Abstract
In this study, we searched the amphibian species Xenopus laevis and Silurana (Xenopus) tropicalis for the presence of genes homologous to mammalian KIRs and avian CHIRs (KRIR family). By experimental and computational procedures, we identified four related ILR (Ig-like receptors) genes in S. tropicalis and three in X. laevis. ILRs encode type I transmembrane receptors with 3–4 Ig-like extracellular domains. All predicted ILR proteins appear to be activating receptors. ILRs have a broad expression pattern, the gene transcripts were found in both lymphoid and non-lymphoid tissues. Phylogenetic analysis shows that the amphibian KRIR family receptors evolved independently from their mammalian and avian counterparts. The only conserved structural element of tetrapod KRIRs is the NxxR motif-containing transmembrane domain that facilitates association with FcR subunit. Our findings suggest that if KRIRs of various vertebrates have any common function at all, such a function is activating rather than inhibitory.
Keywords: genome mining, IgSF, ITAM, paired receptors, evolution, missing self recognition
1. Introduction
Regulation of the immune response in mammals is mediated by various receptors on the surface of the leukocytes. Killer Immunoglobulin-like Receptors or KIRs are among key players in this regulatory network. KIRs have been first discovered as crucial components of the missing self recognition by human NK cells [1]. It has been later demonstrated that mammalian genomes have a large group of genes that are structurally related to the KIR genes but are functionally different from them. In humans, genes of this family, which we will refer to as KIR-Related Ig-like Receptor (KRIR) family, lay within the Leukocyte Receptor Complex (LRC) on chromosome 19. In addition to the KIR genes, there is a large subfamily of genes known as LILR (also named ILT, LIR, MIR), and several singleton genes, such as LAIR-1/2, OSCAR, FCAR, GPVI, NKp46 [2,3]. Except to LAIR2 that is a secreted protein, all members of the family are cell surface receptors. In the KRIR family, we may find pairs of receptors with similar extracellular regions but triggering opposing signaling pathways: either activating or inhibitory. Inhibitory forms of these so-called paired receptors possess ITIM motifs in their cytoplasmic tails and activating forms associate with signaling subunits containing activating ITAM motifs [4].
A variety of functions have been described for the mammalian KRIR family members. The major role of inhibitory KIRs in regulation of immune responses is to couple with MHC class I molecules on target cells and to protect these cells from NK-mediated lysis [1,2]. It has been speculated that activating KIRs may enhance cytotoxic response to cells with unusual or non-self peptides mounted on HLA I molecules [5]. Binding to classical and non-classical MHC class I antigens has been also demonstrated for human inhibitory receptors LILRB1 and LILRB2, along with their mouse counterpart PIR-B [5-8]. LILR and PIR receptors modulate immune response on various cell types [9-12]; NKp46 has been shown to recognize membrane associated heparan sulfate proteoglycans [13]; Human Fc R is a receptor for IgA [14]; GPVI and LAIR1 interact with collagen [15,16]. In addition, some of the family members have been found to recognize pathogen determinants. For example, NKp46 binds influenza virus hemagglutinin [5], whereas LILRB and PIR-B are able to recognize Staphylococcus aureus [12].
Which of these functions is the most ancient for this structural subset of the immunoglobulin superfamily remains unknown. It should be stressed that, in rodents, the function of MHC class I-specific recognition on NK cells is carried out by the Ly49 receptor family that belongs to the C-type lectin superfamily [17,18]. According to recent evidence, marine pinnipeds may use yet another group of molecules for this purpose. In this mammalian species, lineage Ly49 and KIR subfamilies are each represented by single gene that exhibit little polymorphism [19].
More than a hundred KRIR family genes named CHIRs have been identified in chicken [20-23]. Like their mammalian counterparts, CHIRs are subdivided into activating and inhibitory classes. Thus far, the only function known for some of these receptors is to bind to immunoglobulin class Y (IgY) molecules [24,25]. In the bony fish channel catfish (Ictalurus punctatus), Leukocyte Immune-Type Receptors (LITRs) were found to contain Ig-like domains with weak similarity both to the KRIR and the FcR family receptors [26]. MHC class I binding function was proposed for LITRs [27], but no experimental data supporting this assumption are available and LITR ligands remain yet unknown.
This study aimed to get a deeper insight into the structural and functional evolution of KRIRs by analysis of the family genes in the amphibians Xenopus laevis and Silurana tropicalis. The results obtained show that Xenoponidae KRIRs evolved separately from those of birds and mammals. Only activating receptors were found in the family.
2. Materials and methods
Experimental Animals
Adult outbred Xenopus laevis and Silurana tropicalis were obtained from the X. laevis Research Resource for Immunobiology at the University of Rochester Medical Center (www.urmc.rochester.edu/smd/mbi/xenopus/index.htm). All animals were handled under strict laboratory and UCAR regulations. Animals were euthanized with 0.5% Tricaine methanesulfonate (TMS).
RNA extraction, cDNA synthesis, RT-PCR amplification and cloning
Tissue samples were homogenized in 0.8 mL of Trizol reagent (Invitrogen). Total RNA was extracted according to the manufacturer’s protocol. A sample RNA pellet was resuspended in RNase free water and quantified with SmartSpec spectrophotometer (BioRad). 500 ng of quantified total RNA were used to synthesize cDNA with iScript first strand cDNA synthesis kit (BioRad) according to the manufacturer’s protocol. cDNA samples were diluted three times to a final volume of 60 μl before proceeding to PCR amplification. For each PCR reaction (30 μl total volume) 3 μl of 2 mM dNTPs, 3 μl of 10x PCR buffer, 10 pmol of each primer, 2 U of Taq DNA polymerase (Life Technologies), and 1 μl of cDNA were used. Then tubes were set for 30–40 cycles: 45 sec at 95°C, 45 sec at 60–64°C and 30–90 sec at 72°C. Negative controls (without RT) were also performed with same primers to control for genomic DNA contamination. Bands of our interest were re-amplifed with Pfu polymerase (Sibenzyme), cloned into pBluescript (Stratagene) vector and sequenced as described below. The following primers for different exons were used: Xenopus laevis xILR2.1 5′-GACTCTGAATTAAGTGACATGAT-3′ and 5′-GCAGGTTCCCGCTGGATCTCC-3′; xILR2.2 5′-ACTCTAAATTAAGTGACGTCGTG -3′ and 5′-GCAGGTTCATGCTGCATCTGA-3′; Silurana tropicalis sILR1 5′-GAATAGGAGGATCCTGGCTTCT-3′ and 5′-TGGCTCTTCTTGGTATGGCAGT-3′; sILR2 5′-CATCATGCTCAGAGCCTAGTGA-3′ and 5′-TGATGTTGCCTGTGGTGTGATC-3′; sILR3 5′-CCAGCAGGTTCCTACTAGAATG-3′ and 5′-CAAACGGATCTGTGCAGACTC-3′; sILR4 5′-CATGAGTGGCACATACACCTGT-3′ and 5′-AGGATTGCACTGACAGTAACAAC-3′.
cDNA clones and sequencing
S .tropicalis sILR1 L12, L15, M6, M17, M18, H2, H3, H7 and sILR4 B1 clones were obtained through cloning as described above. X. laevis EST cDNA clones IMAGE:4962918, IMAGE:8548181 and IMAGE:8074306 were purchased from the I.M.A.G.E. Consortium [28] through ATCC (USA) or Research Genetics Inc (USA). Both cloned and EST cDNAs were sequenced using an automated fluorescent sequencer ABI-Prizm 3130xl (Applied Biosystems) and submitted to GenBank. Following accession numbers were assigned to: X. laevis xILR1 - AY297107, xILR2.1 - EF431888, xILR2.2 - EF431889; S. tropicalis sILR1 - FJ716717- FJ716724, sILR4 - FJ716725.
Southern blot analysis
Genomic DNA from Xenopus laevis erythrocytes was isolated as described by Sambrook [29] and digested to completion with restriction endonuclease HindIII or PvuII. The digested DNA (10 μg/lane) was separated on 1% agarose gel and transferred onto Zeta-probe nylon membranes (BioRad Laboratories) by the vacuum blotting technique in 0.4 M NaOH. Hybridizations with 32P-labeled probes were performed following the membrane manufacturer’s recommendations in non-stringent conditions (55°C). The probes were PCR amplified fragments coding for the first domain either X. laevis xILR1 (257 bp) or xILR2.1 (210 bp). Following primers were used for probe amplification: xILR1 5′-CATTGCAGATCTTGGCTG-3′ and 5′-TGCGGTCTACTTTCCTCA-3′; xILR2.1 5′-CGACACAATCAGCTTTTTCT-3′ and 5′-CGAACATAGATGTGTTCAGG-3′.
Bioinformatics tools
Nucleotide and amino acid sequences were analyzed using utilities at the NCBI (www.ncbi.nlm.nih.gov), EMBL (www.ebi.ac.uk) and BCM (searchlauncher.bcm.tmc.edu) web sites. Amino acid sequences were aligned using Clustal utilities in the MEGA3 software [30] and shaded manually according to Timberlake classification of aminoacids [31]. The nucleotide and amino acid sequences of known genes were retrieved from GenBank using ENTREZ at the NCBI. The genomic sequences were retrieved from JGI web site (genome.jgi-psf.org). Similarity searches were performed using TBLASTN and BLASTP programs at the NCBI and EMBL sites. The GeneScan program (http://genes.mit.edu/GENSCAN.html) [32] and the Webgene program package (http://www.itb.cnr.it/sun/webgene/) [33] were used for the automated gene structure prediction. The ILR-surrounding genes were identified using the Ensembl (www.ensembl.org) and JGI utilities and were verified by reciprocal sequence comparisons at the NCBI website using the BLASTP program. Phylogenetic analysis was performed with the MEGA3 software [30] using nucleotide sequences of exons and amino acid sequences of domains after alignment with the CLUSTAL option. In certain cases, the CLUSTAL generated alignments were manually corrected. Phylogenetic trees were constructed using the bootstrap and interior branch tests of the Neighbor-joining (NJ) method with p-distances (proportion of differences). Minimum Evolution (ME) trees were essentially the same as the NJ trees in the major branching patterns.
Constructions, transfections and flow cytometry
cDNA regions encoding either an extracellular or EC-TM-Cyt part (with stop-codon at the end) of X. laevis xILR1 [Genbank: AY297107] were cloned using primers containing XmaI and SalI sites and ligated into the pDisplay vector (Invitrogen). The cDNA portion used was 100–900 bp and 100–1090 bp, respectively. Both constructions encoded proteins fused with the N-terminal hemagglutinin (HA)-tag and the first one - also with the C-terminal PDGFR transmembrane. Complete coding region of X. laevis FcRγ [GenBank: AF499689] cDNA was cloned using primers with NheI and ApaI sites and ligated into the pAP-Tag5 vector (GenHunter). The latter construction encoded protein fused with C-terminal c-myc epitope. Eukaryotic 293T cells were transiently transfected with all obtained constructions using Unifectin 56 (IBCH, Moscow, Russia) according to the manufacturer’s protocol. Seventy two hours after they were transfected, the cells were harvested and used for immunocytochemistry and cytometric analysis. For surface staining, transfected cells were washed twice with Wash Buffer (PBS, 1% FCS and 0.1% NaN3). The cells were first incubated with mouse monoclonal 12CA5 anti-HA antibodies (Abcam) in Wash Buffer for 30 min on ice. Cells were then washed three times with cold Wash Buffer and incubated with goat anti-mouse Ig-FITC (BD Bioscience) in Wash Buffer for 30 min on ice. The cells were washed three times with Wash Buffer and analyzed using a microscope Axioscop 2 plus (Carl Zeiss) and FACSAria cytometer (BD Bioscience). For intracellular staining, transfected cells were smeared on glass slides, fixed with acetone and stained for c-myc tagged subunits with anti-c-myc 9E10 monoclonal antibodies (Abcam) and goat anti-mouse IgG-TexasRed (Molecular Probes).
3. Results and discussion
3.1. The amphibian X. laevis possesses a small family of KRIR-related genes - ILR family
The African clawed frog X. laevis is one of the most thoroughly studied non-mammalian immunological model organisms [34]. Abundant molecular data are available for this species - almost 700 thousand cDNA sequences are stored in the EST database (dbEST). We searched this collection using TBLASTN analysis with sequences of various distantly related members of the KRIR family (i.e. KIRs, LILRs, PIRs and CHIRs). As a result, we identified 7 cDNAs similar to KRIR sequences. This similarity was rather weak and it ranged from 25 to 32% identical residues at the amino acid level. However, reciprocal BLASTP search in protein databases invariably indicated CHIRs and mammalian KRIR family proteins as the closest structural homologs of the predicted X. laevis amino-acid sequences. According to their similarity to each other, the identified cDNAs were subdivided into three clusters and a representative of each cluster was obtained from IMAGE consortium and sequenced. The cDNAs appeared to be products of three distinct genes encoding type I transmembrane proteins with similar structure: leader peptide, three Ig-like domains, transmembrane and short cytoplasmic tail. Based on the sequence comparisons, we designated these genes xILR1 (xenopus laevis Immunoglobulin-Like Receptor 1), xILR2.1 and xILR2.2. The deduced amino acid sequence of xILR1 showed less than 20% similarity with other two proteins. xILR2.1 and xILR2.2 share about 70% amino acid residues (Fig. 1).
Fig. 1.
Alignment of Xenopus laevis xILR predicted proteins. Alignment of the KRIR family proteins includes chicken CHIR, human LILRs and GPVI, mouse PIR and OSCAR. Dashes represent gaps introduced for maximal similarity. Conserved and similar amino acid residues are denoted by white letter on the black background and by black letters on grey background, respectively. Similar residues were determined based on amino acid radical polarity and charge [31]. Asterisks designate stop-codons, triangles - conserved cysteine residues. TM domains are boxed.
To estimate the size of the xILR family, we performed Southern blot analysis of X. laevis genomic DNA. Hybridization with a probe encoding the xILR1 domain 1 (xILR1 d1) under non-stringent conditions gave a single strong band in all six individual samples of digested genomic DNA (Fig. 2). Two weak bands were also detected on the same blot. These may be explained either by cross-hybridization with xILR1 d2 and d3 sequences or by the presence of 1–2 xILR1-like functional gene(s) or pseudogenes. The latter possibility is favored by the fact that X. laevis is an allotetraploid species. The probe corresponding to xILR2.1 d1 (86% of similarity with xILR2.2 d1) detected 4–5 hybridizing bands under non-stringent conditions (Fig. 2). Although exact number of xILR genes is to be established, dbEST and Southern blot analyses suggest that the ILR family in X. laevis is relatively small and most probably includes just a few genes per haploid genome.
Fig. 2.
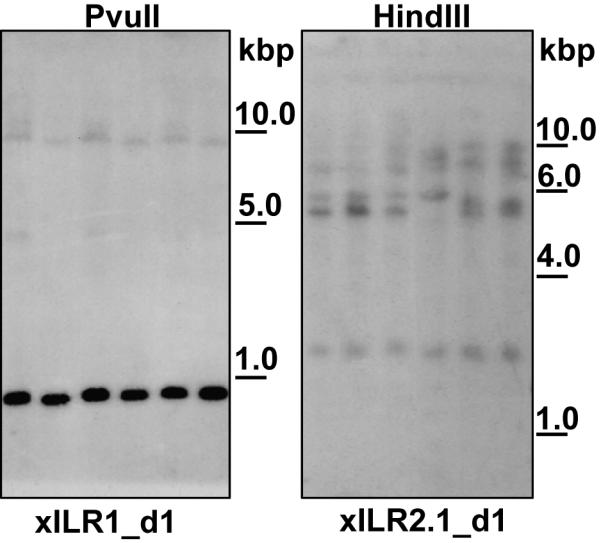
Southern blot analysis of X. laevis genomic DNA digested with HindIII or PvuII, and hybridized with xILR1- or xILR2-specific probes. Six individual samples of genomic DNA were used in each analysis. Molecular weight markers are shown at the right in kilobase pairs.
3.2. The genome of S. tropicalis contains four ILR-related genes
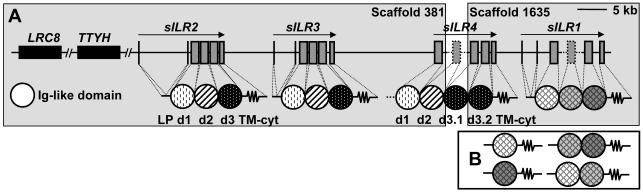
To gain additional information about structure of xILR receptors and their phylogenetic relationships, we studied genome of the diploid amphibian Silurana tropicalis available at JGI [http://genome.jgi-psf.org] and Ensembl [http://www.ensembl.org] sites. The genomic sequences of S. tropicalis were searched for the presence of the KRIR family genes using TBLASTN analysis with sequences of mammalian, chicken and X. laevis KRIR family receptors as probes. In this genomic search, we used a strategy described in our previous analysis of the Xenopus FcR-like genes [35]. Two scaffolds, 381 and 1635, containing exons for Ig-like domains structurally related to those of xILRs, CHIRs, and LILRs were identified in the JGI genome assembly 4.1. Further analysis showed that scaffold 381 contains also TTYH1- and LRC8 (LENG8)-like genes. These two genes are located in the Leukocyte Receptor Complex (LRC) and closely linked to the KRIR family genes in the human and some other mammalian genomes [18].
Automatic and manual gene prediction analyses showed that scaffold 1635 contains a gene highly similar to xILR1. We designated this gene sILR1 (silurana tropicalis Immunoglobulin-Like Receptor 1). sILR1 contains five exons: two mini-exons for the leader peptide (LP), two exons for two extracellular (EC) Ig-like domains and one exon for both the transmembrane (TM) and cytoplasmic (Cyt) parts. The predicted protein lacked one of the Ig-like domains compared to xILR1 but shared with the latter about 70% identity in overlapped parts. To confirm the structure of sILR1, we performed RT-RCR of S. tropicalis spleen total RNA using primers for the second and the last exons of this gene. The RT-PCR yielded three fragments of different size. The largest encoded a transmembrane receptor with 3 EC domains. The exon corresponding to the second domain was absent in all available versions of the S. tropicalis genome assembly (Fig. 3 A). Cloning and sequencing of the smaller size PCR products showed that they correspond to alternative transcripts from the same sILR1 gene. These transcripts encode receptors with either two or one Ig-like domain (Fig. 3 B). Thus, S. tropicalis have one xILR1-similar receptor with three EC domains. However, additional diversity for this receptor may be generated by alternative splicing of the mRNA.
Fig. 3.
Genomic organization of Silurana tropicalis sILR genes. (A) sILR exons encoding Ig-like domains or TM-Cyt portions are designated by grey rectangles, exons for leader peptide (LP) - by vertical lines. The scale for intronic distances is shown at the upper right corner. Dashed lines correspond to sequences missed in the scaffolds but present in raw data files and cDNAs. Ig-like domains of the same phylogenetic type are filled with the identical pattern. Arrows show gene transcription direction. Adjacent non-sILR genes are indicated as black rectangles. Exons encoding sILR4 leader peptide remain unpredicted. (B) Alternative architectures of sILR1-encoded receptors.
Apart from sILR1, the scaffold 381 was found to contain two more xILR-related genes. The TTYH-proximal gene was denoted as sILR2 due to its high similarity to X. laevis xILR2.1 and xILR2.2 (70% identity at the protein level). The TTYH-distal gene was designated sILR3 as its protein product shared only 42% identical residues with that of sILR2. sILR2 and sILR3 genes have genomic organization similar to that of sILR1. They both consist of 6 exons: one for 5′untranslated region (5′UTR) and the first half of LP, one for the second half of LP, three for Ig-like domains and one for TM, Cyt and 3′UTR (Fig. 3, untranslated regions are not indicated). The predicted structure of sILR2 and sILR3 genes was confirmed by comparison with available S. tropicalis EST sequences. It should be noted that the second exon encoding the leader peptide of sILR1, sILR2 or sILR3, is 36 bp long. This is a common feature of the KRIR family genes.
Furthermore, the scaffolds 381 and 1635 were found to contain at their ends orphan exons with nucleotide sequences slightly similar to those of sILR2 and sILR3. We postulated that these two scaffolds include adjacent genomic regions and that the orphan exons are parts of the same gene. We confirmed this by RT-PCR analysis of total RNA from S. tropicalis spleen using a series of primers matching exons from the two scaffolds. The amplification produced a single DNA fragment of 1068 bp, which encoded a protein with four extracellular domains, TM and a long cytoplasmic tail. The exon encoding the second EC domain is absent in all available versions of the S. tropicalis genome assembly. The other parts were completely identical to the sequences found in scaffolds 381 and 1635. We designated this gene as sILR4.
No other KRIR family genes were detected in the S. tropicalis genome. While the initial TBLASTN search produced some additional hits, they were attributed to other subsets of IgSF after subsequent examination. Scaffolds 1009 and 7950 were found to contain two exons completely identical to those of sILR1 (exons encoding d3 domain and TM-Cyt portion). Scaffold 381 contains an additional exon identical to that for the N-terminal domain of sILR4. We suppose that these exons may be either pseudoexons or results of assembly errors. The fact that the sequence identity between all mentioned exons is extended to the flanking non-coding regions and the absence of larger sILR4 cDNA fragments in RT-PCR are in favor of the latter possibility. Partial misassembling is inevitable at the current level of genome sequencing technology. Indeed, additional analysis demonstrated the presence of nucleotide sequences corresponding to the ‘missed’ exons for sILR1 d2 and sILR4 d2 in the Genbank trace archive containing raw data from S. tropicalis genome sequencing project. No sequences were found that could indicate the presence of additional ILR genes. This finding supports our gene models.
Comparisons of deduced amino acid sequences of sILRs showed their relatively weak homology to each other. The highest level of similarity, 45% identical residues, was found between the sILR2 and sILR3 extracellular parts (40–49% when comparing separate domains of homologous types). The sILR4 d1, d2 and d3.1 domains share 35–42% identical amino acid residues with corresponding d1-d3 domains of sILR2 and sILR3. The sILR4 d3.2 domain stands apart with only 21–26% of similarity with sILR4 d3.2 and sILR2/sILR3 d3 domains. The most distant member of the family is sILR1. Its extracellular domains show no more than 26% of identity with those of other sILRs. At the same time, comparisons of xILR1 with sILR1 or xILR2 with sILR2 (X. laevis vs. S. tropicalis homologs) show significantly higher similarity - 68–70% identical residues (Fig. 4).
Fig. 4.
Alignment of X. laevis (xILR) and S. tropicalis (sILR) predicted KRIR family proteins. S. tropicalis amino acid sequences were deduced from genomic sequences and confirmed by EST or cloned cDNAs. Sequences absent in the genomic scaffolds are marked by # sign. Dashes represent gaps introduced for maximal similarity. Conserved and similar amino acid residues are denoted by white letter on the black background and by black letters on grey background, respectively. Similar residues were determined based on radical polarity and charge [31]. Asterisks designate stop codons, triangles - conserved cysteine residues. TMs, switch and ITAM motifs are boxed. X. laevis xILR2.2 was intentionally removed from the alignment due to its high similarity to xILR2.1.
3.3. ILR transcripts have a broad tissue distribution
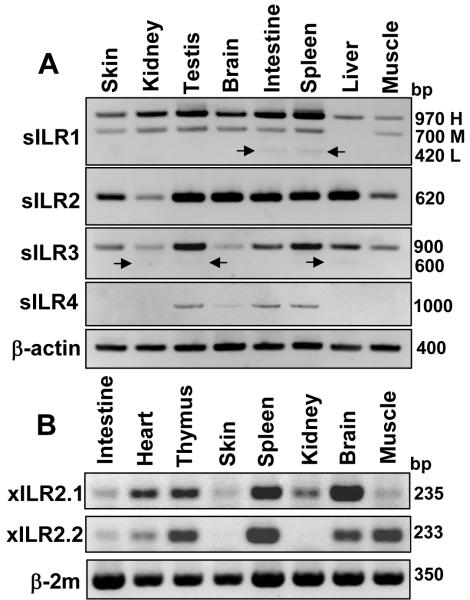
To examine the tissue distribution of sILR transcripts, we performed RT-PCR with primers specific to different exons (L2 and TM-Cyt for sILR1; d1 and TM-Cyt for sILR2, sILR3 and sILR4). sILR1-3 products were detected in all examined S. tropicalis tissues: skin, kidney, testis, brain, intestine, spleen, liver and muscle. Expression of sILR4 was found only in testis, brain, intestine and spleen. RT-PCR showed differential expression and the presence of alternative transcripts for sILR1 and sILR3 (Fig. 5 A). The broad tissue distribution of xILR transcripts was confirmed by RT-PCR analysis of X. laevis tissues (Fig. 5 B). All eight studied X. laevis tissues were found to express xILR2.1, whereas xILR2.2 expression wasn’t detected in kidney and skin (Fig. 5 B). The results of our analyses show that expression and by consequence the functional role of Xenopoidae KRIR family receptors is not restricted to lymphoid tissues. Therefore, it is unlikely that ILR receptors may be functionally equivalent to human KIR and mouse Ly-49 receptors, which are predominantly expressed on NK and NK/T cells.
Fig. 5.
RT-PCR analysis of S. tropicalis sILRs (A) and X. laevis xILR2 (B) expression. Molecular weights are shown at the right edge of gel pictures. Arrows point at weak signals corresponding to alternative splicing variants.
3.4. X. laevis and S. tropicalis KRIR family receptors have features of activating receptors
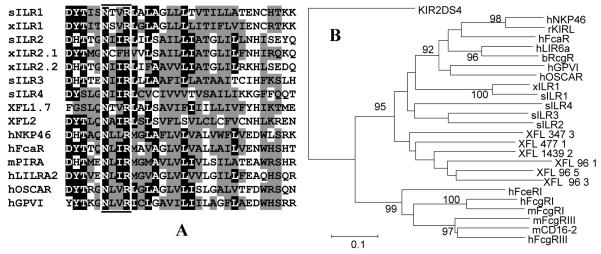
The TM regions of all identified xILR and sILR proteins are homologous to TMs of FcRγ-associating members of the KRIR family, such as NKp46, Fc R, OSCAR, PIR-A, and LILR-A (Fig. 6 A). The characteristic feature of this evolutionary conserved TM domain is the presence of a NxxR motif. Such TM subtype has been recently found in the family of Xenopus FcR-like receptors (XFLs) [35] and in chicken KRIR family receptor ggFcR [36]. Like in mammals [3,37-41], this domain promotes association of frog and chicken proteins with FcRγ [35,36].
Fig. 6.
Alignment of the TM domains belonging to human, Xenopus laevis (xILR) and Silurana tropicalis (sILR) KRIR family receptors. Transmembranes of murine PIR-A and X. laevis XFL receptors are also shown in the alignment. Conserved and similar amino acid residues are denoted by white letter on the black background and by black letters on grey background, respectively. Similar residues were determined based on amino acid radical polarity and charge [31]. NxxR motif is boxed.
Another feature that ILRs share with activating receptors is the use of a single exon for both TM and cytoplasmic tail. This is true even for sILR4 whose intracellular portion includes 80 residues. The presence of tyrosine-based motifs in the cytoplasmic tails of S. tropicalis sILR2 and sILR4 does not contradict the assignment of these proteins as activating receptors. The sILR4 tail contains four tyrosine-based motifs separated by 8–9 residues (Fig. 4). The structural characteristics of these motifs, such as the presence of negatively charged residue at position −3 relative to tyrosine residues resemble those of the ITAM consensus [4]. sILR2 contains a motif similar to the consensus T/SxYxxV/I of the so called ITSM motif [42,43]. In mammals, the latter is known to promote either inhibitory or activating signals depending on various circumstances including the presence of adapter proteins SAP and EAT-2.
To assess the capacity of xILRs to interact with ITAM-bearing subunit, we made constructs encoding X. laevis HA-tagged xILR1 receptor either with its original TM or TM substituted with that of PDGFR. Both HA-xILR1 and HA-xILR1-PDGFRTM receptors were found on the surface of transfected cells in the absence of signal subunits. The Fluorescence Intensity Median for HA-xILR1 was taken as 1.0, and for HA-xILR1-PDGFRTM it reached 1.24. The addition of X. laevis c-myc-tagged FcRγ subunit caused a slight increase in the level of HA-xILR1 surface expression (FIM = 1.21), which nearly reached the level of HA-xILR1-PDGFRTM surface expression (Fig. 7). The experiment was repeated several times and the results were reproducible. These data show that xILR1 doesn’t require FcRγ for surface transport, but the presence of the subunit makes such transport more efficient. The ability to be expressed as monomers is not rare among receptors with NxxR-containing TMs [44-46]. It appears that such ability provide an additional functional flexibility especially in the case of receptors which may possess their own signaling capacities.
Fig. 7.
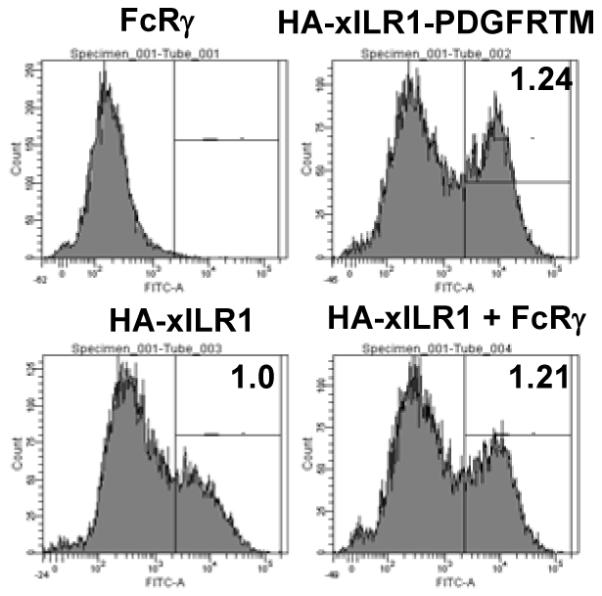
Flow cytometry (FACS) analysis of co-expression of X. laevis HA-xILR1 and FcRγ in 293T eukaryotic cells with α-HA Abs. HA-xILR1 was expressed with original TM domain or with TM substituted to that of PDFGR. FACS results were gated and the Fluorescence Intensity Medians were calculated. xILR1 FIM was taken as 1.0 and relative Medians for other experiments are shown at the top right corner of respective histograms.
3.5. Xenoponidae KRIR family receptors evolved in a species-specific manner
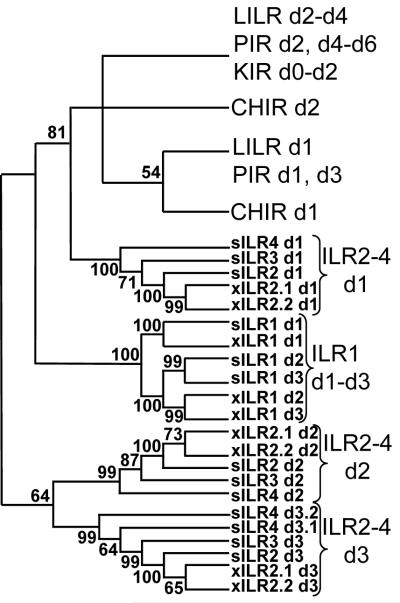
To infer phylogenetic relationships of IRLs with each other and with mammalian and chicken counterparts, we aligned amino acid sequences of their extracellular domains and generated phylogenetic trees using MEGA3 software package. A schematic tree is presented in Fig. 8, for complete tree see Supplementary data. In accordance with the results of sequence comparisons, ILR1 domains form a separate clade with branching pattern suggesting generation of three-domain extracellular region by two consecutive duplications of a single ancestral domain. The second duplication occurred in a species-specific manner after radiation of X.laevis and S.tropicalis. The extracellular regions of ILR2, ILR3 and ILR4 contain three domain subtypes forming three separate clades. In all of them, the xILR 2.1 and 2.2 domains group together demonstrating that these two genes emerged by a X. laevis-specific duplication event. The tree shows that, despite its diverged primary sequence, the sILR4d 3.2 domain clearly belongs to the same subtype as sILR4 d3.1 and sILR2–3 d3. The mammalian and avian KRIR family domains form independent clusters that do not contain ILR domains (Fig. 8 and Supplementary data). Such topology is in agreement with that demonstrated by Nei and collegues [47]. Thus, phylogenetic analysis shows that the evolution of the KRIR family genes in amphibians has been lineage-specific. Separation of ILRs from other known KRIR family genes seems to have occurred early in evolution. This notion is supported by very weak similarity among domains composing extracellular regions of ILRs. The distances among them are comparable to those among ILR domains and their counterparts from ectothermic vertebrates.
Fig. 8.
Phylogenetic analysis of the Ig-like domains of mammalian, avian and amphibian KRIR family proteins. Schematic bootstrapped NJ tree is shown. For the detailed tree see Supplementary data.
We also inferred phylogenetic relationships of the TM domains of the Xenoponidae and mammalian KRIR and FcR family molecules. The generated tree shows that the ILR TMs are closer to those of NKP46, OSCAR and Fc R than to the TM domains of XFLs. However, NxxR-containing TMs group in a statistically supported monophyletic clade separated from TMs of classical FcRs and human KIRs (Fig. 6 B). Altogether these results provide further support for a common origin of the FcR and KRIR families, and a subsequent divergent mode in their evolution. Importantly, the phylogenetic data show that NxxR-containing TM domain has been the most conserved structural element of the FcR/KRIR family during its evolution in tetrapods. This is in disagreement with a model proposing that activating receptors are recurrently generated from inhibitory receptors. Although such model appears to be accurate for KIRs and CHIRs in homeothermic vertebrates [22, 48], it should not be accepted as a common rule for the evolution of paired receptors.
4. Concluding remarks
Comparison of the immune systems of primates and rodents has demonstrated that the principle of functional analogy does not always apply to structurally-related immune genes [49]. The fact that missing-self recognition in human and mouse is performed by proteins belonging to the Ig and C-type lectin superfamilies, respectively, has been one of the first observations of this kind. With progress in studies of non-mammalian species, it is becoming evident that immune-related genes have even larger degree of flexibility and variability than previously thought [50-52]. Our data do not support the view that KRIR’s function was originally related to missing-self recognition through MHC class I binding. Phylogenetic analysis shows that evolution of amphibian, avian and mammalian KRIRs has been completely independent. Furthermore, considering the dramatic differences in the family size and the high degree of divergence of the receptor extracellular regions, one can ask whether KRIRs of various tetrapod lineages have any common function. If the answer is positive, such a function is likely to be determined by the ability to transmit activating signals. Indeed, all identified Xenoponidae ILR proteins have features of activating receptors. The NxxR-containing TM, which is responsible for FcRγ association, is the only conserved structural element of KRIRs in tetrapods. It cannot be excluded, however, that functional evolution of KRIR receptors was independent and that these receptors fulfill various roles in different vertebrate lineages.
Supplementary Material
Acknowledgements
The expert animal husbandry provided by Tina Martin and David Albright is gratefully appreciated. Research was supported by the Russian Fund for Basic Research 05-04-49268 (A.M.N), RAS program “Biodiversity” (A.V.T.), U.S. Civilian Research and Development Foundation RUB1-2837-NO-06 (A.V.T, J.R), NIH R01-CA-108982-02, NIH R24-AI-059830 and NSF MCB-0136536 (J.R.).
Abbreviations and symbols used (in alphabetical order)
- α
alpha
- γ
gamma
- BLAST
Basic Local Alignment Search Tool
- CHIR
Chicken Immunoglobilin-like Receptor
- Cyt
cytoplasmic (tail)
- dbEST
Expressed Sequence Tags Data Base
- EC
extracellular (domain)
- HA
Hemagglutinin
- IgSF
Immunoglobulin Superfamily
- ITAM
Immunoreceptor tyrosine-based activating motif
- ITIM
Immunoreceptor tyrosine-based inhibitory motif
- KIR
Killer-cell Immunoglobulin-like Receptor
- KRIR
KIR-Related Immunoglobilin-like Receptor
- LILR
Leukocyte Immunoglobilin-like Receptor
- LITR
Leukocyte Immune-Type Receptors
- LP
Leader Peptide
- MHC
Major Histocompatibility Complex
- PDGFR
Platelet Derived Growth Factor Receptor
- PIR
Paired Immunoglobilin-like Receptor
- TM
transmembrane (domain)
- ILR
Immunoglobilin-like Receptor
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data. Phylogenetic analysis of the Ig-like domains of mammalian, avian and amphibian KRIR family proteins. Phylogenetic analysis was performed with the MEGA3 software [30] using nucleotide sequences of exons and amino acid sequences of domains after alignment with the CLUSTAL option. Phylogenetic trees were constructed using the bootstrap and interior branch tests of the Neighbor-joining (NJ) method with p-distances (proportion of differences).
Appendix A Supplementary data
Refer to Web version for supplementary material.
References
- [1].Lanier LL. NK cell recognition. Annu Rev Immunol. 2005;23:225–74. doi: 10.1146/annurev.immunol.23.021704.115526. [DOI] [PubMed] [Google Scholar]
- [2].Martin AM, Kulski JK, Witt C, Pontarotti P, Christiansen FT. Leukocyte Ig-like receptor complex (LRC) in mice and men. Trends Immunol. 2002;23(2):81–8. doi: 10.1016/s1471-4906(01)02155-x. [DOI] [PubMed] [Google Scholar]
- [3].Merck E, Gaillard C, Gorman DM, Montero-Julian F, Durand I, Zurawski SM, Menetrier-Caux C, Carra G, Lebecque S, Trinchieri G, Bates EE. OSCAR is an FcRγ-associated receptor that is expressed by myeloid cells and is involved in antigen presentation and activation of human dendritic cells. Blood. 2004;104(5):1386–95. doi: 10.1182/blood-2004-03-0850. [DOI] [PubMed] [Google Scholar]
- [4].Daëron M, Jaeger S, Du Pasquier L, Vivier E. Immunoreceptor tyrosine-based inhibition motifs: a quest in the past and future. Immunol Rev. 2008;224:11–43. doi: 10.1111/j.1600-065X.2008.00666.x. [DOI] [PubMed] [Google Scholar]
- [5].Biassoni R, Cantoni C, Pende D, Sivori S, Parolini S, Vitale M, Bottino C, Moretta A. Human natural killer cell receptors and co-receptors. Immunol Rev. 2001;181:203–14. doi: 10.1034/j.1600-065x.2001.1810117.x. [DOI] [PubMed] [Google Scholar]
- [6].Willcox BE, Thomas LM, Bjorkman PJ. Crystal structure of HLA-A2 bound to LIR-1, a host and viral major histocompatibility complex receptor. Nat Immunol. 2003;4(9):913–9. doi: 10.1038/ni961. [DOI] [PubMed] [Google Scholar]
- [7].Shiroishi M, Kuroki K, Rasubala L, Tsumoto K, Kumagai I, Kurimoto E, Kato K, Kohda D, Maenaka K. Structural basis for recognition of the nonclassical MHC molecule HLA-G by the leukocyte Ig-like receptor B2 (LILRB2/LIR2/ILT4/CD85d) Proc Natl Acad Sci U S A. 2006;103(44):16412–7. doi: 10.1073/pnas.0605228103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Liang S, Baibakov B, Horuzsko A. HLA-G inhibits the functions of murine dendritic cells via the PIR-B immune inhibitory receptor. Eur J Immunol. 2002;32(9):2418–26. doi: 10.1002/1521-4141(200209)32:9<2418::AID-IMMU2418>3.0.CO;2-L. [DOI] [PubMed] [Google Scholar]
- [9].Nakamura A, Kobayashi E, Takai T. Exacerbated graft-versus-host disease in Pirb−/− mice. Nat Immunol. 2004;5(6):623–9. doi: 10.1038/ni1074. [DOI] [PubMed] [Google Scholar]
- [10].Takai T, Ono M. Activating and inhibitory nature of the murine paired immunoglobulin-like receptor family. Immunol Rev. 2001;181:215–22. doi: 10.1034/j.1600-065x.2001.1810118.x. [DOI] [PubMed] [Google Scholar]
- [11].Masuda A, Nakamura A, Maeda T, Sakamoto Y, Takai T. Cis binding between inhibitory receptors and MHC class I can regulate mast cell activation. J Exp Med. 2007;204(4):907–20. doi: 10.1084/jem.20060631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Nakayama M, Underhill DM, Petersen TW, Li B, Kitamura T, Takai T, Aderem A. Paired Ig-like receptors bind to bacteria and shape TLR-mediated cytokine production. J Immunol. 2007;178(7):4250–9. doi: 10.4049/jimmunol.178.7.4250. [DOI] [PubMed] [Google Scholar]
- [13].Hecht ML, Rosental B, Horlacher T, Hershkovitz O, De Paz JL, Noti C, Schauer S, Porgador A, Seeberger PH. Natural cytotoxicity receptors NKp30, NKp44 and NKp46 bind to different heparan sulfate/heparin sequences. J Proteome Res. 2009;8(2):712–20. doi: 10.1021/pr800747c. [DOI] [PubMed] [Google Scholar]
- [14].Wines BD, Hulett MD, Jamieson GP, Trist HM, Spratt JM, Hogarth PM. Identification of residues in the first domain of human Fc alpha receptor essential for interaction with IgA. J Immunol. 1999;162(4):2146–53. [PubMed] [Google Scholar]
- [15].Clemetson JM, Polgar J, Magnenat E, Wells TN, Clemetson KJ. The platelet collagen receptor glycoprotein VI is a member of the immunoglobulin superfamily closely related to FcalphaR and the natural killer receptors. J Biol Chem. 1999;274(41):29019–24. doi: 10.1074/jbc.274.41.29019. [DOI] [PubMed] [Google Scholar]
- [16].Lebbink RJ, de Ruiter T, Adelmeijer J, Brenkman AB, van Helvoort JM, Koch M, Farndale RW, Lisman T, Sonnenberg A, Lenting PJ, Meyaard L. Collagens are functional, high affinity ligands for the inhibitory immune receptor LAIR-1. J Exp Med. 2006;203(6):1419–25. doi: 10.1084/jem.20052554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Yokoyama WM, Plougastel BF. Immune functions encoded by the natural killer gene complex. Nat Rev Immunol. 2003;3(4):304–16. doi: 10.1038/nri1055. [DOI] [PubMed] [Google Scholar]
- [18].Kelley J, Walter L, Trowsdale J. Comparative genomics of natural killer cell receptor gene clusters. PLoS Genet. 2005;1(2):129–39. doi: 10.1371/journal.pgen.0010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Hammond JA, Guethlein LA, Abi-Rached L, Moesta AK, Parham P. Evolution and survival of marine carnivores did not require a diversity of killer cell Ig-like receptors or Ly49 NK cell receptors. J Immunol. 2009;182(6):3618–27. doi: 10.4049/jimmunol.0803026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Dennis G, Jr, Kubagawa H, Cooper MD. Paired Ig-like receptor homologs in birds and mammals share a common ancestor with mammalian Fc receptors. Proc Natl Acad Sci U S A. 2000;97(24):13245–50. doi: 10.1073/pnas.230442897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Viertlboeck BC, Habermann FA, Schmitt R, Groenen MA, Du Pasquier L, Gobel TW. The chicken leukocyte receptor complex: a highly diverse multigene family encoding at least six structurally distinct receptor types. J Immunol. 2005;175(1):385–93. doi: 10.4049/jimmunol.175.1.385. [DOI] [PubMed] [Google Scholar]
- [22].Nikolaidis N, Makalowska I, Chalkia D, Makalowski W, Klein J, Nei M. Origin and evolution of the chicken leukocyte receptor complex. Proc Natl Acad Sci U S A. 2005;102(11):4057–62. doi: 10.1073/pnas.0501040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Laun K, Coggill P, Palmer S, Sims S, Ning Z, Ragoussis J, Volpi E, Wilson N, Beck S, Ziegler A, Volz A. The leukocyte receptor complex in chicken is characterized by massive expansion and diversification of immunoglobulin-like Loci. PLoS Genet. 2006;2(5):e73. doi: 10.1371/journal.pgen.0020073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Viertlboeck BC, Schweinsberg A, Hanczaruk MA, Schmitt R, Du Pasquier L, Herberg FW, Gobel TW. The chicken leukocyte receptor complex encodes a primordial, activating, high-affinity IgY Fc receptor. Proc Natl Acad Sci U S A. 2007;104(28):11718–23. doi: 10.1073/pnas.0702011104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Viertlboeck BC, Schweinsberg S, Schmitt R, Herberg FW, Göbel TW. The chicken leukocyte receptor complex encodes a family of different affinity FcY receptors. J Immunol. 2009;182(11):6985–92. doi: 10.4049/jimmunol.0803060. [DOI] [PubMed] [Google Scholar]
- [26].Stafford JL, Bengten E, Du Pasquier L, McIntosh RD, Quiniou SM, Clem LW, Miller NW, Wilson M. A novel family of diversified immunoregulatory receptors in teleosts is homologous to both mammalian Fc receptors and molecules encoded within the leukocyte receptor complex. Immunogenetics. 2006;58(9):758–73. doi: 10.1007/s00251-006-0134-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Stafford JL, Bengten E, Du Pasquier L, Miller NW, Wilson M. Channel catfish leukocyte immune-type receptors contain a putative MHC class I binding site. Immunogenetics. 2007;59(1):77–91. doi: 10.1007/s00251-006-0169-3. [DOI] [PubMed] [Google Scholar]
- [28].Lennon G, Auffray C, Polymeropoulos M, Soares MB. The I.M.A.G.E. Consortium: an integrated molecular analysis of genomes and their expression. Genomics. 1996;33(1):151–2. doi: 10.1006/geno.1996.0177. [DOI] [PubMed] [Google Scholar]
- [29].Sambrook T, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press; Plainview, NY: 1989. [Google Scholar]
- [30].Kumar S, Tamura K, Nei M. MEGA3. Integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief Bioinform. 2004;5(2):150–63. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- [31].Timberlake KC. Chemistry - 5th Edition. Haper-Collins Publishers Inc; NY: 1992. [Google Scholar]
- [32].Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA. J Mol Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- [33].Milanesi L, D’Angelo D, Rogozin IB. GeneBuilder. interactive in silico prediction of gene structure. Bioinformatics. 1999;15(7–8):612–21. doi: 10.1093/bioinformatics/15.7.612. [DOI] [PubMed] [Google Scholar]
- [34].Robert J, Ohta Y. Comparative and developmental study of the immune system in Xenopus. Dev Dyn. 2009;238(6):1249. doi: 10.1002/dvdy.21891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Guselnikov SV, Ramanayake T, Erilova AY, Mechetina LV, Najakshin AM, Robert J, Taranin AV. The Xenopus FcR family demonstrates continually high diversification of paired receptors in vertebrate evolution. BMC Evol Biol. 2008;8:148. doi: 10.1186/1471-2148-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Viertlboeck BC, Schmitt R, Hanczaruk MA, Crooijmans RP, Groenen MA, Göbel TW. A novel activating chicken IgY FcR is related to leukocyte receptor complex (LRC) genes but is located on a chromosomal region distinct from the LRC and FcR gene clusters. J Immunol. 2009;182(3):1533–40. doi: 10.4049/jimmunol.182.3.1533. [DOI] [PubMed] [Google Scholar]
- [37].van Egmond M, van Vuuren AJ, Morton HC, van Spriel AB, Shen L, Hofhuis FM, Saito T, Mayadas TN, Verbeek JS, van de Winkel JG. Human immunoglobulin A receptor (FcalphaRI, CD89) function in transgenic mice requires both FcR gamma chain and CR3 (CD11b/CD18) Blood. 1999;93(12):4387–94. [PubMed] [Google Scholar]
- [38].Suzuki Y, Ra C, Saito K, Horikoshi S, Hasegawa S, Tsuge T, Okumura K, Tomino Y. Expression and physical association of Fc alpha receptor and Fc receptor gamma chain in human mesangial cells. Nephrol Dial Transplant. 1999;14(5):1117–23. doi: 10.1093/ndt/14.5.1117. [DOI] [PubMed] [Google Scholar]
- [39].Nakajima H, Samaridis J, Angman L, Colonna M. Human myeloid cells express an activating ILT receptor (ILT1) that associates with Fc receptor gamma-chain. J Immunol. 1999;162(1):5–8. [PubMed] [Google Scholar]
- [40].Maeda A, Kurosaki M, Kurosaki T. Paired immunoglobulin-like receptor (PIR)-A is involved in activating mast cells through its association with Fc receptor gamma chain. J Exp Med. 1998;188(5):991–5. doi: 10.1084/jem.188.5.991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Ono M, Yuasa T, Ra C, Takai T. Stimulatory Function of Paired Immunoglobulin-like Receptor-A in Mast Cell Line by Associating with Subunits Common to Fc Receptors. J Biol Chem. 1999;274(42):30288–96. doi: 10.1074/jbc.274.42.30288. [DOI] [PubMed] [Google Scholar]
- [42].Yusa S, Catina TL, Campbel KS. KIR2DL5 can inhibit human NK cell activation via recruitment of Src homology region 2-containing protein tyrosine phosphatase-2 (SHP-2) J Immunol. 2004;172(12):7385–7392. doi: 10.4049/jimmunol.172.12.7385. [DOI] [PubMed] [Google Scholar]
- [43].Torii I, Oka S, Hotomi M, Benjamin WH, Jr, Takai T, Kearney JF, Briles DE, Kubagawa H. PIR-B-deficient mice are susceptible to Salmonella infection. J Immunol. 2008;181(6):4229–4239. doi: 10.4049/jimmunol.181.6.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Morton HC, van den Herik-Oudijk IE, Vossebeld P, Snijders A, Verhoeven AJ, Capel PJ, van de Winkel JG. Functional association between the human myeloid immunoglobulin A Fc receptor (CD89) and FcR gamma chain. Molecular basis for CD89/FcR gamma chain association. J Biol Chem. 1995;270(50):29781–7. doi: 10.1074/jbc.270.50.29781. [DOI] [PubMed] [Google Scholar]
- [45].Taylor LS, McVicar DW. Functional association of FcepsilonRIgamma with arginine(632) of paired immunoglobulin-like receptor (PIR)-A3 in murine macrophages. Blood. 1999;94(5):1790–6. [PubMed] [Google Scholar]
- [46].Berlanga O, Tulasne D, Bori T, Snell DC, Miura Y, Jung S, Moroi M, Frampton J, Watson SP. The Fc receptor gamma-chain is necessary and sufficient to initiate signalling through glycoprotein VI in transfected cells by the snake C-type lectin, convulxin. Eur J Biochem. 2002;269(12):2951–60. doi: 10.1046/j.1432-1033.2002.02969.x. [DOI] [PubMed] [Google Scholar]
- [47].Nikolaidis N, Klein J, Nei M. Origin and evolution of the Ig-like domains present in mammalian leukocyte receptors: insights from chicken, frog, and fish homologues. Immunogenetics. 2005;57(1–2):151–7. doi: 10.1007/s00251-004-0764-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Abi-Rached L, Parham P. Natural selection drives recurrent formation of activating killer cell immunoglobulin-like receptor and Ly49 from inhibitory homologues. J Exp Med. 2005;201(8):1319–32. doi: 10.1084/jem.20042558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Mestas J, Hughes CC. Of mice and not men: differences between mouse and human immunology. J Immunol. 2004;172(5):2731–8. doi: 10.4049/jimmunol.172.5.2731. [DOI] [PubMed] [Google Scholar]
- [50].Alder MN, Rogozin IB, Iyer LM, Glazko GV, Cooper MD, Pancer Z. Diversity and function of adaptive immune receptors in a jawless vertebrate. Science. 2005;310(5756):1970–3. doi: 10.1126/science.1119420. [DOI] [PubMed] [Google Scholar]
- [51].Zhang Q, Zmasek CM, Dishaw LJ, Mueller MG, Ye Y, Litman GW, Godzik A. Novel genes dramatically alter regulatory network topology in amphioxus. Genome Biol. 2008;9(8):R123. doi: 10.1186/gb-2008-9-8-r123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Du Pasquier L. Innate immunity in early chordates and the appearance of adaptive immunity. C R Biol. 2004;327(6):591–601. doi: 10.1016/j.crvi.2004.04.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.