Abstract
Data-independent tandem mass spectrometry isolates and fragments all of the molecular species within a given mass-to-charge window, regardless of whether a precursor ion was detected within the window. For shotgun proteomics on complex protein mixtures, data-independent MS/MS offers certain advantages over the traditional data-dependent MS/MS: identification of low-abundance peptides with insignificant precursor peaks; more direct relative quantification, free of biases caused by competing precursors and dynamic exclusion; and faster throughput due to simultaneous fragmentation of multiple peptides. However, data-independent MS/MS, especially on low-resolution ion-trap instruments, strains standard peptide identification programs, because of less precise knowledge of the peptide precursor mass and large numbers of spectra composed of two or more peptides. Here we describe a computer program called DeMux that deconvolves mixture spectra and improves the peptide identification rate by ~25%. We compare the number of identifications made by data-independent and data-dependent MS/MS at the peptide and protein levels: conventional data-dependent MS/MS makes a greater number of identifications but is less reproducible from run to run.
Introduction
Proteomic samples pose extraordinary analytical challenges. Proteins expressed within the cell have a broad variety of physiochemical properties and a huge range of natural abundances. To overcome the variation in physiochemical properties, methods based on “shotgun” proteomics, digest the proteins to peptides prior to analysis1 2. The peptides in the mixture are then separated by microcapillary liquid chromatography (μLC) and emitted directly into a tandem mass spectrometer. Peptides are isolated and fragmented by tandem mass spectrometry (MS/MS) “on the fly”, and the resulting fragmentation spectra are searched against a sequence database to identify the respective peptide sequences3. This workflow is now standard in most proteomics laboratories, due to a combination of robust commercially available tandem mass spectrometers and well-established database searching software, such as SEQUEST3, Mascot4, and X!Tandem5.
Most shotgun proteomics methods acquire the MS/MS spectra using an approach known as data-dependent acquisition (DDA). With DDA, the mass spectrometer acquires a survey or full-scan mass spectrum of unfragmented precursor ions, followed by tandem mass spectra of individual precursor ions detected in the full scan. For example, “top-five” data-dependent acquisition selects the five most intense peaks in the survey scan for MS/MS, unless a selection is overridden by an inclusion or exclusion list of required and forbidden precursors.
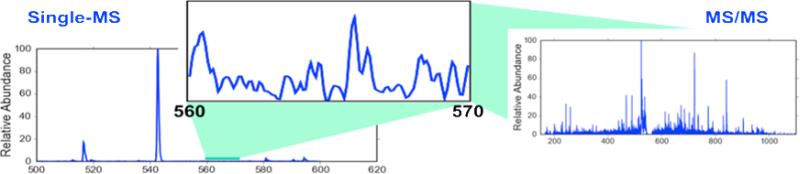
Although DDA is a powerful and versatile strategy, it suffers from several fundamental limitations. First, DDA may compromise the sensitivity of tandem mass spectrometry, because the full-scan mass spectrum used for the selection of the precursor ions typically contains significantly greater chemical background interference than a tandem mass spectrum6. As shown in Figure 1, a peptide could have abundance above the MS/MS detection limit but go unselected for fragmentation because the precursor is masked by background interference.
Figure 1.
Comparison of the signal to noise of data collected using a single stage of mass selection and tandem mass spectrometry. It is possible to identify peptides by MS/MS even in m/z ranges without a detectable precursor ion.
A second limitation of DDA is that the MS/MS spectra are rarely sampled at the optimal portion of the peptide elution profile. An approach known as dynamic exclusion prevents the mass spectrometer from acquiring more than one MS/MS spectrum of the same precursor peak within a short time period – typically 30 seconds. Dynamic exclusion is essential for sensitivity of DDA, because it forces the mass spectrometer to sample peptides other than the most abundant signals at any one point in time. However, an MS/MS spectrum is often acquired at the beginning of the peptide elution, when the signal-to-noise is relatively poor, and then dynamic exclusion prevents the acquisition of a second MS/MS spectrum at the apex of the elution profile. Recently researchers have made efforts to estimate elution profiles in real-time to acquire higher-quality tandem spectra.7
Another related challenge in MS/MS data acquisition is that as many as 15 – 20% of the sampled MS/MS spectra from a complex mixture can contain two or more coeluting molecular species within the isolation window.8 9 10 11 Identifying the peptides in these mixture spectra can be quite difficult, and is beyond the capability of most standard database-search programs. Acquiring MS/MS spectra continuously over time can potentially minimize the challenge posed by coeluting peptides because although coeluting species are present at the same retention time, they rarely have the same elution profile. Thus if MS/MS spectra are acquired continuously, one or more of the peptides could be captured with less coeluting interference, and the mixtures could be computationally resolved.
The most serious limitation of data-dependent acquisition, however, is poor reproducibility. The selection of precursor ions is semi-random, contingent upon elution time, peak intensity rankings, and dynamic exclusion lists, and hence the mass spectrometer acquires MS/MS spectra of different low-abundance peptides in repeated runs of the same material. As many as 30% of the sampled peptides can vary between replicate analyses of the same sample.12 A protein of specific interest, can go undetected if its most detectable peptide misses the DDA list for the few full scans taken during its elution profile, and this chance event may lead to the false conclusion that the protein is absent or decreased in abundance.
A different data acquisition strategy avoids the selection of individual precursors, and instead fragments everything eluting from the column. Versions of this general “shotgun fragmentation” approach include nozzle-skimmer fragmentation13, shotgun CID14, multiplexed tandem mass spectrometry15, and most recently, MSE from Waters Corporation16. Shotgun CID, for example, cycles between no fragmentation and collision-induced dissociation (CID) on alternating scans without precursor isolation. The elution profiles of the fragments are then correlated with the elution profiles of the intact precursors to “deconvolve”, or unmix, the fragments within individual MS/MS spectra. The shotgun-fragmentation approach is appealing because it avoids the random sampling of data-dependent acquisition, and thereby improves the reproducibility of peptide and protein identifications from technical replicates. A downside of this approach is that the fragmentation spectra are not acquired by tandem mass spectrometry, which reduces the chemical background in the product ion spectrum, because only precursor ions within the selected m/z window contribute product ions.
An interesting alternative to both data-dependent acquisition and shotgun fragmentation was reported by Venable et al17. Rather than using a precursor scan to trigger the acquisition of a tandem mass spectrum, Venable et al. reported an approach that steps a relatively wide isolation window (for example, a window of width 10 m/z) across the mass range while acquiring a tandem mass spectrum at each of the 10 m/z increments. Thus, a tandem spectrum is acquired for each m/z range at regular time intervals. The fast duty cycle of the latest generation of linear ion-trap mass spectrometers facilitates 10 to 20 MS/MS scan events within 2 to 4 seconds. The MS/MS spectra are acquired independent of the data in prior scans, and hence Venable et al. named the approach data-independent acquisition (DIA). Automatic gain control on ion trap mass spectrometers further improves the detection of low-abundance molecular species. MS/MS spectra are frequently drawn from m/z ranges that have no strong peaks in the single-MS scan, as shown in Figure 1. Thus data-independent acquisition (DIA) could potentially give both greater sensitivity and greater reproducibility than DDA. DIA should also allow more accurate relative quantification18 than DDA spectral count quantification,12 based on DIA chromatogram alignment and extracted ion currents, as is currently done with label-free differential single-MS profiling.19 By varying the isolation window width and the number of windows, DIA can potentially trade off peptide coverage and quantification accuracy.
To realize these potential benefits, data-independent acquisition requires more powerful data analysis. The approach strains current peptide identification software, both in running time and identification performance, due to high precursor mass uncertainty and complicated mixture spectra containing fragment ions from two or more peptides. In the original implementation, Venable et al. did not develop new data analysis techniques for assigning peptide identifications to the product ion spectra from the wide isolation windows. SEQUEST was used to make (at most) one peptide identification per spectrum.
Here we describe algorithms and software for deconvolving DIA spectra, to identify more than one peptide per spectrum. The same software, called DeMux, also improves m/z accuracy and signal-to-noise by averaging or clustering spectra in adjacent scans. As in the MSE strategy from Waters, or the AMDIS algorithm20 21 for deconvolution of gas chromatography mass spectrometry data, DeMux associates product ion peaks to a single molecular species using the covariance of the elution profiles. However, our software, unlike the proprietary software from Waters, is designed for high-speed, low-resolution ion-trap instruments. We show that deconvolved spectra give approximately 25% better peptide identification rates than unprocessed spectra, when using the ByOnic database search engine22. We minimize the running time associated with the large precursor isolation window using an approach similar to sequence tagging called “lookup peaks” to filter candidate peptides,22 so that only the most promising go on to full scoring. DeMux is available by request from the corresponding author.
Methods
Materials
All reagents were purchased from Sigma-Aldrich unless specified otherwise.
Sample Preparation
Plasma from Dahl Salt Sensitive Rats
Plasma was collected from 15 male Dahl salt sensitive rats. Seven were fed a high-salt diet and developed heart failure and 8 were fed a normal, low-salt diet and were used as healthy controls. Blood was collected by cardiac puncture and placed in EDTA blood collection vials. Plasma was isolated by centrifugation at 1300 × g for 15 min at 4° C, was removed to ensure no cellular contaminants and flash frozen at −80° C until further analysis. All animal procedures were approved by the Institutional Animal Care and Use Committee of The University of Vermont and were conducted in accordance with the Guide for the Care and Use of Laboratory Animals (National Research Council, Washington, DC).
A 10 μL aliquot of each of the unfractionated plasma samples were diluted with 90 μL of ammonium bicarbonate buffer (50 mM, pH 7.8). The samples were reduced with the addition of 2 μL 500 mM dithiothreitol and incubated at 50° C for 30 min. The reduced plasma was then alkylated with 6 μL 500 mM iodoacetamide and incubated at room temperature for 30 min in the dark. To digest the plasma proteins to peptides, 5 μg of trypsin was added to each sample and incubated at 37° C for 4 hours. The digestion was then quenched by the addition of 100 μL of our HPLC buffer A containing 95% water, 4.9% acetonitrile, and 0.1% formic acid. The samples were then centrifuged at 14,000 RPM at 4° C in a table top microcentrifuge (Eppendorf 5417R) to remove insoluble material, and stored at −80° C until analysis.
C. elegans Lysate
C. elegans (Bristol N2 strain) were cultured at 20° C on agarose plates containing E. coli (strain OP50) using standard techniques. Mixed stage worms are washed off the plates with M9 buffer and sucrose floated to remove bacterial contaminants. Worms are then pelleted, washed, resuspended in lysis buffer (310 mM NaF, 3.45 mM NaVO3, 50 mM Tris, 12 mM EDTA, 250 mM NaCl, 140 mM dibasic sodium phosphate pH 7.6), and lysed using immersion sonication. Cell debris and unbroken cells were removed by a low speed spin at 2,000 RPM. The supernatant from the low speed spin was collected and spun again at 14,000 RPM. The supernatant was mixed 1:1 with 0.2% RapiGest in 50 mM NH4HCO3, pH 7.8. The protein sample was then reduced, alkylated, and digested with trypsin as described above for plasma. The resulting peptides were stored at −80° C until analysis by μLC-MS/MS as described below.
Data Acquisition
Fused silica capillary tubing (75 μm I.D.; Polymicro Technologies) was pulled to a tip of ~5 μm at one end and packed with 40 cm of Jupiter Proteo reversed phase chromatography material (Phenomonex, Torrance, CA). The column was then placed in-line with an Agilent 1100 HPLC system and an LTQ ion trap mass spectrometer. The respective digests were loaded onto the microcapillary column from the autosampler as described previously2. The plasma digest was separated using a 45 minute gradient, and the C. elegans digest was separated using a 60 minute gradient. The effluent from the column was electrosprayed into the LTQ using a distal voltage (2.2 kV) applied directly to the solvent. Data-dependent acquisition MS/MS spectra were acquired with a single-MS survey scan triggering 5 MS/MS scans. DDA-triggered precursor ions were isolated using a 2 m/z isolation window and activated with 35% normalized collision energy. The automatic gain control was set to 30,000 and 10,000 charges for MS and MS/MS spectra respectively.
DIA spectra were acquired on the plasma digest by cycling through a fixed number of precursor mass windows of a fixed size. For example, our basic approach used ten individual 10 m/z windows, covering precursors of m/z 500 – 510, 510 – 520, ... 590 – 600. We acquired the mass spectra in profile mode, with the product ion spectra acquired from the low m/z cut-off to 2x the precursor m/z.
For the C. elegans experiment, we investigated three variations of DIA. The first variant used a method with ten scan events with 10 m/z windows. The second variant used a method with ten 5 m/z windows. These ten, 5 m/z windows covered precursors of m/z 500 – 505, 505 – 510, ... 545 – 550. A third variant used a method with ten scan events with 10 m/z windows that were shifted during the elution time. Assuming that the optimal center of the DIA scan events does not remain constant during the μLC-MS/MS analysis, the center of the 100 m/z total window was assessed by the most frequently detected single-MS m/z at each specific retention time based on a prior DDA experiment.
Data Analysis
Problem Statement
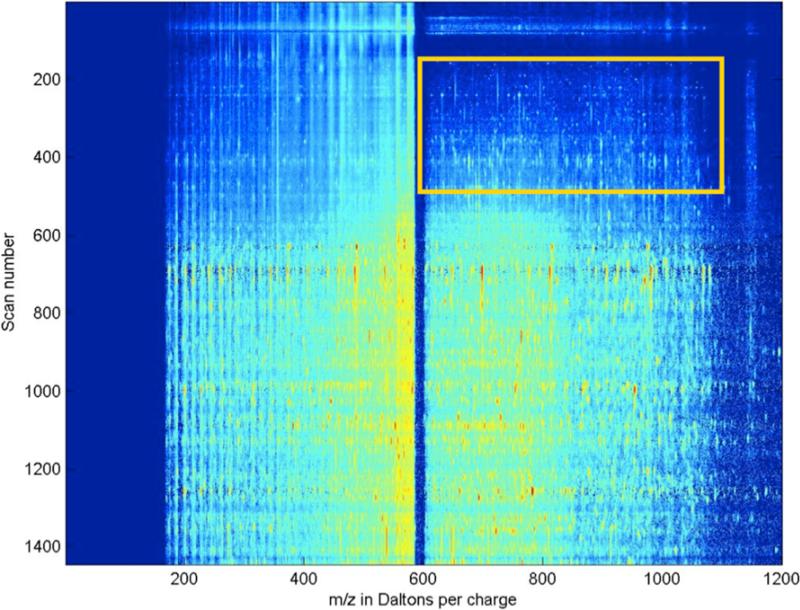
As shown in Figure 2, DIA spectra of the same precursor range, such as m/z 590 – 600, can be formed into a chromatogram showing MS/MS intensities as a function of time. We can consider a tandem-MS chromatogram to be an m × n matrix M with nonnegative entries, where m is the number of time intervals and n is the number of m/z bins. The entry in row i and column j is proportional to the ion current at the i-th time interval and the j-th m/z bin. Our aim is to factor M into a sum of simpler matrices, M = M1 + M2 + ... + Mp, where each matrix Mk contains only a single peptide. If matrix Mk contains a single peptide, then—assuming ideal mass spectra without any chemical or measurement noise—it has a particular form: Mk is the outer product of the peptide's mass spectrum Sk and its elution profile Ek. The mass spectrum Sk is a row vector giving the fraction of the peptide's ion current contributing to fragments at each m/z bin. The elution profile Ek is a column vector giving the ion current of peptide k at each time interval. A typical elution profile Ek has just one nonzero interval with a single maximum, but there are exceptions (Figure 3). For peptide identification we only use the mass spectrum Sk but for quantification we also require Ek.
Figure 2.
Data from data-independent acquisition (DIA) plotted as a heat map. In this heat map, color is proportional to log ion counts. The X-axis is the m/z of the product ions formed from the isolation and activation of molecular species with precursor m/z between 590 and 600. The Y-axis is the scan number. Our DIA strategy cycled through ten of these 10-m/z ranges, completing each cycle in about 2 seconds; a 50-minute LC run gives about 1500 scans for each precursor window. Because the MS/MS spectra were acquired using frequency based activation, all precursor and product ions within the isolation window, in this case m/z 590 – 600, were fragmented to completion. Thus, product ions expected within this relatively large isolation window would not be observed.
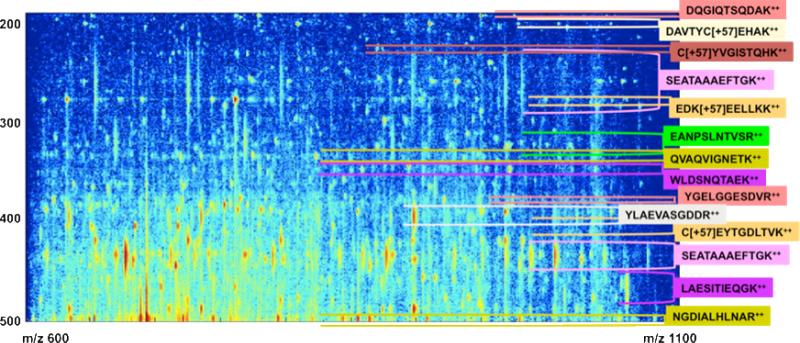
Figure 3.
A blow-up of a narrow region of the heat map shown in Figure 2. Each peptide is represented by a number of fragment ions, which appear as vertical stripes in the chromatogram. Most rows are mixture spectra, crossed by more than one set of vertical stripes. Most peptides elute in a single chromatographic peak, with roughly Gaussian elution profiles spanning 20 – 200 seconds but there are several exceptions. Indeed the abundant peptide SEATAAAEFTGK elutes almost continuously over 300 scans (about 10 minutes), and its intensity has two distinct maxima within this interval.
DeMux Algorithm
DeMux combines peak clustering with time-domain averaging to give approximate answers for the spectra Sk. The algorithm has four major steps:
Conversion of DIA spectra,
Clustering chromatogram columns,
Defining time domain filters, and
Forming synthetic spectra.
Step 1 converts centroided DIA spectra into coarse profile-mode spectra to form chromatograms of manageable size. The coarse spectra have bins of width 1.0005 Daltons. The small fractional part of 0.0005 is included so that, for example, m/z measurements of 1000.49 and 1000.51, which might well represent the same ion, map to the same bin rather than two different bins. This “smart rounding” removes the mass defect characteristic of peptide ions, so that essentially all the singly charged peptide ions with the same integral mass map to the same bin. The chromatogram in Figure 2 is thus a 1440 × 1200 matrix, that is, 1440 mass spectra each containing 1200 m/z bins. Step 1 keeps the original, centroided spectra as well; these will be used in Step 4.
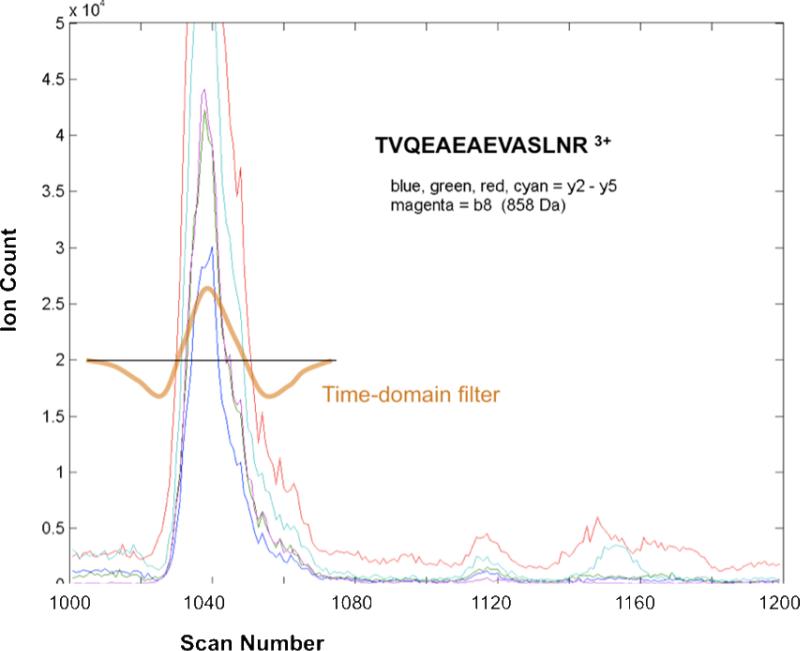
Steps 2 – 4 process the chromatogram in blocks of 100 rows, overlapping by 50%, so that rows 1 – 100 form the first block, 51 – 150 the second block, 101 – 200 the third block, and so forth. The parameter 100 (corresponding to about 200 seconds) is not critical; any number from 50 to 200 works equally well. The number of rows in a block should be large enough that a peptide's intensity is likely to rise and fall within the block, but small enough that many m/z bins are uncontaminated, containing a fragment ion of only one peptide throughout the block. Each block is then processed as follows. Step 2 sorts the columns by decreasing total intensity to form a list C1, C2, ... Cn, where in our case n=1200. The first cluster center is C1. Step 2 finds all the columns with sufficiently high correlation coefficient with C1, and assigns these columns to cluster 1. It then finds the least number j such that Cj is not yet in a cluster; this is the most intense unassigned column. If Cj has sufficiently high intensity, higher than a parameter MinIntensity, it serves as cluster center 2, otherwise the clustering is deemed complete. We then find all the columns with sufficiently high correlation coefficient with Cj and assign these to cluster 2. We proceed in this manner, each time picking the least number j such that Cj is not yet in a cluster, until the intensity of Cj falls below MinIntensity or the number of clusters exceeds a parameter MaxCluster. A column can be assigned to more than one cluster (this happens rarely), but once assigned to a column it cannot act as a cluster center. A correlation coefficient threshold of 0.9 worked well. Our notion of cluster is not intended to capture all the fragment ions from a peptide, but rather it is meant to capture enough ions to form a reasonable elution profile. For intense peptides a single clean column, uncontaminated by fragments of other precursors, is sufficient, and indeed many clusters have only single members. With a strict correlation coefficient threshold like 0.9, the mapping from clusters to peptides is many-to-one, but with a less strict threshold like 0.7, the mapping is almost one-to-one. Figure 4 shows a cluster of five columns for a block of 200 rows.
Figure 4.
Detection of covarying product ions in the time domain. A cluster of 5 highly correlated columns (not yet identified) is used to form a time-domain filter for computing a synthetic spectrum. In this case the 5 columns are fragment ions (y2 – y5 and b8) of the same peptide.
For each cluster c, Step 3 sums all the columns in the cluster to form an approximate elution profile Elute(c); this is a column vector of length equal to the number of rows in a block. Elute(c) is smoothed slightly by averaging each point with its two neighbors, using a 3-point finite-impulse-response filter with weights in the proportions 1, 2, 1. The smoothed elution profile is then used to form another finite-impulse-response filter to extract a synthetic spectrum from the chromatogram. This time-domain filter always has the form of a narrow bell-shaped curve minus a broad bell-shaped curve, as shown in Figure 4, even if the elution profile is noisy and multimodal. More specifically, we set the maximum of the filter to coincide with the maximum point of the elution profile Elute(c). We set the two zero crossings of the filter to coincide with the points on either side of the maximum point where Elute(c) first drops below one-fourth of its maximum height, or if Elute(c) does not drop below one-fourth within that block of spectra, then the end of the block. (Thus the positive part of the time-domain filter may be asymmetrical.) The negative-weight tails each have length equal to the length of the positive part of the filter. The actual weights (for the negative part and each half of the positive part) are drawn from Gaussian distributions, and normalized so that the entire filter integrates to zero.
We explain the algorithm for computing the time-domain filter with an example. Assume Elute(c) is the vector (0, 0, 0, 0, 0.1, 0.5, 0.4, 0.8, 1.7, 1.4, 1.0, 0.8, 0.5, 0.3, 0.2, 0.1, 0, 0, 0, 0)T, with entries representing times 1 to 20. The maximum of Elute(c) is 1.7, attained at time 9. The algorithm sets the zero crossings of the filter to time 7, where Elute(c) has value 0.4 (less than 1.7 / 4), and time 14, where Elute(c) has value 0.3. Thus the positive part of the filter includes times 8 to 13, six points in all. The negative-weight parts of the filter are points 1 to 6 and 15 to 20. Negative weights for points 1 to 20 are computed using the formula for the probability density function of a Gaussian with center 10.5 and standard deviation 5.25, so that the beginning and end of the filter correspond with two standard deviations on either side of the center. Negative weights for points 7 and 14 are “artificially” set to zero. Positive weights for points 8 to 9 are computed using a Gaussian distribution with center 9 and standard deviation 1.0, scaled so that the height at 9 is 1.0; positive weights for points 9 to 13 are computed using a Gaussian distribution with center 9 and standard deviation 4.0, again scaled so that the height at 9 is 1.0. The positive weights for points 8 to 13 are then scaled so that the sum of the positive weights equals the sum of all negative weights (points 1 to 20). Then the final weight for each point j of the time-domain filter is set to the sum of the negative and positive weights for point j.
The time-domain filter is a matched filter, meant to enhance the peaks with a particular elution profile and diminish co-eluting peaks with different elution profiles. The filter also diminishes high-frequency noise, that is, random peaks appearing in only one or two scans. Andreev et al.xxiii published a time-domain filtering method for single-MS chromatograms; their method does not use negative-weight tails so it cannot unmix mixtures.
Using the time-domain filter from Step 3, Step 4 computes a synthetic, or composite, spectrum Synth(c) for each cluster c. Synth(c) is a weighted average of the original centroided spectra, not the coarse profile spectra. Synth(c) contains only those peaks that are found in the centroided spectrum from the maximum-intensity spectrum for cluster c. The intensity of each peak is replaced by a weighted average of the intensities of that “same” peak in adjacent spectra, where the weights are drawn from the time-domain filter. For the example in Figure 4, scans from ~1030 to ~1050 contribute positively to the composite spectrum, and scans from ~1010 to ~1030 and from ~1050 to ~1070 contribute negatively. A peak is considered to be the same peak if it matches within a user-settable tolerance (in this case, 0.25 Thompsons) of the peak in the maximum-intensity spectrum. If a peak ends up with negative intensity, it is dropped from the spectrum. For the example in Figure 4, a fragment peak with uniform intensity over scans 1010 – 1040 will end up with half its original intensity, and one with uniform intensity over scans 1010 – 1070 will end up at zero.
The m/z readings within Synth(c) are also weighted averages. Only the scans with positive weights add their m/z readings to the weighted average, and the weights used for the m/z reading are the ones from the time-domain filter. The rationale here is that the m/z readings from a spectrum drawn at the top of the elution pulse should be more accurate due to better ion counts.
DeMux outputs exactly one synthetic spectrum per cluster. The number of clusters can be adjusted using the parameters MinIntensity and MaxCluster. With a high setting for MinIntensity or a low setting for MaxCluster, DeMux greatly reduces the number of spectra and hence the search time. In all the experiments reported below, we set MinIntensity to zero (meaning no intensity requirement) and MaxCluster to 50, so that DeMux always reduced a 100-spectrum block to 50 synthetic spectra. Because blocks overlapped by 50%, this choice means that the number of output spectra nearly equals the number of input spectra. With such light data reduction, each peptide typically corresponds to several clusters. In the case of multimodal elution profiles, there will be at least one cluster per elution profile maximum.
Database Search
We used ByOnic, a database-search program that uses a small amount of de novo analysis to speed up search times. This speed-up trick is helpful for identification of DIA spectra, because DIA spectra have greater precursor mass uncertainty that do DDA spectra, leading to larger searches. With 10-Thompson DIA windows, the precursor mass is known only to about +/− 10 Daltons for +2 precursors and +/− 15 Da for +3 precursors. To account for measurement inaccuracy and isotope peaks, our ByOnic searches used tolerances of +/− 12 uncertainty for +2 precursors and +/− 18 for +3 precursors.
As a further speed-up, we used two-pass search: the first pass searched only +2 fully tryptic precursors (with any number of missed cleavages), with the fixed modification of carbamidomethylated cysteine (camC) and the variable modifications of pyro-glu of N-terminal Q, E, and camC. The second pass searched a smaller database containing only those proteins (or decoys) with at least one match (of any score) from the first search. The second pass searched both tryptic and semitryptic peptides with the following common variable modifications enabled: oxidized M, deamidated N and Q, pyro-glu N-terminal Q, E, and camC, and alkylated H, R, K, and N-terminus (common artifacts of iodoacetamide treatment). For uniformity, we used two-pass search for both DIA and DDA samples, even though DDA has less need of speed-up. For the rat samples we used a database with ~26,000 protein sequences, and for the C. elegans samples a database with about ~27,500 sequences. In both cases we concatenated a decoy database containing each of the true proteins with residue sequence reversed, to estimate the peptide and protein false discovery rates (FDR).
ByOnic22, like ProbIDtree10, has the capability of identifying more than one peptide per MS/MS spectrum. It does this by removing (“knocking out”) all the identified peaks from a first peptide identification, and then writing out a new MS/MS spectrum that can be searched and scored against the protein database like any other spectrum. We used ByOnic's knockout feature both to increase the number of identifications and to assess the performance of DeMux's deconvolution.
For integrating peptide identifications into protein identifications, we used ByOnic's companion program ComBynexxiv, which is roughly equivalent to ProteinProphetxxv. A Web interface to ByOnic and ComByne is publicly available at http://bio.parc.com/.
Results and Discussion
In brief, we found that ByOnic could identify ~20% more distinct peptides using DeMux and synthetic spectra than it could identify with the original DIA spectra. ByOnic's knockout feature adds another ~10% more distinct peptides. DeMux also improved m/z accuracy. The fragment m/z's in synthetic spectra were slightly more accurate than those from the Max(c) spectrum: the median value of the absolute value of the error (true m/z minus observed m/z) was 0.045 in synthetic spectra and 0.066 in original DIA spectra, centroided by our own in-house software. We also found DIA to be more reproducible than DDA, with ~85% of well-identified peptides found in both LC runs from a pair of technical replicates, as opposed to ~65% for DDA. Despite these improvements, DIA with DeMux and knockouts does not identify as many distinct peptides as a standard DDA strategy. We now give more detailed results on each of the two experiments.
Rat Blood Plasma
We used DIA for an analysis of proteins in a rodent model of heart failure. Each data set contains all the spectra, original or deconvoluted, from a single LC run. Hence we had 60 data sets for 30 LC runs, the data from each run with and without DeMux. On each data set, we performed a two-pass search with modifications as described above on both +2 and +3 precursors. Then we searched knockout spectra for +2 precursors only. (Some 80% of all identifications are +2 peptides, so we decided that searching knockouts for +3 precursors would not be worth the search time.) Each data set took about 12 hours of processor time on a four-core Sun Microsystems computer.
We counted the number of valid identifications at the spectrum, peptide, and protein levels. We called a peptide identification valid if it matched one of the top proteins, that is, a protein ranked higher than the highest-ranking decoy sequence, and its ByOnic score was at least 250 (roughly equivalent to Mascot 25). Thus the filtering by protein was strict but the filtering by score relaxed. We considered the same peptide in two different modification states to be distinct. We called a protein identification valid if it was a true protein, not a decoy, and it ranked higher than the second highest decoy. The number of proteins with a score greater than the second highest scoring decoy is more stable than the number of proteins higher than the highest ranked decoy and corresponds to a 1% to 2% false discovery rate.
As shown in Table 1, the number of identified peptides always increases with the use of DeMux. The number of proteins stays roughly the same, with only slight improvement, typically 1 or 2 more proteins. The use of ByOnic's knockout feature also increases sensitivity. We expected to see that the knockout feature gives a greater percentage increase on the original DIA spectra than on the synthetic spectra, because DeMux separates many mixture spectra, leaving fewer for knockouts. We found, however, that the knockout feature actually gave slightly more improvement on the synthetic spectra than on the original spectra.
Table 1.
Peptide sensitivity increases with the use of DeMux and the use of ByOnic's knockout feature, which attempts to make two identifications per spectrum. The numbers shown are the median, minimum, and maximum over the 30 LC runs on rat blood plasma. The percent improvement is relative to ByOnic without the use of DeMux or knockout spectra. Reproducibility of an LC run is defined as the percent of peptides confidently identified in that run that were confidently re-identified in the run's technical replicate.
| Type of Search | Proteins Med (Min, Max) | Peptides Med (Min, Max) | Percent Improvement Med (Min, Max) | Peptide Reproducibility Med (Min, Max) |
|---|---|---|---|---|
| Original DIA Spectra | 77 (67, 86) | 417 (354, 503) | 0.0 (0.0, 0.0) | 87.0 (80.8, 90.3) |
| Original + Knockout | 77 (68, 85) | 447 (373, 546) | 7.0 (3.4, 8.8) | 88.5 (82.5, 92.5) |
| Synthetic Spectra | 78 (68, 88) | 481 (443, 559) | 22.6 (9.3, 27.0) | 87.7 (83.9, 91.5) |
| Synthetic + Knockout | 79 (68, 91) | 538 (466, 626) | 31.5 (21.8, 40.6) | 88.0 (82.1, 92.7) |
Table 1 also reports the “reproducibility”, meaning the percentage of peptide identifications found in a rat sample that were also found in the sample's technical replicate, that is, Reproducibility(sample i) = (# identifications in i and replicate(i) ) / (# identifications in i). We compared each replicate against the other, that is, percent of replicate 1 found in 2 and vice versa, for a total of 30 comparisons. For our reproducibility statistic, we counted only unmodified peptides from top proteins (ranking above the top decoy) with ByOnic scores at least 350 (approximately equal to a Mascot score of 35); the peptide had to have a high score and be unmodified in both replicates to be counted as observed twice. Our median numbers of ~88% exceed previously reported numbers and our own numbers (see below) for data-dependent acquisition. Indeed not only were peptide identifications highly reproducible between technical replicates but also spectral counts as shown in Figure 5.
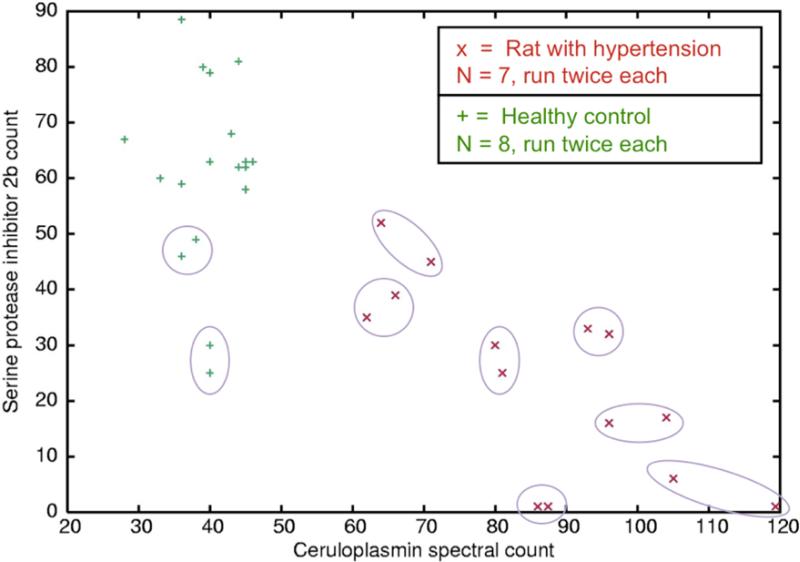
Figure 5.
DIA spectral count on a single protein—ceruloplasmin—gives perfect separation of the hypertensive rats and the controls. Circled pairs show technical replicates; for clarity not all are shown.
DIA's high reproducibility should be advantageous for the analysis for any sample that compares the identification of peptides between groups of samples. Figure 5 shows that spectral counts on a single protein, ceruloplasmin, give perfect separation of the hypertensive and control rats in this model system of heart failure. Many other proteins show some separation power, and we can see that the number of spectra matched to ceruloplasmin is negatively correlated with the number matched to serine protease inhibitor 2b. Ceruloplasmin is an antioxidant, and high levels have been linked previously to heart failure.xxvi xxvii xxviii
For this experiment we used spectral counts on the original DIA spectra for relative quantitation. An alternative would be integration of the area under the extracted ion chromatograms from the DIA MS/MS spectra. Spectral counting on the deconvoluted spectra output by DeMux is unlikely to be quantitative, because the time-domain filtering forms synthetic spectra representing various lengths of time, from as short as ~6 seconds to as long as ~100 seconds in the case of 100-spectra blocks. Thus, DeMux in effect splits DIA data into two sets of spectra: one set better for quantitation and another set better for identification.
We do not know whether spectral count on original DIA spectra scales linearly with actual protein abundance, as it appears to do with DDA's semi-random sampling12. Liu et al.12 explained DDA spectral count with a random sampling model, and hence we are hesitant to make any quantitative claims about spectrum counting with DIA data, because DIA intentionally eliminates randomness in the MS/MS sampling. However, it is reasonable to assume that a larger DIA spectral count indicates greater abundance, because more abundant peptides remain above the instrument's detection limit and the software's “identification threshold” over longer periods of time. Thus, selected proteins such as ceruloplasmin appear to have a difference in abundance between conditions.
Application to a C. elegans lysate
The C. elegans lysate was run with four different data acquisition strategies, (1) DIA with 5-Thompson windows from m/z 500 – 550, (2) DIA with 10-Thompson windows from m/z 500 – 600, (3) DIA with 10-Thompson windows that shift linearly over the course of the LC run, starting at 400 – 500 and ending at 650 – 750, and finally (4) standard top-ten DDA. We collected four technical replicates for each strategy, 16 data sets in all.
As with the rat plasma, on each data set we performed a first-pass database search (+2 precursors only, no variable modifications except pyro-glu at an N-terminal E, Q, or camC) to compile a small protein database. We then performed the second-pass search with variable modifications of oxidized M, deamidated N and Q, pyro-glu N-terminal Q, E, and camC, and alkylated H, R, K, and N-terminus. For DIA data, we also searched knockout spectra, again only for +2 precursors. Table 2, lines 2 and 3, used the same data, but line 2 shows the results of an analysis of the data without using DeMux to produce synthetic spectra (but the analysis did use knockout spectra), to confirm the effectiveness of DeMux on the C. elegans sample. All the other lines of Table 2 report the results of analyses using both synthetic and knockout spectra, as this option gives uniformly better results. As above, we report the number of proteins ranked above the second-highest decoy. For the C. elegans sample, DeMux improved both peptide and protein sensitivity, with improvements ranging from about 15% to 40%. The peptide-level improvement is similar to that for the rat plasma sample, but the protein-level improvement is much better. For reproducibility, we considered all 12 possible ordered pairs of the four technical replicates for each data acquisition strategy, that is, the percent of the peptide identifications from replicate i found in replicate j, for each choice of i and j with 1 ≤ i, j ≤ 4 and i ≠ j, and we report the mean over all pairs. This statistic allows direct comparison of the reproducibility of experiments with any number of technical replicates, for example, the mouse plasma with two replicates and the C. elegans with four replicates. Other statistics, such as the percent of peptides found in all replicates, naturally decrease with the number of replicates. Table 2 shows that the DIA strategies are much more reproducible than the DDA strategies, although not quite as reproducible on the C. elegans lysate as on the mouse blood plasma. DDA is less reproducible partly because it is more sensitive. If we filter the DDA identifications more stringently by requiring a higher ByOnic score, then DDA's reproducibility increases from ~66% to ~73% as its sensitivity falls to DIA level.
Table 2.
Data-independent tandem mass spectrometry gives fewer identifications than standard top-10 data dependent acquisition. Of the four DIA strategies, the best method used ten 10-Thompson windows, shifting to higher m/z later in the elution. DIA with shifting 10-Th windows outperforms DDA by 12% at the protein level and 20% at the peptide level when we limit attention to DDA precursors of m/z 500 – 600. The identification numbers shown are the mean, minimum, and maximum over 4 replicates. The reproducibility percentage is the mean percent of peptides re-identified over all 12 ordered pairs of replicates.
| Type of Data Acquisition | Proteins Mean (Min, Max) | Peptides Mean (Min, Max) | Reproducibility Mean between pairs |
|---|---|---|---|
| DIA, 5-Th windows | 131.75 (127, 137) | 310.5 (295, 327) | 85.0% |
| DIA, 10-Th windows, no DeMux | 170.25 (166, 172) | 335.0 (297, 356) | 82.5% |
| DIA, 10-Th windows | 204.0 (197, 212) | 412.0 (393, 454) | 84.4% |
| DIA, 10-Th windows, shifting | 227.25 (220, 235) | 370.5 (347, 386) | 82.0% |
| DDA, all m/z | 542.25 (476, 640) | 1280.25 (1175, 1423) | 65.4% |
| DDA, m/z 500 - 600 | 201.75 (183, 216) | 316.75 (264, 361) | 66.8% |
As shown in Table 2, DIA does not identify as many peptide and protein identifications as DDA. This smaller number of identifications is not surprising, because DIA collects MS/MS spectra on a smaller range of precursor m/z's, and indeed DIA with 5-Th windows gave the weakest performance of all the trials, as it covers the smallest range of precursors, only m/z 500 – 550. DDA precursors had m/z varying from 402 to 1372. We can also compare the two strategies by considering only the DDA spectra of peptides with precursor m/z in the range 500 – 600, approximately 20% of all DDA tandem spectra. With this limitation, DDA gives fewer unique peptide identifications than DIA, but almost the same number of protein identifications. Additionally, DDA uses only ~3,000 spectra to make its identifications, whereas DIA uses ~15,000. The peptide identifications that are unique to DIA and not DDA include modified forms, especially deamidations, for which the unmodified form is frequently identified by both DIA and DDA. Although these modified peptides have low spectral counts, they inflate the unique peptide count. Thus, the situation shown in Figure 1, in which DIA makes a high-confidence identification of a novel peptide sequence that is undetected in all DDA scans and never selected for MS/MS, was rare in these data.
Conclusions
Identification rate, at either the peptide or protein level, can be expressed as efficiency divided by redundancy, where efficiency is the number of identifications divided by the number of spectra, and redundancy is the total number of identifications divided by the number of unique identifications. The DIA strategy proposed here turned out to have quite acceptable efficiency: ~50% even without the use of knockouts or DeMux, and up to ~75% or more (depending upon the user-settable parameters within DeMux) with knockouts and DeMux. DDA had an efficiency of ~40%. The redundancy of DIA, however, is quite high: each unique peptide is identified an average of ~9 times compared to ~2.4 for standard top-ten DDA with dynamic exclusion.
There is a natural tension between high identification rate, which calls for low redundancy, and accurate relative quantification, which calls for monitoring an eluting species over time. The DIA strategy proposed here can be tuned to various points along the identification/quantification tradeoff. For example, one could cycle through twenty 10-Thompson wide windows, spanning m/z 500 – 700 in ~4 seconds; this would give coarser quantification but superior identification.
Several factors could conceivably be limiting the number of identifications for DIA on an ion-trap instrument. The possible factors include: (1) signal-to-noise ratio drops as the isolation window width increases; (2) the most abundant precursor sets the trap fill time and thus reduces the chance of making a second identification from the same spectrum; (3) DIA spectra are too noisy or complex to identify computationally; and (4) DIA strategies give limited coverage of the m/z range of precursors. The high identification efficiency shown here, and the substantial improvement offered by knockouts and DeMux, argues against factors (1) and (3). The success of knockout spectra argues against factor (2) to some extent, yet we believe that factor (2) remains an important limitation on DIA strategies. Visual inspection of MS/MS chromatograms such as those in Figures 2 and 3 leads us to believe that most DIA spectra contain at least two peptides, so the ~10% improvement due to knockouts and the ~25% improvement due to DeMux reported in Table 1 may still give many fewer identifications than the actual number of eluting peptides. Furthermore, the fact that knockouts contributed ~10% improvement even after DeMux's deconvolution argues that the DIA spectra are multipeptide mixtures that are not so easy to unmix. Finally, we believe factor (4) is overwhelmingly the predominant explanation for DIA's weak performance relative to DDA on the C. elegans benchmark, as evidenced by the fact that 10-Th windows outperformed 5-Th windows, and shifting 10-Th windows outperformed fixed 10-Th windows. This explanation is actually encouraging for DIA on ion-trap instruments, as faster scan speeds in the latest LTQ Velos instrument (Second et al., submitted) will enable broader precursor coverage without excessively compromising quantification.
Even at the current identification rate, the DIA strategy proposed here could be advantageous in certain situations. Our rat plasma experiment suggests that DIA using only a 30 min HPLC gradient would be adequate for monitoring the levels of the abundant plasma proteins, which include many putative markers of disease. This method would offer a faster and more convenient solution than one using single-MS for quantification and tandem-MS for identification, and a more accurate and reproducible solution than one using only data-dependent tandem-MS.
Acknowledgments
The authors acknowledge helpful discussions with John R. Yates from The Scripps Research Institute, Vlad Zabrouskov from ThermoFisher Scientific, and Jesse Canterbury and Jarrett Egertson from the MacCoss Lab. This work was supported in part by NIH grants GM085718 (MB), DK069386 (MJM), and RR011823 (MJM).
References
- 1.MacCoss MJ, Yates JR., III Proteomics: analytical tools and techniques. Curr. Opin. Clin. Nutr. Metab. Care. 2001;4:369–375. doi: 10.1097/00075197-200109000-00006. [DOI] [PubMed] [Google Scholar]
- 2.Wu CC, MacCoss MJ. Shotgun proteomics: tools for the analysis of complex biological systems. Curr. Opin. Mol. Ther. 2002;4:242–250. [PubMed] [Google Scholar]
- 3.Eng JK, McCormack AL, Yates JR., III J. Am. Soc. Mass Spectrom. 1994;5:976–989. doi: 10.1016/1044-0305(94)80016-2. [DOI] [PubMed] [Google Scholar]
- 4.Perkins DN, Pappin DJC, Creasy DM, Cottrell JS. Electrophoresis. 1999;20:3551–3567. doi: 10.1002/(SICI)1522-2683(19991201)20:18<3551::AID-ELPS3551>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 5.Craig R, Beavis RC. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 6.Yost RA, Enke CG. Selected ion fragmentation with a tandem quadrupole mass spectrometer. J. Am. Chem. Soc. 1978;100:2274–2275. [Google Scholar]
- 7.Senko MW, Hemenway E, Hemenway TA. Methods for improved data dependent acquisition. 2007 Nov. 20 United States Patent 7,297,941.
- 8.Hoopmann MR, Finney GL, MacCoss MJ. High-speed data reduction, feature detection, and MS/MS spectrum quality assessment of shotgun proteomics data sets using high-resolution mass spectrometry. Anal. Chem. 2007 doi: 10.1021/ac0700833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Frewen BE, Merrihew GE, Wu CC, Noble WS, MacCoss MJ. Analysis of peptide MS/MS spectra from large-scale proteomics experiments using spectrum libraries. Anal. Chem. 2006;78:5678–5684. doi: 10.1021/ac060279n. [DOI] [PubMed] [Google Scholar]
- 10.Zhang N, et al. ProbIDtree: an automated software program capable of identifying multiple peptides from a single collision-induced dissociation spectrum collected by a tandem mass spectrometer. Proteomics. 2005;5:4096–4106. doi: 10.1002/pmic.200401260. [DOI] [PubMed] [Google Scholar]
- 11.Luethy R, et al. Precursor-ion mass re-estimation improves peptide identification on hybrid instruments. J. Proteome Res. 2006;7:4031–4039. doi: 10.1021/pr800307m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu H, Sadygov RG, Yates JR., III A model for random sampling and estimation of relative protein abundance in shotgun proteomics. Anal. Chem. 2004;76:4193–4201. doi: 10.1021/ac0498563. [DOI] [PubMed] [Google Scholar]
- 13.Hakansson K, Zubarev R, Hakansson P. Combination of nozzle-skimmer fragmentation and partial acid hydrolysis in electrospray ionization time-of-flight mass spectrometry of synthetic peptides. Rapid Comm. Mass Spec. 1998;12:705–711. doi: 10.1002/(SICI)1097-0231(19980615)12:11<705::AID-RCM212>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- 14.Purvine S, Eppel J-T, Yi EC, Goodlett DR. Shotgun collision-induced dissociation of peptides using a time of flight mass analyzer. Proteomics. 2002;3:847–850. doi: 10.1002/pmic.200300362. [DOI] [PubMed] [Google Scholar]
- 15.Page JS, Masselon CD, Smith RD. FTICR mass spectrometry for qualitative and quantitative bioanalyses. Curr. Opin. Biotechnology. 2004;15:3–11. doi: 10.1016/j.copbio.2004.01.002. [DOI] [PubMed] [Google Scholar]
- 16.Plumb RS, Johnson KA, Rainville P, Smith BW, Wilson ID, Castro-Perez JM, Nicholson JK. Rapid Comm. Mass Spectrometry. 2006;20:1989–1994. doi: 10.1002/rcm.2550. [DOI] [PubMed] [Google Scholar]
- 17.Venable JD, Dong MQ, Wohlschegel J, Dillin A, Yates JR., III Automated approach for quantitative analysis of complex peptide mixtures from tandem mass spectra. Nat. Methods. 2004;1:39–45. doi: 10.1038/nmeth705. [DOI] [PubMed] [Google Scholar]
- 18.Turck CW, et al. The association of biomolecular resource facilities proteomics research group 2006 study. Mol. Cellular Proteomics. 2007;6:1291–1298. doi: 10.1074/mcp.M700165-MCP200. [DOI] [PubMed] [Google Scholar]
- 19.Wang W, et al. Quantification of proteins and metabolites by mass spectrometry without isotopic labeling or spiked standards. Analytical Chem. 2003;75:4818–4826. doi: 10.1021/ac026468x. [DOI] [PubMed] [Google Scholar]
- 20.Mallard G, Toropov O. Automatic mass spectral deconvolution and identification software (AMDIS) Chemical Science and Technology Laboratory, National Institute of Standards and Technology; [Google Scholar]
- 21.Stein SE. An integrated method for spectrum extraction and compound identification from gas chromatography/mass spectrometry data. J. Amer. Soc. Mass Spectrometry. 1999;10:770–781. [Google Scholar]
- 22.Bern MW, Cai Y, Goldberg D. Lookup peaks: a hybrid of de novo sequencing and database search for protein identification by tandem mass spectrometry. Anal Chem. 2007;99:1393–1400. doi: 10.1021/ac0617013. [DOI] [PubMed] [Google Scholar]
- xxiii.Andreev VP, Li L, Cao L, Gu Y, Rejtar T, Wu SL, Karger BLJ. Proteome Res. 2007;6:2186–94. doi: 10.1021/pr0606880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- xxiv.Bern M, Goldberg D. Improved ranking functions for protein and modification-site identifications. J. Comp. Biology. 2008;15:705–719. doi: 10.1089/cmb.2007.0119. [DOI] [PubMed] [Google Scholar]
- xxv.Nesvizhskii AI, Keller A, Kolker E. Aebersold. A statistical model for identifying proteins by tandem mass spectrometry. Anal. Chem. 2003;75:4646–4658. doi: 10.1021/ac0341261. [DOI] [PubMed] [Google Scholar]
- xxvi.Reena TR, Annapurna SD, Ushasree B, Pratibha N, Narasimhan C, Soma R. Indian Heart J. 2004;56:72–73. [PubMed] [Google Scholar]
- xxvii.Kim C, Park J, Kim J, Choi C, Kim Y, Chung Y, Lee M, Hong S, Lee K. Metabolism. 2002;7:838–842. doi: 10.1053/meta.2002.33348. [DOI] [PubMed] [Google Scholar]
- xxviii.Garrow TA, Clegg MS, Metzler G, Keen CL. Hypertension. 1991;17:793–797. doi: 10.1161/01.hyp.17.6.793. [DOI] [PubMed] [Google Scholar]