Abstract
CD81 is a tetraspanin cell surface protein that regulates CD19 expression in B lymphocytes and enables hepatitis C virus infection of human cells. Immunohistologic analysis in normal hematopoietic tissue showed strong staining for CD81 in normal germinal center B cells, a cell type in which its increased expression has not been previously recognized. High-dimensional flow cytometry analysis of normal hematopoietic tissue confirmed that among B and T cell subsets, germinal center B cells showed the highest level of CD81 expression. In over 800 neoplastic tissue samples, its expression was also found in a majority of non-Hodgkin lymphomas. Staining for CD81 was rarely seen in multiple myeloma, Hodgkin lymphoma, or myeloid leukemia. In hierarchical cluster analysis of diffuse large B cell lymphoma, staining for CD81 was most similar to other germinal center B cell-associated markers, particularly LMO2. By flow cytometry, CD81 was expressed in diffuse large B cell lymphoma cells independent of the presence or absence of CD10, another germinal center B cell marker. The detection of CD81 in routine biopsy samples and its differential expression in lymphoma subtypes, particularly diffuse large B cell lymphoma, warrants further study to assess CD81 expression and its role in the risk stratification of diffuse large B cell lymphoma patients.
Keywords: CD81, lymphoma, tissue microarray
INTRODUCTION
CD81 is a tetraspanin cell surface protein known to play an important role in multiple cellular interactions by associating with other tetraspanins and partner proteins on the cell membrane [1]. In mature B cells, CD81 regulates CD19 expression and associates with CD19 and CD21 to lower the threshold of B cell activation via the B cell receptor complex [2,3]. Furthermore, the hepatitis C virus is well-known to infect human cells by using CD81 as a cell surface receptor for entry into the cell [4]. The hepatitis C viral envelope glycoprotein E2 binds to CD81 and modulates the properties of CD81. In B lymphocytes, this interaction may help explain the observed epidemiological associations among hepatitis C infection, lymphoproliferative disorders, and non-Hodgkin lymphomas [5]. Binding of E2 to CD81 has been shown to activate naïve B lymphocyte proliferation as well as induce hypermutation of the variable region of immunoglobulin genes in B cells [6,7]. Similarly, ligation of CD81 with the costimulatory molecule CD28 leads to naïve T cell proliferation, which may contribute to the chronic inflammatory environment seen in hepatitis C infection [8].
Previously, gene expression profiling studies of diffuse large B cell lymphoma defined prognostic subgroups within this heterogeneous disease [9,10,11,12]. Subsequently, we described a multivariate model of six genes that predicted survival in diffuse large B cell lymphoma patients [13], the prognostic value of which remained significant in the immunochemotherapy era [14]. Among these six genes, LMO2 expression emerged as the strongest single predictor of superior outcome [13]. We therefore characterized the distribution of the LMO2 protein, whose expression in a germinal center-associated manner was also found to correlate with improved survival in patients with diffuse large B cell lymphoma [15,16]. We also identified CD81 as a potential marker of prognostic significance in patients with diffuse large B cell lymphoma using the supervised principal component method [17]. This identification was accomplished by statistical analysis of multiple diffuse large B cell lymphoma gene profiling studies [9,10,11,12,18], which identified CD81 alongside previously described genes LMO2, MHC class II and BCL6 [13,19]. The potential association of CD81 with LMO2 and other markers relevant to diffuse large B cell lymphoma prognosis further suggests a role for CD81 in lymphoma pathogenesis.
Although the role of CD81 in B cells has been investigated in the context of hepatitis C infection, the tissue distribution pattern of the CD81 protein in hematopoietic tissue has not been previously explored. Given the important role of CD81 in B cell activation and its potential role in diffuse large B cell lymphoma prognosis, we undertook this study to characterize the expression of CD81 protein in normal and neoplastic hematopoietic tissues. We also compared its expression pattern in diffuse large B cell lymphoma cases to other well-characterized germinal center and non-germinal center markers.
MATERIALS AND METHODS
Tissue samples
Formalin-fixed paraffin-embedded tissue samples of normal and neoplastic hematolymphoid cases were obtained from the archives of the Departments of Pathology, Stanford University Medical Center, Stanford, California. Institutional Review Board approval was obtained for these studies. The cases were studied by immunohistochemistry on tissue microarrays. Whole sections were also evaluated to confirm findings seen on the tissue microarray sections, including cases of follicular lymphoma, diffuse large B cell lymphoma, and small lymphocytic lymphoma/chronic lymphocytic leukemia, the latter with known IgH V hypermutation status. Hematolymphoid neoplasia were classified according to the current World Health Organization scheme [20].
Fresh tonsil, thymus, normal bone marrow, and bone marrow involved by plasma cell myeloma were also obtained from Stanford University Medical Center. Portions of the tissue were frozen and cryostat sections were obtained for immunohistochemistry. In addition, fresh tonsil, thymus, and bone marrow samples were washed twice with phosphate-buffered saline, then once with RPMI1640 media containing 10% fetal calf serum. Single cell suspensions were prepared by passing the tissue through nylon mesh and collecting the cells in phosphate-buffered saline. Cells were used directly or washed and frozen in fetal calf serum containing 10% dimethyl sulfoxide.
Single cell suspensions were also prepared from tumor biopsies from 8 diffuse large B cell lymphoma patients at the Norwegian Radium Hospital between 1988 and 1992 and stored in liquid nitrogen in cryotubes as described previously [21]. The biopsies were obtained at the time of diagnosis, prior to any treatment. Each individual cryotube was thawed, pelleted and then resuspended in RPMI media with 10% fetal bovine serum at 5 - 10 × 106 cells per milliliter. Thawed cells were allowed to rest at 37 °Celsius for 15 minutes in a 5% carbon dioxide tissue culture incubator.
Immunohistochemistry
Two primary antibodies directed against CD81 (clones 1D6 and JS81) were tested and optimized on frozen and formalin-fixed paraffin-embedded normal human tonsil tissue. The reactivity patterns of both clones were similar and clone 1D6 was used for the remainder of the studies on formalin-fixed paraffin-embedded tissue and tissue microarrays. Serial 4 micrometer-thick sections from paraffin-embedded conventional tissue and tissue microarray blocks were deparaffinized in xylene and hydrated in a series of graded alcohols. Heat-induced antigen retrieval was carried out by microwave pretreatment in citric acid buffer (10 millimolar, pH 6.0, for 10 minutes). Clone 1D6 was used at a concentration of 20 micrograms per milliliter. Detection was carried out using the DAKO Envision method (DAKO Corporation, Carpinteria, California). The cut-off of staining in greater than 30% of lymphoma cells was assigned a positive score. This cut-off was based on the need for using a non-ambiguous threshold for scoring tissue microarrays and has been used in similar characterization studies on hematopoetic neoplasms [15]. The cut-off does not reflect differences in staining intensity between normal and neoplastic tissue or among different diagnoses and was chosen before correlation with other immunohistologic markers. All tissue microarray slides were scored independently by four pathologists (RL, YN, DG, AW), and discordant scores were resolved by joint review on a multi-headed microscope. Materials and methods for LMO2, HGAL, BCL6, CD10, BCL2 and MUM1/IRF4 immunostaining have been described previously [22]. Double immunofluorescence labeling with CD19, CD163, Ki67, and CD3 was also performed as previously described [23]. Antibodies to CD3 and PAX5 were purchased from DAKO Corporation.
High-dimensional (11-color) flow cytometry
Fluorescent conjugated monoclonal antibodies against CD3, CD10, CD19, CD20, CD38, CD81, IgD, IgM and isotype controls were purchased from BD Biosciences (San Diego, California) (Table 1). Single cell suspensions of normal tonsil, thymus, and bone marrow, along with biopsies of diffuse large B cell lymphoma and multiple myeloma, were stained with a cocktail of fluorochrome-conjugated antibodies. Propidium iodide was added to all samples before data collection to identify dead cells. High-dimensional flow cytometry data was collected on a LSRII FACS instrument (BD Biosciences, San Jose, CA).
Table 1. Antibodies used for flow cytometry.
| Antigen | Clone | Fluorochrome |
|---|---|---|
| CD3 | SP34-2, UCHT1 | Pacific Blue |
| CD10 | HI10a | Cy7PE |
| CD19 | SJ25C1 | AmCyan |
| CD20 | L27 | Cy7APC, |
| PerCPCy5.5 | ||
| CD38 | HIT2 | PerCPCy5.5 |
| CD81 | JS81 | APC |
| IgD | IA6-2 | PE |
| IgM | G20-127 | FITC |
Data analysis and visualization
The stained lymphoma tissue microarray slides were scanned and stored as high resolution images using an automated scanner (Bacus Laboratories, Inc., Slide Scanner, Lombard, Illinois). The “Deconvoluter” algorithm (custom WBS macro, Excel, Microsoft, Redmond, Washington) with appropriate layout for use in the Cluster software was used for hierarchical clustering to integrate all immunohistologic staining results as previously described [24]. Positive staining is represented as red, lack of staining as green, and non-interpretable staining as black.
For flow cytometry data, FLOWJO (TreeStar, San Carlos, California) and Cytobank (http://www.cytobank.org) software was used for fluorescence compensation and analysis. Data were depicted as dot plots, or histograms displaying fluorescence intensity plotted against cell numbers/fluorescent intensity interval with a total of 256 intervals per parameter. Based on IgD and CD38 expression in the tonsil preparation, CD19+ B cells were resolved into naïve (IgD+CD38-), pre-germinal center (IgD+CD38+), germinal center (IgD-CD38+) and memory (IgD-CD38-) B cells and the expression of CD10, CD20 and CD81 was analyzed in these subsets.
RESULTS
CD81 protein is expressed in normal hematopoietic tissue and is present at high levels in germinal center B cells
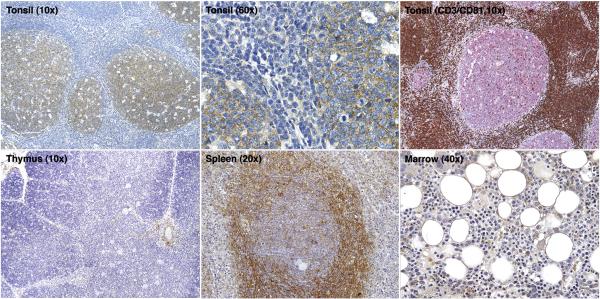
In formalin-fixed paraffin-embedded normal tonsils and lymph nodes, CD81 protein was highly expressed in germinal center lymphocytes with strong membrane and cytoplasmic staining. No staining was seen in mantle zones, marginal zones, or interfollicular areas (Figure 1). Double immunofluorescence microscopy showed that CD81 expression co-localized in germinal centers with CD19 and Ki67 expression but not with CD3-labeled T cells or CD163-labeled macrophages. In formalin-fixed paraffin-embedded normal thymi, CD81 staining highlighted rare scattered lymphocytes in the cortex and medulla. In normal spleen, CD81 staining was found in the marginal zones as well as in germinal centers. In normal bone marrow, only scattered lymphocytes showed positive staining. Erythroid and myeloid precursors and megakaryocytes lacked staining. Examples of staining in normal tonsil, thymus, spleen, and bone marrow are shown in Figure 1. In frozen cryostat sections of normal tonsils, CD81 staining was seen in all B cell and paracortical T cell zones, although the intensity of staining in the germinal centers was higher.
Figure 1. Immunohistochemical staining for CD81 in normal hematopoietic tissue.
Low and high magnification images of normal tonsil sections show cytoplasmic CD81 staining within germinal centers. Double immunofluorescence labeling on tonsil tissue shows CD81 (red) labeling germinal center B cells and CD3 labeling germinal center T cells and interfollicular T cells (brown). Normal thymus shows rare, scattered CD81-positive lymphocytes in the cortex and medulla. Normal spleen shows CD81 staining in germinal centers and marginal zones. Normal bone marrow shows only rare CD81-positive lymphocytes.
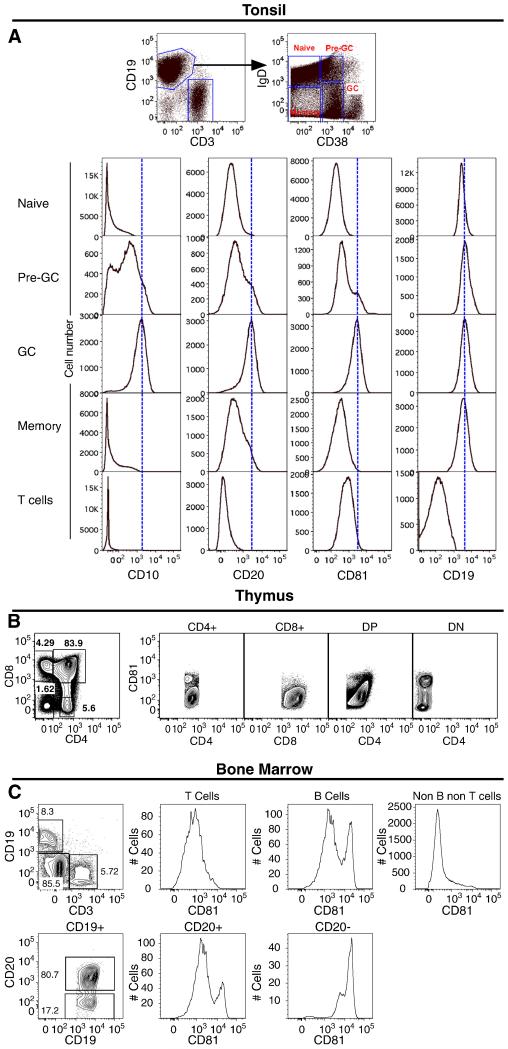
To further explore the pattern of CD81 expression in normal lymphoid subpopulations, flow cytometry was done on normal tonsil, thymus, and bone marrow. In flow cytometry analysis of normal tonsil, B cells were divided into subsets based on IgD and CD38 expression, with expression of CD10 in CD19+IgD-CD38+ cells identifying germinal center B cells. All B cell subsets showed homogeneous CD19 expression (Figure 2A). In contrast, the expression of CD81 was variable and germinal center B cells had the highest level of CD81 expression. We also noticed that these B cells expressed significantly higher amounts of CD20 compared to other subsets (naïve, pre-germinal center and memory). Flow cytometry of normal thymus showed a homogeneous level of CD81 expression in T cell subsets with the exception of CD4-CD8-double negative T cells which showed increased expression of CD81 (Figure 2B). In the bone marrow, CD19+CD20+ mature B cells expressed lower levels of CD81 than CD19+CD20- early B cell progenitors, which expressed the highest CD81 levels (Figure 2C). In addition, bone marrow erythroid and myeloid precursors (non B, non T cells) lacked expression of CD81. Given that T cells in tonsil, thymus and bone marrow also express significant amounts of CD81, a major difference exists in the detection of CD81 by immunohistochemistry in that immunohistochemistry detects only the highest expression levels of CD81 in comparison to flow cytometry.
Figure 2. Flow cytometry of normal tonsil, thymus and bone marrow.
(A) Based on IgD and CD38 expression, B cells (CD19+CD3-) from normal tonsil were resolved into naïve (IgD+CD38-), pre-germinal center (IgD+CD38+), germinal center (IgD- CD38+) and memory (IgD-CD38-) B cells (top right panel, dot plot). The expression of CD10, CD20 and CD81 in these subsets is shown, with germinal center B cells expressing the highest levels of CD81. T cells (CD3+CD19-CD10-CD20-) also demonstrated significant amounts of CD81 expression. (B) In the normal thymus, CD4+, CD8+, and CD4+CD8+ double positive T-cells show homogenous expression of CD81 whereas CD4-CD8- double negative T-cells show a higher level of CD81 expression. (C) In normal bone marrow cells, CD19+CD20- early B cell progenitors show the highest level of CD81 expression, whereas CD19+CD20+ mature B cells and T-cells show lower levels of expression and other hematopoietic marrow precursors (non B non T cells) lack CD81 expression.
CD81 protein is expressed in the majority of non-Hodgkin lymphoma but rarely in Hodgkin lymphoma, plasma cell myeloma and myeloid leukemia
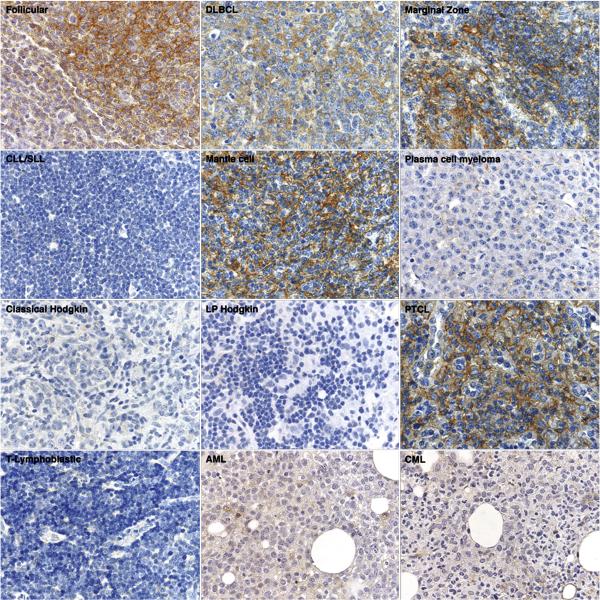
The results of immunohistochemical staining in hematolymphoid neoplasia are summarized in Table 2, with specific examples shown in Figure 3. In addition, digital images of all original CD81-stained tissue microarray cores of the non-Hodgkin and Hodgkin lymphomas tested in this paper are shown on the following freely-accessible website: http://tma.stanford.edu/tma_portal/CD81/. Among B cell lymphomas, CD81 staining was positive in the majority of diffuse large B cell lymphomas (123/196), follicular lymphomas of all three histologic grades (139/164), marginal zone lymphomas of all types (23/27), mantle cell lymphomas (12/18), mediastinal large B cell lymphomas (5/8), lymphoplasmacytic lymphomas (4/5), precursor B-lymphoblastic lymphomas (5/9) and Burkitt lymphomas (2/2). Examination of conventional sections of follicular lymphomas showed that CD81 staining was found in germinal centers as well as in scattered interfollicular B cells. Only a minority of small lymphocytic lymphoma/chronic lymphocytic leukemia (10/43) showed staining for CD81. In a subset of 10 cases with known IgH V hypermutation status, two of five hypermutation positive cases stained positively for CD81. Similarly, two of five hypermutation negative cases of small lymphocytic lymphoma/chronic lymphocytic leukemia also stained positive for CD81.
Table 2. Immunohistologic analysis of CD81 protein expression in hematolymphoid neoplasia.
| Lymphoma Subtype | Total Positive* | % Positive |
|---|---|---|
| B cell Lymphoma [N=462] | ||
| Follicular Lymphoma | 139/164 | 85% |
| Grade 1 | 35/41 | 85% |
| Grade 2 | 48/53 | 91% |
| Grade 3 | 56/70 | 80% |
| Diffuse Large B cell Lymphoma | 123/196 | 63% |
| Mediastinal Large B cell Lymphoma | 5/8 | 63% |
| Burkitt Lymphoma | 2/2 | 100% |
| Extranodal Marginal Zone Lymphoma | 14/17 | 82% |
| Splenic Marginal Zone Lymphoma | 5/5 | 100% |
| Nodal Marginal Zone Lymphoma | 4/5 | 80% |
| Mantle Cell Lymphoma | 12/18 | 67% |
| Small Lymphocytic Lymphoma/Chronic | 10/43 | 23% |
| Lymphocytic Leukemia | ||
| Lymphoplasmacytic Lymphoma | 4/5 | 80% |
| Precursor B-Lymphoblastic Lymphoma | 5/9 | 56% |
| T cell Lymphoma [N=129] | ||
| Precursor T-Lymphoblastic Lymphoma | 4/10 | 40% |
| Peripheral T cell Lymphoma | 11/14 | 79% |
| Anaplastic Large Cell Lymphoma | 4/8 | 50% |
| Angioimmunoblastic T cell Lymphoma | 2/3 | 67% |
| NK Lymphoma | 9/94 | 10% |
| Plasma Cell Neoplasms [N=116] | ||
| Multiple Myeloma | 13/101 | 13% |
| Plasma Cell Leukemia | 0/10 | 0% |
| Monoclonal Gammopathy of | 1/5 | 20% |
| Undetermined Significance | ||
| Hodgkin Lymphoma [N=98] | ||
| Lymphocyte Predominant | 3/18 | 17% |
| Classical Hodgkin | 2/80 | 3% |
| Myeloid Leukemia [N=10] | ||
| Acute Myeloid Leukemia | 0/9 | 0% |
| Chronic Myeloid Leukemia | 0/1 | 0% |
CD81 immunostaining was similar in intensity to normal germinal center B cells and was localized to the cytoplasm in all hematopoietic neoplasms tested. Cases were scored positive if >30% of lymphoma cells stained for CD81.
Figure 3. Immunohistologic staining for CD81 in hematolymphoid neoplasia.
Representative examples of CD81 immunostaining in lymphomas (60x magnification) show CD81 expression in follicular lymphoma, diffuse large B cell lymphoma, marginal zone lymphoma, mantle cell lymphoma, and peripheral T cell lymphoma. CD81 staining was absent in small lymphocytic lymphoma/chronic lymphocytic leukemia, plasma cell myeloma, Hodgkin lymphoma, T-lymphoblastic lymphoma, acute myeloid leukemia, and chronic myeloid leukemia.
Among T- and NK-cell lymphomas, CD81 staining was present in many peripheral T cell lymphomas (11/14), angioimmunoblastic T cell lymphomas (2/3), and anaplastic large cell lymphomas (4/8), but only in a minority of precursor T-lymphoblastic lymphomas (4/10) and NK lymphomas (9/94). Cases of plasma cell myeloma (13/101) and Hodgkin lymphomas of all types (5/98) showed CD81 staining only in rare cases.
Among myeloid leukemias, CD81 staining was lacking in immature blasts in all acute myeloid leukemias (0/9), including those with multilineage dysplasia and monocytic, erythroid and megakaryocytic differentiation. Similarly, a case of chronic myeloid leukemia, chronic phase, lacked staining for CD81.
CD81 protein expression correlates best with LMO2 and other germinal center markers
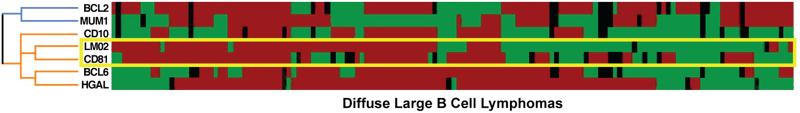
Because statistical analysis of multiple diffuse large B cell lymphoma gene expression profiling studies had identified a potential role for CD81 in diffuse large B cell lymphoma patients (Tibshirani R, unpublished observations), we further characterized the expression of CD81 protein in diffuse large B cell lymphoma samples. CD81 staining was present in 123 of 196 cases of diffuse large B cell lymphoma (63%). The staining intensity was similar to that seen in normal germinal center B cells. In 143 of these cases, CD81 expression was compared to the expression of six additional markers, LMO2, HGAL, BCL6, CD10, BCL2 and MUM1/IRF4, documented in previous work by our group and others (Table 2) [15,22,25,26]. As seen in Figure 4, hierarchical cluster analysis demonstrated that CD81 expression correlated most closely with the germinal center-specific marker LMO2 and secondarily with other germinal center-associated markers HGAL, BCL6, and CD10. It did not correlate with the non-germinal center markers, MUM1/IRF4 or BCL2.
Figure 4. Hierarchical cluster analysis of immunohistologic data.
The expression patterns of seven proteins CD81, LMO2, HGAL, CD10, BCL6, MUM1/IRF4 (MUM1) and BCL2 in 143 cases of diffuse large B cell lymphoma are shown. Positive staining is indicated in red, lack of staining in green, and uninformative data in black. CD81 protein expression is clustered on the same branch of the dendrogram as the germinal center protein LMO2. Both CD81 and LMO2 cluster together with other germinal center proteins, HGAL, BCL6, and CD10 (colored orange), and away from non-germinal center proteins, MUM1 and BCL2 (colored blue).
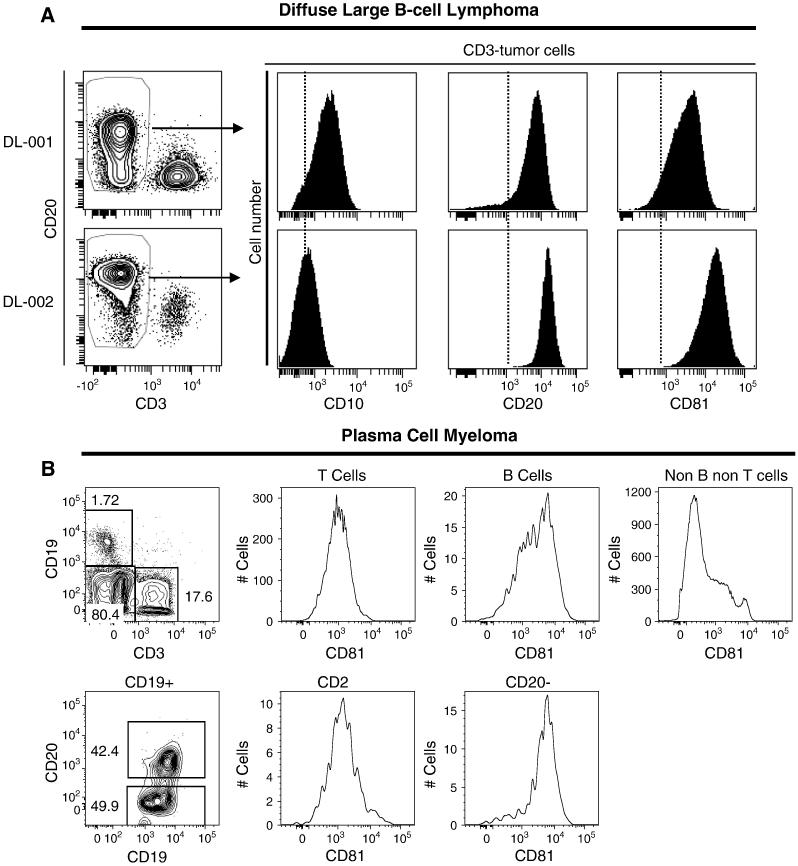
We also measured CD81 and CD10 expression in cell suspensions from diffuse large B cell lymphoma tumor specimens by flow cytometry analysis. All tumor cells showed high levels of CD81 expression, independent of the level of CD10 expression (Figure 5A). In contrast, flow cytometry of plasma cell myeloma showed diminished expression of CD81 in myeloma cells (Figure 5B), which is in keeping with the observation that only a small minority of plasma cell myelomas stained for CD81 on formalin-fixed paraffin-embedded tissue.
Figure 5. Flow cytometry of diffuse large B cell lymphoma and plasma cell myeloma.
(A) Flow cytometry analysis was done for CD10, CD20 and CD81 expression in 2 individual diffuse large B cell lymphoma patient samples, DL-01 and DL-02. Shown is a dot plot of CD20 expression and CD3 expression gated on live cells, along with histograms for CD10, CD20 and CD81 expression gated on CD3-negative tumor cells. CD81 expression was high in all cases, regardless of CD10 expression. The data are representative of 8 individuals. (B) Flow cytometry of plasma cell myeloma involving the bone marrow shows increased expression of CD81 in plasma cells (CD20-negative).
DISCUSSION
CD81 is a widely expressed tetraspanin cell surface protein known to play an important role in synapse formation between B and T cells [27]. Although its role in B cell development and activation, hepatitis C viral entry, and T-cell proliferation have been well studied, its distribution in human hematopoietic tissue has been largely unexplored. For the first time, the current study provides a comprehensive characterization of its expression pattern at the protein level in normal and neoplastic hematolymphoid tissue. High levels of CD81 protein expression were detected in germinal center B cells, a finding that was previously unrecognized. In addition, our data show that there is differential expression of this protein in human lymphomas and that some subtypes of B cell lymphomas express CD81 protein at high levels. Furthermore, high-dimensional flow cytometry data suggests that CD81 expression is regulated developmentally in a similar nature to CD10, being high in both early B cell progenitors in the bone marrow and in the germinal center.
In formalin-fixed paraffin-embedded sections of normal hematopoietic tissue, there was strong membrane and cytoplasmic staining of CD81 in germinal center B cells but not in other lymphoid or non-lymphoid compartments. The high levels of staining for CD81 in germinal center B cells was best appreciated in formalin-fixed paraffin-embedded tissue where the staining in other B- and T-cell subsets was diminished or absent. The staining was distinctly localized to germinal center B cells and reproducible across multiple rounds of staining with both anti-CD81 clones that were tested in this study. In addition, double labeling of CD81 with CD19, CD3, and the proliferation marker Ki-67, also unequivocally showed that the staining in formalin-fixed paraffin-embedded tissue was localized to germinal center B cells. This pattern of expression was further confirmed by frozen section immunohistochemistry as well as flow cytometry studies, where although less intense staining was appreciated in other B- and T-cell compartments, the most intense expression of CD81 was seen in germinal center B cells. The most likely explanation for the difference in staining between fresh and formalin-fixed paraffin-embedded tissue is that only a high level of expression of CD81 protein is detected by formalin-fixed paraffin-embedded -based immunohistologic methods. This finding is not unusual as differences in preservation and denaturation (and therefore detection) of proteins due to the processing steps involved in generating formalin-fixed paraffin-embedded sections is well known. However, the importance of the previously unrecognized finding of high levels of expression of CD81 protein in germinal center B cells will likely encourage future investigations of the role of CD81 in the specialized germinal center niche.
In formalin-fixed paraffin-embedded normal bone marrow, CD81 was not detected by immunohistochemical staining in any of the hematopoietic lineages, and its expression was also lacking in all myeloid leukemias tested (including erythroid and megakaryocytic types). Flow cytometry of bone marrow lymphoid cells also showed that early B-cell progenitors expressed the highest level of CD81 and confirmed the lack of CD81 expression in other hematopoietic precursors. In this regard, the differential expression of CD81 in lymphoid cell types in the bone marrow could potentially be exploited for diagnostic purposes in separating lymphoid from non-lymphoid acute and chronic leukemias. This separation may be particularly useful in minimally differentiated leukemias where other known immunophenotypic markers or cytogenetic abnormalities may be lacking. However, further studies are needed to formally show its utility for this purpose.
The ability to detect CD81 protein in routinely processed lymphoma tissue samples is also likely to facilitate investigations into its role in HCV-associated lymphomagenesis. Our findings show differential expression of CD81 protein in human lymphoma subtypes. Staining for CD81 was detected in the majority of all non-Hodgkin lymphomas tested except for small lymphocytic lymphoma/chronic lymphocytic leukemia, plasma cell myeloma, precursor T-lymphoblastic lymphoma and NK lymphoma. Follicular, marginal zone and lymphoplasmacytic lymphoma, three of the most common non-Hodgkin lymphomas encountered in patients with hepatitis C infection [28], showed the highest rates (≥80%) of CD81 expression among all lymphoma subtypes. In plasma cell myeloma and Hodgkin lymphoma, only a small subset of cases showed CD81 expression, and these lymphomas show a much less frequent association with hepatitis C infection. These findings, although only observational, may warrant further study into the relationships among CD81, hepatitis C, and lymphomagenesis.
The identification of CD81 as a potential marker of prognostic importance in diffuse large B cell lymphoma patients by statistical methodology and the strong germinal center-associated expression pattern of the CD81 protein, prompted us to further explore its expression in diffuse large B cell lymphoma subtypes that we had previously characterized using germinal center B cell markers. Hierarchical cluster analysis of immunohistologic data demonstrated that CD81 is aligned most closely with LMO2, another germinal center B cell-associated marker whose expression both at the gene and protein levels in patients with diffuse large B cell lymphoma is associated with a superior overall and progression-free survival [13,14,16]. Together, CD81 and LMO2 cluster with other germinal center markers HGAL, BCL6, and CD10 and away from non-germinal center markers BCL2 and MUM1/IRF4. It is however worthy to note that like LMO2 and HGAL [15,22], CD81 expression was also found in a significant number of diffuse large B cell lymphoma cases (30/66) that by immunophenotypic algorithms alone would have been characterized as belonging to the non-germinal center subgroup. As these discordant findings have already been noted in other germinal center markers, current models to distinguish germinal center- from non-germinal center-derived diffuse large B cell lymphomas may benefit from further study and optimization using newer markers such as CD81.
Flow cytometry on diffuse large B cell lymphoma cases demonstrated that CD81 expression, often seen at high levels, is independent of the expression of another germinal center marker, CD10. Again, such differences in expression between germinal center markers suggest that current multivariate models of diffuse large B cell lymphoma survival may need to be expanded to create more precise prognostic subcategories of patients. Multiple recent studies have shown that the addition of the anti-CD20 antibody rituximab to anthracycline-based chemotherapy significantly improves the overall survival of patients with diffuse large B cell lymphoma [29,30,31,32]. Thus, multivariate prognostic models should be revalidated in patients treated with the new standard immunochemotherapy regimen. Given that gene expression profiling studies have shown that a germinal center-specific gene expression signature is associated with an improved clinical outcome, it is of interest to characterize new markers that aid in identifying a germinal center phenotype. Our findings that the CD81 protein is expressed at high levels in the germinal center and that the protein expression patterns of CD81 and LMO2 in diffuse large B cell lymphoma tumor samples are closely related provide compelling arguments to assess the relationship of CD81 expression to clinical outcome in a well-characterized cohort of diffuse large B cell lymphoma patients treated with immunochemotherapy.
In conclusion, we have characterized the expression of CD81 protein in normal and neoplastic hematopoietic tissues and have demonstrated that it is expressed at high levels in normal germinal center B cells and in subtypes of non-Hodgkin lymphomas. In diffuse large B cell lymphoma cases, CD81 aligns closely with LMO2 and other germinal center markers and may aid in future categorizations of germinal center-like diffuse large B cell lymphoma patients. Further work is warranted to assess the usefulness of CD81 expression in the risk stratification of diffuse large B cell lymphoma patients.
Acknowledgments
Support: This work was partly funded by NIH CA34233, CA33399, the Leukemia and Lymphoma Society SCORE grant. RL is an American Cancer Society Clinical Research Professor and JHM is a Norwegian Cancer Society Fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Levy S, Shoham T. The tetraspanin web modulates immune-signaling complexes. Nat Rev Immunol. 2005;5:136–148. doi: 10.1038/nri1548. [DOI] [PubMed] [Google Scholar]
- 2.Shoham T, Rajapaksa R, Boucheix C, Rubinstein E, Poe JC, Tedder TF, Levy S. The tetraspanin CD81 regulates the expression of CD19 during B cell development in a postendoplasmic reticulum compartment. J Immunol. 2003;171:4062–4072. doi: 10.4049/jimmunol.171.8.4062. [DOI] [PubMed] [Google Scholar]
- 3.Tedder TF, Inaoki M, Sato S. The CD19-CD21 complex regulates signal transduction thresholds governing humoral immunity and autoimmunity. Immunity. 1997;6:107–118. doi: 10.1016/s1074-7613(00)80418-5. [DOI] [PubMed] [Google Scholar]
- 4.Pileri P, Uematsu Y, Campagnoli S, Galli G, Falugi F, Petracca R, Weiner AJ, Houghton M, Rosa D, Grandi G, Abrignani S. Binding of hepatitis C virus to CD81. Science. 1998;282:938–941. doi: 10.1126/science.282.5390.938. [DOI] [PubMed] [Google Scholar]
- 5.Weng WK, Levy S. Hepatitis C virus (HCV) and lymphomagenesis. Leuk Lymphoma. 2003;44:1113–1120. doi: 10.1080/1042819031000076972. [DOI] [PubMed] [Google Scholar]
- 6.Rosa D, Saletti G, De Gregorio E, Zorat F, Comar C, D’Oro U, Nuti S, Houghton M, Barnaba V, Pozzato G, Abrignani S. Activation of naïve B lymphocytes via CD81, a pathogenic mechanism for hepatitis C virus-associated B lymphocyte disorders. Proc Natl Acad Sci USA. 2005;102:18544–18549. doi: 10.1073/pnas.0509402102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Machida K, Cheng KT, Pavio N, Sung VM, Lai MM. Hepatitis C virus E2-CD81 interaction induces hypermutation of the immunoglobulin gene in B cells. J Virol. 2005;79:8079–8089. doi: 10.1128/JVI.79.13.8079-8089.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serra A, Nuti S, Tavarini S, Sammicheli C, Rosa D, Saletti G, Soldaini E, Abrignani S, Wack A. Coligation of the hepatitis C virus receptor CD81 with CD28 primes naïve T lymphocytes to acquire type 2 effector function. J Immunol. 2008;181:174–185. doi: 10.4049/jimmunol.181.1.174. [DOI] [PubMed] [Google Scholar]
- 9.Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, Powell JI, Yang L, Marti GE, Moore T, Hudson J, Jr, Lu L, Lewis DB, Tibshirani R, Sherlock G, Chan WC, Greiner TC, Weisenburger DD, Armitage JO, Warnke R, Levy R, Wilson W, Grever MR, Byrd JC, Botstein D, Brown PO, Staudt LM. Distinct types of diffuse large B cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- 10.Shipp MA, Ross KN, Tamayo P, Weng AP, Kutok JL, Aguiar RC, Gaasenbeek M, Angelo M, Reich M, Pinkus GS, Ray TS, Koval MA, Last KW, Norton A, Lister TA, Mesirov J, Neuberg DS, Lander ES, Aster JC, Golub TR. Diffuse large B cell lymphoma outcome prediction by gene-expression profiling and supervised machine learning. Nat Med. 2002;8:68–74. doi: 10.1038/nm0102-68. [DOI] [PubMed] [Google Scholar]
- 11.Monti S, Savage KJ, Kutok JL, Feuerhake F, Kurtin P, Mihm M, Wu B, Pasqualucci L, Neuberg D, Aguiar RC, Dal Cin P, Ladd C, Pinkus GS, Salles G, Harris NL, Dalla-Favera R, Habermann TM, Aster JC, Golub TR, Shipp MA. Molecular profiling of diffuse large B cell lymphoma identifies robust subtypes including one characterized by host inflammatory response. Blood. 2005;105:1851–1861. doi: 10.1182/blood-2004-07-2947. [DOI] [PubMed] [Google Scholar]
- 12.Rosenwald A, Wright G, Chan WC, Connors JM, Campo E, Fisher RI, Gascoyne RD, Muller-Hermelink HK, Smeland EB, Giltnane JM, Hurt EM, Zhao H, Averett L, Yang L, Wilson WH, Jaffe ES, Simon R, Klausner RD, Powell J, Duffey PL, Longo DL, Greiner TC, Weisenburger DD, Sanger WG, Dave BJ, Lynch JC, Vose J, Armitage JO, Montserrat E, López-Guillermo A, Grogan TM, Miller TP, LeBlanc M, Ott G, Kvaloy S, Delabie J, Holte H, Krajci P, Stokke T, Staudt LM, Lymphoma/Leukemia Molecular Profiling Project The use of molecular profiling to predict survival after chemotherapy for diffuse large B cell lymphoma. N Engl J Med. 2002;346:1937–1947. doi: 10.1056/NEJMoa012914. [DOI] [PubMed] [Google Scholar]
- 13.Lossos IS, Czerwinski DK, Alizadeh AA, Wechser MA, Tibshirani R, Botstein D, Levy R. Prediction of survival in diffuse large-B cell lymphoma based on the expression of six genes. N Engl J Med. 2004;350:1828–1837. doi: 10.1056/NEJMoa032520. [DOI] [PubMed] [Google Scholar]
- 14.Malumbres R, Chen J, Tibshirani R, Johnson NA, Sehn LH, Natkunam Y, Briones J, Advani R, Connors JM, Byrne GE, Levy R, Gascoyne RD, Lossos IS. Paraffin-based 6-gene model predicts outcome in diffuse large B-cell lymphoma patients treated with R-CHOP. Blood. 2008;111:5509–5514. doi: 10.1182/blood-2008-02-136374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Natkunam Y, Zhao S, Mason DY, Chen J, Taidi B, Jones M, Hammer AS, Hamilton Dutoit S, Lossos IS, Levy R. The oncoprotein LMO2 is expressed in normal germinal-center B cells and in human B cell lymphomas. Blood. 2007;109:1636–1642. doi: 10.1182/blood-2006-08-039024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Natkunam Y, Farinha P, Hsi ED, Hans CP, Tibshirani R, Sehn LH, Connors JM, Gratzinger D, Rosado M, Zhao S, Pohlman B, Wongchaowart N, Bast M, Avigdor A, Schiby G, Nagler A, Byrne GE, Levy R, Gascoyne RD, Lossos IS. LMO2 protein expression predicts survival in patients with diffuse large B cell lymphoma treated with anthracycline-based chemotherapy with and without rituximab. J Clin Oncol. 2008;26:447–454. doi: 10.1200/JCO.2007.13.0690. [DOI] [PubMed] [Google Scholar]
- 17.Bair E, Tibshirani R. Semi-supervised methods to predict patient survival from gene expression data. PLoS Biol. 2004;2:E108. doi: 10.1371/journal.pbio.0020108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hummel M, Bentink S, Berger H, Klapper W, Wessendorf S, Barth TF, Bernd HW, Cogliatti SB, Dierlamm J, Feller AC, Hansmann ML, Haralambieva E, Harder L, Hasenclever D, Kühn M, Lenze D, Lichter P, Martin-Subero JI, Möller P, Müller-Hermelink HK, Ott G, Parwaresch RM, Pott C, Rosenwald A, Rosolowski M, Schwaenen C, Stürzenhofecker B, Szczepanowski M, Trautmann H, Wacker HH, Spang R, Loeffler M, Trümper L, Stein H, Siebert R, Molecular Mechanisms in Malignant Lymphomas Network Project of the Deutsche Krebshilfe A biologic definition of Burkitt’s lymphoma from transcriptional and genomic profiling. N Engl J Med. 2006;354:2419–2430. doi: 10.1056/NEJMoa055351. [DOI] [PubMed] [Google Scholar]
- 19.Lossos IS, Morgensztern D. Prognostic biomarkers in diffuse large B cell lymphoma. J Clin Oncol. 2006;24:995–1007. doi: 10.1200/JCO.2005.02.4786. [DOI] [PubMed] [Google Scholar]
- 20.Swerdlow SH, Campo E, Harris NL, Jaffe ES, Pileri SA, Stein H, Thiele J, Vardiman JW. WHO classification of tumours of haematopoietic and lymphoid tissue. IARC Press; Lyon: 2008. [Google Scholar]
- 21.Stokke T, Galteland E, Holte H, Smedshammer L, Suo Z, Smeland EB, Børresen-Dale AL, DeAngelis P, Steen HB. Oncogenic aberrations in the p53 pathway are associated with a high S phase fraction and poor patient survival in B cell non-Hodgkin’s lymphoma. Int J Cancer. 2000;89:313–324. doi: 10.1002/1097-0215(20000720)89:4<313::aid-ijc1>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 22.Natkunam Y, Lossos IS, Taidi B, Zhao S, Lu X, Ding F, Hammer AS, Marafioti T, Byrne GE, Jr, Levy S, Warnke RA, Levy R. Expression of the human germinal center-associated lymphoma (HGAL) protein, a new marker of germinal center B cell derivation. Blood. 2005;105:3979–3986. doi: 10.1182/blood-2004-08-3112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mason DY, Micklem K, Jones M. Double immunofluorescence labelling of routinely processed paraffin sections. J Pathol. 2000;191:452–461. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH665>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 24.Marinelli RJ, Montgomery K, Liu CL, Shah NH, Prapong W, Nitzberg M, Zachariah ZK, Sherlock GJ, Natkunam Y, West RB, Rijn MV, Brown PO, Ball CA. The Stanford tissue microarray database. Nucleic Acids Res. 2008;36:D871–7. doi: 10.1093/nar/gkm861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hans CP, Weisenburger DD, Greiner TC, Gascoyne RD, Delabie J, Ott G, Müller-Hermelink HK, Campo E, Braziel RM, Jaffe ES, Pan Z, Farinha P, Smith LM, Falini B, Banham AH, Rosenwald A, Staudt LM, Connors JM, Armitage JO, Chan WC. Confirmation of the molecular classification of diffuse large B cell lymphoma by immunohistochemistry using a tissue microarray. Blood. 2004;103:275–282. doi: 10.1182/blood-2003-05-1545. [DOI] [PubMed] [Google Scholar]
- 26.Natkunam Y, Warnke RA, Montgomery K, Falini B, van De Rijn M. Analysis of MUM1/IRF4 protein expression using tissue microarrays and immunohistochemistry. Mod Pathol. 2001;14:686–694. doi: 10.1038/modpathol.3880373. [DOI] [PubMed] [Google Scholar]
- 27.Mittelbrunn M, Yáñez-Mó M, Sancho D, Ursa A, Sánchez-Madrid F. Cutting edge: dynamic redistribution of tetraspanin CD81 at the central zone of the immune synapse in both T lymphocytes and APC. J Immunol. 2002;15(169):6691–6695. doi: 10.4049/jimmunol.169.12.6691. [DOI] [PubMed] [Google Scholar]
- 28.Viswanatha DS, Dogan A. Hepatitis C virus and lymphoma. J Clin Pathol. 2007;60:1378–1383. doi: 10.1136/jcp.2007.051870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coiffier B, Lepage E, Briere J, Herbrecht R, Tilly H, Bouabdallah R, Morel P, Van Den Neste E, Salles G, Gaulard P, Reyes F, Lederlin P, Gisselbrecht C. CHOP chemotherapy plus rituximab compared with CHOP alone in elderly patients with diffuse large B cell lymphoma. N Engl J Med. 2002;346:235–242. doi: 10.1056/NEJMoa011795. [DOI] [PubMed] [Google Scholar]
- 30.Habermann TM, Weller EA, Morrison VA, Gascoyne RD, Cassileth PA, Cohn JB, Dakhil SR, Woda B, Fisher RI, Peterson BA, Horning SJ. Rituximab-CHOP versus CHOP alone or with maintenance rituximab in older patients with diffuse large B cell lymphoma. J Clin Oncol. 2006;24:3121–3127. doi: 10.1200/JCO.2005.05.1003. [DOI] [PubMed] [Google Scholar]
- 31.Pfreundschuh M, Trümper L, Osterborg A, Pettengell R, Trneny M, Imrie K, Ma D, Gill D, Walewski J, Zinzani PL, Stahel R, Kvaloy S, Shpilberg O, Jaeger U, Hansen M, Lehtinen T, López-Guillermo A, Corrado C, Scheliga A, Milpied N, Mendila M, Rashford M, Kuhnt E, Loeffler M, MabThera International Trial Group CHOP-like chemotherapy plus rituximab versus CHOP-like chemotherapy alone in young patients with good-prognosis diffuse large B cell lymphoma: a randomised controlled trial by the MabThera International Trial (MInT) group. Lancet Oncol. 2006;7:379–391. doi: 10.1016/S1470-2045(06)70664-7. [DOI] [PubMed] [Google Scholar]
- 32.Sehn LH, Donaldson J, Chhanabhai M, Fitzgerald C, Gill K, Klasa R, MacPherson N, O’Reilly S, Spinelli JJ, Sutherland J, Wilson KS, Gascoyne RD, Connors JM. Introduction of combined CHOP plus rituximab therapy dramatically improved outcome of diffuse large B cell lymphoma in British Columbia. J Clin Oncol. 2005;23:5027–5033. doi: 10.1200/JCO.2005.09.137. [DOI] [PubMed] [Google Scholar]