Abstract
Apolipoprotein A5 (APOA5) and lipoprotein lipase (LPL) proteins interact functionally to regulate lipid metabolism, and single nucleotide polymorphisms (SNPs) for each gene have also been associated independently with obesity risk. Evaluating gene combinations may be more effective than single SNP analyses in identifying genetic risk, but insufficient minor allele frequency (MAF) often limits evaluations of potential epistatic relationships. Populations with multiple ancestral admixtures may provide unique opportunities for evaluating genetic interactions. We examined relationships between LPL m107 (rs1800590) and APOA5 S19W (rs3135506) and lipid and anthropometric measures in Caribbean origin Hispanics (n=1019, aged 45–5 years) living in the Boston metropolitan area. Significant interaction terms between LPL m107 and APOA5 S19W were observed for BMI (P=0.003) and waist circumference (P=0.019). Higher BMI (P=0.001), waist (P=0.011) and hip (P=0.026) were observed in minor allele (G) carriers for LPL m107 who also carried the APOA5 S19W minor allele (G). Additionally, extreme obesity (BMI≥40 kg/m2) risk was higher (OR=4.02; 95% CI:1.81–.91; global P=0.008) for minor allele carriers for both SNPs (LPL TG+GG, APOA5 CG+GG) compared to major allele carriers for both SNPs. In summary, we identified significant interactions for APOA5 S19W and LPL m107 for obesity in Caribbean Hispanics. Population-specific MAFs increase the difficulties of replicating gene-gene interactions, but may support the hypothesis that combinations of frequencies in selected genes could heighten obesity susceptibility in a given population. Analyses of gene-gene interactions may improve understanding of genetically-based obesity risk, and underscore the need for further study of groups with multiple ancestral admixtures.
Introduction
Obesity is a complex, multifactorial condition for which heritability estimates vary widely, and this variability could be related to the large number of potential genetic contributors. Hundreds of obesity candidate loci have been proposed, and genome-wide association studies continue to identify new loci, but each individual locus may account for only a small percentage of the variability in populations (1). Analytic approaches which evaluate combinations of gene variants may improve our understanding of the genetic factors which modulate obesity variation and may help to clarify disparate results in genetic association studies (2).
Lipoprotein lipase (LPL), an important regulator of lipid and energy metabolism, has been explored as a potential genetic contributor to plasma lipids and obesity. LPL is the rate-limiting enzyme for hydrolysis of circulating triglycerides (TG) that comprise chylomicrons and very low density lipoproteins (VLDL), thereby regulating fatty acids supplied for oxidation in skeletal muscle or for storage in adipose tissue (3). Associations between LPL genetic variants and plasma lipids are well-documented (4) and the regulatory function of LPL in fatty acid partitioning and energy metabolism supports a possible additional role in obesity risk. Genetic studies linking LPL variants to obesity or altered adipose tissue distribution have reported gene-gene interactions between LPL and beta adrenergic receptor (ADR) variants or associations involving LPL haplotypes (5–7). In addition, the LPL promoter single nucleotide polymorphism (SNP), LPL m107 (−93 T/G), has been reported to be associated with obesity in Asian Indians in South India (8).
Apolipoprotein A5 (APOA5) is another key regulator of TG metabolism and its genetic associations with plasma lipids and cardiovascular disease risk have been replicated in multiple populations (9–12). One mechanism by which APOA5 regulates plasma lipids is via activation of proteoglycan-bound lipoprotein lipase to augment TG hydrolysis (13). Overexpression of APOA5 has been shown to increase LPL activity in an animal model (14). A second mechanism by which APOA5 has been hypothesized to modulate plasma TG is via inhibition of VLDL synthesis (15). Associations between individual APOA5 SNPs and obesity, as well as lipids, have been reported (10,16) but in another study the association between APOA5 −1131 T>C and obesity became apparent only when an interaction was considered, that of dietary fat (17).
Interactions between APOA5 and additional genetic factors, such as LPL, may be relevant to modulation of plasma lipids as well as obesity. Demonstrated functional relationships between LPL and APOA5 proteins in the modulation of plasma lipids lend further support to an analytic approach based on gene-gene interactions. For example, the APOA5 A139X variant is associated with altered LPL mass and activity in hyperchylomicronemia (18). Further, human carriers of either APOA5 S16W or APOA5 m1123 have been demonstrated to exhibit lower LPL activity in vivo following an intravenous fat tolerance test (19). Multiple novel APOA5 variants have been shown to exert variable effects on LPL activation, resulting in varying degrees of impaired lipolysis and hypertriglyceridemia in people (20).
With respect to the obesity phenotype, as opposed to hypertriglyceridemia, potential epistasis between APOA5 and LPL is unexplored. We based selection of specific APOA5 and LPL SNPs on reported phenotypic associations and also on the presence of adequate minor allele frequencies (MAF) in our population. APOA5 is a strong determinant of plasma TG in many ethnic groups (12,21) but the frequency of SNP APOA5 S16W (56C>G) is higher in Hispanic Americans compared to non-Hispanic Whites and African Americans (21). The LPL SNP m107 (−93 T/G) is of interest in part due to its reported association with increased obesity risk (8), but also because of earlier work reporting its ethnically variable modulation of plasma lipids (22,23). MAF for the LPL m107 SNP is highly variable across populations, ranging from undetectable in Chinese and 0.017 in non-Hispanic Whites, to 0.764 in Black South Africans (23). Caribbean Hispanics are characterized by admixture from European, Taíno American Indian and African ancestries, and this provides sufficiently high MAF of both APOA5 S16W and LPL m107 SNPS. In this unique population, we are able to evaluate epistatic relationships which would require much larger sample sizes in other populations. Therefore, in the current study, we examined whether LPL m107 and APOA5 S16W interact to modulate obesity and plasma lipids in Caribbean origin Hispanics.
Methods
Study Design and Subjects
Participants were recruited for a prospective two-year cohort study of men and women of Puerto Rican origin aged 45–74 years and living in the Boston, Massachusetts metropolitan area. Interviews to collect baseline demographic information, medical history, anthropometric and dietary data were conducted between 2004 and 2007 by trained bilingual staff. The Institutional Review Board at Tufts University/New England Medical Center approved the protocol of the current study. Anthropometric data including height, weight, and waist and hip circumference were measured in duplicate consistent with the technique used by the National Health and Nutrition Surveys (NHANES).
Laboratory Methods
Blood was collected for biochemical analyses and genetic analysis. Plasma was separated within four hours in a refrigerated centrifuge and then stored at −70° C. High density lipoprotein cholesterol (HDL-C) was analyzed using EDTA plasma with the enzymatic endpoint reaction with Olympus HDL reagents (OSR6156) and TG were analyzed using EDTA plasma with Olympus TG reagents (OSR6033). HDL-C and TG measurements were performed using the Olympus AU400e (Olympus America Inc., Melville, NY).
Genetic analysis
Genomic DNA was isolated from peripheral blood lymphocytes by standard methods. The SNPs LPL m107 (rs1800590) and APOA5 S16W (rs3135506) were genotyped using the ABI Prism TaqMan multiplex system (Applied Biosystems, Foster City, California).
Population ancestry admixture
The study participants reflect three ancestral populations, European, Taíno American Indian and African, which may be represented in varying proportions for each individual. One hundred ancestral markers were chosen to reduce confounding associated with population stratification. The programs STRUCTURE 2.2 (24) and IAE3CI (25) were used to calculate three admixture values for each subject and similar results were achieved (26). Multivariate regression analysis models incorporate adjustment for population admixture through the addition of two of the three admixture variables as covariates.
Statistical Analyses
All continuous variables were examined for normal distribution. Triglycerides were log transformed in order to normalize the distribution. The relationship between the individual SNPs LPL m107 and APOA5 S16W, and anthropometric measures and plasma lipids were evaluated by analysis of variance techniques. Interactions between the polymorphisms and anthropometric measures or plasma lipids were tested in a multivariate interaction model, with control for potential confounders including age, sex, current alcohol use, current smoking, diabetes, antilipemic medication, energy intake, physical activity and ancestry admixture. Combinations of genotypes were constructed as major-minor allele pairs to evaluate associations of genotype combination with differences in anthropometric measures and lipids. Logistic regression models were fitted to estimate risk (OR and 95% CI) of obesity (BMI ≥30 kg/m2) and extreme obesity (BMI ≥40 kg/m2) associated with the presence of one or two genetic variants, as compared with the absence of the genetic variants. SAS (Version 9.1 for Windows) was used to analyze data. A P value of 0.05 was considered statistically significant.
Results
Demographic, biochemical, anthropometric and genotypic data are presented in Table 1. Genotype frequencies for the APOA5 S19W SNP did not deviate from Hardy-Weinberg equilibrium (HWE) expectations but LPL m107 deviated from expectations (P=0.001). Minor allele frequency (MAF) was 0.128 for LPL m107 and 0.104 for the APOA5 S19W SNP. Anthropometric measures did not differ according to LPL m107 genotype, but HDL-C was significantly lower in carriers of the minor allele (P=0.013; Table 2). No differences in anthropometric measures were observed for APOA5 S19W, but TG was significantly higher in carriers of the minor allele (P<0.0001; Table 3). Multivariate adjustment for potential confounders included age, sex, current smoking, current alcohol, physical activity, diabetes, antilipemic medication, energy intake and admixture. Lipid measures were also adjusted by waist circumference and hormone use in women.
Table 1.
Demographic, biochemical, lifestyle and genotypic characteristics.1
| Age,y | 57.8±7.3 |
| Female,n (%) | 714(71.9) |
| BMI,kg/m2 | 32.2±6.8 |
| Obese,n (%)2 | 574(59%) |
| Extreme obesity, n(%)3 | 119(12%) |
| Waist,cm | 102±15 |
| Hip,cm | 110±14 |
| HDL-C, mmol/L | 1.16±0.33 |
| Triglyceride, mmol/L | 1.86±1.4 |
| Total energy intake (kcal) | 2106±861 |
| Diabetes, n(%)4 | 408(41.2) |
| Antilipemic medication, n(%) | 383(38.6) |
| Current alcohol use, n(%) | 379(38.4) |
| Current smoker, n(%) | 227(23.1) |
| LPL m107, n(%) | |
| TT | 787(77.2) |
| TG | 204 (20) |
| GG | 28(2.8) |
| APOA5 s19w, n(%) | |
| CC | 810(80) |
| CG | 193(19.1) |
| GG | 9(0.9) |
Values are mean±SD or n(%)
Obesity=BMI ≥30 mg/kg2
Extreme obesity=BMI ≥40 mg/kg2
Diabetes=Fasting glucose≥126 mg/dL or taking antiglycemic medication.
Table 2.
Associations between LPL m107 genotype, anthropometric measures and lipids in Caribbean Hispanics.
| TT (n=787) | TG+GG (n=232) | P value | |
|---|---|---|---|
| BMI(mg/kg2)1 | 30.8±0.3 | 31.5±0.5 | 0.161 |
| Waist(cm)1 | 101±1 | 103±1 | 0.250 |
| Hip(cm)1 | 107±1 | 109±1 | 0.147 |
| HDL-C(mmol/L)2 | 1.21±0.04 | 1.15±0.04 | 0.013 |
| Triglycerides(mmol/L)2 | 1.80±0.01 | 1.84±0.01 | 0.698 |
Meansadjusted for age, sex, alcohol, smoking, diabetes, antilipemic medication, physical activity, energy, admixture.
Meansadjusted for age, sex, alcohol, smoking, diabetes, antilipemic medication, physical activity, admixture, hormone use(women), waist.
Table 3.
Associations between APOA5 S19W genotype, anthropometric measures and lipids in Caribbean Hispanics.
| CC (n=810) | GC+GG (n=202) | P value | |
|---|---|---|---|
| BMI (mg/kg2)1 | 30.9±0.3 | 31.3±0.5 | 0.451 |
| Waist (cm)1 | 102±1 | 102±1 | 0.894 |
| Hip (cm)1 | 107±1 | 108±1 | 0.811 |
| HDL-C(mmolL)2 | 1.20±0.04 | 1.18±0.04 | 0.333 |
| Triglycerides(mmol/L)2 | 1.71±0.01 | 2.05±0.01 | <0.0001 |
Meansadjusted for age, sex, alcohol, smoking, diabetes, antilipemic medication, physical activity, energy, admixture.
Meansadjusted for age, sex, alcohol, smoking, diabetes, antilipemic medication, physical activity, admixture, hormone use(women), waist.
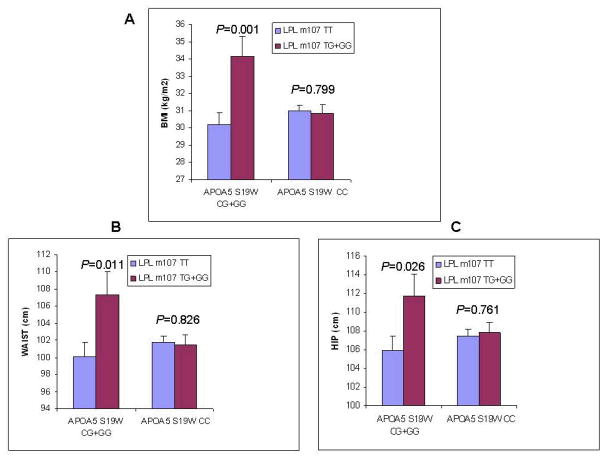
Genotype combinations for the two SNPs were constructed to evaluate interactions between the genes for lipids and anthropometric measures. No significant interactions were observed between LPL m107 and APOA5 S19W for plasma lipids (data not shown). Significant interaction terms between LPL m107 and APOA5 S19W were observed for BMI (P=0.003; Figure 1; panel A) and waist circumference (P=0.019; Figure 1; panel B) and marginally significant interaction was observed for hip circumference (P=0.054; Figure 1; panel C). Significantly higher BMI was observed in subjects with the minor allele for LPL m107 (G) who also carried the APOA5 S19W minor allele (G) (Figure 1; panel A; P = 0.001). Significantly higher waist circumference was observed in subjects with the minor allele for LPL m107 (G) who also carried the APOA5 S19W minor allele (G) (Figure 1; panel B; P = 0.011). Similarly, significantly higher hip circumference was present in subjects with the G allele for LPL m107 who were also carriers of APOA5 S19W minor allele (G) (Figure 1; panel C; P = 0.026). Differences for BMI, waist and hip were not observed for LPL m107 genotype in subjects who lacked the APOA5 S19W minor allele (G). Total energy intake did not differ according to genotype combination (data not shown). We also evaluated obesity measures and genotype combinations using a second model in which diabetes was not included as a confounder, but this did not alter significant relationships for gene-gene interactions or genotype-associated differences.
Figure 1.
Anthropometric measures by LPL m107/APOA5 S19W genotype combination. (A)BMI (kg/m2);(B)Waist;(C)Hip. Means adjusted for age, sex, smoking, alcohol, antilipemic medication, physical activity, diabetes, energy, admixture. Error bars indicate standard error of means.
Examination of the relationship between LPL m107 and APOA5 S19W genotype combination and extreme obesity (BMI ≥40 kg/m2) revealed an increased frequency of extreme obesity for carriers of the minor allele for both SNPs (LPL TG+GG and APOA5 CG+GG; Table 4). Subjects with extreme obesity for whom genotype data are available for both SNPs are categorized by combined genotype (n=113). Compared to the reference group of subjects carrying major alleles for both SNPs, the presence of both LPL TG+GG and APOA5 CG+GG alleles was associated with an odds ratio (OR) of 4.02 for extreme obesity (95% CI of 1.81 to 8.91; global P=0.008). The same multivariate adjustments used for the previous analyses were applied.
Table 4.
| BMI≥40,n(%) | OR | 95% CI Lower | Upper | P value | |
|---|---|---|---|---|---|
| APOA5 S19W CC/LPL m107 TT | 64(11.1) | 11 | |||
| APOA5 S19W CC/LPL m107 TG+GG | 20(11.9) | 1.24 | 0.70 | 2.19 | 0.469 |
| APOA5 S19WCG+GG/LPL m107 TT | 17(12.1) | 1.02 | 0.55 | 1.89 | 0.946 |
| APOA5 S19W CG+GG/LPL m107 TG+GG | 12(26.7) | 4.02 | 1.81 | 8.91 | 0.0006 |
Reference genotype combination
Maximum likelihood estimates, global genotype effect: Chi square statistic=11.96, 3 DF, P=0.008
Multivariate adjustment for age, sex, alcohol, smoking, diabetes, antilipemic medication, physical activity, energy, admixture.
Discussion
We have observed an interaction between APOA5 S19W and LPL m107 in which the presence of minor alleles for both SNPs was associated with significantly greater measures of obesity, including extreme obesity, compared to subjects with one or no minor alleles. Significant associations for obesity were not observed for either of the individual SNPs, although HDL-C and TG differed according to LPL m107 and APOA5 genotype, respectively. Previous studies have implicated various SNPs of LPL and APOA5, either individually or in combination with other genes, for obesity and plasma lipids, but interaction between these two genes for obesity has not been reported previously.
Evidence for LPL as a regulator of energy balance and potential modulator of obesity is extensive, although inconclusive. One previous study reported an increased risk of obesity associated with the individual SNP LPL m107 in a population of Asian Indians (8), but the tendency towards an association with obesity for this SNP alone did not reach significance in the current study. Consideration of LPL as a potential modulator of obesity is based on the role of LPL in regulating the supply of TG-derived fatty acids to skeletal muscle for oxidation or to adipose for storage (3). LPL sensitivity to energy balance is reflected in its tissue-specific responses to feeding, fasting and exercise (3,27). The effect of weight loss on adipose LPL activity varies among individuals, and may predict subsequent weight regain. In the 6 months following a hypocaloric intervention, human subjects whose adipose LPL had decreased during the intervention demonstrated less weight regain than those whose LPL had increased during the intervention, suggesting that LPL may play a regulatory role in ongoing weight maintenance (28). Whether this variability in LPL response to energy deficit is related to genetic variants, or to additional factors acting to regulate LPL, is unknown. In another study of adipose LPL and obesity, leptin-deficient (ob/ob) mice lacking adipose LPL but expressing skeletal muscle LPL demonstrated reduced weight and fat mass compared to ob/ob mice which did not lack LPL, although the apparent resistance to obesity conferred by adipose LPL knockout was not observed in non-ob/ob mice (29). Animal studies demonstrating that the LPL activator NO-1886 (ibrolipim) is associated with decreased fat accumulation and increased uncoupling protein 3 (UCP3) mRNA in high-fat-induced obese animals suggest a role for LPL in energy balance, possibly through stimulation of increased fatty acid oxidation by skeletal muscle (30).
Collectively, the preceding physiologic studies provide a framework by which LPL may modulate obesity, but do not address the question of whether single SNP genetic variation in LPL is sufficient to influence obesity or whether variation in other regulators of energy and lipid metabolism is also required. Obesity association studies for LPL often report interactions between LPL and other potential obesity candidates, such as ADR and glucocorticoid receptor variants, and suggest an interplay between LPL and other genetic factors in modulating obesity (6,7). These results support observations in our population, in which neither LPL nor APOA5 variants alone were associated with obesity, but in which the presence of both minor alleles was associated with higher anthropometric measures.
In order to better characterize the nature of the increased measures of obesity associated with genotype, we also examined the risk of extreme obesity (BMI ≥ 40 kg/m2) by genotype combination. Extreme obesity prevalence was 12% in this Caribbean-origin Hispanic population, which exceeds estimates for prevalence in Hispanic Americans of unspecified origin, aged 50–79 years (31). While obesity risk (BMI ≥ 30 kg/m2) did not differ according to genotype combination, the OR of extreme obesity was four times greater in subjects carrying both minor alleles. This observation represents a potentially important clinical finding, as BMI≥40 kg/m2 is associated with more co-morbidities and higher mortality than less severe obesity (31).
In addition to the interactions observed for obesity, APOA5 S16W was associated with higher TG in our population. An association between APOA5 and altered TG is consistent with previous studies in other groups (32). While we detected no association between LPL m107 and TG, previous studies have produced conflicting results in which the LPL m107 minor allele has been associated with unchanged, higher or with lower TG in different ethnic groups (23, 33). One source of apparent conflict in associations for LPL m107 and lipids may be the differential LD patterns between LPL m107 and other LPL SNPs. For example, LD between LPL D9N and m107 is strong in non-Hispanic Whites and the D9N SNP has been shown to be associated with high TG (23, 34). LD between LPL m107 and LPL D9N is weak in Africans, and in the current study of subjects with African as well as European admixture, weak LD between LPL D9N and LPL m107 may account for the lack of an association for LPL m107 with TG. Alternatively, APOA5 may be a more important modulator of plasma TG in this population, and may modulate TG via inhibition of VLDL synthesis in addition to its interaction with LPL (15).
Previous studies examining LPL m107 and HDL-C are similarly mixed. While we observed a significant association for LPL m107 minor allele with lower HDL-C, an earlier study reported no differences in HDL-C for LPL m107/D9N haplotypes in a small, combined African-American/Hispanic American population (35). Similarly, no associations between LPL m107 and plasma lipids were reported for Asian Indians (8). However, lower HDL-C was associated with the LPL m107 minor allele in two non-Hispanic White populations (34, 36). Ethnic-specific LD patterns may underlie differential associations for HDL-C as well as for TG. Alternatively, the lower HDL-C observed in LPL m107 carriers in the current study may be related to the non-significant tendency for increased obesity and increased abdominal obesity in minor allele carriers.
Several limitations for the current study must be addressed. One question relating to LPL m107 is the deviation from HWE observed in the current population, because deviation may indicate genotyping error. Laboratory procedures in place to detect error include blinded no-template controls and DNA sample replicates. Further, genotype clusters were re-checked visually to confirm that genotyping error did not occur. Genotype error rate was estimated at less than 0.5%. Previous studies which have evaluated HWE have not reported deviation for this SNP; however, HWE analysis has not been reported in all studies. In one previous study for which LPL m107 allele frequencies were reported but HWE not evaluated, we calculated a slight deviation from HWE [P=0.03] (22). HWE deviation may reflect population substructure or selection, or may support an association between genotype and disease (37). A second consideration is the relatively small number of subjects (n=47) carrying minor alleles for both SNPs. Ideally, a larger population with a greater number of minor allele carriers would be preferable.
In summary, we observed an interaction between LPL m107 and APOA5 S19W for obesity in Caribbean origin Hispanics. Replication is critical for validating association and interaction studies, and the observations reported for a single population in the current study must therefore be considered hypothesis-generating rather than definitive. However, replication of gene-gene interactions may be more difficult than replication of single SNP studies due to the requirement for adequate MAF for both SNPs. LPL m107 MAF is highly variable, ranging from 0.76 in South African Blacks (23), 0.28 in African Americans (35), 0.128 in the current population, 0.05–0.06 in Asian Indians (8), and 0.017 in non-Hispanic Whites (23). In addition, groups in which LPL m107 frequency may be sufficient for testing, such as Asian Indians, may exhibit a relatively lower MAF (0.03) for APOA5 S19W, which is substantially less than that reported for the current Caribbean origin Hispanic population (0.104) or other Hispanic populations [0.15](21). Population differences in allelic frequencies increase the difficulties of replicating gene-gene interactions, but also support the hypothesis that combinations of allelic frequencies in multiple genes may heighten genetic susceptibility for disease risk in a given population, such as Hispanics of Caribbean origin. Increasing understanding of gene-gene interaction may provide a more realistic picture of genetically-based obesity risk, and underscores the need for further study of groups with multiple ancestral admixtures.
Acknowledgments
Supported by the National Institutes of Health, National Institute on Aging, Grant Number 5P01AG023394 and NIH/NHLBI grant number HL54776 and NIH/NIDDK DK075030 and contracts 53-K06-5-10 and 58–1950-9–001 from the U.S. Department of Agriculture Research Service. C. Smith is supported by T32 DK007651-19.
Footnotes
Disclosure
The authors have no conflicts of interest to disclose.
References
- 1.Perusse L, Rankinen T, Zuberi A, et al. The human obesity gene map: the 2004 update. Obes Res. 2005;13(3):381–490. doi: 10.1038/oby.2005.50. [DOI] [PubMed] [Google Scholar]
- 2.Bouchard L, Tremblay A, Bouchard C, Perusse L. Contribution of several candidate gene polymorphisms in the determination of adiposity changes: results from the Quebec Family Study. Int J Obes. 2007;31(6):891–899. doi: 10.1038/sj.ijo.0803542. [DOI] [PubMed] [Google Scholar]
- 3.Lithell H, Boberg J, Hellsing K, Lundqvist G, Vessby B. Lipoprotein-lipase activity in human skeletal muscle and adipose tissue in the fasting and the fed states. Atherosclerosis. 1978;30(1):89–94. doi: 10.1016/0021-9150(78)90155-7. [DOI] [PubMed] [Google Scholar]
- 4.Chen W, Srinivasan SR, Elkasabany A, Ellsworth DL, Boerwinkle E, Berenson GS. Influence of lipoprotein lipase serine 447 stop polymorphism on tracking of triglycerides and HDL cholesterol from childhood to adulthood and familial risk of coronary artery disease: the Bogalusa heart study. Atherosclerosis. 2001;159(2):367–373. doi: 10.1016/s0021-9150(01)00508-1. [DOI] [PubMed] [Google Scholar]
- 5.Goodarzi MO, Taylor KD, Guo X, et al. Haplotypes in the lipoprotein lipase gene influence fasting insulin and discovery of a new risk haplotype. J Clin Endocrinol Metab. 2007;92(1):293–296. doi: 10.1210/jc.2006-1195. [DOI] [PubMed] [Google Scholar]
- 6.Li S, Chen W, Srinivasan SR, Boerwinkle E, Berenson GS. Influence of lipoprotein lipase gene Ser447Stop and beta1-adrenergic receptor gene Arg389Gly polymorphisms and their interaction on obesity from childhood to adulthood: the Bogalusa Heart Study. Int J Obes. 2006;30(8):1183–1188. doi: 10.1038/sj.ijo.0803281. [DOI] [PubMed] [Google Scholar]
- 7.Ukkola O, Perusse L, Chagnon YC, Despres JP, Bouchard C. Interactions among the glucocorticoid receptor, lipoprotein lipase and adrenergic receptor genes and abdominal fat in the Quebec Family Study. Int J Obes Relat Metab Disord. 2001;25(9):1332–1339. doi: 10.1038/sj.ijo.0801735. [DOI] [PubMed] [Google Scholar]
- 8.Radha V, Vimaleswaran KS, Ayyappa KA, Mohan V. Association of lipoprotein lipase gene polymorphisms with obesity and type 2 diabetes in an Asian Indian population. Int J Obes. 2007;31(6):913–918. doi: 10.1038/sj.ijo.0803547. [DOI] [PubMed] [Google Scholar]
- 9.Chandak GR, Ward KJ, Yajnik CS, et al. Triglyceride associated polymorphisms of the APOA5 gene have very different allele frequencies in Pune, India compared to Europeans. BMC Med Genet. 2006:7–76. doi: 10.1186/1471-2350-7-76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Elosua R, Cupples LA, Fox CS, et al. Association between well-characterized lipoprotein-related genetic variants and carotid intimal medial thickness and stenosis: The Framingham Heart Study. Atherosclerosis. 2006;189(1):222–228. doi: 10.1016/j.atherosclerosis.2005.12.005. [DOI] [PubMed] [Google Scholar]
- 11.Hubacek JA, Skodova Z, Adamkova V, Lanska V, Poledne R. The influence of APOAV polymorphisms (T-1131>C and S19>W) on plasma triglyceride levels and risk of myocardial infarction. Clin Genet. 2004;65(2):126–130. doi: 10.1111/j.0009-9163.2004.00199.x. [DOI] [PubMed] [Google Scholar]
- 12.Lai CQ, Tai ES, Tan CE, et al. The APOA5 locus is a strong determinant of plasma triglyceride concentrations across ethnic groups in Singapore. J Lipid Res. 2003;44(12):2365–2373. doi: 10.1194/jlr.M300251-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Merkel M, Loeffler B, Kluger M, et al. Apolipoprotein AV accelerates plasma hydrolysis of triglyceride-rich lipoproteins by interaction with proteoglycan-bound lipoprotein lipase. J Biol Chem. 2005;280(22):21553–21560. doi: 10.1074/jbc.M411412200. [DOI] [PubMed] [Google Scholar]
- 14.Fruchart-Najib J, Bauge E, Niculescu LS, et al. Mechanism of triglyceride lowering in mice expressing human apolipoprotein A5. Biochem Biophys Res Commun. 2004;319(2):397–404. doi: 10.1016/j.bbrc.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 15.Weinberg RB, Cook VR, Beckstead JA, et al. Structure and interfacial properties of human apolipoprotein A-V. J Biol Chem. 2003;278:34438–34444. doi: 10.1074/jbc.M303784200. [DOI] [PubMed] [Google Scholar]
- 16.Niculescu LS, Fruchart-Najib J, Fruchart JC, Sima A. Apolipoprotein A-V gene polymorphisms in subjects with metabolic syndrome. Clin Chem Lab Med. 2007;45(9):1133–1139. doi: 10.1515/CCLM.2007.257. [DOI] [PubMed] [Google Scholar]
- 17.Corella D, Lai CQ, Demissie S, et al. APOA5 gene variation modulates the effects of dietary fat intake on body mass index and obesity risk in the Framingham Heart Study. J Mol Med. 2007;85:119–128. doi: 10.1007/s00109-006-0147-0. [DOI] [PubMed] [Google Scholar]
- 18.Marcais C, Verges B, Charriere S, et al. Apoa5 Q139X truncation predisposes to late-onset hyperchylomicronemia due to lipoprotein lipase impairment. J Clin Invest. 2005;115(10):2862–2869. doi: 10.1172/JCI24471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kovar J, Adamkova V. Lipoprotein lipase activity determined in vivo is lower in carriers of apolipoprotein A-V gene variants 19W and −1131C. Physiol Res. 2008;57(4):555–561. doi: 10.33549/physiolres.931245. [DOI] [PubMed] [Google Scholar]
- 20.Dorfmeister B, Zeng WW, Dichlberger A, et al. Effects of six APOA5 variants, identified in patients with severe hypertriglyceridemia, on in vitro lipoprotein lipase activity and receptor binding. Arterioscler Thromb Vasc Biol. 2008;28(10):1866–1871. doi: 10.1161/ATVBAHA.108.172866. [DOI] [PubMed] [Google Scholar]
- 21.Pennacchio LA, Olivier M, Hubacek JA, Krauss RM, Rubin EM, Cohen JC. Two independent apolipoprotein A5 haplotypes influence human plasma triglyceride levels. Hum Mol Genet. 2002;11(24):3031–3038. doi: 10.1093/hmg/11.24.3031. [DOI] [PubMed] [Google Scholar]
- 22.Hall S, Chu G, Miller G, et al. A common mutation in the lipoprotein lipase gene promoter, −93T/G, is associated with lower plasma triglyceride levels and increased promoter activity in vitro. Arterioscler Thromb Vasc Biol. 1997;17(10):1969–1976. doi: 10.1161/01.atv.17.10.1969. [DOI] [PubMed] [Google Scholar]
- 23.Ehrenborg E, Clee SM, Pimstone SN, et al. Ethnic variation and in vivo effects of the −93t-->g promoter variant in the lipoprotein lipase gene. Arterioscler Thromb Vasc Biol. 1997;17(11):2672–2678. doi: 10.1161/01.atv.17.11.2672. [DOI] [PubMed] [Google Scholar]
- 24.Falush D, Stephens M, Pritchard JK. Inference of population structure using multilocus genotype data: linked loci and correlated allele frequencies. Genetics. 2003;164:1567–1587. doi: 10.1093/genetics/164.4.1567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tsai HJ, Choudhry S, Naqvi M, Rodriguez-Cintron W, Burchard EG, Ziv E. Comparison of three methods to estimate genetic ancestry and control for stratification in genetic association studies among admixed populations. Hum Genet. 2005;118(3–4):424–433. doi: 10.1007/s00439-005-0067-z. [DOI] [PubMed] [Google Scholar]
- 26.Lai CQ, Tucker KL, Choudry S, et al. Hum Genet. 2008 [Epub ahead of print]. Population admixture associated with disease prevalence in the Boston Puerto Rican health study. [Google Scholar]
- 27.Ruge T, Svensson M, Eriksson JW, Olivecrona G. Tissue-specific regulation of lipoprotein lipase in humans: effects of fasting. Eur J Clin Invest. 2005;35(3):194–200. doi: 10.1111/j.1365-2362.2005.01470.x. [DOI] [PubMed] [Google Scholar]
- 28.Nicklas BJ, Rogus EM, Berman DM, Dennis KE, Goldberg AP. Responses of adipose tissue lipoprotein lipase to weight loss affect lipid levels and weight regain in women. Am J Physiol Endocrinol Metab. 2000;79(5):E1012–1019. doi: 10.1152/ajpendo.2000.279.5.E1012. [DOI] [PubMed] [Google Scholar]
- 29.Weinstock PH, Levak Frank S, Hudgins LC, et al. Lipoprotein lipase controls fatty acid entry into adipose tissue, but fat mass is preserved by endogenous synthesis in mice deficient in adipose tissue lipoprotein lipase. Proc Natl Acad Sci USA. 1997;94:10261–10266. doi: 10.1073/pnas.94.19.10261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kusunoki M, Tsutsumi K, Iwata K, et al. NO-1886 (ibrolipim), a lipoprotein lipase activator, increases the expression of uncoupling protein 3 in skeletal muscle and suppresses fat accumulation in high-fat diet-induced obesity in rats. Metabolism. 2005;54(12):1587–1592. doi: 10.1016/j.metabol.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 31.McTigue K, Larson JC, Valoski A, et al. Mortality and cardiac and vascular outcomes in extremely obese women. JAMA. 2006;296(1):79–86. doi: 10.1001/jama.296.1.79. [DOI] [PubMed] [Google Scholar]
- 32.Lai CQ, Demissie S, Cupples LA, et al. Influence of the APOA5 locus on plasma triglyceride, lipoprotein subclasses, and CVD risk in the Framingham Heart Study. J Lipid Res. 2004;45(11):2096–105. doi: 10.1194/jlr.M400192-JLR200. [DOI] [PubMed] [Google Scholar]
- 33.Hall S, Talmud PJ, Cook DG, et al. Frequency and allelic association of common variants in the lipoprotein lipase gene in different ethnic groups: the Wandsworth Heart and Stroke Study. Genet Epidemiol. 2000;18(3):203–216. doi: 10.1002/(SICI)1098-2272(200003)18:3<203::AID-GEPI2>3.0.CO;2-I. [DOI] [PubMed] [Google Scholar]
- 34.Samuels ME, Forbey KC, Reid JE, et al. Identification of a common variant in the lipoprotein lipase gene in a large Utah kindred ascertained for coronary heart disease: the −93G/D9N variant predisposes to low HDL-C/high triglycerides. Clin Genet. 2001;59(2):88–98. doi: 10.1034/j.1399-0004.2001.590205.x. [DOI] [PubMed] [Google Scholar]
- 35.Talmud PJ, Hall S, Holleran S, Ramakrishnan R, Ginsberg HN, Humphries SE. LPL promoter −93T/G transition influences fasting and postprandial plasma triglycerides response in African-Americans and Hispanics. J Lipid Res. 1998;39(6):1189–1196. [PubMed] [Google Scholar]
- 36.Kastelein JJ, Groenemeyer BE, Hallman DM, et al. The Asn9 variant of lipoprotein lipase is associated with the −93G promoter mutation and an increased risk of coronary artery disease. The Regress Study Group. Clin Genet. 1998;53(1):27–33. doi: 10.1034/j.1399-0004.1998.531530106.x. [DOI] [PubMed] [Google Scholar]
- 37.Wang J, Shete S. A test for genetic association that incorporates information about deviation from Hardy-Weinberg proportions in cases. Am J Hum Genet. 2008;83(1):53–63. doi: 10.1016/j.ajhg.2008.06.010. [DOI] [PMC free article] [PubMed] [Google Scholar]