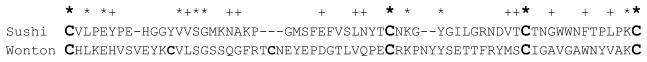Abstract
Pathogen recognition and rapid initiation of defense responses are essential for the survival of host insects. In Manduca sexta, hemolymph proteinase-14 precursor (proHP14) senses non-self presence and triggers a branched serine proteinase pathway which leads to prophenoloxidase activation and melanin formation around the invading organisms. To understand functions of individual domains in HP14, we have produced a series of HP14 domains and truncation mutants and studied their interactions with microbial polysaccharides and β-1,3-glucan recognition protein-1 (βGRP1) – a biosensor for fungal and bacterial infection. These include: the low-density lipoprotein receptor class A repeats 1–5 (LDL1–5), Sushi domain, Wonton domain, and proteinase catalytic domain of HP14, as well as proHP14 missing 1~4 LDL repeats (ΔLDL1, ΔLDL12, ΔLDL1–3 and ΔLDL1–4). LDL1–5, Sushi, and Wonton domains specifically recognized Lys-type PG, whereas the latter two also bound βGRP1. Wonton in addition bound to lipopolysaccharide (LPS), lipoteichoic acid (LTA), and meso-diaminopimelic acid (DAP)-type peptidoglycan (PG). The four N-terminally truncated proHP14 (ΔLx) further confirmed specific interactions with LPS, LTA, DAP-PG, Lys-PG, laminarin, and βGRP1. These binding data suggest a broad specificity of proHP14 in pattern recognition. Its role in mediating immune responses is anticipated to be influenced by other plasma factors and surface structures of invading pathogens.
Keywords: insect immunity, pattern recognition, phenoloxidase, melanization, serine proteinase pathway, hemolymph protein
1. Introduction
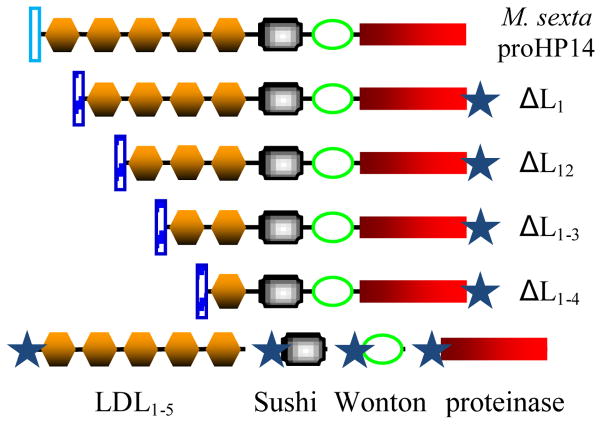
Innate immunity is critically important for the survival and wellbeing of insects in environments abundant in pathogenic microorganisms. This defense system is composed of factors that recognize/immobilize microbes, relay signals outside and inside immune tissues/cells, kill the invading organisms, and regulate immune mechanisms [1–4]. Some of the cellular and humoral mechanisms are mediated by an extracellular serine proteinase network which generates spätzle, phenoloxidase (PO), and plasmatocyte spreading peptide via limited proteolysis [5,6]. Among the network components discovered so far, Manduca sexta hemolymph proteinase-14 (HP14) and its orthologs in other insects are unique in three ways: 1) they contain 4–5 low-density lipoprotein receptor class A (LDLA or simply LDL) repeats, a Sushi domain, a Wonton domain, and a carboxyl-terminal serine proteinase domain (PD) (Fig. 1), 2) they directly or indirectly recognize pathogen surface molecules such as bacterial peptidoglycans (PGs) and fungal β-1,3-glucan, and 3) they autoactivate and trigger the serine proteinase network [7–10]. Interaction of recombinant M. sexta proHP14 with Lys-type PG resulted in its proteolytic processing, and supplementing hemolymph with the proHP14 greatly enhanced prophenoloxidase (proPO) activation in response to M. luteus [7]. ProHP14 purified from hemolymph of M. sexta larvae injected with bacteria was converted to a two-chain active form after incubation with β-1,3-glucan and M. sexta βGRP2 [8]. Such autoactivated HP14 greatly elevated PO activity in the larval plasma. HP14 activates proHP21 to HP21, HP21 converts proPAP2/3 to PAP2/3, and PAP2/3 generates active PO in the presence of a high Mr complex of clip-domain serine proteinase homolog-1 and -2 (SPH1 and SPH2) [11,12].
Fig. 1. Domain structure of M. sexta proHP14 (top), its deletion mutants (middle) and domain regions (bottom).
The vertical bars (open and hatched) represent M. sexta proHP14 and honeybee melittin signal peptides, respectively. The hexahistidine tag is denoted by a star.
In order to investigate roles of the amino-terminal putative regulatory domains in M. sexta HP14, we produced different regions of the zymogen and studied interactions of the recombinant proteins with microbial cell wall components. In this paper, we report binding properties of individual domains or regions in proHP14 and their associations with M. sexta βGRP1 [13], a protein similar to βGRP2 which recognizes fungi and bacteria [14]. Implications of the broad binding spectrum of binding are also discussed.
2. Methods and materials
2.1. Construction of expression plasmids for producing proHP14 domain regions in Escherichia coli
Full-length cDNA for M. sexta HP14 was used as template for amplification of the four segments with the primer pairs listed in Table S1. The 25 μl reaction contained 2 ng template, 10 pmol of each primer, and 2.5 U Advantage cDNA polymerase mix (Clontech). The thermal cycling conditions were 35 cycles of 94°C, 30s; 50°C, 30s; 68°C, 60s, followed by 3 min of incubation at 68°C. Following gel purification, the PCR products were cloned into pGEM-T (Promega) and the transformants were examined for correct restriction digestion patterns, insert sizes, and nucleotide sequences. The cDNA segments, retrieved by digestion with NcoI and BamHI/SphI, were inserted to the same sites of plasmid H6pQE60 [15] to generate plasmids LDL1–5/H6pQE60, Sushi/H6pQE60, Wonton/H6pQE60, and PD/H6pQE60. The transformants were examined for induced expression of recombinant proteins at expected sizes by SDS-polyacrylamide gel electrophoresis (SDS-PAGE) followed by immunoblot analysis using 1:2000 diluted anti-HP14 serum as the first antibody.
2.2. Prokaryotic expression, purification, antibody raising, and renaturation of the four domain regions
The domain regions of M. sexta proHP14 were individually produced using E. coli JM109 harboring the recombinant plasmids according to Wang et al [16]. The hexahistidine-tagged proteins from 500 ml of the cultures were purified on a nickel-nitrilotriacetic acid (NTA) agarose column under denaturing condition. The affinity-purified proteins were resolved by 10% SDS-PAGE, and the gel slices containing LDL1–5, Sushi, Wonton, and PD (0.4 mg each) were used as antigens to generate four region-specific rabbit polyclonal antisera (Cocalico Biologicals Inc.). An aliquot of each protein was renatured by dialysis against 50 mM Tris-HCl (pH 8.0), 3 mM reduced glutathione, 1 mM oxidized glutathione, and 0.5 M arginine for 16 h at 4°C and then 20 mM Tris-HCl (pH 7.5), 50 mM NaCl for 8 h at 4°C, and then centrifuged at 15,000g for 10 min at 4°C. The supernatants (10 μl) were analyzed by 15% SDS-polyacrylamide gel electrophoresis (SDS-PAGE) and visualized by Coomassie Blue staining.
2.3. Preparation of expression constructs for producing N-terminally truncated proHP14 in baculovirus-insect cell system
For PCR amplification of the cDNA fragments, each 50 μl-reaction contained 5 ng full-length proHP14 cDNA, 20 pmol of each primer, and 5 U Advantage cDNA polymerase mix (Clontech). The thermal cycling conditions were 35 cycles of 94°C, 20s; 50°C, 30s; 68°C, 90s, followed by 10 min of incubation at 68°C. The gel-purified PCR products were cloned into pGEM-T and plasmids from the resulting transformants were sequenced entirely to ensure error-free inserts. The cDNA segments, retrieved by digestion with EcoRI and XhoI, were inserted to the same sites of plasmid pMFH6 [17] to generate plasmids ΔL1/pMFH6, ΔL12/pMFH6, ΔL1–3/pMFH6, and ΔL1–4/pMFH6. The modified Bac-to-Bac vector allowed the recombinant proteins to be synthesized under the control of polyhedrin promoter, secreted into the medium using the honeybee mellitin signal peptide, and purified on a Ni-NTA column via the carboxyl-terminal hexahistidine tag.
2.4. Baculovirus generation, insect cell infection, and protein isolation
In vivo transposition of the expression cassette in ΔLx/pMFH6, selection of bacterial colonies carrying recombinant bacmids, and isolation of bacmid DNA were performed according to the manufacturer’s protocols (Invitrogen Life Technologies). The initial viral stocks were separately obtained by transfecting Spodoptera frugiperda Sf21 cells with a bacmid DNA-CellFECTIN mixture, and their titers were improved through serial infections [18]. The V5 viral stocks, containing the highest levels of baculoviruses (1~2×108), were stored at −70°C for further experiments. Sf21 cells (at 2.4×106 cells/ml) in 1.0 L of Sf-900™ III serum-free medium (Invitrogen Life Technologies) were separately infected with the baculovirus stocks at a multiplicity of infection of 10 and grown at 27°C for 84 h with gentle agitation (100 rpm). After the cells were removed by centrifugation at 5,000g for 10 min, a 50 mL-aliquot of the supernatant was diluted with an equal volume of 1 mM benzamidine and gently mixed with 6.0 ml dextran sulfate (DS)-Sepharose CL-6B beads equilibrated in buffer A (0.01% Tween-20, 1 mM benzamidine, 10 mM potassium phosphate, pH 6.4) on ice for 1 h. The suspension was loaded into a column, washed with 30 ml buffer A, and eluted with a linear gradient of 0–1 M NaCl in buffer A at 1.0 ml/min for 30 min. Following SDS-PAGE and immunoblot analysis, fractions containing the recombinant proteins were pooled, supplemented with 2 mM MgCl2, and loaded to a 5-ml Concanavalin A-Sepharose (GE Healthcare Life Sciences) column. The column was washed with 30 ml buffer B (0.01% Tween-20, 0.5 M NaCl, 20 mM Tris-HCl, pH 7.5) and eluted with 30 ml 0.4 M methyl-α-D-mannopyranoside in buffer B. The recombinant protein fractions were combined and loaded onto a 1-ml Ni2+-NTA agarose column equilibrated with 10 mM imidazole in buffer C (1 mM benzamidine, 0.3 M NaCl, 50 mM sodium phosphate, pH 8.0). Bound proteins were eluted with a linear gradient of 10–250 mM imidazole in buffer C at 0.5 ml/min for 40 min. Fractions containing the mutant proteins were stored at −80°C prior to binding assays.
2.5. Enzyme-linked immunosorbent assay (ELISA)-based binding assays
Laminarin from Laminaria digitata (Sigma L9634), lipopolysaccharide from E. coli O127: B8 (Sigma L3129), lipoteichoic acid from Bacillus subtilis (Sigma L3265), DAP-type PG from E. coli (InvivoGen), or Lys-type PG from Staphylococcus aureus (InvivoGen) were individually dissolved in H2O at 40 μg/ml. These samples (2.0 μg) were applied to a 96-well microplate and air dried overnight at room temperature. The plate was incubated at 60°C for 30 min to fix the ligands, and the wells were blocked with 200 μl of 1 mg/ml bovine serum albumin (BSA) in TBS (137 mM NaCl, 3 mM KCl, 25 mM Tris-HCl, pH 7.6) at 37°C for 2 h. After a washing step (200 μl TBS, 3 min each time for 4 times), each recombinant protein (100 ng in 50 μl TBS containing 0.1 mg/ml BSA) was added to the wells and incubated for 3 h at room temperature. A competition binding assay was performed to test whether the binding was a result of specific interaction between the protein and ligand: the protein (100 ng) was first incubated with the ligand (20 μg) in solution for 30 min at room temperature, TBS containing 0.1 mg/ml BSA was then added to a final volume of 50 μl, and the mixture was then incubated for 3 h at room temperature in a well coated with the same ligand. After washing, 100 μl of 1:1000 diluted proHP14 antiserum in TBS containing 0.1 mg/ml BSA was incubated with the bound antigen for 2 h at 37°C. Following a washing step, 100 μl of 1:1500 diluted goat anti-rabbit IgG conjugated to alkaline phosphatase (Bio-Rad) in TBS containing 0.1 mg/ml BSA was added and incubated for 2 h at 37°C. Then the wells were rinsed four times with TBS and once with 0.5 mM MgCl2, 10 mM diethanolamine. Aliquots of 50 μl of p-nitrophenyl phosphate (1.0 mg/ml in the diethanolamine buffer) were added to the wells and absorbance at 405 nm was monitored in the kinetic mode using a microplate reader (Molecular Devices).
2.6. SDS-PAGE analysis of the mutant proteins binding to curdlan
Five μl of LDL1–5, Sushi, Wonton, ΔL1, ΔL12, ΔL1–3, or ΔL1–4 was mixed with 15 μl, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM benzamidine and 50 μl curdlan pellet (500 μg) on ice for 1 h. After brief centrifugation, the original protein, supernatant, and pellet were subjected to sample buffer treatment, SDS-PAGE, and immunoblot analysis using specific antibodies.
2.7. Purification of M. sexta βGRP1 from plasma of naïve larvae
A frozen hemolymph sample (40 ml) was fractionated with 15–38% saturation of ammonium sulfate, and the fraction was dialyzed against HT buffer (pH 6.8, 10 mM potassium phosphate, 0.5 M NaCl) supplemented with 0.001% 1-phenyl-2-thiourea and 1 mM benzamidine (2.0 L for 8 h, twice). After centrifugation at 15,000×g for 30 min, the cleared supernatant was applied to a hydroxylapatite column (2.5 cm i.d. × 7 cm, Bio-Rad) equilibrated in HT buffer. Following a washing step with 100 ml HT buffer, bound proteins were eluted at 0.4 ml/min for 6.25 h with a linear gradient of 10–150 mM potassium phosphate (pH 6.8), 0.5 M NaCl. Guided by immunoblot analysis using its antibodies (Ma and Kanost, 2000), βGRP1 fractions were pooled, concentrated, and resolved on a Sephacryl S100-HR column equilibrated with 20 mM Tris-HCl, 0.5 M NaCl, pH 7.6. The active fractions from the gel filtration column were combined, supplemented with 1 mM CaCl2 and MgCl2, and loaded onto a concanavalin A-Sepharose column (5.0 ml). The flow-through and washing fractions were combined, diluted with eight volumes of S buffer (50 mM sodium acetate, pH 4.8), and then loaded onto an UNO™ S6 column using a BioLogic DuoFlow System (Bio-Rad). Following washing, bound proteins were eluted with a linear gradient of 0–500 mM NaCl in S buffer at 1 min/min for 1 h. The purified βGRP1 was stored at −80°C before use.
2.8. Autoactivation of M. sexta proHP14 triggered by insoluble β-1,3-glucan and βGRP1
Purified βGRP1 (20 ng, 1 μl), curdlan (10 μg, 1 μl), proHP14 from plasma (200 ng, 10 μl) [8], CaCl2 (100 mM, 1 μl), and 20 mM Tris-HCl, pH 8.0, 20 mM NaCl (1 μl) were incubated at 37°C for 1 h. To test the contribution of curdlan and βGRP1 in proHP14 autoactivation, one or two of them were replaced by the same volumes of the buffer. The reaction mixtures were separated by 10% SDS-PAGE under reducing condition and visualized by silver staining to assess cleavage.
2.9. Association of M. sexta βGRP1 with microbial surface molecules or purified HP14 mutants
An ELISA-based binding assay was performed to study the association of βGRP1 with the immobilized cell wall components. The first antibody was 1:1000 diluted βGRP1 antiserum. To examine its association with the HP14 protein constructs, renatured recombinant βGRP1 [13] (2.0 ng/well) was immobilized on a 96-well plate to interact with LDL1–5, Sushi or Wonton (12.5 pmol each), ΔL1, ΔL12, ΔL1–3 or ΔL1–4 (100 ng each). Other conditions were the same as described in section 2.5.
3. Results
3.1. Expression, purification, and binding properties of the domain regions in M. sexta HP14
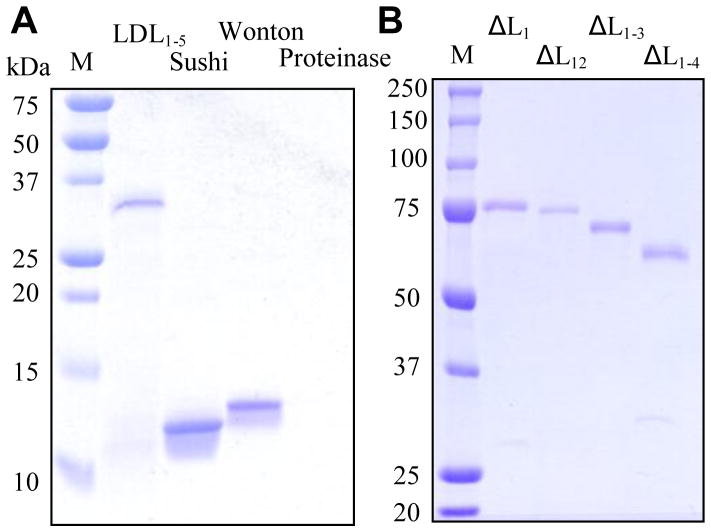
We constructed four plasmids to produce recombinant LDL1–5, Sushi domain, Wonton domain, and proteinase domain (PD) in E. coli (Fig. 1). All the domain regions were insoluble and, therefore, purified under the denaturing condition. From 1 L of the cultures, we obtained 1.3 mg LDL1–5, 3.8 mg Sushi, 6.6 mg Wonton, and 3.6 mg PD, part of which was used as antigen to generate region-specific antibodies. The rest of the proteins were subjected to renaturation. During dialysis, PD completely precipitated while the other three remained soluble (Fig. 2A). As judged from the stained gel, LDL1–5, Sushi, and Wonton were essentially pure and they migrated to positions consistent with their expected molecular masses.
Fig. 2. SDS-polyacrylamide gel electrophoretic analysis of purified domain regions (A) and deletion mutants (B) of M. sexta proHP14.
M, molecular weight markers with their positions and sizes indicated. Note that proteinase domain (PD) did not show up on the gel since it was completely insoluble after denaturation and renaturation.
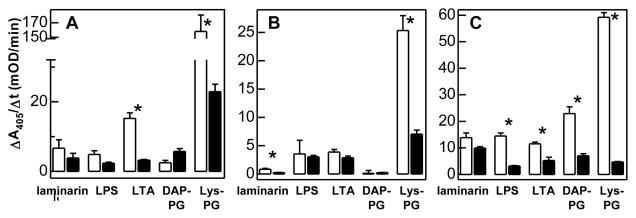
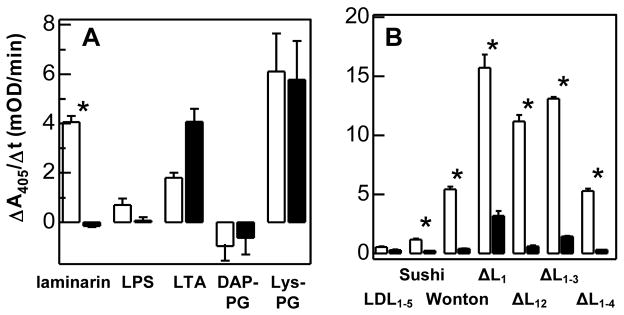
LDL1–5 specifically associated with lipoteichoic acid (LTA) and Lys-type PG. Binding to the Lys-type PG was ten fold more than binding to LTA (Fig. 3). Sushi domain also bound the Lys-type PG isolated from S. aureus. Wonton domain specifically associated with Lys- and DAP-type PGs, and the amount of binding was higher than that with lipopolysaccharide (LPS) or LTA.
Fig. 3. Binding of LDL1–5 (A), Sushi (B), and Wonton (C) to soluble microbial cell wall components.
As described in Methods and Materials, each purified domain region was preincubated with buffer (control, white bar) or a ligand (competition, black bar) and then added to wells that contain the immobilized ligand. The associated protein was incubated with specific first antibodies and enzyme-linked secondary antibodies. The alkaline phosphatase activities were measured and plotted as mean ± SEM (n = 3). The statistical significance between control and competition groups was analyzed by two-sample unpaired t-test. * indicates total binding is significantly (p<0.05) larger than binding after preincubation with a large excess of the ligands.
3.2. Isolation and characterization of the truncation mutants of M. sexta proHP14
To investigate the contribution of individual LDLA repeats in binding to Lys-type PG and LTA, we generated four recombinant baculoviruses that produced mutants with the first 1, 2, 3, and 4 LDL repeats removed from the intact proHP14 (Fig. 1). These N-terminally truncated proteins also serve as standards to verify some of the results obtained using the renatured domain regions. The deletion mutants were secreted into the culture media in a soluble form, which were enriched by cationic exchange chromatography and purified by nickel affinity chromatography. From 50 mL of the media, 130 μg ΔL1, 50 μg ΔL12, 200 μg ΔL1–3, and 60 μg ΔL1–4 were obtained. The purified proteins ran as single bands to their anticipated positions (Fig. 2B).
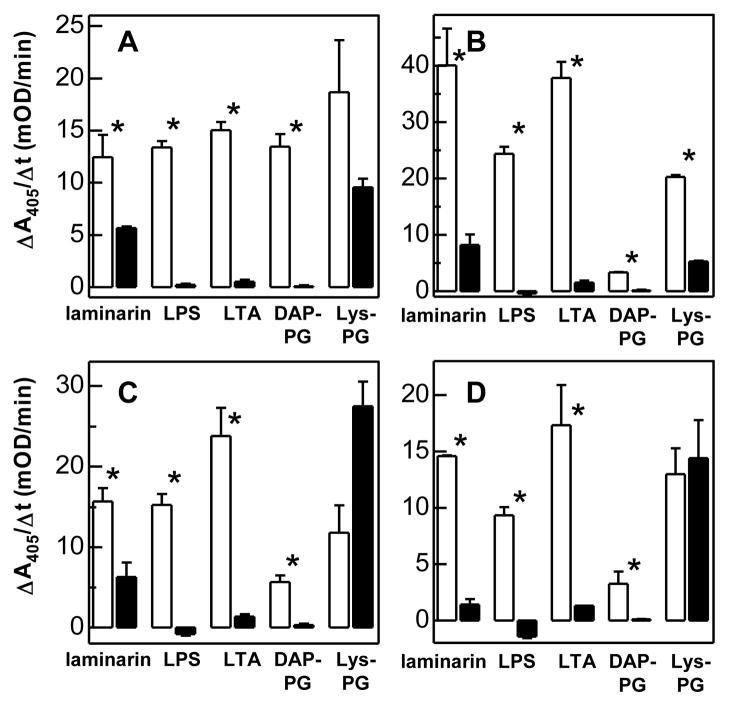
ΔL1 specifically associated with laminarin, LPS, LTA, and DAP-PG (Fig. 4A). Its binding to Lys-type PG was not statistically significant (p = 0.147). Removing the second LDLA repeat led to an increase in binding for all ligands except for the DAP-type PG from E. coli (Fig. 4B). The association with Lys-type PG became specific. The binding of ΔL1–3 to all ligands decreased except for DAP-PG (Fig. 4C). The association of Lys-type PG with ΔL1–3 or ΔL1–4 was nonspecific, and there was no major difference between ΔL1–3 and ΔL1–4 in binding amount or specificity with the other ligands (Fig. 4D).
Fig. 4. Binding of ΔL1 (A), ΔL12 (B), ΔL1–3 (C), and ΔL1–4 (D) to soluble microbial cell wall components.
See Fig. 3 legend.
Therefore, LDL1–5 and Wonton appeared to be responsible for specific interaction between LTA and proHP14 (Fig. 3, A and B), and the binding was modulated by the number of LDLA repeats (Fig. 4). Wonton domain specifically bound to LPS (Fig. 3B), and maximum binding occurred when the first two LDLA repeats were removed from the amino-terminus (Fig. 4). Specific association between Sushi domain and laminarin (Fig. 3C) became greater when LDLA repeats, Wonton, and PD were present in the same protein. Again, the binding was at highest level with ΔL12 (Fig. 4B). The specific association between Wonton domain and DAP-type PG remained unchanged, but its level was modulated by the LDLA region (Fig. 3B and Fig. 4). In contrast, the specific binding of Lys-type PG to LDL1–5, Sushi, or Wonton (Fig. 3) did not increase the specific association with the deletion mutants: ΔL12 was the only one that exhibited binding specificity (Fig. 4B).
3.3. Association of HP14 domains and truncation mutants with curdlan and βGRP1
While the ELISA-based binding assays provided quantitative data on how different domain regions or deletion mutants associate with immobilized ligands, we further tested direct binding of these proteins to curdlan, an insoluble β-1,3-glucan recognized by β-1,3-glucan recognition protein-2 (βGRP2) and if they caused proHP14 autoactivation [8]. We detected that LDL1–5, Sushi, or Wonton associated with curdlan to different extents, but the binding was not as complete as with the N-terminally truncated proteins (Fig. 5), suggesting that a combination of individual domain regions enhanced the total binding. The enhanced bindings were, however, weak, as washing the curdlan pellet with buffer led to a complete loss of signal in the bound fractions (data not shown).
Fig. 5. Binding of the purified domain regions and deletion mutants of M. sexta proHP14 to insoluble β-1,3-glucan.
Five μl of LDL1–5, Sushi, or Wonton (500 ng each) (top panels), ΔL1, ΔL12, ΔL1–3, or ΔL1–4 (200 ng each) (bottom panel) was mixed with 15 μl, 20 mM Tris-HCl, pH 8.0, 500 mM NaCl, 1 mM benzamidine and 50 μl curdlan pellet (500 μg) on ice for 1 h. After brief centrifugation, the original protein (5 μl with 15 μl H2O and 5 μl, 5×SDS sample buffer), supernatant (20 μl with 5 μl, 5×SDS sample buffer), and pellet (with 20 μl, 2×SDS) were subjected to heat treatment, 10% SDS-PAGE, and immunoblot analysis using specific antibodies. Immunoblot analysis using HP14 first antibody and goat-anti-rabbit IgG conjugated to alkaline phosphatase. 7.5% SDS-PAGE. Lane 1, total protein before binding; lane 2, unbound protein in the supernatant; lane 3, bound protein directly eluted from curdlan.
We then investigated the association among proHP14, curdlan, and β-1,3-glucan recognition protein-1 (βGRP1). After βGRP1 had been incubated with proHP14 isolated from induced plasma, there was no change in electrophoretic mobility of the proenzyme (Fig. 6). Also, no mobility change was detected after curdlan alone was mixed with the purified proHP14. However, when all three components were present at the same time, 75 kDa proHP14 was processed into 45 kDa regulatory domain and 30 kDa catalytic domain. The same phenomenon was observed after mixing proHP14, βGRP2 and curdlan [8], indicating that binding of the fungal cell wall component by its recognition protein βGRP2 (or βGRP1 in this case) and their interaction with proHP14 led to the autoactivation of proHP14. ELISA-based binding assay demonstrated that βGRP1 specifically recognized laminarin but not other microbial polysaccharides tested (Fig. 7A). βGRP1 also bound to Sushi or Wonton domain (Fig. 7B). Although the association between LDL1–5 and βGRP1 was low and nonspecific, the co-presence of PD and at least LDL45 significantly increased the binding.
Fig. 6. Autoactivation of M. sexta proHP14 in the presence of curdlan and βGRP1.
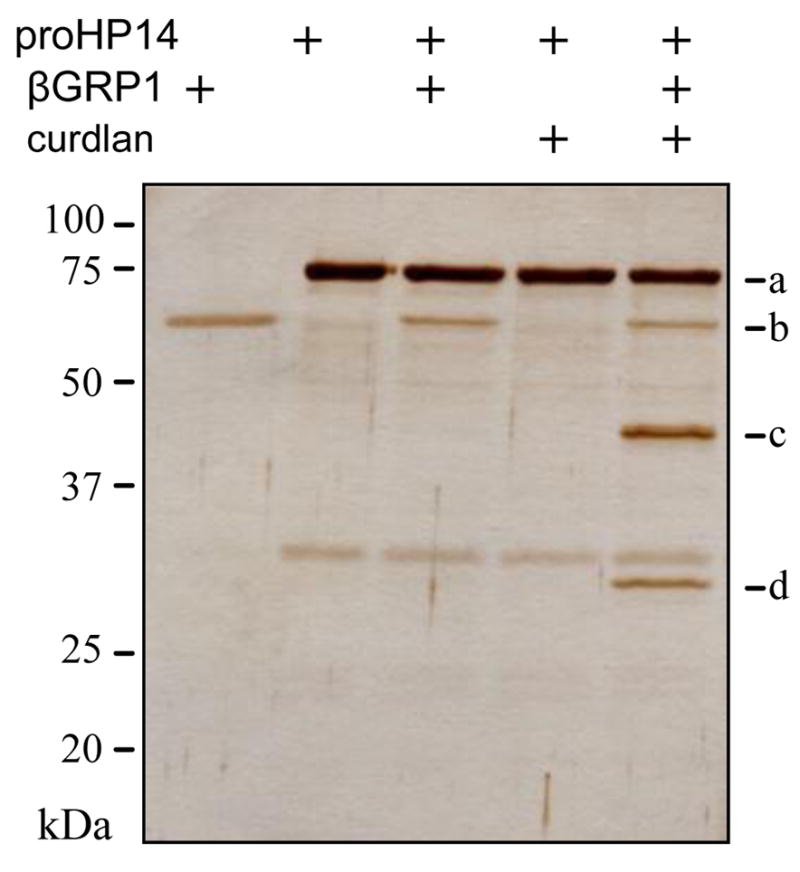
The purified proHP14 was incubated with curdlan and βGRP1 at 37°C for 1 h. After being treated with SDS sample buffer containing dithiothreitol, the reaction mixture and controls were subjected to 10% SDS-PAGE and silver staining. Sizes and positions of the molecular weight markers are indicated on the left. a, 75 kDa proHP14; b, βGRP1; c, 45 kDa HP14 heavy chain; d, 30 kDa HP14 light chain.
Fig. 7. Specific recognition of laminarin by βGRP1 (A) and binding of βGRP1 to domain regions or truncation mutants (B) of M. sexta proHP14.
See Fig. 3 legend.
4. Discussion
Innate immunity is vital for multicellular organisms to fend off invading pathogens. In order to distinguish self from non-self, this system employs a limited number of proteins to recognize surface features present in a multitude of microbes but not in host tissues or cells. By binding to common structures of bacteria or fungi, the so-called “pattern recognition receptors” form clusters on the foreign surfaces to recruit signaling molecules and downstream effectors that immobilize and kill pathogens [4,5,19]. In this study, we have examined the binding properties of LDL1–5, Sushi, and Wonton domains in M. sexta HP14 and their interaction with βGRP1.
The renatured LDL1–5, Sushi, and Wonton domains expressed in E. coli were soluble (Fig. 1), indicating that the proteins are folded. Since the three-dimensional structure of M. sexta HP14 is unknown, we are not able to confirm whether the recombinant proteins adopt the same folds as their counterparts in the natural proteins. Nevertheless, their specific binding to PGs, laminarin, curdlan, and βGRP1 (Figs. 3, 5, and 7) suggests correct folding because, otherwise, the renatured proteins are not expected to have similar binding properties manifested by proHP14 [7,8].
As summarized in Table 1, the binding patterns for different ligands and mutant proteins are complex. Specific binding of laminarin, LPS, or LTA to the truncated proHP14s occurred. Sushi domain might contribute specificity to the laminarin binding, whereas Wonton domain seems to be responsible for the specific interaction with LPS and (in part) LTA. The specificity for the binding of Wonton domain and DAP-type PG did not change but the degree of binding was greatly influenced by the LDL repeats (Figs. 3C and 4). The most intriguing result came from the assays using Lys-type PG: although LDL1–5, Sushi and Wonton each specifically bound to the ligand (Fig. 3), the binding specificity was lost in all of the deletion mutants except for ΔL12 (Fig. 4). Perhaps, interactions among the specific binding sites in Sushi, Wonton, and LDLA repeats led to lower and less specific association of the truncated proHP14 with Lys-type PG. Since such interactions did not seem to negatively impact the specific association of Sushi or Wonton with the other ligands (Table 1), we suggest that Lys-type PG itself contributed to the reduction in binding and specificity. The structural differences between DAP- and Lys-type PGs are critical for understanding the binding property changes.
Table 1.
Summary of the associations between M. sexta proHP14 mutants and microbial cell wall components a
| LDL1–5 | Sushi | Wonton | ΔL1 | ΔL12 | ΔL1–3 | ΔL1–4 | |
|---|---|---|---|---|---|---|---|
| Laminarin | − | +/lb | − | + | +/hb | + | + |
| lipopolysaccharide | − | −/l | + | + | +/h | + | + |
| lipoteichoic acid | + | −/l | + | + | +/h | +/h | + |
| DAP-type peptidoglycan | −/l | −/l | +/h | + | +/l | + | +/l |
| Lys-type peptidoglycan | +/h | +/h | +/h | − | +/h | − | − |
+, specific binding; −, no significant difference in the competition binding assay.
low (l) and high (h) degree of association (< 5.0 and > 20.0 mOD/min for binding without a competitor, respectively).
Some of the binding data must be cautiously evaluated. For instance, we could not explain why association became significantly higher after of ΔL1–3 had been pre-incubated with an excess amount of Lys-type PG (Fig. 4C). While the increase was statistically insignificant in the case of ΔL1–4 and Lys-type PG (Fig. 4D), neither do we understand the significant increase in binding of βGRP1 to LTA after blocking with excess the ligand (Fig. 7A). Additionally, although renatured LDL1–5, Sushi, and Wonton from E. coli were soluble, they may or may not adopt the same fold as those in the native proteins from the insect cells. Further binding analysis using the domain regions from the baculovirus expression system could be useful for verifying the data presented herein (Fig. 3).
Wonton domain appears to be critical for specific binding to bacterial cell wall components (Table 1). BLASTP search of GenBank indicates it is most similar in sequence to Lonomia oblique serine proteinase-1 (60% identity; 73% similarity, and 2 gaps). Like M. sexta HP14, the L. oblique enzyme contains a Sushi domain before its Wonton domain. The Sushi domains are 37% identical and 48% similar, significantly less conserved than the Wonton domains. The binding of HP14 Wonton domain to LPS, LTA, and two types of PGs may demand a higher degree of structural conservation. Interestingly, Wonton and Sushi domains seem to be evolutionarily related – in M. sexta HP14, their sequences are 17.6% identical and 35.2% similar (Fig. 8). As a matter of fact, the Lonomia protein is predicted to contain two Sushi domains, the second of which contains two extra Cys residues located in the same positions of the HP14 Wonton domain (data not shown). Although the LDLA repeats modulate the degree and specificity of proHP14’s binding to the microbial surface molecules, their role seems to be less important than the Wonton domain. L. oblique serine proteinase-1 has only one LDLA repeat between the signal peptide and (first) Sushi domain, suggesting that the LDLA is partially dispensable.
Fig. 8. Sequence alignment of the Sushi and Wonton domains in M. sexta HP14.
The sequences are manually aligned to show identical (*) and similar (+) residues. Cys residues are in bold, and the ones conserved in Sushi domains form two disulfide bonds (Cys-1 and Cys-3; Cys-2 and Cys-4) [22].
As demonstrated previously [8] and in this study (Fig. 6), specific binding of proHP14 to the microbial polysaccharides by itself was insufficient for its autoactivation. Additional protein such as βGRP1 or βGRP2 is also required. Both βGRPs specifically recognized soluble or insoluble β-1,3-glucan (Fig. 7A; Wang and Jiang, unpublished data), which enhanced the binding specificity of LDL1–5, Sushi, and Wonton (Fig. 3). The added specificity, passed onto proHP14 through specific protein-protein interactions (Fig. 7B), ensures that initiation of the M. sexta proPO activation system occurs only at the site of infection. In the beetle Tenebrio molitor, autoactivation of the proHP14 ortholog is triggered by PGRP-SA and GNBP1 that recognize Lys-type PG [20].
Binding site analysis of the horseshoe crab coagulation factor G demonstrated that β-1,3-glucan recognition was mediated by the two xylanase Z-like domains in the α subunit [21]. Factor G and HP14 dramatically differ in their quaternary structure and domain constitution, but the presence of two Sushi-like domains in HP14 is reminiscent of the tandem repeats in Factor G. Increase in binding specificity and avidity through multiple domains or via interaction with other pattern recognition receptors (e.g. βGRPs) may be an important factor for triggering localized innate immune responses.
Supplementary Material
Acknowledgments
This work was supported by National Institutes of Health Grants GM58634. We would like to thank Drs. Michael Kanost, Jack Dillwith, and Andrew Mort for their helpful comments on the manuscript. This article was approved for publication by the Director of Oklahoma Agricultural Experimental Station and supported in part under project OKLO2450.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Gillespie JP, Kanost MR, Trenczek T. Biological mediators of insect immunity. Ann Rev Entomol. 1997;42:611–43. doi: 10.1146/annurev.ento.42.1.611. [DOI] [PubMed] [Google Scholar]
- 2.Lavine MD, Strand MR. Insect hemocytes and their role in immunity. Insect Biochem Mol Biol. 2002;32:1295–309. doi: 10.1016/s0965-1748(02)00092-9. [DOI] [PubMed] [Google Scholar]
- 3.Nappi AJ, Christensen BM. Melanogenesis and associated cytotoxic reactions: applications to insect innate immunity. Insect Biochem Mol Biol. 2005;35:443–59. doi: 10.1016/j.ibmb.2005.01.014. [DOI] [PubMed] [Google Scholar]
- 4.Lemaitre B, Hoffmann J. The host defense of Drosophila melanogaster. Ann Rev Immunol. 2007;25:697–743. doi: 10.1146/annurev.immunol.25.022106.141615. [DOI] [PubMed] [Google Scholar]
- 5.Jiang H. The biochemical basis of antimicrobial responses in Manduca sexta. Insect Sci. 2008;15:53–66. [Google Scholar]
- 6.Cerenius L, Lee BL, Söderhäll K. The proPO-system: pros and cons for its role in invertebrate immunity. Trends Immunol. 2008;29:263–71. doi: 10.1016/j.it.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Ji C, Wang Y, Guo X, Hartson S, Jiang H. A pattern recognition serine proteinase triggers the prophenoloxidase activation cascade in the tobacco hornworm, Manduca sexta. J Biol Chem. 2004;279:34101–6. doi: 10.1074/jbc.M404584200. [DOI] [PubMed] [Google Scholar]
- 8.Wang Y, Jiang H. Interaction of β-1,3-glucan with its recognition protein activates hemolymph proteinase 14, an initiation enzyme of the prophenoloxidase activation system in Manduca sexta. J Biol Chem. 2006;281:9271–8. doi: 10.1074/jbc.M513797200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim CH, Kim SJ, Kan H, Kwon HM, Roh KB, Jiang R, Yang Y, Park JW, Lee HH, Ha NC, Kang HJ, Nonaka M, Söderhäll K, Lee BL. A three-step proteolytic cascade mediates the activation of the peptidoglycan-induced toll pathway in an insect. J Biol Chem. 2008;283:7599–607. doi: 10.1074/jbc.M710216200. [DOI] [PubMed] [Google Scholar]
- 10.Buchon N, Poidevin M, Kwon HM, Guillou A, Sottas V, Lee BL, Lemaitre B. A single modular serine protease integrates signals from pattern-recognition receptors upstream of the Drosophila Toll pathway. Proc Natl Acad Sci USA. 2009;106:12442–7. doi: 10.1073/pnas.0901924106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y, Jiang H. Reconstitution of a branch of Manduca sexta prophenoloxidase activation cascade in vitro: Snake-like hemolymph proteinase 21 cleaved by HP14 activates prophenoloxidase-activating proteinase-2 precursor. Insect Biochem Mol Biol. 2007;37:1015–25. doi: 10.1016/j.ibmb.2007.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gorman MJ, Wang Y, Jiang H, Kanost MR. Manduca sexta hemolymph proteinase 21 activates prophenoloxidase activating proteinase 3 in an insect innate immune response proteinase cascade. J Biol Chem. 2007;282:11742–9. doi: 10.1074/jbc.M611243200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ma C, Kanost MR. A β-1,3-glucan recognition protein from an insect, Manduca sexta, agglutinates microorganisms and activates the phenoloxidase cascade. J Biol Chem. 2000;275:7505–14. doi: 10.1074/jbc.275.11.7505. [DOI] [PubMed] [Google Scholar]
- 14.Jiang H, Ma C, Lu Z, Kanost MR. β-1,3-glucan recognition protein-2 (βGRP-2) from Manduca sexta: an acute-phase protein that binds β-1,3-glucan and lipoteichoic acid to aggregate fungi and bacteria. Insect Biochem Mol Biol. 2004;34:89–100. doi: 10.1016/j.ibmb.2003.09.006. [DOI] [PubMed] [Google Scholar]
- 15.Lee E, Linder ME, Gilman AG. Expression of G-protein alpha subunits in Escherichia coli. Methods Enzymol. 1994;237:146–63. doi: 10.1016/s0076-6879(94)37059-1. [DOI] [PubMed] [Google Scholar]
- 16.Wang Y, Jiang H, Kanost MR. Expression and purification of Manduca sexta prophenoloxidase-activating proteinase precursor (proPAP) from baculovirus-infected insect cells. Protein Exp Purif. 2004;23:328–37. doi: 10.1006/prep.2001.1517. [DOI] [PubMed] [Google Scholar]
- 17.Lu Z, Jiang H. Expression of Manduca sexta serine proteinase homolog precursors in insect cells and their proteolytic activation. Insect Biochem Mol Biol. 2008;38:89–98. doi: 10.1016/j.ibmb.2007.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ji C, Wang Y, Ross J, Jiang H. Expression and in vitro activation of Manduca sexta prophenoloxidase-activating proteinase-2 precursor (proPAP-2) from baculovirus-infected insect cells. Protein Exp Purif. 2003;29:235–43. doi: 10.1016/s1046-5928(03)00020-2. [DOI] [PubMed] [Google Scholar]
- 19.Zhao P, Li J, Wang Y, Jiang H. Broad-spectrum antimicrobial activity of the reactive compounds generated in vitro by Manduca sexta phenoloxidase. Insect Biochem Mol Biol. 2007;37:952–9. doi: 10.1016/j.ibmb.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Park JW, Kim CH, Kim JH, Je BR, Roh KB, Kim SJ, Lee HH, Ryu JH, Lim JH, Oh BH, Lee WJ, Ha NC, Lee BL. Clustering of peptidoglycan recognition protein-SA is required for sensing lysine-type peptidoglycan in insects. Proc Natl Acad Sci USA. 2007;104:6602–7. doi: 10.1073/pnas.0610924104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Takaki Y, Seki N, Kawabata SI, Iwanaga S, Muta T. Duplicated binding sites for (1,3)-beta-D-glucan in the horseshoe crab coagulation factor G: implications for a molecular basis of the pattern recognition in innate immunity. J Biol Chem. 2002;277:14281–7. doi: 10.1074/jbc.M200177200. [DOI] [PubMed] [Google Scholar]
- 22.Wei XQ, Orchardson M, Gracie JA, Leung BP, Gao BM, Guan H, Niedbala W, Paterson GK, McInnes IB, Liew FY. The Sushi domain of soluble IL-15 receptor alpha is essential for binding IL-15 and inhibiting inflammatory and allogenic responses in vitro and in vivo. J Immunol. 2001;167:277–82. doi: 10.4049/jimmunol.167.1.277. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.