Abstract
Pig-tailed macaques (Macaca nemestrina) provide important animal models in biomedical research, but utility of this species for HIV and other disease pathogenesis research is limited by incomplete knowledge of major histocompatibility complex (MHC) class I genetics. Here, we describe comprehensive MHC class I genotyping of 24 pig-tailed macaques, using pyrosequencing to evaluate a 367 bp cDNA-PCR amplicon spanning the highly polymorphic peptide-binding region of MHC class I transcripts. We detected 29 previously described Mane transcripts, 90 novel class I sequences, and eight shared MHC class IB haplotypes. We used this genotyping data to inform full-length MHC class I cDNA allele discovery, characterizing 66 novel full-length transcripts. These new full-length sequences nearly triple the number of Mane-B cDNA sequences previously characterized. The comprehensive genotypes and full-length Mane transcripts described herein add value to pig-tailed macaques as model organisms in biomedical research; furthermore, the coordinated method for MHC genotyping and allele discovery is extensible to other less well-characterized nonhuman primate species.
Keywords: Macaca nemestrina, pig-tailed macaques, MHC, pyrosequencing
Introduction
Nonhuman primates are important animal models for biodefense, transplant immunology, and infectious disease research (Gardner & Luciw 2008; Berger et al. 2009; Patterson & Carrion 2005). In particular, macaque infection with pathogenic strains of simian immunodeficiency virus (SIV) or chimeric SIV/HIV (SHIV) serves as the primary model system for understanding HIV pathogenesis (Pratt et al. 2006; Baroncelli et al. 2008; Valentine & Watkins 2008). Major histocompatibility complex (MHC) class I-restricted CD8+ T cell responses are crucial in determining the host adaptive immune response against infection by viruses like HIV/SIV (Goulder & Watkins 2008). However, characterizing CD8+ T cell responses requires detailed knowledge of MHC class I alleles present in infected macaques. MHC class I genotyping in macaques is complicated by the fact that macaque MHC class I loci have undergone a complex series of segmental duplications—genomic sequencing of the MHC region shows that at least 22 functional MHC class I genes are encoded in both rhesus (Daza-Vamenta et al. 2004) and cynomolgus macaques (Watanabe et al. 2007).
Indian rhesus macaques (Macaca mulatta) have historically been the preferred macaque population for modeling infectious disease; as such, they are to date the most well-characterized population in regards to known MHC class I allele sequences (Baroncelli et al. 2008). In this population, sequences for over 600 MHC classical and nonclassical class I alleles have been at least partially characterized (www.ebi.ac.uk/imgt/mhc; Robinson et al. 2003). While cynomolgus macaques (Macaca fasicularis) provide alternative models for biomedical research, pig-tailed macaques (Macaca nemestrina) are emerging as important models for HIV infection. Significantly, pig-tailed macaques can be infected not only with SIV/SHIV (Buch et al. 2002; Polacino et al. 2008), but also with minimally modified HIV-1. Unlike rhesus and cynomolgus macaques, in which the functional TRIM5α protein is a barrier to HIV-1 replication, pig-tailed macaques express a nonfunctional TRIM5α variant that makes them susceptible to HIV-1 infection (Brennan et al. 2007; Igarashi et al. 2007; Brennan et al. 2008; Newman et al. 2008). The recent success in challenging pig-tailed macaques with minimally modified HIV-1 containing only SIV-derived Vif sequences (Hatziioannou et al. 2009) suggests that pig-tailed macaques will become more widely used in HIV pathogenesis and vaccine research.
MHC class I alleles can both influence the course of infection following viral challenge and confound interpretation of vaccination effects (Yant et al. 2006; Florese et al. 2008; Loffredo et al. 2008; Sauermann et al. 2008; Mee et al. 2009). Thorough investigation of cellular immune responses and correlates of protection against infection in pig-tailed macaques is hindered, however, by limited knowledge of MHC class I genetics. Only 28 Mane-A and 22 Mane-B cDNA sequences have been partially or fully described (www.ebi.ac.uk/imgt/mhc; Robinson et al., 2003). Restriction of an SIV epitope has been defined for a single allele, Mane-A1*08401 (previously known as Mane-A*10, accession numbers AY557348, DQ916064, and EF010518), expression of which has been correlated to lower viral loads following challenge with SIVmac239 (Smith et al. 2005; Mankowski et al. 2008).
Recently, we introduced cDNA amplicon Roche/454 pyrosequencing as a method to rapidly determine MHC class I transcript profiles in macaques (Wiseman et al. 2009). Taking advantage of the high throughput and sensitivity of GS-FLX pyrosequencing, we sequenced a 190 base pair (bp) cDNA amplicon spanning a portion of the highly polymorphic peptide-binding region of MHC class I transcripts. In a cohort of twelve pig-tailed macaques, we detected twenty-four previously described Mane-A and Mane-B sequences or lineages, along with 98 putative novel Mane-A and Mane-B-like sequences (Wiseman et al. 2009). This preponderance of putative novel sequences indicated that the existing Mane class I allele database is incomplete.
Here, we describe amplicon pyrosequencing for MHC class I genotyping in pig-tailed macaques using a 367 bp cDNA amplicon that encodes the MHC class I peptide-binding domain. This amplicon provides improved resolution of closely related class I sequences, more clearly illuminating shared sequences among animals and independent cohorts; additionally, spanning intron two of the MHC transcript eliminates the possibility of genomic DNA contamination. The comprehensive genotypes we obtained by this method elucidated MHC class I diversity within individual animals and also highlighted the need to characterize full-length Mane sequences to confirm that these novel cDNA amplicon sequences represent functional MHC class I transcripts.
Taking advantage of amplicon pyrosequencing data to pre-screen and prioritize individual animals for allele discovery by cDNA cloning and Sanger sequencing, we characterized 66 novel Mane sequences and extended the known sequences for five previously characterized transcripts. This full-length characterization of novel Mane sequences provides a necessary confirmation of the diversity of novel sequences identified by amplicon pyrosequencing. More importantly, elucidation of these novel full-length sequences, adds value to the pig-tailed macaque as a model organism in biomedical research; these full-length cDNA sequences can serve as reagents for a variety of immunological assays that will aid in investigating mechanisms underlying protective MHC class I-restricted immune responses in pig-tailed macaques.
Methods and Materials
Pig-tailed macaque samples
We genotyped the MHC class I region by 367 bp amplicon pyrosequencing in twenty-four pigtailed macaques from two distinct breeding centers. Cellular RNA and genomic DNA for twelve macaques (PT029-PT040) were provided by investigators at Johns Hopkins University (Baltimore, MD); RNA, peripheral blood mononuclear cells (PBMC), T cells, or bone marrow samples were provided for an independent cohort of twelve pig-tailed macaques (PT044-PT055) from the Fred Hutchinson Cancer Research Center (Seattle, WA). We obtained full-length cDNA sequences from twenty-four pig-tailed macaques for which we had comprehensive pyrosequencing genotypes. Cellular RNA for PT029-PT040, macaques genotyped in this report by 367 bp amplicon pyrosequencing, was provided by Johns Hopkins University researchers. The other twelve pig-tailed macaques used here for allele discovery were previously genotyped by 190 bp amplicon pyrosequencing (Wiseman et al. 2009): PBMC samples from nine of these pig-tailed macaques (PT020-PT028) were obtained from the University of Pennsylvania (Philadelphia, PA), while cellular RNA from PT009, PT010, and PT019 was obtained from Johns Hopkins University. All animals were cared for according to the regulations and guidelines of the Institutional Care and Use Committees at their respective institutions.
Preparation and pyrosequencing of 367 bp amplicons
Samples were prepared as described previously (Wiseman et al. 2009). If necessary, RNA was isolated using the MagNA Pure LC RNA Isolation Kit (Roche Applied Sciences, Indianapolis, IN). RNA was reverse transcribed to cDNA using the Superscript™III First-Strand Synthesis System (Invitrogen, Carlsbad, CA). We generated PCR amplicons from cDNA using high-fidelity Phusion polymerase (New England Biolabs, Ipswich, MA). For the 367 bp amplicon we used the previously described exon two forward primer, SBT190F, paired with a reverse primer, SBT367R (5′-TCCCACTTSCGCTGGGT-3′), that binds a conserved region of exon three. Forward and reverse PCR primer pairs contained one of twelve Multiplex Identifier (MID) tags, unique ten bp sequences annealed to the 5′ end of the primer, along with GS-A or GS-B adaptor sequences required for emulsion PCR: seventeen bp sequences annealed to the 5′ end of the MID tag-primer oligonucleotide (Supplemental Table 1). The total amplicon length including adaptor and MID sequences is 421 bp.
We generated primary amplicons from cDNA using the following PCR program on an MJ Research Tetrad Thermocycler (Bio-Rad Laboratories, Hercules, CA): initial denaturation, 98°C for 3 min; amplification over 23 cycles of 98°C for 5 s, 60°C for 1 s, 72°C for 20 s; final extension, 72°C for 5 min. Aliquots from each reaction were run on a FlashGel DNA cassette (Lonza Walkersville Inc., Walkersville, MD) to check for sufficient amplification; if necessary, the reaction was put back on the thermocycler for 3–6 additional PCR cycles to generate sufficient product. PCR products were then separated using a 1% agarose gel in 1X TAE buffer, purified using the MinElute Gel Extraction Kit (Qiagen, Valencia, CA), and quantified with a Qubit Quantitation Platform using the Quant-iT dsDNA HS Assay fluorescence kit (Invitrogen). We normalized amplicons to equimolar concentrations and then pooled samples, confirming purity of amplicons using a 2100 BioAnalyzer (Agilent Technologies, Santa Clara, CA).
The emulsion PCR, bead recovery, and pyrosequencing steps were performed following the manufacturer’s GS FLX protocols at the University of Illinois – Urbana Champaign Sequencing Center. The two 367 bp amplicon pools were sequenced in two independent instrument runs on 1/16th regions of a 70×75 PicoTiterPlate.
Data Analysis
We associated sequence reads with individual animals by binning high quality reads according to MID tag. Because the sequence of the 367 bp amplicon exceeds the maximum read length for the GS-FLX instrument, we first assembled forward and reverse reads, averaging about 250 bp in length, into 100% identical unidirectional contigs using the SeqMan Pro assembler (DNASTAR, Madison, WI). We further examined only sequences that assembled into contigs of two or more identical reads. Following assembly of reads into unidirectional contigs, we performed manual editing as described in Wiseman et al. (2009) to remove pyrosequencing-associated artifacts (chiefly insertions or deletions in homopolymers) and to identify single base and primer mismatches introduced during the amplification processes. An increased frequency of insertion/deletion-type pyrosequencing artifacts resulted from inclusion of a conserved guanine homopolymer region, at the beginning of exon 3, in the 367 bp product for a subset of class I sequences; as a result, a somewhat higher percentage of total sequencing reads were identified as artifacts following manual editing than was observed with 190 bp amplicon pyrosequencing. We then used CodonCode Aligner (Dedham, MA) software to assemble forward and reverse contigs into bidirectional reads based on 100% identity in the overlapping region of approximately 120 bases. Requiring 100% identity over the entire overlapping region was sufficient to unambiguously pair forward and reverse reads for almost all sequences within an individual animal. In a small minority of cases, a definitive 367 bp sequence could not be identified because contigs of distinct forward reads associated with a single contig of reverse reads, or vice-versa; these ambiguous sequences were excluded in our final data analysis. All sequences we analyzed subsequently represent spliced mRNA transcripts as the presence of any genomic contamination from the second intron of MHC class I genes would be evident in the assembled bidirectional sequences. To minimize the likelihood of erroneously identifying sequencing artifacts as novel class I sequences, we required at least two identical sequencing reads in each orientation, for a minimum of four reads, to consider a putative novel class I sequence as present within an animal; we determined this to be an appropriate limit of detection given the stringent artifact analysis and manual assembly of bidirectional reads to ensure that only unambiguously paired forward and reverse unidirectional contigs were included in the data presented.
Confirmation of initial 367 bp amplicon pyrosequencing results by microsatellite analysis
We performed microsatellite analysis for PT029-PT040, the first pool of macaques genotyped by pyrosequencing of the 367 bp amplicon. As previously described (Wiseman et al. 2007; Karl et al. 2008), genomic DNA served as template for PCR using a panel of 16 microsatellite markers that span the five Mb MHC region in macaques. Sizes of the resulting fluorescently labeled products were determined by capillary electrophoresis. We scored peaks using Data Acquisition and Data Analysis Software (Van Mierlo Software Co., Einhoven, the Netherlands). All markers gave a single peak value per haplotype, with the exception of the P03-193435 marker in the MHC class IB region that yielded a variable number of peaks per haplotype.
cDNA cloning and sequencing to characterize full-length MHC class IB alleles
We selected two cohorts, of twelve pig-tailed macaques each, for allele discovery; comprehensive genotyping data from amplicon pyrosequencing existed for both cohorts (Wiseman et al. 2009; this report). The MHC class I cDNA cloning and Sanger sequencing method follows the protocol described by Karl et al. (2009). RNA was isolated and cDNA was generated, as described above. We performed PCR using primers specific for the untranslated regions of MHC class IB alleles to preferentially amplify full-length MHC class IB cDNAs. The sense primer for cDNA-PCR, 5′MHC_UTR_CY1_MIDx (5′-AGAGTCTCCTCAGACGCCGAG-3′), was tagged with MID sequences as previously described in Karl et al. (2009); the antisense primer, 3′MHC_UTR_CY1-MIDx (5′-GGCTGTCTCTCCACCTCCTCAC-3′), was likewise tagged with MID sequences (Supplemental Table 1). PCR for amplification of MHC class I cDNA sequences, ligation and transformation of purified product into chemically competent E. coli, and preparation of plasmid DNA was done as described by Karl et al. (2009). Where the concentration of cDNA-PCR product allowed, we ligated multiple, uniquely MID-tagged samples into vector as a pooled, equimolar sample. At least 96 colonies were isolated per transformation. We did Sanger sequencing, as previously described (Karl et al. 2009), of all clones using a single primer to identify potentially novel cDNA clones; we used the T7 sequencing primer for pooled samples or the 5′Refstrand_v2 primer for individual samples. Sequence analysis was performed using CodonCode Aligner and Lasergene (DNASTAR) software. We sequenced novel class I transcripts detected in at least three clones by single-pass sequencing with a total of five primers, as described by Campbell et al. (2009); for the full-length cDNAs characterized we obtained overlapping sequence coverage for an average of 1221 nucleotides between the 5′ and 3′ UTR primers (Supplemental Table 1).
The 71 full-length sequences described in this report are available in GenBank under the following accession numbers: FJ875218-FJ875276, GQ274880, GQ153465, GQ153484, GQ153471, GQ274894, GQ274896, GQ153467, GQ153511, GQ281749, GQ153468, GQ274887, GQ274890. They were also submitted to the IMGT/MHC Non-human Primate Immuno Polymorphism Database-MHC (IPD) for official nomenclature assignments (Robinson et al. 2003). Nomenclature for pig-tailed macaque MHC class I sequences has been recently updated based on homology to rhesus macaques and the most recent IPD designations for previously described cDNAs are given in Supplemental Table 2.
Results
Comprehensive MHC class I genotyping by amplicon pyrosequencing
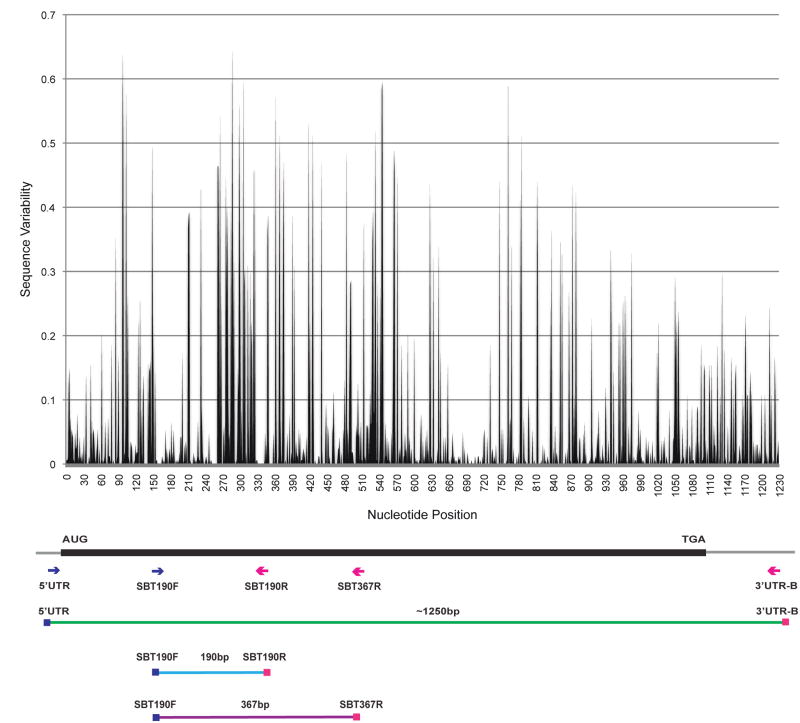
The sequences encoding the peptide-binding domain of MHC class I proteins are highly polymorphic; therefore, obtaining partial sequence coverage of this region of class I transcripts allows us to distinguish specific sequences or lineages. Utilizing amplicon pyrosequencing for sequence-based typing applications is a method to rapidly and comprehensively genotype the MHC class I transcripts of macaques used for biomedical research. Although originally designed based on alignment of known rhesus and cynomolgus macaque MHC class I sequences, the 367 bp primers employed here for pyrosequencing effectively amplify pig-tailed macaque MHC class I transcripts as well, binding highly conserved regions of MHC class I transcripts that flank regions of great nucleotide variability in the peptide-binding region (Figure 1).
Fig. 1.
Schematic of ~1.2 kb MHC class I product and PCR primers. Approximate primer positions are indicated. SBT190F was paired with SBT190R to generate the 190 bp amplicon (Wiseman et al. 2009) and with SBT367R for 367 bp amplicon for pyrosequencing. We used UTR primers to preferentially amplify full-length MHC class IB cDNAs. Relative nucleotide variability along the length of the transcript is calculated based on alignment, using MUSCLE (Edgar 2004), of previously known pig-tailed macaque MHC class I sequences, as well as the novel full-length pig-tailed macaque MHC class I sequences described here.
We evaluated an average of 856 sequencing reads for each of the twenty-four pig-tailed macaques genotyped by 367 bp amplicon pyrosequencing. We distinguished approximately fourteen distinct Mane sequences per animal, with a minimum of two Mane-A and six Mane-B transcripts identified in each animal (Figure 2). A total of 119 unique Mane sequences were distinguished. We detected sixteen previously described Mane-A sequences, but only ten known Mane-B sequences. Putative novel MHC class I sequences predominated; we observed twenty-one Mane-A and sixty-eight Mane-B sequences previously unreported. In addition, three previously characterized and one novel Mane-I sequences were identified. We determined that approximately 82% of Mane sequences, including the novel cDNA sequences described in this report, are uniquely resolved within the sequence of this amplicon. Interesting to those investigating the protective effects of Mane-A1*08401 against SIV disease progression, the 367 bp amplicon allows resolution of Mane-A1*08401 from closely related sequence variants. Within the amplicon sequence, Mane-A1*08401 is distinct from Mane-A1*08402 and also from two putative novel variants that we detected: Mn-A*nov013 and Mn-A*nov030, which differ by one and three nucleotide substitutions, respectively, from Mane-A1*08401 in the peptide-binding region.
Fig. 2.
Amplicon pyrosequencing MHC class I genotypes for 24 pig-tailed macaques. Samples from two cohorts of macaques were obtained from distinct breeding centers and genotyped with the 367 bp amplicon in two separate GS-FLX instrument runs: PT029-PT040 in the first pool and PT044-PT055 in the second. Abundance of Mane sequences is given as a percentage of the total reads analyzed per animal, following removal of pyrosequencing artifacts. Where multiple sequences are noted, the class I genotype is ambiguous due to sequence identity within the 367 bp region examined. The frequency column to the far right indicates the number of animals in which each transcript was observed. Names of novel class I transcripts for which full-length sequences are here described are in bold; all transcripts in bold are novel unless (FL) indicates that previously published sequence has been extended. GenBank accession numbers for full-length novel sequences are given in Table 2; partial sequences for identified “nov” transcripts were also submitted to GenBank (Supplemental Table 3).
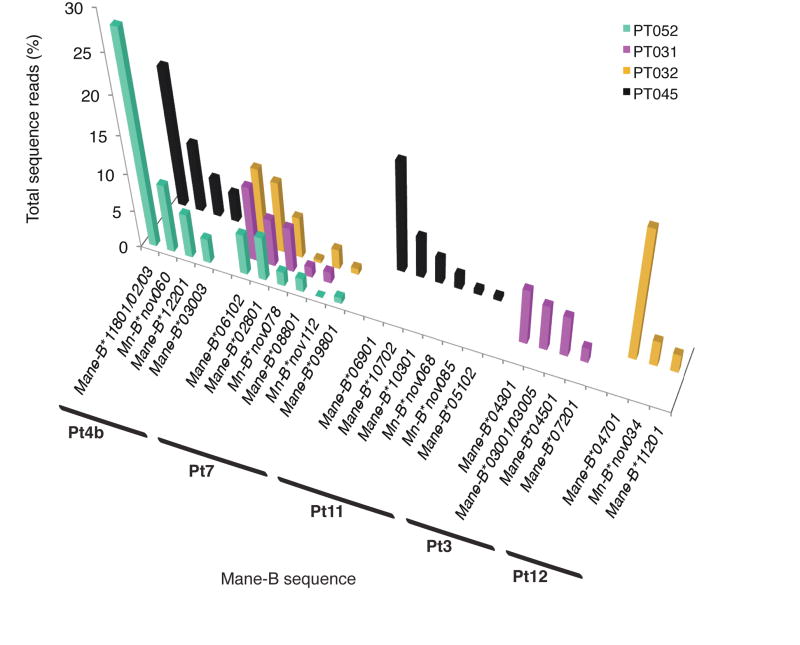
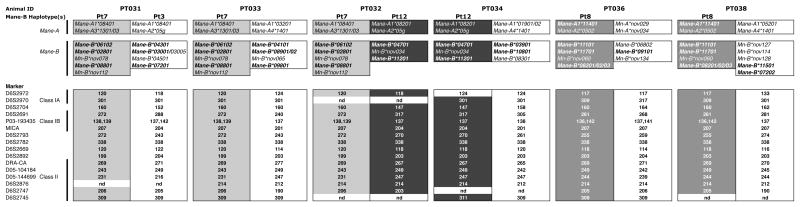
We observed widespread sharing of MHC class I sequences within and between the independent pig-tailed macaque cohorts that were genotyped. Although pedigree data was not available for most animals, we deduced putative Mane-B haplotypes on the basis that particular combinations of three or more MHC class IB sequences were observed in two or more macaques with similar profiles of transcript abundance. In the twenty-four pig-tailed macaques reported here, we observed eight shared Mane-B haplotypes (Figure 2); together with our previous amplicon pyrosequencing study (Wiseman et al 2009) we inferred a total of twelve distinct Mane-B haplotypes (Table 1). The similar transcript profiles we observed among animals deduced to share a haplotype suggest that genotyping by amplicon pyrosequencing offers a semi-quantitative measure of relative transcript levels. This reproducibility in shared transcript profiles is exemplified in Figure 3 for two Mane-B haplotypes (Pt4b and Pt7) that shared among two and three animals, respectively, from distinct breeding centers. The correspondence between shared Mane-B haplotypes deduced by microsatellite analysis and amplicon pyrosequencing data for PT029-PT040 serves as confirmation of inferred haplotypes within this cohort, validating the predictions made based on sequence sharing among two or more animals (Figure 4). Partial breeding records available for PT031, PT032, PT033, and PT034 were also consistent with the haplotype segregation inferred by both microsatellite analysis and amplicon pyrosequencing genotypes; this provides further support for the notion that deduced Mane-B haplotypes based on observations of shared sequences and transcript profiles in two or more animals are not simply chance arrangements. Finally, for the animals illustrated in Figure 4, microsatellite profiles suggest sharing of extended MHC haplotypes despite the predicted amplicon pyrosequencing haplotypes being determined based only on shared Mane-B transcripts.
Table 1.
Identification of twelve deduced Mane-B haplotypes in pig-tailed macaques1
| Haplotype | Mane-B sequences observed2 |
|---|---|
| Pt1 | Mane-B*01502, Mane-B*06802, Mane-B*03002, Mane-B*08901, Mn-B*nov004 |
| Pt2 | Mn-B*nov006, Mane-B*05701, Mane-B*07201, Mn-B*nov009, Mane-B*05102, Mane-B*07901, Mn-B*nov012, Mn-B*nov013 |
| Pt3 | Mane-B*04301, Mane-B*03001/03005, Mane-B*04501, {Mane-B*07201} |
| Pt4a3 | Mane-B*11801/02/03, Mn-B*nov060, Mane-B*12201, Mane-B*08201/02/03, Mane-B*07901, Mane-B*03501 |
| Pt4b | Mane-B*11801/02/03, Mn-B*nov060, Mane-B*12201, Mane-B*03003 |
| Pt5 | Mane-B*01702, Mane-B*03901, Mane-B*10801, Mn-B*nov016, Mane-B*08801, Mn-B*nov023, Mn-B*nov087 |
| Pt6 | Mane-B*05201, Mane-B*05801, Mane-B*11501, Mn-B*nov080 |
| Pt7 | Mane-B*02801, Mane-B*06102, Mn-B*nov078, Mane-B*08801, {Mn-B*nov112}, {Mane-B*09801} |
| Pt8 | Mane-B*11101, Mane-B*11701, Mn-B*nov060, Mane-B*08201/02/03, Mane-B*05101 |
| Pt9 | Mn-B*nov142, Mn-B*nov057, Mane-B*08901/02, Mane-B*07202 |
| Pt10 | Mane-B*06802, Mane-B*01501/03, Mane-B*06401 |
| Pt11 | Mane-B*06901, Mane-B*10702, {Mane-B*05102} |
| Pt12 | Mane-B*04701, Mn-B*nov034, Mane-B*11201 |
Haplotypes were deduced based on combinations of Mane-B sequences shared among multiple animals. Haplotypes Pt1–Pt6 were originally identified based on pyrosequencing of a 190 bp amplicon (see Supplemental Figure 4 in Wiseman et al. 2009).
Named sequences in bold represent full-length cDNAs characterized here. “Mn-B*nov” indicates that only partial sequence is available. Sequences denoted in brackets {} were observed in a majority but not in all animals predicted to share a given haplotype.
Two variants of haplotype Pt4 were observed: Both Pt4a and Pt4b were detected in multiple animals from different breeding centers. The distinguishing sequences between the “a” and “b” variants were amplified to similar degrees by both 190 and 367 bp amplicon primer pairs.
Fig. 3.
MHC class IB transcript profiles of animals inferred to share Mane-B haplotypes. Similarity in relative sequence abundance profiles among animals inferred to share haplotypes suggests comparable levels of expression for these transcripts. The Pt7 and Pt4b haplotypes were observed in multiple animals from both cohorts genotyped. The Pt4b haplotype is a variant of haplotype Pt4a originally observed in a separate cohort using the 190 bp genotyping amplicon (Wiseman et al. 2009). Haplotypes Pt11, Pt3, and Pt12 were also observed in multiple animals from distinct cohorts (Fig. 2).
Fig. 4.
Microsatellite analysis confirms shared haplotypes identified by amplicon pyrosequencing. Based on combinations of shared Mane-B sequences observed in the genotyping data, PT031, PT032, and PT033 were inferred to share a common haplotype designated Pt7. PT032 also shares haplotype Pt12 with PT034. These findings are consistent with partial breeding colony records available for these animals indicating that they share a common sire and/or grandsire. Pt8 appears to be conserved between different breeding centers, since these animals were derived from unrelated parents and originated from distinct breeding colonies. Sequences associated with both haplotypes present in each animal are inferred based on amplicon pyrosequencing results; only shared Mane-B sequences have been designated with haplotype names. Novel full-length cDNA sequences characterized in this report are indicated in bold. Failed amplification resulted in no data, indicated by “nd.”
Full-length MHC class IB allele discovery
MHC class I genotyping by amplicon pyrosequencing indicated that most novel MHC class I sequences in pig-tailed are Mane-B sequences; therefore, we focused our allele discovery effort to characterize novel Mane-B transcripts. We characterized 66 novel, full length MHC class I cDNA sequences. In addition, we obtained full-length cDNA sequences for five previously reported Mane-B and Mane-I transcripts, extending each of these known sequences by at least 100 bp to obtain sequences inclusive of both start and stop codons (Table 2). We prioritized cDNA cloning and sequencing for macaques inferred by amplicon pyrosequencing to share Mane-B haplotypes or express highly abundant novel Mane-B transcripts. We did not observe any full-length cDNA sequences that were not detected by amplicon pyrosequencing and identification of the novel Mane-B transcripts strongly correlated to the relative abundance determined by amplicon pyrosequencing. Thirty-five of the full-length Mane-B transcripts were sequences we observed at frequencies greater than 5% of sequence reads obtained per animal in the genotyping experiment. We observed twenty-one of these novel Mane-B transcripts at intermediate levels, between 1% and 5% of analyzed pyrosequencing reads. In contrast, we only characterized four of the full-length novel sequences that were detected at less than 1% of the total pyrosequencing reads per animal.
Table 2.
71 full-length MHC class I cDNA sequences identified in pig-tailed macaques1
| IPD Name | Working Name | Representative Animal(s) | GenBank Accession No. | BLASTP Hit(s) |
|---|---|---|---|---|
| Mane-A1*11401 | Mn-A*nov014 | PT038 | GQ274880 | |
| Mane-B*00701 | Mn-B*nov025 | PT019 | FJ875228 | Mamu B*00704(α1/α2) |
| Mane-B*01101 | Mn-B*nov054 | PT023 | FJ875239 | Mafa-B*04501(α1/α2) |
| Mane-B*01502 | Mn-B*nov001 | PT028 | FJ875219 | Mafa-B*03702 |
| Mane-B*01503 | Mane-B*01301(FL) | PT019 | FJ875254 | |
| Mane-B*01703 | Mn-B*nov096 | PT034, PT039 | FJ875257 | |
| Mane-B*02401 | Mn-B*nov048 | PT019 | FJ875234 | |
| Mane-B*02801 | Mn-B*nov103 | PT031 | FJ875264 | Mamu-B*028024/Mafa-B*03501(α1/α2) |
| Mane-B*03001 | Mane-B*09(FL) | PT022 | FJ875273 | Mane-B*015/009 |
| Mane-B*03002 | Mn-B*nov002 | PT009, PT023 | FJ875220 | Mafa-B*03802 |
| Mane-B*03003 | Mn-B*nov059 | PT028 | FJ875241 | Mafa-B*03801/02/Mamu-B*03001/02/03(α1/α2) |
| Mane-B*03004 | Mn-B*nov098 | PT040 | FJ875259 | Mafa-B*03401 |
| Mane-B*03501 | Mn-B*nov017 | PT019 | FJ875225 | |
| Mane-B*03901 | Mn-B*nov014 | PT026, PT028 | FJ875224 | |
| Mane-B*04101 | Mn-B*nov131 | PT033 | GQ274896 | |
| Mane-B*04301 | Mn-B*nov071 | PT022, PT023 | FJ875247 | |
| Mane-B*04701 | Mn-B*nov063 | PT010 | FJ875244 | Mamu-B*04701 |
| Mane-B*05101 | Mn-B*nov106 | PT036 | FJ875267 | Mafa-B*09602(α1/α2) |
| Mane-B*05102 | Mn-B*nov010 | PT026 | GQ153467 | |
| Mane-B*05201 | Mn-B*nov081 | PT024, PT027 | FJ875249 | Mamu-B*05201(α1/α2) |
| Mane-B*05601 | Mn-B*nov052 | PT026 | FJ875237 | Mamu-B*05602 |
| Mane-B*05701 | Mn-B*nov007 | PT009 | GQ153465 | Mafa-B*05402 |
| Mane-B*05801 | Mn-B*nov079 | PT025, PT027 | GQ153511 | Mamu-B*05802 |
| Mane-B*06101 | Mn-B*nov050 | PT024 | FJ875235 | Mafa-B*01501(α1/α2) |
| Mane-B*06102 | Mn-B*nov102 | PT031 | FJ875263 | |
| Mane-B*06401 | Mn-B*nov035 | PT039 | GQ153484 | |
| Mane-B*06901 | Mn-B*nov097 | PT037 | FJ875258 | |
| Mane-B*07201 | Mn-B*nov008 | PT009 | FJ875223 | |
| Mane-B*07202 | Mn-B*nov101 | PT031, PT037 | FJ875262 | |
| Mane-B*07203 | Mn-B*nov128 | PT038 | GQ274894 | |
| Mane-B*07801 | Mn-B*nov066 | PT026, PT019 | FJ875246 | Mamu-B*05002/Mafa-B*11301(α1/α2) |
| Mane-B*07802 | Mn-B*nov083 | PT025, PT027 | FJ875250 | |
| Mane-B*07901 | Mn-B*nov011 | PT026 | GQ153468 | Mamu-B*0790101/0102 |
| Mane-B*08201 | Mn-B*nov061 | PT024 | FJ875242 | |
| Mane-B*08202 | Mn-B*nov092 | PT026 | FJ875253 | |
| Mane-B*08203 | Mn-B*nov136 | PT022 | FJ875269 | |
| Mane-B*08302 | Mn-B*nov099 | PT020, PT036 | FJ875260 | |
| Mane-B*08801 | Mn-B*nov019 | PT019 | FJ875226 | |
| Mane-B*08901 | Mn-B*nov003 | PT023, PT027 | FJ875221 | |
| Mane-B*08902 | Mn-B*nov119 | PT033 | GQ274890 | Mamu-B*08901/Mafa-B*08601(α1/α2) |
| Mane-B*09101 | Mn-B*nov100 | PT036 | FJ875261 | |
| Mane-B*09801 | Mn-B*nov056 | PT031 | FJ875240 | |
| Mane-B*10301 | Mn-B*nov026 | PT022 | FJ875229 | Mafa-B*10301 |
| Mane-B*10302 | Mn-B*nov051 | PT023 | FJ875236 | |
| Mane-B*10401 | Mn-B*nov005 | PT009 | FJ875222 | |
| Mane-B*10402 | Mn-B*nov031 | PT026 | FJ875231 | Mafa-B*04401 |
| Mane-B*10403 | Mn-B*nov072 | PT028 | FJ875248 | |
| Mane-B*10701 | Mn-B*nov036 | PT026 | FJ875232 | |
| Mane-B*10702 | Mn-B*nov095 | PT037 | FJ875256 | |
| Mane-B*10801 | Mn-B*nov015 | PT025 | GQ153471 | |
| Mane-B*11001 | Mn-B*nov094 | PT039 | FJ875255 | |
| Mane-B*11101 | Mn-B*nov107 | PT036 | FJ875268 | |
| Mane-B*11201 | Mn-B*nov064 | PT032 | FJ875245 | |
| Mane-B*11301 | Mn-B*nov104 | PT036 | FJ875265 | |
| Mane-B*11501 | Mn-A*nov012 | PT038 | FJ875218 | |
| Mane-B*11601 | Mn-B*nov053 | PT037 | FJ875238 | |
| Mane-B*11701 | Mn-B*nov105 | PT036 | FJ875266 | Mafa-B*08501/02(α1/α2) |
| Mane-B*11801 | Mn-B*nov030 | PT022 | FJ875230 | |
| Mane-B*11802 | Mn-B*nov062 | PT028 | FJ875243 | |
| Mane-B*11803 | Mn-B*nov091 | PT026 | FJ875252 | |
| Mane-B*11901 | Mn-B*nov090 | PT026, PT028 | FJ875251 | |
| Mane-B*11901del | Mn-B*nov082 | PT026, PT028 | GQ281749 | |
| Mane-B*12001 | Mn-B*nov037 | PT019 | FJ875233 | |
| Mane-B*12002 | Mn-B*nov108 | PT029 | GQ274887 | |
| Mane-B*12101 | Mn-B*nov024 | PT019 | FJ875227 | |
| Mane-I*010102 | Mane-I*010102(FL) | PT027 | FJ875274 | |
| Mane-I*0104 | Mane-I*0104(FL) | PT019 | FJ875275 | |
| Mane-I*0107 | Mane-I*0107(FL) | PT031 | FJ875276 | |
| Mane-I*010802 | Mn-I*nov003 | PT037 | FJ875270 | |
| Mane-I*010803 | Mn-I*nov004 | PT019 | FJ875271 | |
| Mane-I*0111 | Mn-I*nov006 | PT035 | FJ875272 |
IPD name, working sequence name, GenBank accession number, and reference animal(s) are listed for each sequence. If 100% identity was found between MHC class IB transcripts by BLASTP analysis, the Mafa/Mamu hit is shown; (α1/α2) indicates that 100% identity exists in the peptide-binding region only. cDNAs designated (FL) in working name extend previously known allele sequences by at least 100 bp.
Given the limited number of MHC class IB alleles characterized previously in pig-tailed macaques, we compared these novel Mane-B transcripts to the more extensively characterized Mamu and Mafa sequences. The official nomenclature (Robinson et al. 2003) assigned to our novel Mane-B sequences suggests that we identified fifteen lineage groups, each consisting of two or more Mane-B transcripts (Table 2). At least three unique cDNA sequences were characterized for five Mane-B lineages. To illuminate possible structural or functional similarities to other known macaque MHC sequences, we performed BLASTP analysis using conceptual translations for these Mane-B transcripts. We analyzed similarity across species for the predicted protein products of these full-length novel Mane-B transcripts (averaging 362 amino acids), as well as for the peptide-binding region encoded by exons two and three (predicted to be 182 amino acids). Over a third of predicted proteins encoded by the novel Mane-B sequences characterized in this report have 100% amino acid identity within the peptide-binding region to previously described rhesus and cynomolgus macaque gene products (Table 2). Eight novel pig-tailed macaque MHC class I gene products are amino acid identical to complete cynomolgus macaque predicted proteins; four others have 100% identity to complete rhesus macaque proteins.
Discussion
Amplicon pyrosequencing and full-length cDNA sequencing as complementary methods
The coordinated use of amplicon pyrosequencing with cDNA cloning and Sanger sequencing to characterize novel MHC class I transcripts offers advantages for genotyping and allele discovery in species for which our knowledge of MHC genetics is limited. Firstly, full-length characterization of novel Mane sequences originally identified by amplicon pyrosequencing confirms the authenticity of the novel amplicon sequences as functional class I transcripts. While a major concern for pyrosequencing-based genotyping remains the relatively high incidence of sequencing artifacts, the concordance of the described cloning and sequencing results with the MHC class I transcript profiles generated by amplicon pyrosequencing is an important confirmation that amplicon pyrosequencing, when employed with appropriately stringent data analysis, is a reliable method to rapidly genotype macaques. Adding to this, use of the 367 bp amplicon spanning intron two of the MHC transcript provides assurance that analyzed sequence reads represent spliced mRNAs and not genomic DNA sequence. The second advantage of this combined method is in using comprehensive genotypes to guide full-length allele discovery. Use of the 367 bp genotyping amplicon provides sufficient sequence resolution to aid in the unique identification of closely related sequence variants, adding sensitivity to our ability to detect novel Mane sequences. This genotyping data enabled us to target specific loci and focus allele discovery efforts primarily on animals predicted to carry shared haplotypes or highly abundant novel sequences.
Applications and limitations of MHC class I genotyping by amplicon pyrosequencing
The genotyping data generated by amplicon pyrosequencing is useful for a variety of applications. Comprehensive MHC class I genotyping in cohorts of pig-tailed macaques can simplify interpretation of experimental results by illuminating shared and unique haplotypes or individual transcripts. This may be particularly relevant in examining immune responses made against specific infectious agents and in interpretation of vaccination results, as genetic correlates to disease susceptibility and resistance are poorly understood in pig-tailed macaques. MHC class I genotyping by amplicon pyrosequencing also holds promise for design and management of breeding colonies. Because the MHC class I region recombines rarely during meiosis (Penedo et al. 2005), inheritance of known haplotypes can be readily traced from parents to offspring; availability of comprehensive genotypes for breeding sires and dams allows for simplified MHC class I genotyping of offspring by alternative techniques, such as microsatellite analysis.
While this deep sequencing method provides a comprehensive genotype overview, utility of the genotyping data is more limited in species for which few MHC class I sequences have been characterized. The short amplicon sequences obtained by pyrosequencing do not provide template material suitable for further investigation of the structure and function of novel MHC class I transcripts. Thus, full-length cDNA cloning and sequencing to characterize novel MHC class I sequences remains of great importance to researchers investigating MHC class I-restricted immune responses in less-well characterized nonhuman primate species. Additionally, the presence of certain MHC class I transcripts are likely masked due to either low expression or mismatches to the amplification primers used in pyrosequencing. Although all of the novel full-length cDNAs that we characterized were detected by amplicon pyrosequencing, four of our novel sequences were originally detected at relatively low abundance. Two of these (Mane-B*09801, Mane-B*05102) were detected in four or more distinct animals and may be commonly expressed at low transcript levels. In contrast, the full-length sequences of Mane-B*01101 and Mane-B*03004 revealed mismatches under the forward pyrosequencing primer which may have caused underrepresentation in our amplicon pyrosequencing results. Augmenting the library of full-length cDNA sequences enables us to fine-tune our amplicon pyrosequencing primers to capture a wider diversity of MHC class I sequences specifically in pig-tailed macaques.
Characterization of novel Mane transcripts
MHC class I transcripts are largely distinct even among geographically distinct populations of the same species (Karl et al. 2008; Campbell et al. 2009); the data described here, however, makes a case for conservation of common classical MHC class I transcripts and lineage groups across species. The predicted amino acid sequences of the Mane transcripts described in this report exhibit a high degree of homology to MHC transcripts characterized in rhesus and cynomolgus macaques, despite the fact that pig-tailed macaques belong to a distinct evolutionary clade (silenus) that diverged over five million years ago from the fasicularis clade which gave rise to both rhesus and cynomolgus macaques (Tosi et al. 2000; Deinard & Smith 2001; Li et al. 2009). Previously, the most notable example of conserved MHC class I protein sequences between pig-tailed macaques and the more closely related rhesus and cynomolgus macaques was the observed sequence homology of nonclassical MHC class I sequences (Lafont et al. 2003; Lafont et al. 2004). Among the novel Mane-B transcripts characterized here, twenty are 100% identical in the peptide binding region to Mamu or Mafa gene products, and twelve of these transcripts are identical to their Mamu or Mafa homologues throughout the complete protein translation.
While such similarity may not be unexpected given the similar susceptibility of these macaque species to certain infectious diseases, it has not previously been possible to compare such a diversity of Mane-B sequences with MHC class IB sequences expressed in other macaques. One striking similarity among species is the apparent evolutionary conservation of a sequence lineage known to give rise to alternatively spliced transcriptional variants. We sequenced two novel Mane-B*11901 transcripts, one of which appears to be an alternatively sliced variant encoding a truncated protein. Sequence homology between Mane-B*11901 and rhesus and cynomolgus macaque transcripts known to have splicing variants (Mamu-B*07402, Mafa-B*03901) makes it likely that this alternative splice site is conserved in all three species. Furthermore, certain abundantly expressed MHC class I haplotypes may be conserved across species, a possibility exemplified by the combination of Mane-B*03003 and Mn-B*nov060 on the Pt4b haplotype which was detected in a third of the pig-tailed macaques described in this report. Mane-B*03003 is 100% identical in the peptide binding region to the gene product of Mamu-B*03003 (Table 2), while Mn-B*nov060, for which sequence covers only the peptide-binding region, is 100% amino acid identical to Mamu-B*02702 (Supplemental Table 3); these two rhesus macaque transcripts are linked on recently described Indian rhesus macaque haplotypes (Sauermann et al. 2008; Wiseman et al. 2009).
The importance of amino acid homology in class I sequences from distinct species may be more fully understood by considering the amino acid homology that exists between two novel MHC class IB sequences characterized here and two MHC alleles previously shown to be protective in SIV infection. Mane-B*01703 is 99% similar at the amino acid level to Mamu-B*01701, and Mane-B*04701 has 100% identity to the Mamu-B*04701 transcript. Mamu-B*01701 is correlated with decreased viral loads following infection with SIVmac239 (Yant et al., 2006), while haplotypes containing Mamu-B*04701 are similarly correlated to slow disease progression (Sauermann et al. 2008). Mane-B*01703 differs from Mamu-B*01701 by three amino acids, however these residues are located at the start of the α2 domain in positions predicted to be highly variable (Parham et al. 1988). Additionally, recent studies show that even disparate MHC class IB transcripts can present highly similar peptides (Loffredo et al. 2009). In the case of Mane-B*04701, identical to the protein product of Mamu-B*04701, this transcript is identified here as a component of a shared haplotype, designated Pt12 (Figures 2 & 3, Table 1). While functional studies are required to determine if the role of Mane-B*04701 or Mane-B*01703 during SIV infection of pig-tailed macaques is similar to the protective role of the homologue rhesus macaque alleles, the degree of identity remains striking. Furthermore, in the case of Mane-B*04701, existence of a corresponding haplotype makes it possible to consider more than single-allele effects on disease progression.
Advancing pig-tailed macaques as model organisms for biomedical research
Using the coordinated approach of amplicon pyrosequencing to generate comprehensive genotypes with full-length cDNA cloning and sequencing for allele discovery, we have characterized twelve distinct Mane-B haplotypes and obtained full-length sequence for 66 novel, as well as five previously described, Mane transcripts. Previously, only 50 classical MHC class I cDNAs and 10 nonclassical MHC class I cDNAs had been characterized in pig-tailed macaques (Lafont et al. 2003; Smith et al. 2005; Lafont et al. 2004; Pratt et al. 2006; Lafont et al. 2007); thus, this report represents a significant increase in our knowledge of the MHC genetics in this important animal for HIV and other infectious disease research. The comprehensive genotypes generated by amplicon pyrosequencing have broad applications for biomedical research using pig-tailed macaques, while the full-length characterization of these novel alleles makes it possible to generate reagents necessary for functional immunological studies in pig-tailed macaques, such as MHC class I transferrants to determine CD8+ T cell restriction and tetramer constructs for sensitive detection of specific CD8+ T cell populations. Additional improvements in the use of pyrosequencing for MHC class I genotyping, as well as continued full-length sequencing of potential novel alleles, make the pig-tailed macaque an increasingly valuable animal model for biomedical studies, including HIV vaccine development.
Supplementary Material
Acknowledgments
This work was funded in part by: National Center for Research Resources (NCRR) R24 RR02174, NCRR P51 RR000167, National Institute of Allergy and Infectious Disease (NIAID) HHSN266200400088C/N01-AI-40088, and NIAID R21 AI068488. The pig-tailed macaque samples utilized in these studies were graciously provided by investigators at Johns Hopkins University (Drs. Susan Queen, Joseph Mankowski, and Robert Adams), the Fred Hutchinson Cancer Research Center (Dr. Carolina Berger), and the University of Pennsylvania (Drs. James Hoxie and Patricia Fultz). Experiments using macaque samples from Johns Hopkins University were funded in part by NCRR P40 RR019995. We would also like to acknowledge Nel Otting and Natasja de Groot with the Immuno Polymorphism Database for assigning official nomenclature to the submitted full-length sequences, as well as the members of the O’Connor laboratory at the University of Wisconsin for providing helpful discussion and critical commentary on this manuscript.
References
- Baroncelli S, Negri DR, Michelini Z, Cara A. Macaca mulatta, fascicularis and nemestrina in AIDS vaccine development. Expert Rev Vaccines. 2008;7:1419–1434. doi: 10.1586/14760584.7.9.1419. [DOI] [PubMed] [Google Scholar]
- Berger C, Turtle CJ, Jensen MC, Riddell SR. Adoptive transfer of virus-specific and tumor-specific T cell immunity. Curr Opin Immunol. 2009;21:224–232. doi: 10.1016/j.coi.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan G, Kozyrev Y, Kodama T, Hu SL. Novel TRIM5 isoforms expressed by Macaca nemestrina. J Virol. 2007;81:12210–12217. doi: 10.1128/JVI.02499-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brennan G, Kozyrev Y, Hu SL. TRIMCyp expression in Old World primates Macaca nemestrina and Macaca fascicularis. Proc Natl Acad Sci USA. 2008;105:3569–3574. doi: 10.1073/pnas.0709511105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buch SJ, Villinger F, Pinson D, Hou Y, Adany I, Li Z, et al. Innate differences between simian-human immunodeficiency virus (SHIV)(KU-2)-infected rhesus and pigtailed macaques in development of neurological disease. Virol. 2002;295:54–62. doi: 10.1006/viro.2001.1369. [DOI] [PubMed] [Google Scholar]
- Campbell KJ, Detmer AM, Karl JA, Wiseman RW, Blasky AJ, Hughes AL, et al. Characterization of 47 MHC class I sequences in Filipino cynomolgus macaques. Immunogenetics. 2009;61:177–187. doi: 10.1007/s00251-008-0351-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daza-Vamenta R, Glusman G, Rowen L, Guthrie B, Geraghty DE. Genetic divergence of the rhesus macaque major histocompatibility complex. Genome Res. 2004;14:1501–1515. doi: 10.1101/gr.2134504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deinard A, Smith DG. Phylogenetic relationships among the macaques: evidence from the nuclear locus NRAMP1. J Hum Evol. 2001;41:45–59. doi: 10.1006/jhev.2001.0480. [DOI] [PubMed] [Google Scholar]
- Edgar Robert C. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–97. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florese RH, Wiseman RW, Venzon D, Karl JA, Demberg T, Larsen K, et al. Comparative study of Tat vaccine regimens in Mauritian cynomolgus and Indian rhesus macaques: Influence of Mauritian MHC haplotypes on susceptibility/resistance to SHIV89.6P infection. Vaccine. 2008;26:3312–3321. doi: 10.1016/j.vaccine.2008.03.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner MB, Luciw PA. Macaque models of human infectious disease. ILAR J/Natl Res Council, Inst Lab Anim Resour. 2008;49:220–255. doi: 10.1093/ilar.49.2.220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulder PJ, Watkins DI. Impact of MHC class I diversity on immune control of immunodeficiency virus replication. Nat Rev Immunol. 2008;8:619–630. doi: 10.1038/nri2357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatziioannou T, Ambrose Z, Chung NP, Piatak M, Jr, Yuan F, Trubey CM, et al. A macaque model of HIV-1 infection. Proc Natl Acad Sci USA. 2009;106:4425–4429. doi: 10.1073/pnas.0812587106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Igarashi T, Iyengar R, Byrum RA, Buckler-White A, Dewar RL, Buckler CE, et al. Human immunodeficiency virus type 1 derivative with 7% simian immunodeficiency virus genetic content is able to establish infections in pig-tailed macaques. J Virol. 2007;81:11549–11552. doi: 10.1128/JVI.00960-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl JA, Wiseman RW, Campbell KJ, Blasky AJ, Hughes AL, Ferguson B, et al. Identification of MHC class I sequences in Chinese-origin rhesus macaques. Immunogenetics. 2008;60:37–46. doi: 10.1007/s00251-007-0267-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karl JA, Wiseman RW, O’Connor DH. Cost-effective sequence-based nonhuman primate MHC class I genotyping from RNA. Methods. 2009 doi: 10.1016/j.ymeth.2009.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafont BAP, Buckler-White A, Plishka R, Buckler C, Martin MA. Characterization of Pig-Tailed Macaque Classical MHC Class I Genes: Implications for MHC Evolution and Antigen Presentation in Macaques. J Immunol. 2003;171:875–885. doi: 10.4049/jimmunol.171.2.875. [DOI] [PubMed] [Google Scholar]
- Lafont BA, Buckler-White A, Plishka R, Buckler C, Martin MA. Pig-tailed macaques (Macaca nemestrina) possess six MHC-E families that are conserved among macaque species: implication for their binding to natural killer receptor variants. Immunogenetics. 2004;56:142–154. doi: 10.1007/s00251-004-0663-4. [DOI] [PubMed] [Google Scholar]
- Lafont BA, McGraw CM, Stukes SA, Buckler-White A, Plishka RJ, Byrum RA, et al. The locus encoding an oligomorphic family of MHC-A alleles (Mane-A*06/Mamu-A*05) is present at high frequency in several macaque species. Immunogenetics. 2007;59:211–223. doi: 10.1007/s00251-007-0190-1. [DOI] [PubMed] [Google Scholar]
- Li J, Han K, Xing J, Kim HS, Rogers J, Ryder OA, et al. Phylogeny of the macaques (Cercopithecidae: Macaca) based on Alu elements. Gene. 2009 doi: 10.1016/j.gene.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Bean AT, Beal DR, Leon EJ, May GE, Piaskowski SM, et al. Patterns of CD8+ immunodominance may influence the ability of Mamu-B*08-positive macaques to naturally control simian immunodeficiency virus SIVmac239 replication. J Virol. 2008;82:1723–1738. doi: 10.1128/JVI.02084-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loffredo JT, Sidney J, Bean AT, Beal DR, Bardet W, Wahl A, et al. Two MHC class I molecules associated with elite control of immunodeficiency virus replication, Mamu-B*08 and HLA-B*2705, bind peptides with sequence similarity. J Immunol. 2009;182:7763–7775. doi: 10.4049/jimmunol.0900111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mankowski JL, Queen SE, Fernandez CS, Tarwater PM, Karper JM, Adams RJ, et al. Natural host genetic resistance to lentiviral CNS disease: a neuroprotective MHC class I allele in SIV-infected macaques. PloS One. 2008;3:e3603. doi: 10.1371/journal.pone.0003603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mee ET, Berry N, Ham C, Sauermann U, Maggiorella MT, Martinon F, et al. Mhc haplotype H6 is associated with sustained control of SIVmac251 infection in Mauritian cynomolgus macaques. Immunogenetics. 2009;61:327–39. doi: 10.1007/s00251-009-0369-8. [DOI] [PubMed] [Google Scholar]
- Newman RM, Hall L, Kirmaier A, Pozzi LA, Pery E, Farzan M, et al. Evolution of a TRIM5-CypA splice isoform in old world monkeys. PLoS Pathog. 2008;4:e1000003. doi: 10.1371/journal.ppat.1000003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parham P, Lomen CE, Lawlor DA, Ways JP, Holmes N, Coppin HL, et al. Nature of polymorphism in HLA-A, -B, and -C molecules. Proc Natl Acad Sci USA. 1988;85:4005–4009. doi: 10.1073/pnas.85.11.4005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson JL, Carrion R., Jr Demand for nonhuman primate resources in the age of biodefense. ILAR J. 2005;46:15–22. doi: 10.1093/ilar.46.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penedo MC, Bontrop RE, Heijmans CM, Otting N, Noort R, Rouweler AJ, et al. Microsatellite typing of the rhesus macaque MHC region. Immunogenetics. 2005;57:198–209. doi: 10.1007/s00251-005-0787-1. [DOI] [PubMed] [Google Scholar]
- Polacino P, Larsen K, Galmin L, Suschak J, Kraft Z, Stamatatos L, et al. Differential pathogenicity of SHIV infection in pig-tailed and rhesus macaques. J Med Primatol. 2008;37(Suppl 2):13–23. doi: 10.1111/j.1600-0684.2008.00325.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt BF, O’Connor DH, Lafont BA, Mankowski JL, Fernandez CS, Triastuti R, et al. MHC class I allele frequencies in pigtail macaques of diverse origin. Immunogenetics. 2006;58:995–1001. doi: 10.1007/s00251-006-0164-8. [DOI] [PubMed] [Google Scholar]
- Robinson J, Waller MJ, Parham P, de Groot N, Bontrop R, Kennedy LJ, et al. IMGT/HLA and IMGT/MHC: sequence databases for the study of the major histocompatibility complex. Nucleic Acids Res. 2003;31:311–314. doi: 10.1093/nar/gkg070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sauermann U, Siddiqui R, Suh YS, Platzer M, Leuchte N, Meyer H, et al. MHC class I haplotypes associated with survival time in simian immunodeficiency virus (SIV)-infected rhesus macaques. Genes Immun. 2008;9:69–80. doi: 10.1038/sj.gene.6364448. [DOI] [PubMed] [Google Scholar]
- Smith MZ, Dale CJ, De Rose R, Stratov I, Fernandez CS, Brooks AG, et al. Analysis of pigtail macaque major histocompatibility complex class I molecules presenting immunodominant simian immunodeficiency virus epitopes. J Virol. 2005;79:684–695. doi: 10.1128/JVI.79.2.684-695.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tosi AJ, Morales JC, Melnick DJ. Comparison of Y chromosome and mtDNA phylogenies leads to unique inferences of macaque evolutionary history. Mol Phylogenet Evol. 2000;17:133–144. doi: 10.1006/mpev.2000.0834. [DOI] [PubMed] [Google Scholar]
- Valentine LE, Watkins DI. Relevance of studying T cell responses in SIV-infected rhesus macaques. Trends Microbiol. 2008;16:605–611. doi: 10.1016/j.tim.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe A, Shiina T, Shimizu S, Hosomichi K, Yanagiya K, Kita YF, et al. A BAC-based contig map of the cynomolgus macaque (Macaca fascicularis) major histocompatibility complex genomic region. Genomics. 2007;89:402–412. doi: 10.1016/j.ygeno.2006.11.002. [DOI] [PubMed] [Google Scholar]
- Wiseman RW, Wojcechowskyj JA, Greene JM, Blasky AJ, Gopon T, Soma T, et al. Simian immunodeficiency virus SIVmac239 infection of major histocompatibility complex-identical cynomolgus macaques from Mauritius. J Virol. 2007;81:349–361. doi: 10.1128/JVI.01841-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiseman RW, Karl JA, Bimber BN, O’Leary CE, Lank SM, Tuscher JJ, et al. MHC genotyping with massively parallel pyrosequencing. Nat Med. 2009 doi: 10.1038/nm.2038. (in press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yant LJ, Friedrich TC, Johnson RC, May GE, Maness NJ, Enz AM, et al. The high-frequency major histocompatibility complex class I allele Mamu-B*17 is associated with control of simian immunodeficiency virus SIVmac239 replication. J Virol. 2006;80:5074–5077. doi: 10.1128/JVI.80.10.5074-5077.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.