoid signaling pathways in association with the variable effect that hGR gene mutations/polymorphisms might have on glucocorticoid signal transduction, indicates that alterations in hGR action may have important implications for many critical biological processes, such as the behavioral and physiologic responses to stress, the immune and inflammatory reaction, the process of sleep, as well as basic functions, such as growth and reproduction. Research studies on the molecular mechanisms of hGR action enhance our understanding of several conditions associated with glucocorticoid resistance or hypersensitivity and underscore the importance of cellular and molecular signaling pathways in maintaining homeostasis and preserving normal physiology.
Acknowledgements
Literary work of this article was funded by the EU-European Social Fund, the Greek Ministry of Development-General Secretariat of Research and Technology, and the Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health, Bethesda, Maryland, 20892, USA,
References
- 1.Clark JK, Schrader WT, O'Malley BW. Mechanism of steroid hormones. In: Wilson JD, Foster DW, editors. Williams Textbook of Endocrinology. Philadelphia: WB Sanders Co.; 1992. pp. 35–90. [Google Scholar]
- 2.Chrousos GP, Charmandari E, Kino T. Glucocorticoid action networks–an introduction to systems biology. J. Clin. Endocrinol. Metab. 2004;89:563–564. doi: 10.1210/jc.2003-032026. [DOI] [PubMed] [Google Scholar]
- 3.Chrousos GP. The glucocorticoid receptor gene, longevity, and the complex disorders of Western societies. Am. J. Med. 2004;117:204–207. doi: 10.1016/j.amjmed.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 4.Galon J, Franchimont D, Hiroi N, Frey G, Boettner A, Ehrhart-Bornstein M, O'Shea JJ, Chrousos GP, Bornstein SR. Gene profiling reveals unknown enhancing and suppressive actions of glucocorticoids on immune cells. FASEB J. 2002;16(1):61–71. doi: 10.1096/fj.01-0245com. [DOI] [PubMed] [Google Scholar]
- 5.Zhou J, Cidlowski JA. The human glucocorticoid receptor: one gene, multiple proteins and diverse responses. Steroids. 2005;70(5–7):407–417. doi: 10.1016/j.steroids.2005.02.006. [DOI] [PubMed] [Google Scholar]
- 6.Duma D, Jewell CM, Cidlowski JA. Multiple glucocorticoid receptor isoforms and mechanisms of post-translational modification. J Steroid Biochem Mol Biol. 2006;102(1–5):11–21. doi: 10.1016/j.jsbmb.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 7.Thornton JW. Evolution of vertebrate steroid receptors from an ancestral estrogen receptor by ligand exploitation and serial genome expansions. Proc Natl Acad Sci U S A. 2001;98(10):5671–5676. doi: 10.1073/pnas.091553298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Thornton JW, Need E, Crews D. Resurrecting the ancestral steroid receptor: ancient origin of estrogen signaling. Science. 2003;301(5640):1714–1747. doi: 10.1126/science.1086185. [DOI] [PubMed] [Google Scholar]
- 9.Thornton JW, DeSalle R. Gene family evolution and homology: genomics meets phylogenetics. Annu Rev Genomics Hum Genet. 2000;1:41–73. doi: 10.1146/annurev.genom.1.1.41. [DOI] [PubMed] [Google Scholar]
- 10.Suga H, Hoshiyama D, Kuraku S, Katoh K, Kubokawa K, Miyata T. Protein tyrosine kinase cDNAs from amphioxus, hagfish, and lamprey: isoform duplications around the divergence of cyclostomes and gnathostomes. J Mol Evol. 1999;49(5):601–608. doi: 10.1007/pl00006581. [DOI] [PubMed] [Google Scholar]
- 11.Bridgham JT, Carroll SM, Thornton JW. Evolution of hormone-receptor complexity by molecular exploitation. Science. 2006;312(5770):97–101. doi: 10.1126/science.1123348. [DOI] [PubMed] [Google Scholar]
- 12.Ortlund EA, Bridgham JT, Redinbo MR, Thornton JW. Crystal structure of an ancient protein: evolution by conformational epistasis. Science. 2007;317(5844):1544–1548. doi: 10.1126/science.1142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hollenberg SM, Weinberger C, Ong ES, Cerelli G, Oro A, Lebo R, Thompson EB, Rosenfeld MG, Evans RM. Primary structure and expression of a functional human glucocorticoid receptor cDNA. Nature. 1985;318:635–641. doi: 10.1038/318635a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kino T, Manoli I, Kelkar S, Wang Y, Su YA, Chrousos GP. Glucocorticoid receptor (GR) β has intrinsic, Gα-independent transcriptional activity. Biochem Biophys Res Commun. 2009;381(4):671–675. doi: 10.1016/j.bbrc.2009.02.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Oakley RH, Jewell CM, Yudt MR, Bofetiado DM, Cidlowski JA. The dominant negative activity of the human glucocorticoid receptor β isoform. Specificity and mechanisms of action. J Biol Chem. 1999;274(39):27857–27866. doi: 10.1074/jbc.274.39.27857. [DOI] [PubMed] [Google Scholar]
- 16.Bledsoe RK, Montana VG, Stanley TB, Delves CJ, Apolito CJ, McKee DD, Consler TG, Parks DJ, Stewart EL, Willson TM, Lambert MH, Moore JT, Pearce KH, Xu HE. Crystal structure of the glucocorticoid receptor ligand binding domain reveals a novel mode of receptor dimerization and coactivator recognition. Cell. 2002;110:93–105. doi: 10.1016/s0092-8674(02)00817-6. [DOI] [PubMed] [Google Scholar]
- 17.Tanenbaum DM, Wang Y, Williams SP, Sigler PB. Crystallographic comparison of the estrogen and progesterone receptor's ligand binding domains. Proc Natl Acad Sci U S A. 1998;95:5998–6003. doi: 10.1073/pnas.95.11.5998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kauppi B, Jakob C, Farnegardh M, Yang J, Ahola H, Alarcon M, Calles K, Engstrom O, Harlan J, Muchmore S, Ramqvist AK, Thorell S, Ohman L, Greer J, Gustafsson JA, Carlstedt-Duke J, Carlquist M. The three-dimensional structures of antagonistic and agonistic forms of the glucocorticoid receptor ligand-binding domain: RU-486 induces a transconformation that leads to active antagonism. J Biol Chem. 2003;278(25):22748–22754. doi: 10.1074/jbc.M212711200. [DOI] [PubMed] [Google Scholar]
- 19.Zhang S, Liang X, Danielsen M. Role of the C terminus of the glucocorticoid receptor in hormone binding and agonist/antagonist discrimination. Mol Endocrinol. 1996;10(1):24–34. doi: 10.1210/mend.10.1.8838142. [DOI] [PubMed] [Google Scholar]
- 20.Lu NZ, Cidlowski JA. Translational regulatory mechanisms generate N-terminal glucocorticoid receptor isoforms with unique transcriptional target genes. Mol Cell. 2005;18:331–342. doi: 10.1016/j.molcel.2005.03.025. [DOI] [PubMed] [Google Scholar]
- 21.Breslin MB, Geng D, Vedeckis WV. Multiple promoters exist in the human GR gene, one of which is activated by glucocorticoids. Mol Endocrinol. 2001;15:1381–1395. doi: 10.1210/mend.15.8.0696. [DOI] [PubMed] [Google Scholar]
- 22.Chrousos GP, Kino T. Intracellular glucocorticoid signaling: a formerly simple system turns stochastic. Sci STKE. 2005;2005(304):pe48. doi: 10.1126/stke.3042005pe48. [DOI] [PubMed] [Google Scholar]
- 23.Pratt WB. The role of heat shock proteins in regulating the function, folding, and trafficking of the glucocorticoid receptor. J Biol Chem. 1993;268(29):21455–21458. [PubMed] [Google Scholar]
- 24.Terry LJ, Shows EB, Wente SR. Crossing the nuclear envelope: hierarchical regulation of nucleocytoplasmic transport. Science. 2007;318(5855):1412–1416. doi: 10.1126/science.1142204. [DOI] [PubMed] [Google Scholar]
- 25.Bamberger CM, Schulte HM, Chrousos GP. Molecular determinants of glucocorticoid receptor function and tissue sensitivity to glucocorticoids. Endocr Rev. 1996;17(3):245–261. doi: 10.1210/edrv-17-3-245. [DOI] [PubMed] [Google Scholar]
- 26.Schaaf MJ, Cidlowski JA. Molecular mechanisms of glucocorticoid action and resistance. J Steroid Biochem Mol Biol. 2002;83(1–5):37–48. doi: 10.1016/s0960-0760(02)00263-7. [DOI] [PubMed] [Google Scholar]
- 27.Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, Herrlich P. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62(6):1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- 28.Scheinman RI, Gualberto A, Jewell CM, Cidlowski JA, Baldwin AS., Jr Characterization of mechanisms involved in transrepression of NF-κB by activated glucocorticoid receptors. Mol Cell Biol. 1995;15(2):943–953. doi: 10.1128/mcb.15.2.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kino T, Chrousos GP. Tissue-specific glucocorticoid resistance-hypersensitivity syndromes: multifactorial states of clinical importance. J Allergy Clin Immunol. 2002;109(4):609–613. doi: 10.1067/mai.2002.123708. [DOI] [PubMed] [Google Scholar]
- 30.Liu J, DeFranco DB. Protracted nuclear export of glucocorticoid receptor limits its turnover and does not require the exportin 1/CRM1-directed nuclear export pathway. Mol Endocrinol. 2000;14(1):40–51. doi: 10.1210/mend.14.1.0398. [DOI] [PubMed] [Google Scholar]
- 31.Carrigan A, Walther RF, Salem HA, Wu D, Atlas E, Lefebvre YA, Haché RJ. An active nuclear retention signal in the glucocorticoid receptor functions as a strong inducer of transcriptional activation. J Biol Chem. 2007;282(15):10963–10971. doi: 10.1074/jbc.M602931200. [DOI] [PubMed] [Google Scholar]
- 32.Lieberman BA, Bona BJ, Edwards DP, Nordeen SK. The constitution of a progesterone response element. Mol Endocrinol. 1993;7:515–527. doi: 10.1210/mend.7.4.8388996. [DOI] [PubMed] [Google Scholar]
- 33.Beato M, Sanchez-Pacheco A. Interaction of steroid hormone receptors with the transcription initiation complex. Endocr Rev. 1996;17:587–609. doi: 10.1210/edrv-17-6-587. [DOI] [PubMed] [Google Scholar]
- 34.McKenna NJ, Lanz RB, O'Malley BW. Nuclear receptor coregulators: cellular and molecular biology. Endocr Rev. 1999;20(3):321–344. doi: 10.1210/edrv.20.3.0366. [DOI] [PubMed] [Google Scholar]
- 35.McKenna NJ, Xu J, Nawaz Z, Tsai SY, Tsai MJ, O'Malley BW. Nuclear receptor coactivators: multiple enzymes, multiple complexes, multiple functions. J Steroid Biochem Mol Biol. 1999;69:3–12. doi: 10.1016/s0960-0760(98)00144-7. [DOI] [PubMed] [Google Scholar]
- 36.McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- 37.Auboeuf D, Honig A, Berget SM, O'Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 38.Heery DM, Kalkhoven E, Hoare S, Parker MG. A signature motif in transcriptional coactivators mediates binding to nuclear receptors. Nature. 1997;387(6634):733–736. doi: 10.1038/42750. [DOI] [PubMed] [Google Scholar]
- 39.Reichardt HM, Kaestner KH, Tuckermann J, Kretz O, Wessely O, Bock R, Gass P, Schmid W, Herrlich P, Angel P, Schutz G. DNA binding of the glucocorticoid receptor is not essential for survival. Cell. 1998;93:531–541. doi: 10.1016/s0092-8674(00)81183-6. [DOI] [PubMed] [Google Scholar]
- 40.Cole TJ, Blendy JA, Monaghan AP, Krieglstein K, Schmid W, Aguzzi A, Fantuzzi G, Hummler E, Unsicker K, Schutz G. Targeted disruption of the glucocorticoid receptor gene blocks adrenergic chromaffin cell development and severely retards lung maturation. Genes Dev. 1995;9:1608–1621. doi: 10.1101/gad.9.13.1608. [DOI] [PubMed] [Google Scholar]
- 41.Reichardt HM, Tuckermann JP, Gottlicher M, Vujic M, Weih F, Angel P, Herrlich P, Schutz G. Repression of inflammatory responses in the absence of DNA binding by the glucocorticoid receptor. EMBO J. 2001;20:7168–7173. doi: 10.1093/emboj/20.24.7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Karin M, Chang L. AP-1-glucocorticoid receptor crosstalk taken to a higher level. J Endocrinol. 2001;169:447–451. doi: 10.1677/joe.0.1690447. [DOI] [PubMed] [Google Scholar]
- 43.Barnes PJ, Karin M. Nuclear factor-κB: a pivotal transcription factor in chronic inflammatory diseases. N Engl J Med. 1997;336:1066–1071. doi: 10.1056/NEJM199704103361506. [DOI] [PubMed] [Google Scholar]
- 44.Didonato JA, Saatcioglu F, Karin M. Molecular mechanisms of immunosuppression and anti-inflammatory activities by glucocorticoids. Am J Respir Crit Care Med. 1996;154:S11–S15. doi: 10.1164/ajrccm/154.2_Pt_2.S11. [DOI] [PubMed] [Google Scholar]
- 45.Ismaili N, Garabedian MJ. Modulation of glucocorticoid receptor function via phosphorylation. Ann N Y Acad Sci. 2004;1024:86–101. doi: 10.1196/annals.1321.007. [DOI] [PubMed] [Google Scholar]
- 46.Orti E, Hu LM, Munck A. Kinetics of glucocorticoid receptor phosphorylation in intact cells. Evidence for hormone-induced hyperphosphorylation after activation and recycling of hyperphosphorylated receptors. J Biol Chem. 1993;268:7779–7784. [PubMed] [Google Scholar]
- 47.Wang Z, Frederick J, Garabedian MJ. Deciphering the phosphorylation "code" of the glucocorticoid receptor in vivo. J Biol Chem. 2002;277:26573–26580. doi: 10.1074/jbc.M110530200. [DOI] [PubMed] [Google Scholar]
- 48.Miller AL, Webb MS, Copik AJ, Wang Y, Johnson BH, Kumar R, Thompson EB. p38 Mitogen-activated protein kinase (MAPK) is a key mediator in glucocorticoid-induced apoptosis of lymphoid cells: correlation between p38 MAPK activation and site-specific phosphorylation of the human glucocorticoid receptor at serine 211. Mol Endocrinol. 2005;19:1569–1583. doi: 10.1210/me.2004-0528. [DOI] [PubMed] [Google Scholar]
- 49.Kino T, Ichijo T, Amin ND, Kesavapany S, Wang Y, Kim N, Rao S, Player A, Zheng YL, Garabedian MJ, Kawasaki E, Pant HC, Chrousos GP. Cyclin-dependent kinase 5 differentially regulates the transcriptional activity of the glucocorticoid receptor through phosphorylation: clinical implications for the nervous system response to glucocorticoids and stress. Mol Endocrinol. 2007;21(7):1552–1568. doi: 10.1210/me.2006-0345. [DOI] [PubMed] [Google Scholar]
- 50.Rogatsky I, Waase CL, Garabedian MJ. Phosphorylation and inhibition of rat glucocorticoid receptor transcriptional activation by glycogen synthase kinase-3 (GSK-3). Species-specific differences between human and rat glucocorticoid receptor signaling as revealed through GSK-3 phosphorylation. J Biol Chem. 1998;273(23):14315–14321. doi: 10.1074/jbc.273.23.14315. [DOI] [PubMed] [Google Scholar]
- 51.Galliher-Beckley AJ, Williams JG, Collins JB, Cidlowski JA. Glycogen synthase kinase 3β-mediated serine phosphorylation of the human glucocorticoid receptor redirects gene expression profiles. Mol Cell Biol. 2008;28(24):7309–7322. doi: 10.1128/MCB.00808-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Rogatsky I, Logan SK, Garabedian MJ. Antagonism of glucocorticoid receptor transcriptional activation by the c-Jun N-terminal kinase. Proc Natl Acad Sci U S A. 1998;95(5):2050–2055. doi: 10.1073/pnas.95.5.2050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Itoh M, Adachi M, Yasui H, Takekawa M, Tanaka H, Imai K. Nuclear export of glucocorticoid receptor is enhanced by c-Jun N-terminal kinase-mediated phosphorylation. Mol Endocrinol. 2002;16(10):2382–2392. doi: 10.1210/me.2002-0144. [DOI] [PubMed] [Google Scholar]
- 54.Dennis AP, O'Malley BW. Rush hour at the promoter: how the ubiquitin-proteasome pathway polices the traffic flow of nuclear receptor-dependent transcription. J Steroid Biochem Mol Biol. 2005;93:139–151. doi: 10.1016/j.jsbmb.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 55.Kinyamu HK, Chen J, Archer TK. Linking the ubiquitin-proteasome pathway to chromatin remodeling/modification by nuclear receptors. J Mol Endocrinol. 2005;34:281–297. doi: 10.1677/jme.1.01680. [DOI] [PubMed] [Google Scholar]
- 56.Gillette TG, Gonzalez F, Delahodde A, Johnston SA, Kodadek T. Physical and functional association of RNA polymerase II and the proteasome. Proc Natl Acad Sci USA. 2004;101:5904–5909. doi: 10.1073/pnas.0305411101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Deroo BJ, Rentsch C, Sampath S, Young J, DeFranco DB, Archer TK. Proteasomal inhibition enhances glucocorticoid receptor transactivation and alters its subnuclear trafficking. Mol Cell Biol. 2002;22(12):4113–4123. doi: 10.1128/MCB.22.12.4113-4123.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kino T, Liou SH, Charmandari E, Chrousos GP. Glucocorticoid receptor mutants demonstrate increased motility inside the nucleus of living cells: time of fluorescence recovery after photobleaching (FRAP) is an integrated measure of receptor function. Mol Med. 2004;10(7–12):80–88. doi: 10.2119/2005-00026.Kino. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ito K, Yamamura S, Essilfie-Quaye S, Cosio B, Ito M, Barnes PJ, Adcock IM. Histone deacetylase 2-mediated deacetylation of the glucocorticoid receptor enables NF-κB suppression. J Exp Med. 2006;203(1):7–13. doi: 10.1084/jem.20050466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Nader N, Chrousos GP, Kino T. Circadian rhythm transcription factor CLOCK regulates the transcriptional activity of the glucocorticoid receptor by acetylating its hinge region lysine cluster: potential physiological implications. FASEB J. 2009 doi: 10.1096/fj.08-117697. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Le Drean Y, Mincheneau N, Le Goff P, Michel D. Potentiation of glucocorticoid receptor transcriptional activity by sumoylation. Endocrinology. 2002;143(9):3482–3489. doi: 10.1210/en.2002-220135. [DOI] [PubMed] [Google Scholar]
- 62.Göttlicher M, Heck S, Doucas V, Wade E, Kullmann M, Cato AC, Evans RM, Herrlich P. Interaction of the Ubc9 human homologue with c-Jun and with the glucocorticoid receptor. Steroids. 1996;61(4):257–262. doi: 10.1016/0039-128x(96)00032-3. [DOI] [PubMed] [Google Scholar]
- 63.Cho S, Kagan BL, Blackford JA, Jr, Szapary D, Simons SS., Jr Glucocorticoid receptor ligand binding domain is sufficient for the modulation of glucocorticoid induction properties by homologous receptors, coactivator transcription intermediary factor 2, and Ubc9. Mol Endocrinol. 2005;19(2):290–311. doi: 10.1210/me.2004-0134. [DOI] [PubMed] [Google Scholar]
- 64.Tian S, Poukka H, Palvimo JJ, Jänne OA. Small ubiquitin-related modifier-1 (SUMO-1) modification of the glucocorticoid receptor. Biochem J. 2002;367(Pt 3):907–911. doi: 10.1042/BJ20021085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Holmstrom S, Van Antwerp ME, Iñiguez-Lluhi JA. Direct and distinguishable inhibitory roles for SUMO isoforms in the control of transcriptional synergy. Proc Natl Acad Sci U S A. 2003;100(26):15758–15763. doi: 10.1073/pnas.2136933100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kino T, Chrousos GP. Glucocorticoid and mineralocorticoid resistance/hypersensitivity syndromes. J Endocrinol. 2001;169(3):437–445. doi: 10.1677/joe.0.1690437. [DOI] [PubMed] [Google Scholar]
- 67.Chrousos GP. Hormone Resistance and Hypersensitivity States. In: Chrousos GP, Olefsky JM, Samols E, editors. Modern Endocrinology Series. Philadelphia, PA: Lippincott, Williams & Wilkins; 2002. p. 542. [Google Scholar]
- 68.Kino T, De Martino MU, Charmandari E, Mirani M, Chrousos GP. Tissue glucocorticoid resistance/hypersensitivity syndromes. J Steroid Biochem Mol Biol. 2003;85(2–5):457–467. doi: 10.1016/s0960-0760(03)00218-8. [DOI] [PubMed] [Google Scholar]
- 69.Chrousos GP, Kino T. Glucocorticoid action networks and complex psychiatric and/or somatic disorders. Stress. 2007;10(2):213–219. doi: 10.1080/10253890701292119. [DOI] [PubMed] [Google Scholar]
- 70.Vingerhoeds AC, Thijssen JH, Schwarz F. Spontaneous hypercortisolism without Cushing's syndrome. J Clin Endocrinol Metab. 1976;43(5):1128–1133. doi: 10.1210/jcem-43-5-1128. [DOI] [PubMed] [Google Scholar]
- 71.Chrousos GP, Vingerhoeds A, Brandon D, Eil C, Pugeat M, DeVroede M, Loriaux DL, Lipsett MB. Primary cortisol resistance in man. A glucocorticoid receptor-mediated disease. J Clin Invest. 1982;69(6):1261–1269. doi: 10.1172/JCI110565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chrousos GP, Detera-Wadleigh SD, Karl M. Syndromes of glucocorticoid resistance. Ann Intern Med. 1993;119(11):1113–1124. doi: 10.7326/0003-4819-119-11-199312010-00009. [DOI] [PubMed] [Google Scholar]
- 73.Kino T, Vottero A, Charmandari E, Chrousos GP. Familial/sporadic glucocorticoid resistance syndrome and hypertension. Ann N Y Acad Sci. 2002;970:101–111. doi: 10.1111/j.1749-6632.2002.tb04416.x. [DOI] [PubMed] [Google Scholar]
- 74.Charmandari E, Kino T, Chrousos GP. Familial/sporadic glucocorticoid resistance: clinical phenotype and molecular mechanisms. Ann N Y Acad Sci. 2004;1024:168–181. doi: 10.1196/annals.1321.014. [DOI] [PubMed] [Google Scholar]
- 75.Charmandari E, Kino T. Novel causes of generalized glucocorticoid resistance. Horm Metab Res. 2007;39(6):445–450. doi: 10.1055/s-2007-980196. [DOI] [PubMed] [Google Scholar]
- 76.Charmandari E, Kino T, Ichijo T, Chrousos GP. Generalized glucocorticoid resistance: clinical aspects, molecular mechanisms, and implications of a rare genetic disorder. J Clin Endocrinol Metab. 2008;93(5):1563–1572. doi: 10.1210/jc.2008-0040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Karl M, Lamberts SW, Koper JW, Katz DA, Huizenga NE, Kino T, Haddad BR, Hughes MR, Chrousos GP. Cushing's disease preceded by generalized glucocorticoid resistance: clinical consequences of a novel, dominant-negative glucocorticoid receptor mutation. Proc Assoc Am Physicians. 1996;108(4):296–307. [PubMed] [Google Scholar]
- 78.Hurley DM, Accili D, Stratakis CA, Karl M, Vamvakopoulos N, Rorer E, Constantine K, Taylor SI, Chrousos GP. Point mutation causing a single amino acid substitution in the hormone binding domain of the glucocorticoid receptor in familial glucocorticoid resistance. J Clin Invest. 1991;87(2):680–686. doi: 10.1172/JCI115046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Karl M, Lamberts SW, Detera-Wadleigh SD, Encio IJ, Stratakis CA, Hurley DM, Accili D, Chrousos GP. Familial glucocorticoid resistance caused by a splice site deletion in the human glucocorticoid receptor gene. J Clin Endocrinol Metab. 1993;76(3):683–689. doi: 10.1210/jcem.76.3.8445027. [DOI] [PubMed] [Google Scholar]
- 80.Malchoff DM, Brufsky A, Reardon G, McDermott P, Javier EC, Bergh CH, Rowe D, Malchoff CD. A mutation of the glucocorticoid receptor in primary cortisol resistance. J Clin Invest. 1993;91(5):1918–1925. doi: 10.1172/JCI116410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kino T, Stauber RH, Resau JH, Pavlakis GN, Chrousos GP. Pathologic human GR mutant has a transdominant negative effect on the wild-type GR by inhibiting its translocation into the nucleus: importance of the ligand-binding domain for intracellular GR trafficking. J Clin Endocrinol Metab. 2001;86(11):5600–5608. doi: 10.1210/jcem.86.11.8017. [DOI] [PubMed] [Google Scholar]
- 82.Ruiz M, Lind U, Gafvels M, Eggertsen G, Carlstedt-Duke J, Nilsson L, Holtmann M, Stierna P, Wikstrom AC, Werner S. Characterization of two novel mutations in the glucocorticoid receptor gene in patients with primary cortisol resistance. Clin Endocrinol (Oxf) 2001;55(3):363–371. doi: 10.1046/j.1365-2265.2001.01323.x. [DOI] [PubMed] [Google Scholar]
- 83.Mendonca BB, Leite MV, de Castro M, Kino T, Elias LL, Bachega TA, Arnhold IJ, Chrousos GP, Latronico AC. Female pseudohermaphroditism caused by a novel homozygous missense mutation of the GR gene. J Clin Endocrinol Metab. 2002;87(4):1805–1809. doi: 10.1210/jcem.87.4.8379. [DOI] [PubMed] [Google Scholar]
- 84.Vottero A, Kino T, Combe H, Lecomte P, Chrousos GP. A novel, C-terminal dominant negative mutation of the GR causes familial glucocorticoid resistance through abnormal interactions with p160 steroid receptor coactivators. J Clin Endocrinol Metab. 2002;87(6):2658–2667. doi: 10.1210/jcem.87.6.8520. [DOI] [PubMed] [Google Scholar]
- 85.Charmandari E, Kino T, Vottero A, Souvatzoglou E, Bhattacharyya N, Chrousos GP. Natural glucocorticoid receptor mutants causing generalized glucocorticoid resistance: Molecular genotype, genetic transmission and clinical phenotype. J Clin Endocrinol Metab. 2004;89(4):1939–1949. doi: 10.1210/jc.2003-030450. [DOI] [PubMed] [Google Scholar]
- 86.Charmandari E, Raji A, Kino T, Ichijo T, Tiulpakov A, Zachman K, Chrousos GP. A novel point mutation in the ligand-binding domain (LBD) of the human glucocorticoid receptor (hGR) causing generalized glucocorticoid resistance: the importance of the C terminus of hGR LBD in conferring transactivational activity. J Clin Endocrinol Metab. 2005;90(6):3696–3705. doi: 10.1210/jc.2004-1920. [DOI] [PubMed] [Google Scholar]
- 87.Charmandari E, Kino T, Ichijo T, Zachman K, Alatsatianos A, Chrousos GP. Functional characterization of the natural human glucocorticoid receptor (hGR) mutants hGRαR477H and hGRαG679S associated with generalized glucocorticoid resistance. J Clin Endocrinol Metab. 2006;91(4):1535–1543. doi: 10.1210/jc.2005-1893. [DOI] [PubMed] [Google Scholar]
- 88.Charmandari E, Kino T, Ichijo T, Jubiz W, Mejia L, Zachman K, Chrousos GP. A novel point mutation in helix 11 of the ligand-binding domain of the human glucocorticoid receptor gene causing generalized glucocorticoid resistance. J Clin Endocrinol Metab. 2007;92(10):3986–3990. doi: 10.1210/jc.2006-2830. [DOI] [PubMed] [Google Scholar]
- 89.Savory JG, Hsu B, Laquian IR, Giffin W, Reich T, Hache RJ, Lefebvre YA. Discrimination between NL1- and NL2-mediated nuclear localization of the glucocorticoid receptor. Mol Cell Biol. 1999;19(2):1025–1037. doi: 10.1128/mcb.19.2.1025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Picard D, Yamamoto KR. Two signals mediate hormone-dependent nuclear localization of the glucocorticoid receptor. EMBO J. 1987;6(11):3333–3340. doi: 10.1002/j.1460-2075.1987.tb02654.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Qi M, Hamilton BJ, DeFranco D. v-mos oncoproteins affect the nuclear retention and reutilization of glucocorticoid receptors. Mol Endocrinol. 1989;3(8):1279–1288. doi: 10.1210/mend-3-8-1279. [DOI] [PubMed] [Google Scholar]
- 92.Wikstrom AC, Bakke O, Okret S, Bronnegard M, Gustafsson JA. Intracellular localization of the glucocorticoid receptor: evidence for cytoplasmic and nuclear localization. Endocrinology. 1987;120(4):1232–1242. doi: 10.1210/endo-120-4-1232. [DOI] [PubMed] [Google Scholar]
- 93.Ding XF, Anderson CM, Ma H, Hong H, Uht RM, Kushner PJ, Stallcup MR. Nuclear receptor-binding sites of coactivators glucocorticoid receptor interacting protein 1 (GRIP1) and steroid receptor coactivator 1 (SRC-1): multiple motifs with different binding specificities. Mol Endocrinol. 1998;12(2):302–313. doi: 10.1210/mend.12.2.0065. [DOI] [PubMed] [Google Scholar]
- 94.Hong H, Kohli K, Garabedian MJ, Stallcup MR. GRIP1, a transcriptional coactivator for the AF-2 transactivation domain of steroid, thyroid, retinoid, and vitamin D receptors. Mol Cell Biol. 1997;17(5):2735–2744. doi: 10.1128/mcb.17.5.2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hong H, Kohli K, Trivedi A, Johnson DL, Stallcup MR. GRIP1, a novel mouse protein that serves as a transcriptional coactivator in yeast for the hormone binding domains of steroid receptors. Proc Natl Acad Sci U S A. 1996;93(10):4948–4952. doi: 10.1073/pnas.93.10.4948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Lamberts SW, Huizenga AT, de Lange P, de Jong FH, Koper JW. Clinical aspects of glucocorticoid sensitivity. Steroids. 1996;61(4):157–160. doi: 10.1016/0039-128x(96)00005-0. [DOI] [PubMed] [Google Scholar]
- 97.Smit P, Russcher H, de Jong FH, Brinkmann AO, Lamberts SW, Koper JW. Differential regulation of synthetic glucocorticoids on gene expression levels of glucocorticoid-induced leucine zipper and interleukin-2. J Clin Endocrinol Metab. 2005;90(5):2994–3000. doi: 10.1210/jc.2004-2298. [DOI] [PubMed] [Google Scholar]
- 98.van Rossum EF, Lamberts SW. Polymorphisms in the glucocorticoid receptor gene and their associations with metabolic parameters and body composition. Recent Prog Horm Res. 2004;59:333–357. doi: 10.1210/rp.59.1.333. [DOI] [PubMed] [Google Scholar]
- 99.Koper JW, Stolk RP, de Lange P, Huizenga NA, Molijn GJ, Pols HA, Grobbee DE, Karl M, de Jong FH, Brinkmann AO, Lamberts SW. Lack of association between five polymorphisms in the human glucocorticoid receptor gene and glucocorticoid resistance. Hum Genet. 1997;99(5):663–668. doi: 10.1007/s004390050425. [DOI] [PubMed] [Google Scholar]
- 100.Huizenga NA, Koper JW, De Lange P, Pols HA, Stolk RP, Burger H, Grobbee DE, Brinkmann AO, De Jong FH, Lamberts SW. A polymorphism in the glucocorticoid receptor gene may be associated with and increased sensitivity to glucocorticoids in vivo. J Clin Endocrinol Metab. 1998;83(1):144–151. doi: 10.1210/jcem.83.1.4490. [DOI] [PubMed] [Google Scholar]
- 101.Lin RC, Wang XL, Dalziel B, Caterson ID, Morris BJ. Association of obesity, but not diabetes or hypertension, with glucocorticoid receptor N363S variant. Obes Res. 2003;11(6):802–808. doi: 10.1038/oby.2003.111. [DOI] [PubMed] [Google Scholar]
- 102.Di Blasio AM, van Rossum EF, Maestrini S, Berselli ME, Tagliaferri M, Podestà F, Koper JW, Liuzzi A, Lamberts SW. The relation between two polymorphisms in the glucocorticoid receptor gene and body mass index, blood pressure and cholesterol in obese patients. Clin Endocrinol (Oxf) 2003;59(1):68–74. doi: 10.1046/j.1365-2265.2003.01798.x. [DOI] [PubMed] [Google Scholar]
- 103.Roussel R, Reis AF, Dubois-Laforgue D, Bellanné-Chantelot C, Timsit J, Velho G. The N363S polymorphism in the glucocorticoid receptor gene is associated with overweight in subjects with type 2 diabetes mellitus. Clin Endocrinol (Oxf) 2003;59(2):237–241. doi: 10.1046/j.1365-2265.2003.01831.x. [DOI] [PubMed] [Google Scholar]
- 104.Dobson MG, Redfern CP, Unwin N, Weaver JU. The N363S polymorphism of the glucocorticoid receptor: potential contribution to central obesity in men and lack of association with other risk factors for coronary heart disease and diabetes mellitus. J Clin Endocrinol Metab. 2001;86(5):2270–2274. doi: 10.1210/jcem.86.5.7465. [DOI] [PubMed] [Google Scholar]
- 105.Lin RC, Wang XL, Morris BJ. Association of coronary artery disease with glucocorticoid receptor N363S variant. Hypertension. 2003;41(3):404–407. doi: 10.1161/01.HYP.0000055342.40301.DC. [DOI] [PubMed] [Google Scholar]
- 106.Buemann B, Vohl MC, Chagnon M, Chagnon YC, Gagnon J, Pérusse L, Dionne F, Després JP, Tremblay A, Nadeau A, Bouchard C. Abdominal visceral fat is associated with a BclI restriction fragment length polymorphism at the glucocorticoid receptor gene locus. Obes Res. 1997 May;5(3):186–192. doi: 10.1002/j.1550-8528.1997.tb00292.x. [DOI] [PubMed] [Google Scholar]
- 107.Rosmond R, Chagnon YC, Holm G, Chagnon M, Pérusse L, Lindell K, Carlsson B, Bouchard C, Björntorp P. A glucocorticoid receptor gene marker is associated with abdominal obesity, leptin, and dysregulation of the hypothalamic-pituitary-adrenal axis. Obes Res. 2000;8(3):211–218. doi: 10.1038/oby.2000.24. [DOI] [PubMed] [Google Scholar]
- 108.Ukkola O, Pérusse L, Chagnon YC, Després JP, Bouchard C. Interactions among the glucocorticoid receptor, lipoprotein lipase and adrenergic receptor genes and abdominal fat in the Québec Family Study. Int J Obes Relat Metab Disord. 2001;25(9):1332–1339. doi: 10.1038/sj.ijo.0801735. [DOI] [PubMed] [Google Scholar]
- 109.Weaver JU, Hitman GA, Kopelman PG. An association between a Bc1I restriction fragment length polymorphism of the glucocorticoid receptor locus and hyperinsulinaemia in obese women. J Mol Endocrinol. 1992;9(3):295–300. doi: 10.1677/jme.0.0090295. [DOI] [PubMed] [Google Scholar]
- 110.van Rossum EF, Koper JW, van den Beld AW, Uitterlinden AG, Arp P, Ester W, Janssen JA, Brinkmann AO, de Jong FH, Grobbee DE, Pols HA, Lamberts SW. Identification of the BclI polymorphism in the glucocorticoid receptor gene: association with sensitivity to glucocorticoids in vivo and body mass index. Clin Endocrinol (Oxf) 2003;59(5):585–592. doi: 10.1046/j.1365-2265.2003.01888.x. [DOI] [PubMed] [Google Scholar]
- 111.Russcher H, Smit P, van den Akker EL, van Rossum EF, Brinkmann AO, de Jong FH, Lamberts SW, Koper JW. Two polymorphisms in the glucocorticoid receptor gene directly affect glucocorticoid-regulated gene expression. J Clin Endocrinol Metab. 2005;90(10):5804–5810. doi: 10.1210/jc.2005-0646. [DOI] [PubMed] [Google Scholar]
- 112.van Rossum EF, Koper JW, Huizenga NA, Uitterlinden AG, Janssen JA, Brinkmann AO, Grobbee DE, de Jong FH, van Duyn CM, Pols HA, Lamberts SW. A polymorphism in the glucocorticoid receptor gene, which decreases sensitivity to glucocorticoids in vivo, is associated with low insulin and cholesterol levels. Diabetes. 2002;51(10):3128–3134. doi: 10.2337/diabetes.51.10.3128. [DOI] [PubMed] [Google Scholar]
- 113.van Rossum EF, Feelders RA, van den Beld AW, Uitterlinden AG, Janssen JA, Ester W, Brinkmann AO, Grobbee DE, de Jong FH, Pols HA, Koper JW, Lamberts SW. Association of the ER22/23EK polymorphism in the glucocorticoid receptor gene with survival and C-reactive protein levels in elderly men. Am J Med. 2004;117(3):158–162. doi: 10.1016/j.amjmed.2004.01.027. [DOI] [PubMed] [Google Scholar]
- 114.van Rossum EF, Voorhoeve PG, te Velde SJ, Koper JW, Delemarre-van de Waal HA, Kemper HC, Lamberts SW. The ER22/23EK polymorphism in the glucocorticoid receptor gene is associated with a beneficial body composition and muscle strength in young adults. J Clin Endocrinol Metab. 2004;89(8):4004–4009. doi: 10.1210/jc.2003-031422. [DOI] [PubMed] [Google Scholar]
- 115.Russcher H, van Rossum EF, de Jong FH, Brinkmann AO, Lamberts SW, Koper JW. Increased expression of the glucocorticoid receptor-A translational isoform as a result of the ER22/23EK polymorphism. Mol Endocrinol. 2005;19(7):1687–1696. doi: 10.1210/me.2004-0467. [DOI] [PubMed] [Google Scholar]
- 116.Detera-Wadleigh SD, Encio IJ, Rollins DY, Coffman D, Wiesch D. A TthIII1 polymorphism on the 5' flanking region of the glucocorticoid receptor gene (GRL) Nucleic Acids Res. 1991;19(8):1960. doi: 10.1093/nar/19.8.1960-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.van Rossum EF, Roks PH, de Jong FH, Brinkmann AO, Pols HA, Koper JW, Lamberts SW. Characterization of a promoter polymorphism in the glucocorticoid receptor gene and its relationship to three other polymorphisms. Clin Endocrinol (Oxf) 2004;61(5):573–581. doi: 10.1111/j.1365-2265.2004.02132.x. [DOI] [PubMed] [Google Scholar]
- 118.Charmandari E, Ichijo T, Jubiz W, Baid S, Zachman K, Chrousos GP, Kino T. A novel point mutation in the amino terminal domain of the human glucocorticoid receptor (hGR) gene enhancing hGR-mediated gene expression. J Clin Endocrinol Metab. 2008;93(12):4963–4968. doi: 10.1210/jc.2008-0892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Syed AA, Irving JA, Redfern CP, Hall AG, Unwin NC, White M, Bhopal RS, Weaver JU. Association of glucocorticoid receptor polymorphism A3669G in exon 9beta with reduced central adiposity in women. Obesity (Silver Spring) 2006;14(5):759–764. doi: 10.1038/oby.2006.86. [DOI] [PubMed] [Google Scholar]
ADDDITIONAL REFERENCES
- Brown GM, Grota LJ, Penney DP, Reichlin S. Pituitary–adrenal function in the squirrel monkey. Endocrinology. 1970;86:519–529. doi: 10.1210/endo-86-3-519. [DOI] [PubMed] [Google Scholar]
- Davies TH, Ning YM, Sanchez ER. A new first step in activation of steroid receptors: hormone-induced switching of FKBP51 and FKBP52 immunophilins. J. Biol. Chem. 2002;277:4597–4600. doi: 10.1074/jbc.C100531200. [DOI] [PubMed] [Google Scholar]
- Denny WB, Valentine DL, Reynolds PD, Smith DF, Scammell JG. Squirrel monkey immunophilin FKBP51 is a potent inhibitor of glucocorticoid receptor binding. Endocrinology. 2000;141:4107–4113. doi: 10.1210/endo.141.11.7785. Full Text via CrossRef | View Record in Scopus | Cited By in Scopus (70) [DOI] [PubMed] [Google Scholar]
- Gottlicher M, Heck S, Herrlich P. Transcriptional crosstalk, the second mode of steroid hormone receptor action. J Mol Med. 1998;76:480–489. doi: 10.1007/s001090050242. [DOI] [PubMed] [Google Scholar]
- Jonat C, Rahmsdorf HJ, Park KK, Cato AC, Gebel S, Ponta H, Herrlich P. Antitumor promotion and antiinflammation: down-modulation of AP-1 (Fos/Jun) activity by glucocorticoid hormone. Cell. 1990;62:1189–1204. doi: 10.1016/0092-8674(90)90395-u. [DOI] [PubMed] [Google Scholar]
- Heck S, Kullmann M, Gast A, Ponta H, Rahmsdorf HJ, Herrlich P, Cato AC. A distinct modulating domain in glucocorticoid receptor monomers in the repression of activity of the transcription factor AP-1. EMBO J. 1994;13:4087–4095. doi: 10.1002/j.1460-2075.1994.tb06726.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konig H, Ponta H, Rahmsdorf HJ, Herrlich P. Interference between pathway-specific transcription factors: glucocorticoids antagonize phorbol ester-induced AP-1 activity without altering AP-1 site occupation in vivo. EMBO J. 1992;11:2241–2246. doi: 10.1002/j.1460-2075.1992.tb05283.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKay LI, Cidlowski JA. Molecular control of immune/inflammatory responses: interactions between nuclear factor-kappa B and steroid receptor-signaling pathways. Endocr Rev. 1999;20:435–459. doi: 10.1210/edrv.20.4.0375. [DOI] [PubMed] [Google Scholar]
- Schule R, Rangarajan P, Kliewer S, Ransone LJ, Bolado J, Yang N, Verma IM, Evans RM. Functional antagonism between oncoprotein c-Jun and the glucocorticoid receptor. Cell. 1990;62:1217–1226. doi: 10.1016/0092-8674(90)90397-w. [DOI] [PubMed] [Google Scholar]
- Stocklin E, Wissler M, Gouilleux F, Groner B. Functional interactions between Stat5 and the glucocorticoid receptor. Nature. 1996;383:726–728. doi: 10.1038/383726a0. [DOI] [PubMed] [Google Scholar]