Abstract
In the present study, we investigated the relationship between the KIR loci and the genes encoding their HLA ligands and genetic susceptibility to Crohn’s disease (CD). Analyses of the interactions between KIR3DL1, KIR2DL1, KIR2DL2 and KIR2DL3 with their respective HLA ligands indicate that there is a protective effect for KIR2DL2 in the absence of its HLA ligand C1. Given that KIR2DL2 and KIR2DL3 segregate as alleles, we compared their genotypic distributions to expectations under Hardy-Weinberg Equilibrium (HWE) with regard to the HLA ligand C1 status. While all the genotypic distributions conform to expectations under HWE in controls, in C2 ligand homozygous cases there is significant deviation from HWE, with a reduction of KIR2DL2, KIR2DL3 heterozygotes. KIR2DL2, KIR2DL3 heterozygosity is the only genotypic combination that confers protection from CD. In addition to the protective effect (OR = 0.44, CI = 0.22–0.87; p = 0.018) observed in C2 ligand homozygotes, the KIR2DL2, KIR2DL3 genotype is predisposing (OR = 1.34, CI = 1.03–4.53; p = 0.031) in the presence of C1 ligand. A test for trend of HLA class I C ligand group genotypes with KIR2DL2, KIR2DL3 heterozygosity in cases and controls indicates that C1, C2 ligand group heterozygotes have an intermediate effect on predisposition. These results show for the first time that disease susceptibility may be related to heterozygosity at a specific KIR locus, and that HLA ligand genotype influences the relative effect of the KIR genotype.
Keywords: Crohn’s disease, KIR, HLA, heterozygosity, ligand
INTRODUCTION
The inflammatory bowel diseases (IBD) predominately consist of two idiopathic clinical entities, Crohn’s disease (CD) and ulcerative colitis (UC). The majority of patients with IBD characteristically exhibit relapsing, chronic, intestinal inflammation although there can be great variability in disease sub-phenotype (reviewed in (Targan 2003; Targen SR 2003; Taylor 2007). Patients with UC or CD can usually be distinguished by clinical and histopathologic characteristics. Patients with CD characteristically present with diarrhea and abdominal pain, and inflammation may occur anywhere in the GI tract, with the commonest sites affected being the ileum and colon. Chronic inflammation of the colonic and rectal mucosa with relapses and remissions of rectal bleeding characterize UC.
Association studies carried out by our lab and many others over the last several years have identified a number of susceptibility genes for IBD, and have shown that the innate immune system and the regulation of the Th1/Th2 balance are important in IBD pathophysiology. While several specific microbes have been suggested to play a role in CD pathogenesis, including measles virus and Mycobacteria paratuberculosis, the current consensus is that these conditions occur through an inappropriate immune response to the host’s commensal flora. Both our group and others have reported associations between IBD subgroups and the serum expression of antibodies to various microbial antigens including anti-Saccharomyces cerevisiae (ASCA, oligomannan)(Sandborn et al. 2001; Vermeire and Wild 2001), anti-Escherichia coli outer membrane porin C (Landers et al. 2002), anti-CD-related bacterial sequence from Pseudomonas fluorescens (Wei et al. 2002), and anti-flagellin (CBir1)(Lodes et al. 2004). We have observed that expression of serum antibodies to more than one of these antigens is associated with a more aggressive CD phenotype or sub-type (Mow et al. 2004), and that expression of the ASCA antibody, anti-ompC and anti-CIBR antibodies are familial (Mei 2006; Sutton et al. 2000; Takedatsu 2009) and are linked and associated with regions of the human genome (McGovern et al. 2009). These observations are consistent with the hypothesis that genetic variants generate defects in both the innate and in the adaptive immune system, leading to an imbalance between the regulatory cell and T-cell population, resulting in loss of tolerance to microbial antigens, expression of antibodies to these antigens, and more severe disease (Devlin 2007). Intestinal immunity relies on the balance between effector cells and regulatory cells and thus between responsiveness and tolerance. Interactions between the intestinal microflora, altered innate immune function and dysregulation of Th1 balance result in an imbalance in regulatory T and B cells that lead to an enhanced Th1 response characteristic of aggressive CD.
The importance of NK cells in early defense against infection suggests that KIR diversity has evolved as a consequence of repeated selection by pathogens acting on human NK response (Trowsdale 2001; Trowsdale and Parham 2004; Vilches and Parham 2002). Evidence suggests the KIR genes also evolved rapidly through recombination, in response to pathogen-driven selection (Canavez et al. 2001; Guethlein et al. 2002; Khakoo et al. 2000; Mager et al. 2001; Moretta et al. 2002; Rajalingam et al. 2001; Shilling et al. 1998; Trowsdale 2001; Trowsdale et al. 2001; Vilches and Parham 2002). Others have suggested that KIR are co-evolving with HLA, in which changes in HLA class I frequencies due to morbidity from infectious disease effects the evolution, via maintenance or expansion, of KIR repertoires to interact advantageously with HLA class I molecules (Khakoo et al. 2000). Other reports indicate that this diversity may also play a role in autoimmune disease and specifically IBD, with reported associations of KIR with several different autoimmune disorders, including ulcerative colitis (Jones et al. 2006), psoriatic arthritis (Nelson et al. 2004), scleroderma (Momot et al. 2004), sarcoidosis (Mizuki et al. 2000) and type I diabetes (van der Slik et al. 2003). In the majority of autoimmune disorders analyzed, an increase in the frequency of the stimulatory locus KIR2DS2 and/or an overall increase in the frequency of activating KIR, or an imbalance of KIR and their HLA class I ligands was observed in patients. Furthermore recent data have also suggested that a number of immune mediated conditions ‘share’ other susceptibility loci such as the IL23R (Burton et al. 2007; Cargill et al. 2007; Duerr et al. 2006). The KIR complex is located under a peak of linkage for CD (van Heel et al. 2003), and IBD has previously been associated with genetic variation at the HLA (Taylor 2007; Trachtenberg et al. 2000), and recent work has suggested an association of Crohn’s disease with KIR in combination with the HLA ligand (Zhang et al. 2008). In the present study, we investigated the relationship between the KIR loci and their HLA ligands with protection from or susceptibility to Crohn’s disease.
METHODS
Subjects
The study cohort consists of 1306 adult Caucasian CD patients, consisting of equal numbers of men and women. Of these, 512 are of self-reported Ashkenazi Jewish origin, while the remaining 794 reported as non-Jewish. The control population consists of 299 adult Caucasian individuals from the same geographic area as the disease cohort, of whom 96 are Ashkenazi Jewish and 203 non-Jewish origin, and also consisting of equal numbers of women and men. All individuals in the control population have no personal or family history of autoimmune disease.
KIR genotyping
To genotype the KIR loci in this CD cohort, we utilized our high throughput single nucleotide polymorphism (SNP)-based KIR genotyping assay developed using the SEQUENOM™ (San Diego, CA) MALDI-TOF mass spectrometer platform (Houtchens et al. 2007), which we have modified to improve efficiency and accuracy, including all known alleles at the time. The assay types for the presence or absence of 16 KIR loci and common alleles, including KIR2DL1*004, KIR2DS4*003/004/006/007 (form with truncated protein products which are not expressed on the cell surface (Middleton et al. 2007) and KIR2DS4*001 (expressed and capable of being membrane-bound). Briefly, 38 capture primer pairs (Table 1) are used to capture the region surrounding the SNPs to be queried, and 39 homogenous mass extend (HME) primers are used to differentiate individual SNP patterns for the 16 KIR genes on the MALDI-TOF platform. These assays were run using the KIR sequence alignment in the Immuno Polymorphism Database (IPD; http://www.ebi.ac.uk/ipd/kir) using version 12.19.06. Quality control measures included: 1) 4 negative and 4 positive (previously characterized and incorporating various combinations of all loci) controls per microchip; 2) 11% of samples (N=188) were blinded and typed in duplicate, yielding discrepancy rates of <0.02% discrepancy between typings.
Table 1.
Primer sequences for KIR analysis using MALDI-TOF.
| Assay Name | Forward Capture Primer | Reverse Capture Primer | hME Primer |
|---|---|---|---|
| 2DL1.2DS1.D1.S.tri | ACGTTGGATGAAGGCCAACTTCTCCATCA | ACGTTGGATGGTGAGTAACAGAACCGTAGC | GGTCCCTGCCAGGTCTTGC |
| 2DL1.D1.Gb | ACGTTGGATGACTTCTCCATCAGTCGCATGAC | ACGTTGGATGATGATCACGATGTCCAGAGG | GTCACTGGGAGCTGACA |
| 2DL1.D2.G.no004 | ACGTTGGATGCAGGGCCCAAGGTCAACG | ACGTTGGATGGACTTTGACCACTCGTAT | ATGCTTCGGCTCTTTCC |
| 2DL1.no005.2DL2.004.TC.S | ACGTTGGATGGTAATGGACCAAGAGTCTGC | ACGTTGGATGCGGGCCGAGGAGTACCTACCT | CGCTATTCGCTGTTCTGTT |
| 2DL2.001.2.3.D1.G | ACGTTGGATGCCTGCAATGTTGGTCAGATG | ACGTTGGATGGGAGCTGACAACTGATAGGG | CATGATGGGGTCTCCAA |
| 2DL2.004.TC.G | ACGTTGGATGGTAATGGACCAAGAGTCTGC | ACGTTGGATGGGCCGAGGAGTACCTACCT | GAAACAGAACAGCGAATA |
| 2DL2.D2.S | ACGTTGGATGGAGCTCCTATGACATGTACC | ACGTTGGATGGCCTGGAATGTTCCGTTGACCTTG | CCCTGCAGAGAACCTAC |
| 2DL3.2DL2.D1.S | ACGTTGGATGGAGTCCACAGAAAACCTTCCCTCC | ACGTTGGATGAGTGTCCTTAAACTTCCCTTCTC | CTTCTGATTTCACCAGG |
| 2DL3.TC.S.INT | ACGTTGGATGGTAACCCCAGACACCTGCATG | ACGTTGGATGCTGCTTCGTGAGACTTACTT | TCTCCTTCATCGCTGGTGCT |
| 2DL4.DO.G | ACGTTGGATGCCCTGAGCTCTACAACAGAA | ACGTTGGATGTGCCGACCACTCAGTGGG | TGGAACAGTTTCCTCAT |
| 2DL4.TC.G | ACGTTGGATGAGGTGACATACGCACAGTTG | ACGTTGGATGATCTGTTGAGGGTCTCTTGC | CACAGTTGGATCACTGC |
| 2DL5.D2.G | ACGTTGGATGGACTTTCCTCTGGGCCCTG | ACGTTGGATGTGACAGAAACAAGCAGTGGG | CCACGGAGGGACCTACA |
| 2DL5.TC.G | ACGTTGGATGCAAGACCCTCAGGAGGTGAC | ACGTTGGATGCTTGGGCCTCTGAGAAGGG | CACTGCGTTTTCACACAGA |
| 2DL5sub1 | ACGTTGGATGAGGACAAGCCCTTGCTGTCT | ACGTTGGATGCAAGACGAGAGCGACACA | GTCCTCCTCGAGGCACCACAG |
| 2DL5sub2 | ACGTTGGATGCCCTGAGCTCTACAACAA | ACGTTGGATGGACATGAGTCCTCTGACCTG | CGCTCCCCCATTGAGTGGTC |
| 2DL5sub3 | ACGTTGGATGCCCTGAGCTCTACAACAA | ACGTTGGATGGACATGAGTCCTCTGACCTG | GCAACCCCCTGGTGATC |
| 2DL5sub4 | ACGTTGGATGCCACGGAGGGACCTACAC | ACGTTGGATGGTGACAGAAACAAGCAGTGG | GTGAGTCATGGAGAGAGC |
| 2DL5sub5 | ACGTTGGATGGATCTTGGCTTAGCATTTGG | ACGTTGGATGCTGCGTTTTCACACAGAC | CTTCTCAGAGGCCCAAG |
| 2DL5sub6 | ACGTTGGATGTAAGGTGGCGCCTCCTTCTC | ACGTTGGATGCAAGACGAGAGCGACACA | AGCAAGGGCTTGTCCTG |
| 2DP1.D0.G | ACGTTGGATGGGGTTTAACAACTTCAGTCTGT | ACGTTGGATGTGTGCTGGGGTCACAGGGCC | ATTCTGTTGTAGAGCTCAG |
| 2DS1.D2.G | ACGTTGGATGATTCCAGGCCAACTTTCCTC | ACGTTGGATGATGGAGAGTCACGGAAAGAG | CATCTGTAGGTCCCTCC |
| 2DS2.D1.G | ACGTTGGATGCCTGCAATGTTGGTCAGATG | ACGTTGGATGAGAAGTTGGCCTTGGAGACC | GCACAGAGAGGGGAAGT |
| 2DS2.D2.G | ACGTTGGATGGTCTATATGAGAAACCTTC | ACGTTGGATGGGACAAGGTCACGCTCTCTC | CACGCTCTCTCCTGCCA |
| 2DS3.D1.G | ACGTTGGATGAAGGCCAACTTCTCCATCGG | ACGTTGGATGCTGTGATCACGATGTCCAG | CACTCCCCCTATCAGTT |
| 2DS3.D2.S | ACGTTGGATGAGGTCAACGGAACATTCCAGGCCG | ACGTTGGATGAAGAGCCGAAGCATCTGTAG | CATCTGTAGGTTCCTCC |
| 2DS4.D1.G | ACGTTGGATGAGAGACAGTCATCCTGCAATG | ACGTTGGATGATGGAGAAGTTGGCCTTGGA | GAAGTGCTCAAACATGACATC |
| 2DS4.D2.S | ACGTTGGATGGCATCAACGGAACATTCCAGGCC | ACGTTGGATGGAGCTCTGTGACGGAAACAA | TCGGCTCTTTCCGTGAC |
| 2DS4del.sub | ACGTTGGATGCGGTTCAGGCAGGAGAGAAT | ACGTTGGATGTTGACCACTCGTAGGGAGC | CCTTGTCCTGCAGCTCC |
| 2DS5.D1.G | ACGTTGGATGACACTTTGCGCCTCATTGGAG | ACGTTGGATGGTGAGTAACAGAACCGTAG | GACCGATGGAGAAGTTG |
| 2DS5.D2.G | ACGTTGGATGAGGCCCATGAACGTAGGCTCC | ACGTTGGATGAAGAGCCGAAGCATCTGTAG | CTCCGTGGGTGGCAGGG |
| 3DL1.TC.S | ACGTTGGATGCACTGCGTTTTCACACAGAG | ACGTTGGATGATGGGCAGGAGACAACTTTG | AGGCCCAAGACACCCCC |
| 3DL2.D1.G | ACGTTGGATGAAGGCCAACTTCTCCATCGG | ACGTTGGATGTGGGAGCTGACAACTGATAG | GTGAGGAACAGAACCATA |
| 3DL2.TC.G | ACGTTGGATGGATGAACAAGACCCTCAGGAGGTG | ACGTTGGATGTACACGCTGGTATCTGTT | GCCTCTGAGAAGGGCGA |
| 3DL3.D1.G | ACGTTGGATGGATCACTGAGGACCCCTTGC | ACGTTGGATGTCATGGGACCCATGGAATAG | AATAGTTGACCTGGGAACCC |
| 3DL3.D2.G | ACGTTGGATGAGAATGTGACCTTGTCCTGC | ACGTTGGATGCAGTGAGCCTAAGTTCACCG | GGATAGATGGTAAATGTCAAA |
| 3DP1.D2.G2 | ACGTTGGATGTGGGAAACCTTCTCTCTCAGCC | ACGTTGGATGGAGCTGCAGGACAAGGTCAC | CTCTCTCAGCCCAGCCG |
| 3DS1.3DL1.D1.S | ACGTTGGATGCAAGGCCAATTTCTCCATCG | ACGTTGGATGGGGAGCTGACAACTGATAGG | CTGTAGGTCCCTGCAAGGGCA |
| 3DS1.DO.S | ACGTTGGATGTGTGTAGTTCCCTGCATGTG | ACGTTGGATGTCATGCTATACAAAGAAGAC | AGGGCTCATGTTGAAGC |
| 3DS1.TC.S.INT | ACGTTGGATGAACTGCTATGATTAGCTTC | ACGTTGGATGGATGAAGGAGAAAGAAGAGGAGGA | GAATGTGCAGGTGTCTG |
HLA C1, C2 ligand groups and HLA Bw4 ligand group assays
Assays were developed using SEQUENOM™ AssayDesigner software for homogeneous MassEXTEND (hME) reactions and the primer extension products were analyzed on the MALDI-TOF mass spectrometer using SEQUENOM’S TyperAnalyzer v3.3.
The HLA-C alleles are classified as C1 or C2 KIR ligand groups, depending upon two amino acid (aa) positions encoded in exon 2. The C1 ligand group contains serine (AGC) at aa77 and asparagine (AAC) at aa80, while the C2 ligand group encodes asparagine (AAC) and lysine (AAA) at those positions. HLA-B alleles fall into two broad groups, Bw4 or Bw6, depending upon the presence of either glycine or arginine at aa83, respectively; only Bw4 is a KIR ligand for KIR3DL receptors. Amplicons containing exon 2 of HLA-C or HLA-B genes were produced using PCR primers designed in regions previously used for locus-specific amplification of HLA class I genes (Cereb et al. 1995).
Using SEQUENOM’S AssayDesigner v3.0.1, extend primers were designed to extend through the polymorphic positions encoding aa77 (AGC or AAC) and aa80 (AAC or AAA) on the HLA-C gene amplicon and aa83 (GGC or CGC) on the HLA-B gene amplicon (Table 2). The extend primers for HLA-C aa77 and 80 were multiplexed to run in one well. PCR reactions contained 2ng of DNA in a total volume of 5 µl in a 384-well format. Amplification conditions were modified from the original Sequenom protocols. All reactions contained 1.26X HotStarTaq buffer (Qiagen), 500µM dNTPs (BioLine), 600nM forward and reverse primers, 1.6mM MgCl2, 10% glycerol (HLA-C amplifications) or 15% glycerol (HLA-B amplifications) and 0.15 units HotStarTaq (Qiagen). Samples were cycled as follows: 1 cycle at 95°C for 15min; 45 cycles at 95°C for 20sec, 72°C for 1min; 1 cycle at 72°C for 3min and a hold at 4 °C.
Table 2.
Primer sequences for HLA-B (Bw4 ligand, Bw6) and HLA-C (C1, C2) ligand group analysis using MALDI-TOF.
| Primer | Sequence | Region |
|---|---|---|
| HLACP1 | ACG TTG GAT GGG AGG GAA DCG GCC TCT GSG GA | PCR HLA-C upstream primer |
| HLACP2 | ACG TTG GAT GGC CCC RGG CCG GGG TCA CTC AC | PCR HLA-C, HLA-B downstream primer |
| HLAPBforw | ACG TTG GAT GGG GAG GAG MGA GGG GAC CGC AG | PCR HLA-B upstream primer |
| hlaC80 | GGT TGT AGT AGC CGC GCA G | Extension primer HLA-C aa80 |
| hlaC77 | ACA GGC TGA CCG AGT GA | Extension primer HLA-C aa77 |
| hlaB83hME | GCT CTG GTT GTA GTA GC | Extension primer HLA-B aa83 |
Notes: The first 10 bp of the PCR primers are primer tags (ACGTTGGATG), specified by the SEQUENOM™ AssayDesigner. D = A, G, or T; S = G or C; R =A or G; M = A or C
After a shrimp alkaline phosphatase (SAP) step to neutralize unincorporated dNTPs, both extend primers for the HLA-C positions 77 and 80 were added to the PCR reactions, along with SEQUENOM’S CGT(ddC, ddG, ddT and dA) termination mix and enzyme. The extension primer hlaB83forw and the AGT termination mix (ddA, ddG, ddT, and dC) were added to the HLA–B amplicon wells. Thermocycling of the reactions resulted in the addition of nucleotides through the SNP site. After desalting, the products were spotted onto SpectroCHIPs; spectra were acquired using the SEQUENOM™ MassARRAY Compact and analyzed. The readout for for individuals homozygous for the C1 ligand is G (homozygous GG) at aa77 and C (homozygous CC) at aa80; the readout for individuals homozygous for the C2 ligand is A (homozygous AA) at aa77 and A (homozygous AA) at aa80. Individuals heterozygous for C1 and C2 will show GA at aa77 and CA at aa80. The readout for the HLA Bw4 and Bw6 assay is C (homozygous CC) for individuals that carry only Bw4 alleles, G (homozygous GG) for individuals with only Bw6 alleles, and CG for those individuals that carry both an Bw4 and a Bw6 allele (Table 3).
Table 3.
Expected HLA-C (C1, C2 ligand groups) and HLA-B (Bw4 ligand and Bw6) SNP nucleotide calls for homozygous and heterozygous samples using the HLA ligand MALDI-TOF assay.
| C1 Homozygous |
C2 Homozygous |
C1/C2 Heterozygous |
Bw4 Homozygous |
Bw6 Homozygous |
Bw4/Bw6 Heterozygous |
|
|---|---|---|---|---|---|---|
| HLA-C aa77 |
G (homozygous GG) |
A (homozygous AA) |
GA (heterozygous) |
|||
| HLA-C aa80 |
C (homozygous CC) |
A (homozygous AA) |
CA (heterozygous) |
|||
| HLA-B aa83 |
C (homozygous CC) |
G (homozygous GG) |
CG (heterozygous) |
Statistical analysis
Association analyses and test for fit to Hardy-Weinberg Equilibrium (HWE) proportions were performed using contingency table testing and a standard Chi-square measure. All p- values are uncorrected. Logistic regression was performed in R using the logit function in the VGAM package (Team 2008). A test for trend was accomplished using the Cochran-Armitage method (Agresti 2002). All association testing assumed an additive model.
RESULTS
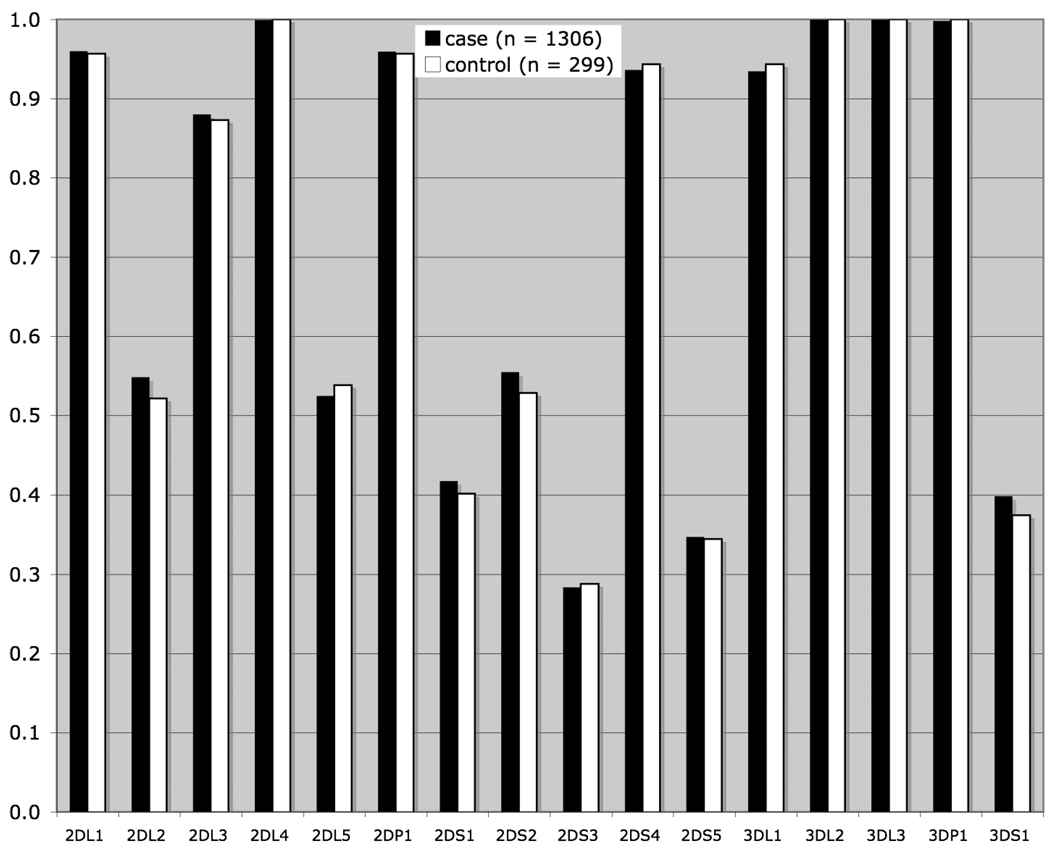
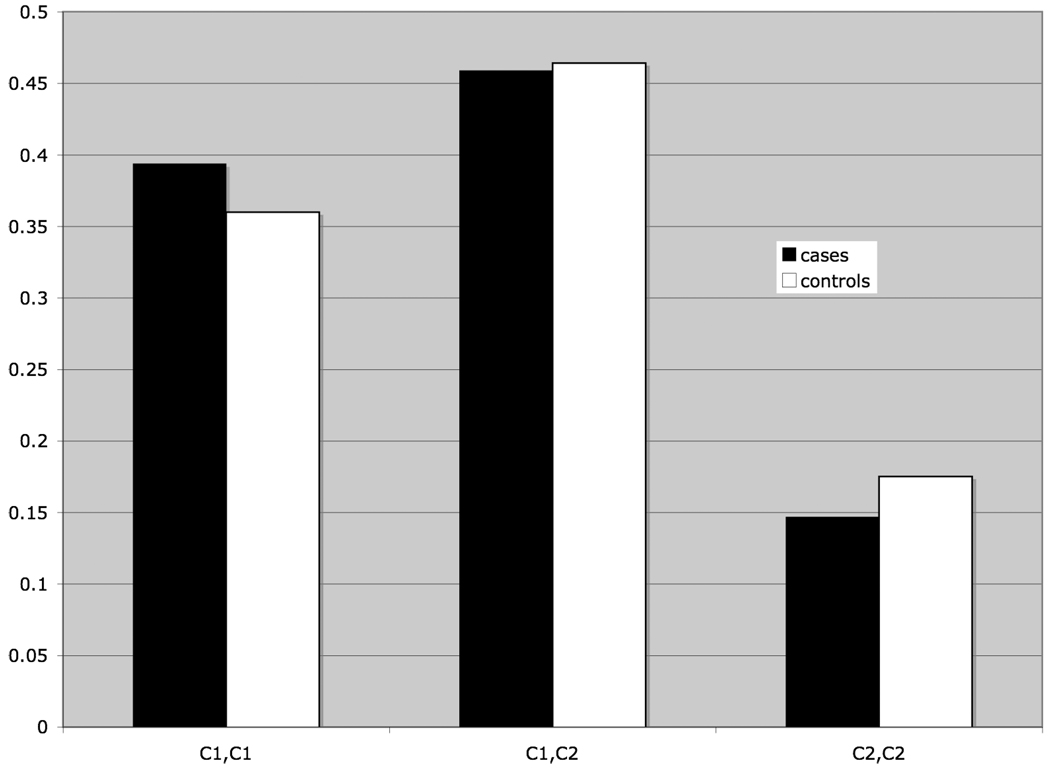
Frequency distributions for the KIR loci and HLA ligands were tested for heterogeneity between Ashkenazi Jewish and non-Jewish populations; no significant heterogeneity was observed, allowing us to analyze the case and control populations as a whole, thereby maximizing statistical power for this study. The frequency distribution of the sixteen KIR loci typed in patients and controls is shown in Figure 1 No significant differences between patient and control samples are observed. Likewise, there are no significant differences between patients and controls in the frequencies of the HLA C1 and C2 ligand genotypes (Figure 2) or for the Bw4 ligand (data not shown), although a slight trend is observed toward a higher frequency of HLA C1 ligand homozygosity in patients and a decrease in C2 ligand homozygosity
Fig 1.
Fig 2.
Analysis of the interactions between inhibitory KIR loci and their HLA class I ligands (KIR2DL1 and its ligand C2; KIR2DL2 with C1; KIR2DL3 with C1; KIR3DL1 with Bw4) indicated a protective effect (p <0.05) for KIR2DL2 in the absence of its ligand C1. No other receptor-ligand pair is significantly associated with disease. The frequency distribution for the possible receptor-ligand combinations between KIR2DL2 and C1 is given in Table 4. While the overall frequency of the associated KIR-HLA genotype combination is relatively low, the magnitude of the deviation from the expected value for the control population (n = 22) is nearly 50%, with an enrichment of KIR2DL2 in the presence of C2 homozygosity. No significant differences are observed for the combined receptor-ligand genotypes between the Jewish and non-Jewish populations.
Table 4.
Carrier frequencies for KIR2DL2 and HLA-C ligand combinations in CD cases and controls.
| KIR2DL2 | C1 Ligand | Case frequency (n) |
Control frequency (n) |
p-value | OR (95% CI) |
|---|---|---|---|---|---|
| present | present | 0.476 (614) | 0.418 (124) | ns | --- |
| present | absent | 0.067 (87) | 0.101 (30) | 0.046 | 0.64 (0.42 – 0.99) |
| absent | present | 0.376 (486) | 0.407 (121) | ns | --- |
| absent | absent | 0.081 (104) | 0.074 (22) | ns | --- |
Note: C1 ligand absent = C2 ligand homozygosity.
C1 ligand present = C1 ligand homozygosity (C1/C1) or C1 heterozygosity (C1/C2).
Given that KIR2DL2 and KIR2DL3 segregate as alleles, analysis of their fit to expected distributions under Hardy-Weinberg Equilibrium (HWE) with regard to the HLA-C ligand was investigated (Table 5). Testing of fit to expectations under HWE can often reveal evidence of disease associations. Given that we had previously identified, upon conditioning on the presence of its HLA ligand, C1, an association KIR 2DL2, we sought to investigate whether genotypic distributions for KIR2DL2 and KIR2DL3 conformed to expectations under HWE when the ligand interaction was taken into account. While all the genotypic distributions conform to expectations under HWE in controls both with and without the C1 ligand, in cases without C1 (i.e. C2 homozygous), there is a significant (p =0.002) deviation from HWE, with a reduction of heterozygotes in this group. Although not statistically significant, a similar trend is observed when the Jewish and non-Jewish populations are analyzed separately (data not shown). A potential exists whereby subdividing the population based on the HLA–C data might create a population substructure resulting in skewed frequency distributions for the KIR, but this should be evident in the control population. However, deviations from HWE were observed only within the patient population, suggesting a role in disease predisposition.
Table 5.
Observed and expected values for KIR2DL2, KIR2DL3 and HLA ligand genotypic distributions under HWE.
| KIR genotype: | KIR2DL2, KIR2DL2 | KIR2DL2, KIR2DL3 | KIR2DL3, KIR2DL3 | |||
|---|---|---|---|---|---|---|
| C ligand genotype | ||||||
| Cases: | 0bs/exp | 0bs/exp | 0bs/exp | p-value | ||
| C1, C1 | 48/51 | 229/222 | 237/240 | ns | ||
| C1, C2 | 81/77 | 268/276 | 250/246 | ns | ||
| C2, C2 | 27/18 | 63/81 | 101/92 | 0.002 | ||
| Controls: | ||||||
| C1, C1 | 15/11 | 39/47 | 53/49 | ns | ||
| C1, C2 | 17/14 | 55/61 | 68/65 | ns | ||
| C2, C2 | 6/6 | 24/24 | 22/22 | ns | ||
Comparing the association of KIR2DL2 and KIR2DL3 homozygotes and heterozygotes in the presence and absence of HLA ligand (Table 6) confirms that it is only the heterozygote combination which is important in protection from CD. In addition to the protective effect (OR = 0.44, CI = 0.22–0.87; p = 0.018) accorded to this genotype in the absence of the HLA-C1 ligand (C2 homozygosity), in the presence of the ligand, the KIR2DL2, KIR2DL3 heterozygote genotype is predisposing in the presence of C1 ligand (C1 homozygosity or C1, C2 heterozygosity; OR = 1.34, CI = 1.03–4.53; p = 0.031). Given that this analysis is a follow-up to the finding with regard to deviations from HWE, no correction for multiple tests was applied to the p- values. This result is confirmed in a logistic regression model in which interaction terms are included for the KIR genotype as well as the HLA ligand (p < 0.05). Furthermore, a test for trend of the C ligand genotypes with KIR2DL2, KIR2DL3 heterozygosity indicates an intermediate effect of C1/C2 heterozygotes, such that C2 has a moderating protective effect over the C1 susceptibility effect. Accordingly, in the presence of C2 ligand only, the KIR2DL2, KIR2DL3 heterozygote combination is protective.
Table 6.
KIR2DL2, KIR2DL3 genotypic distributions stratified by HLA C1 ligand status.
| KIR genotype | HLA C1 ligand | case freq (n) | control freq (n) | p-value | OR (95% CI) |
|---|---|---|---|---|---|
| 2DL2,2DL2 | present | 0.099 (128) | 0.108 (32) | ns | --- |
| 2DL2,2DL2 | absent | 0.021 (27) | 0.020 (2) | ns | --- |
| 2DL2,2DL3 | present | 0.376 (486) | 0.310 (92) | 0.031 | 1.34 (1.02– 1.76) |
| 2DL2,2DL3 | absent | 0.046 (60) | 0.081 (24) | 0.017 | 0.55 (0.34– 0.91) |
| 2DL3,2DL3 | present | 0.376 (486) | 0.407 (121) | ns | --- |
| 2DL3,2DL3 | absent | 0.081 (104) | 0.074 (22) | ns | --- |
DISCUSSION
A substantial body of evidence is accumulating which shows an association of the KIR genes and their HLA ligands in autoimmune diseases (Carrington and Martin 2006; Khakoo and Carrington 2006). Notably, the most commonly associated KIR genes cited are the inhibitory KIR2DL2 and/or the stimulatory KIR2DS2, which are in near complete linkage disequilibrium. Here we present results which show that susceptibility to CD is mediated in part by KIR2DL2, KIR2DL3 heterozygosity, and that the direction of the association (i.e. predisposing vs. protective) is related to HLA–C ligand status. While previous studies have shown a hierarchical effect of the stimulatory and inhibitory KIR in autoimmune disease (Nelson et al. 2004), these results show for the first time that disease susceptibility may be related to heterozygosity at a specific KIR locus, and that HLA ligand genotype determines the relative effect of the KIR genotype. It is important to note that due to the high level of linkage disequilibrium between KIR2DL2 and KIR2DS2, it is impossible to distinguish their effects in this analysis. Thus, any association with KIR2DL2 and/or KIR2DS2 may be attributable to either or both of these genes. While the B haplotype associated KIR2DL2 segregates as an allele of KIR2DL3 (which is associated with the A haplotype), KIR2DS2 has no corresponding allelic partner. KIR2DL2/KIR2DS2 serve as markers for the centromeric portion of the B haplotype. In this study, individuals with a KIR genotype that includes KIR2DL2/KIR2DS2 have three stimulatory KIR genes on average (not including KIR2DS4, which is found on both the A and B haplotypes and may be of a common truncated variant) compared to only one stimulatory KIR gene for those without these loci. It is conceivable that the observed KIR2DL2/KIR2DS2 association is merely a feature of a more stimulatory KIR genotype. However, there is no evidence in this study that the overall number of stimulatory and/or inhibitory KIR is related to disease status. Analysis on the basis of KIR haplotype status also indicated no significant relationship with the KIR inhibitory (A/A) haplotype versus the stimulatory (B/x) haplotype (any haplotypic combination which has between one and four more stimulatory KIR genes than the A/A haplotype, as defined in http://www.ebi.ac.uk/ipd/kir) and disease. Another important consideration is the pattern of LD whereby 90% of individuals who bear KIR2DL3 also possess KIR2DL1, compared to only two-thirds of those who do not have KIR2DL3. Given that KIR2DL1 binds HLA ligand C2, it is possible that this very strong inhibitory interaction could account in part for the protective effect observed for the KIR2DL2, KIR2DL3 heterozygous combination in C2 homozygotes, as well as the moderation of the predisposing effect observed in C1, C2 heterozygotes.
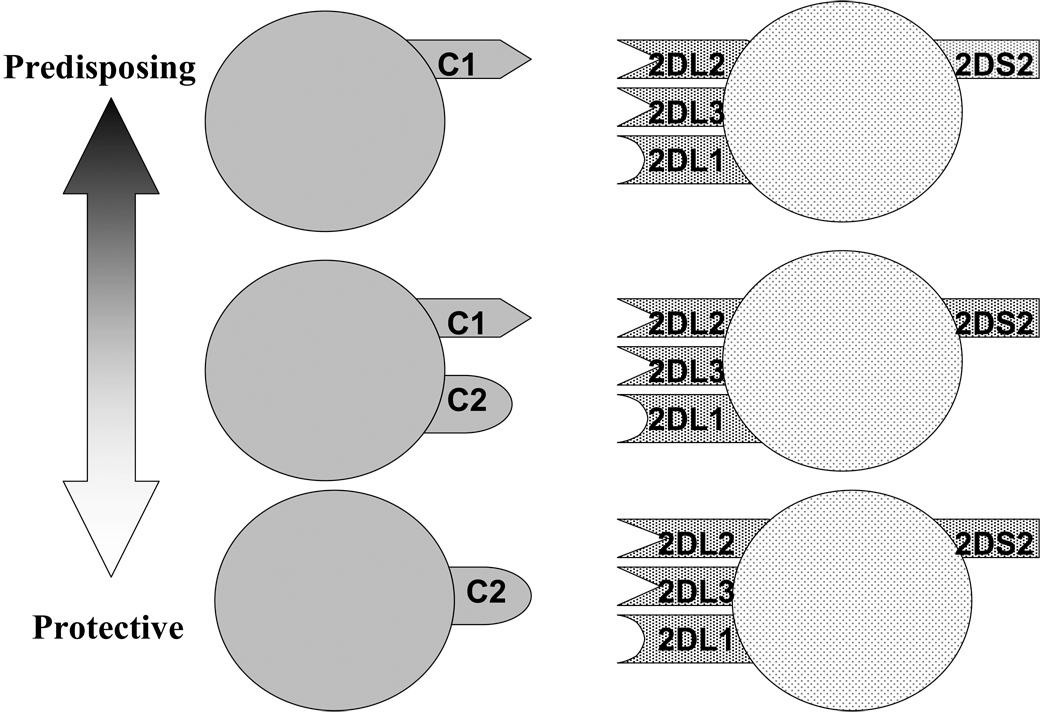
A model illustrating these various interactions and their potential impact upon disease susceptibility is shown in Figure 3. The finding that heterozygosity for the KIR2DL2 and KIR2DL3 genes may be important in disease may reflect the particularly delicate balance in immune responsiveness in the CD patient gut. Interaction of the inhibitory KIR with the HLA ligand functions not only to inhibit NK cell activation, but also serves to prime the particular NK cell for activation in the absence of self (Kim et al. 2005; Kim et al. 2008). The variegated expression pattern of the KIR gene products (Young and Uhrberg 2002) may produce KIR2DL2, KIR2DL3 heterozygous NK cells in individuals whose only inhibitory receptor is the weakly inhibitory KIR2DL3, resulting in a strong activation potential. The presence of the activating KIR2DS2 renders the cell potentially more reactive. Alternatively, in the absence of HLA ligand C1, one might imagine that NK cell activation will be strongly inhibited (except in the case of down regulation of HLA class I on the target cells), as only the strongly inhibitory KIR2DL1 bearing cells will be primed for response. A microbial trigger in CD could become the tipping point between NK cell activation sufficient to keep infection in check versus an ultimately pathological immune response resulting in ongoing inflammation in the gut. Too little NK cell activation therefore, and one risks viral pathology, while too much activation may trigger an augmented inflammatory response and disease. The KIR2DL2/KIR3DL3 heterozygote combination in association with C2 ligand may therefore be the intermediate “sweetspot” of NK activation potential with regard to CD. These potential scenarios for the action of NK cells in CD are likely to occur in the context of other risk factors, both environmental and genetic. Nonetheless, these findings demonstrate a complex role for the KIR genes in CD, and support the general notion that any examination of the impact of the KIR genes in disease must consider the possibility of multiple receptors and ligands acting in concert.
Fig 3.
ACKNOWLEDGMENTS
This study was supported by a National Institutes of Health research grant no. U01 AI067068 (JAH, MBL, KS, HAE and EAT). Assay development at Children’s Hospital Oakland Research Institute was supported by National Institutes of Health grant no. R21 A-03-107. Sample collection and clinical identification at Cedars Sinai Medical Center was supported by research grants AI 067068, IBD Program Project Grant grant DK 046763 (KDT, LM, TH, DPBM and JIR), General Clinical Research Center grant MO1 RR00069, Diabetes Endocrinology Research Center grant DK 063491 and the Cedars-Sinai Board of Governors' in Medical Genetics (JIR). The authors would also like to thank Kristine Munir for her administrative assistance.
REFERENCES
- Agresti A. Categorical Data Analysis. Second Edition. Wiley; 2002. [Google Scholar]
- Burton PR, Clayton DG, Cardon LR, Craddock N, Deloukas P, Duncanson A, Kwiatkowski DP, McCarthy MI, Ouwehand WH, Samani NJ, Todd JA, Donnelly P, Barrett JC, Davison D, Easton D, Evans DM, Leung HT, Marchini JL, Morris AP, Spencer CC, Tobin MD, Attwood AP, Boorman JP, Cant B, Everson U, Hussey JM, Jolley JD, Knight AS, Koch K, Meech E, Nutland S, Prowse CV, Stevens HE, Taylor NC, Walters GR, Walker NM, Watkins NA, Winzer T, Jones RW, McArdle WL, Ring SM, Strachan DP, Pembrey M, Breen G, St Clair D, Caesar S, Gordon-Smith K, Jones L, Fraser C, Green EK, Grozeva D, Hamshere ML, Holmans PA, Jones IR, Kirov G, Moskivina V, Nikolov I, O'Donovan MC, Owen MJ, Collier DA, Elkin A, Farmer A, Williamson R, McGuffin P, Young AH, Ferrier IN, Ball SG, Balmforth AJ, Barrett JH, Bishop TD, Iles MM, Maqbool A, Yuldasheva N, Hall AS, Braund PS, Dixon RJ, Mangino M, Stevens S, Thompson JR, Bredin F, Tremelling M, Parkes M, Drummond H, Lees CW, Nimmo ER, Satsangi J, Fisher SA, Forbes A, Lewis CM, Onnie CM, Prescott NJ, Sanderson J, Matthew CG, Barbour J, Mohiuddin MK, Todhunter CE, Mansfield JC, Ahmad T, Cummings FR, Jewell DP, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canavez F, Young NT, Guethlein LA, Rajalingam R, Khakoo SI, Shum BP, Parham P. Comparison of chimpanzee and human leukocyte Ig-like receptor genes reveals framework and rapidly evolving genes. J Immunol. 2001;167:5786–5794. doi: 10.4049/jimmunol.167.10.5786. [DOI] [PubMed] [Google Scholar]
- Cargill M, Schrodi SJ, Chang M, Garcia VE, Brandon R, Callis KP, Matsunami N, Ardlie KG, Civello D, Catanese JJ, Leong DU, Panko JM, McAllister LB, Hansen CB, Papenfuss J, Prescott SM, White TJ, Leppert MF, Krueger GG, Begovich AB. A large-scale genetic association study confirms IL12B and leads to the identification of IL23R as psoriasis-risk genes. Am J Hum Genet. 2007;80:273–290. doi: 10.1086/511051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carrington M, Martin MP. The impact of variation at the KIR gene cluster on human disease. Curr Top Microbiol Immunol. 2006;298:225–257. doi: 10.1007/3-540-27743-9_12. [DOI] [PubMed] [Google Scholar]
- Cereb N, Maye P, Lee S, Kong Y, Yang SY. Locus-specific amplification of HLA class I genes from genomic DNA: locus-specific sequences in the first and third introns of HLA-A, -B, and -C alleles. Tissue Antigens. 1995;45:1–11. doi: 10.1111/j.1399-0039.1995.tb02408.x. [DOI] [PubMed] [Google Scholar]
- Devlin SM, Yang H, Ippoliti A, Taylor KD, Landers CJ, Su X, Abreu MT, Papadakis K, Vasiliauskas E, Melmed G, Fleshner P, Mei L, Rotter JI, Targan SR. NOD2/CARD15 variants are significantly associated with sero-reactivity to microbial antigens in patients with Crohn's disease and thair unaffected relatives. Gastroenterology. 2007;132:576–586. doi: 10.1053/j.gastro.2006.11.013. [DOI] [PubMed] [Google Scholar]
- Duerr RH, Taylor KD, Brant SR, Rioux JD, Silverberg MS, Daly MJ, Steinhart AH, Abraham C, Regueiro M, Griffiths A, Dassopoulos T, Bitton A, Yang H, Targan S, Datta LW, Kistner EO, Schumm LP, Lee AT, Gregersen PK, Barmada MM, Rotter JI, Nicolae DL, Cho JH. A genome-wide association study identifies IL23R as an inflammatory bowel disease gene. Science. 2006;314:1461–1463. doi: 10.1126/science.1135245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guethlein LA, Flodin LR, Adams EJ, Parham P. NK cell receptors of the orangutan (Pongo pygmaeus): a pivotal species for tracking the coevolution of killer cell Ig-like receptors with MHC-C. J Immunol. 2002;169:220–229. doi: 10.4049/jimmunol.169.1.220. [DOI] [PubMed] [Google Scholar]
- Houtchens KA, Nichols RJ, Ladner MB, Boal HE, Sollars C, Geraghty DE, Davis LM, Parham P, Trachtenberg EA. High-throughput killer cell immunoglobulin-like receptor genotyping by MALDI-TOF mass spectrometry with discovery of novel alleles. Immunogenetics. 2007;59:525–537. doi: 10.1007/s00251-007-0222-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DC, Edgar RS, Ahmad T, Cummings JR, Jewell DP, Trowsdale J, Young NT. Killer Ig-like receptor (KIR) genotype and HLA ligand combinations in ulcerative colitis susceptibility. Genes Immun. 2006;7:576–582. doi: 10.1038/sj.gene.6364333. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Carrington M. KIR and disease: a model system or system of models? Immunol Rev. 2006;214:186–201. doi: 10.1111/j.1600-065X.2006.00459.x. [DOI] [PubMed] [Google Scholar]
- Khakoo SI, Rajalingam R, Shum BP, Weidenbach K, Flodin L, Muir DG, Canavez F, Cooper SL, Valiante NM, Lanier LL, Parham P. Rapid evolution of NK cell receptor systems demonstrated by comparison of chimpanzees and humans. Immunity. 2000;12:687–698. doi: 10.1016/s1074-7613(00)80219-8. [DOI] [PubMed] [Google Scholar]
- Kim S, Poursine-Laurent J, Truscott SM, Lybarger L, Song YJ, Yang L, French AR, Sunwoo JB, Lemieux S, Hansen TH, Yokoyama WM. Licensing of natural killer cells by host major histocompatibility complex class I molecules. Nature. 2005;436:709–713. doi: 10.1038/nature03847. [DOI] [PubMed] [Google Scholar]
- Kim S, Sunwoo JB, Yang L, Choi T, Song YJ, French AR, Vlahiotis A, Piccirillo JF, Cella M, Colonna M, Mohanakumar T, Hsu KC, Dupont B, Yokoyama WM. HLA alleles determine differences in human natural killer cell responsiveness and potency. Proc Natl Acad Sci U S A. 2008;105:3053–3058. doi: 10.1073/pnas.0712229105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Landers CJ, Cohavy O, Misra R, Yang H, Lin YC, Braun J, Targan SR. Selected loss of tolerance evidenced by Crohn's disease-associated immune responses to auto- and microbial antigens. Gastroenterology. 2002;123:689–699. doi: 10.1053/gast.2002.35379. [DOI] [PubMed] [Google Scholar]
- Lodes MJ, Cong Y, Elson CO, Mohamath R, Landers CJ, Targan SR, Fort M, Hershberg RM. Bacterial flagellin is a dominant antigen in Crohn disease. J Clin Invest. 2004;113:1296–1306. doi: 10.1172/JCI20295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mager DL, McQueen KL, Wee V, Freeman JD. Evolution of natural killer cell receptors: coexistence of functional Ly49 and KIR genes in baboons. Curr Biol. 2001;11:626–630. doi: 10.1016/s0960-9822(01)00148-8. [DOI] [PubMed] [Google Scholar]
- McGovern DP, Taylor KD, Landers C, Derkowski C, Dutridge D, Dubinsky M, Ippoliti A, Vasiliauskas E, Mei L, Mengesha E, King L, Pressman S, Targan SR, Rotter JI. MAGI2 genetic variation and inflammatory bowel disease. Inflamm Bowel Dis. 2009;15:75–83. doi: 10.1002/ibd.20611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mei L, Targan SR, Landers C, Dutridge D, Ippoloti A, Vasiliauskas EA, Papadakis KA, Fleshner P, Rotter JI, Yang H. Familial expression of anti-Escherichia coli outer membrane porin C (OmpC) in relatives of patients with Crohn’s disease. Gastroenterology. 2006;130:1078–1085. doi: 10.1053/j.gastro.2006.02.013. [DOI] [PubMed] [Google Scholar]
- Middleton D, Gonzalez A, Gilmore PM. Studies on the Expression of the Deleted KIR2DS4*003 Gene Product and Distribution of KIR2DS4 Deleted and Nondeleted Versions in Different Populations. Hum Immunol. 2007;68:128–134. doi: 10.1016/j.humimm.2006.12.007. [DOI] [PubMed] [Google Scholar]
- Mizuki M, Eklund A, Grunewald J. Altered expression of natural killer cell inhibitory receptors (KIRs) on T cells in bronchoalveolar lavage fluid and peripheral blood of sarcoidosis patients. Sarcoidosis Vasc Diffuse Lung Dis. 2000;17:54–59. [PubMed] [Google Scholar]
- Momot T, Koch S, Hunzelmann N, Krieg T, Ulbricht K, Schmidt RE, Witte T. Association of killer cell immunoglobulin-like receptors with scleroderma. Arthritis Rheum. 2004;50:1561–1565. doi: 10.1002/art.20216. [DOI] [PubMed] [Google Scholar]
- Moretta A, Bottino C, Mingari MC, Biassoni R, Moretta L. What is a natural killer cell? Nat Immunol. 2002;3:6–8. doi: 10.1038/ni0102-6. [DOI] [PubMed] [Google Scholar]
- Mow WS, Lo SK, Targan SR, Dubinsky MC, Treyzon L, Abreu-Martin MT, Papadakis KA, Vasiliauskas EA. Initial experience with wireless capsule enteroscopy in the diagnosis and management of inflammatory bowel disease. Clin Gastroenterol Hepatol. 2004;2:31–40. doi: 10.1016/s1542-3565(03)00289-1. [DOI] [PubMed] [Google Scholar]
- Nelson GW, Martin MP, Gladman D, Wade J, Trowsdale J, Carrington M. Cutting edge: heterozygote advantage in autoimmune disease: hierarchy of protection/susceptibility conferred by HLA and killer Ig-like receptor combinations in psoriatic arthritis. J. Immunol. 2004;173:4273–4276. doi: 10.4049/jimmunol.173.7.4273. [DOI] [PubMed] [Google Scholar]
- Rajalingam R, Hong M, Adams EJ, Shum BP, Guethlein LA, Parham P. Short KIR haplotypes in pygmy chimpanzee (Bonobo) resemble the conserved framework of diverse human KIR haplotypes. J Exp Med. 2001;193:135–146. doi: 10.1084/jem.193.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandborn WJ, Loftus EV, Jr, Colombel JF, Fleming KA, Seibold F, Homburger HA, Sendid B, Chapman RW, Tremaine WJ, Kaul DK, Wallace J, Harmsen WS, Zinsmeister AR, Targan SR. Evaluation of serologic disease markers in a population-based cohort of patients with ulcerative colitis and Crohn's disease. Inflamm Bowel Dis. 2001;7:192–201. doi: 10.1097/00054725-200108000-00003. [DOI] [PubMed] [Google Scholar]
- Shilling HG, Lienert-Weidenbach K, Valiante NM, Uhrberg M, Parham P. Evidence for recombination as a mechanism for KIR diversification. Immunogenetics. 1998;48:413–416. doi: 10.1007/s002510050453. [DOI] [PubMed] [Google Scholar]
- Sutton CL, Yang H, Li Z, Rotter JI, Targan SR, Braun J. Familial expression of anti-Saccharomyces cerevisiae mannan antibodies in affected and unaffected relatives of patients with Crohn's disease. Gut. 2000;46:58–63. doi: 10.1136/gut.46.1.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takedatsu H, Taylor KD, Mei L, McGovern DPB, Landers CJ, Gonsky R, Cong Y, Vasiliauskas EA, Ippoliti A, Elson CO, Rotter JI, Targan SR. Linkage of CD-related serological phenotypes: NFKB1 haplotypes are associated with anti-CBir1 and ASCA, and show reduced NF-kB activation. Gut. 2009;58:60–67. doi: 10.1136/gut.2008.156422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Targan SSF, Karp LC. Inflammatory Bowel Disease: From Bench to Bedside. Kluwer Academic Publishers; 2003. [Google Scholar]
- Targen SR SF, Karp LC. Inflammatory Bowel Disease: From Bench to Bedside. Kluwer Academic Publishers; 2003. [Google Scholar]
- Taylor KD, Yang H, Rotter JI. Emery and Rimoin’s Principles and Practice of Medical Genetics. London: Churchill Livingstone; 2007. Inflammatory bowel disease; pp. 1549–1582. [Google Scholar]
- Team RDC. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2008. [Google Scholar]
- Trachtenberg E, Yang H, Hayes E, Vinson M, Li S, Targan S, Tyan D, Erlich H, Rotter J. HLA class II haplotype associations with Inflammatory Bowel Disease in Jewish (Ashkenazi) and non-Jewish caucasian populations. Human Immunology. 2000;61:326–333. doi: 10.1016/s0198-8859(99)00134-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trowsdale J. Genetic and functional relationships between MHC and NK receptor genes. Immunity. 2001;15:363–374. doi: 10.1016/s1074-7613(01)00197-2. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Barten R, Haude A, Stewart CA, Beck S, Wilson MJ. The genomic context of natural killer receptor extended gene families. Immunol Rev. 2001;181:20–38. doi: 10.1034/j.1600-065x.2001.1810102.x. [DOI] [PubMed] [Google Scholar]
- Trowsdale J, Parham P. Mini-review: defense strategies and immunity-related genes. Eur J Immunol. 2004;34:7–17. doi: 10.1002/eji.200324693. [DOI] [PubMed] [Google Scholar]
- van der Slik AR, Roeleman PC, Verduijn W, Jan Bruining G, Roep BO, Giphart MJ. KIR in type 1 diabetes: Disparate distribution of activating and inhibitory natural killer cell receptors in patients versus HLA-matched control subjects. Diabetes. 2003;52:2639–2642. doi: 10.2337/diabetes.52.10.2639. [DOI] [PubMed] [Google Scholar]
- van Heel DA, Dechairo BM, Dawson G, McGovern DP, Negoro K, Carey AH, Cardon LR, Mackay I, Jewell DP, Lench NJ. The IBD6 Crohn's disease locus demonstrates complex interactions with CARD15 and IBD5 disease-associated variants. Hum Mol Genet. 2003;12:2569–2575. doi: 10.1093/hmg/ddg281. [DOI] [PubMed] [Google Scholar]
- Vermeire S, Wild G. RE: Development of an assay for antibodies to Saccharomyces cerevisiae: Easy, cheap and specific for Crohn's disease. Can J Gastroenterol. 2001;15:841–842. [PubMed] [Google Scholar]
- Vilches C, Parham P. KIR: Diverse, rapidly evolving receptors of innate and adaptive immunity. Annu Rev Immunol. 2002;20:217–251. doi: 10.1146/annurev.immunol.20.092501.134942. [DOI] [PubMed] [Google Scholar]
- Wei B, Huang T, Dalwadi H, Sutton CL, Bruckner D, Braun J. Pseudomonas fluorescens encodes the Crohn's disease-associated I2 sequence and T-cell superantigen. Infect Immun. 2002;70:6567–6575. doi: 10.1128/IAI.70.12.6567-6575.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young NT, Uhrberg M. KIR expression shapes cytotoxic repertoires: a developmental program of survival. Trends Immunol. 2002;23:71–75. doi: 10.1016/s1471-4906(01)02113-5. [DOI] [PubMed] [Google Scholar]
- Zhang HX, Li JC, Liu ZJ. [Relationship between expression of inhibitory killer cell immunoglobulin-like receptor HLA-Cw ligand and susceptibility to inflammatory bowel disease] Zhonghua Yi Xue Za Zhi. 2008;88:3108–3111. [PubMed] [Google Scholar]