Abstract
Extracts of fifty-seven newly isolated strains of dinoflagellates and raphidophytes were screened for protein phosphatase (PP2A) inhibition. Five strains, identified by rDNA sequence analysis as Prorocentrum rhathymum, tested positive and the presence of okadaic acid was confirmed in one strain by HPLC-MS/MS and by HPLC with fluorescence detection and HPLC-MS of the okadaic acid ADAM derivative. Quantitation of the ADAM derivative indicated that the concentration of okadaic acid in the culture medium is 0.153 μg/L.
Keywords: Prorocentrum rhathymum, okadaic acid, diarrheic shellfish poisoning (DSP) toxins, LC-MS, ADAM derivative, HPLC-FLD
The suite of marine toxins that includes okadaic (OA) acid and the dinophysistoxins (DTX) is collectively known as DSP (diarrheic shellfish poisoning) toxins. The acute symptoms of DSP include diarrhea, nausea, vomiting and abdominal pain. Outbreaks have been documented worldwide and are associated with the consumption of mussels, scallops, or clams tainted with OA, its analogs or derivatives. (Gestal-Otero, 2000). OA has been isolated from several dinoflagellates in the genera Prorocentrum and Dinophysis, including the species P. lima (Yasumoto et al., 1987), P. hoffmanianum (Murakami et al., 1982), P. concavum (Dickey et al., 1990), P. maculosum (Zhou and Fritz, 1993), P. belizeanum (Morton et al., 1998), P. faustiae (Morton 1998), P. arenarium (Ten-Hage et al., 2000), D. acuta (Draisci et al., 1998) and D. fortii (Murata et al., 1982). In addition to OA, several related polyethers were isolated from these dinoflagellates, including the dinophysis toxins, DTX-1 and DTX-2 which differ in the location and number of methyl substituents (Murata et al., 1982; Yasumoto, et al., 1985; Hu et al., 1993). OA, DTX-1 and DTX-2 are inhibitors of protein phosphatases PP1 and PP2A (Dounay and Forsyth, 2002).
We recently reported the isolation of over fifty strains of dinoflagellates by viable high speed single-cell sorting (Sinigalliano et al., 2009). Five of these strains (6–9 and 25) matched most closely with P. rhathymum in a BLAST (Altschul et al., 1990) analysis of their large-subunit ribosomal genes (Scorzetti et al., 2009) and tested positive for protein phosphatase (PP2A) inhibition in preliminary screening. The presence of OA was confirmed in one strain by HPLC with fluorescent detection of the ADAM (9-anthryldiazomethane) derivative (Quilliam et al., 1998) and by HPLC-MS and MS/MS experiments.
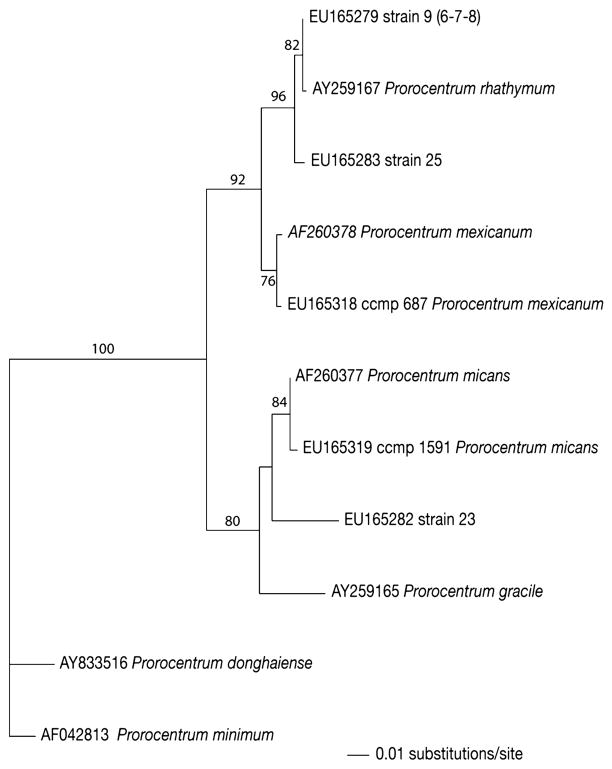
Fifty-seven field strains were sequenced in the D1/D2 region of the LSU rDNA and compared with GenBank sequence data (Scorzetti et al., 2009). Dinoflagellate and raphidophyte genera identified included Akashiwo, Amphidinium, Heterocapsa, Cachonina, Chatonella, Coolia, Fragilidinium, Heterosigma, Karlodinium, Karenia, Protoceratium, Scripsciella and Prorocentrum. Field strains 6, 7, 8, 9, 23, 25 and identified strains CCMP687 (Prorocentrum mexicanum) and CCMP1591 (Prorocentrum micans) clustered within a clade of the genus Prorocentrum (Fig. 1). The sequences for strains 6, 7, 8 and 9 were identical to each other and matched with two sequences (AY259166 and AY259167) of P. rhathymum (Pearce and Hallegraeff unpublished), which are the only two D1/D2 sequences of that species in GenBank. Strain 6 was deposited in the Provasoli-Guillard National Center for culture of Marine Phytoplankton as strain CCCMP2933 and sequenced in the ITS regions (Ferrel and Beaton unpublished). The ITS sequence (EU927561) showed only 2 mismatches from other ITS sequences present in the GenBank database: EU244464 (Rial et al., unpublished) and FJ155840 (Caillaud et al., 2009), thus confirming the identification as P. rhathymum. Base pair differences at two locations, particularly in the highly variable ITS region, are considered to be insignificant and may be due to technical difficulties in comparing data from different laboratories (D’Onofrio et al 1999). Strain 23 was more than 20 bp different from its closest match, AF260377, P. micans. The sequence of the identified strain of P. mexicanum agreed with the GenBank strain AF260378 (Daugbjerg et al., 2000). After its initial description in 1979 by Loeblich, the P. rhathymum taxonomy was dissolved and reclassified as P. mexicanum (Faust, 1990; Steidinger, 1983). Later the species was reinstated as a unique organism (Cortes-Altamirano, 2003). Our sequence data agrees with the concept that P. mexicanum and P. rhathymum are separate species. Strain 25 clustered with P. rhathymum and differed from that species by 6 bp in the D2 region. It has a distinct position in the clade and demonstrates significant microscopic and macroscopic differences from P. rhathymum, which indicates that strain 25 may represent a new taxon or species within the genus. Other members of the clade include P. micans, P. gracile and strain 23, which may also represent a new species in the genus. Studies among micro-eukaryotes (D’Onofrio et al, 1999; Scorzetti et al, 2002; Montresor et al 2003) have recognized that strains with identical sequences are members of a single species and that sequence differences at the magnitude exhibited by strains 23 and 25 indicate phylogenetically distinct genotypes.
Fig. 1.
LSU D1D2 rRNA phylogenetic tree comprising studied strains and related GenBank sequences. The tree was constructed with likelihood analysis in heuristic search (stepwise-addition option with 1000 replicates). Bootstrap values were reported on branches when higher than 50%. Sequences for Prorocentrum minimum and Prorocentrum donghiae were used as outgroup.
Solid phase C18 extracts of the culture medium of fifty-seven strains of dinoflagellates and raphidophytes were screened for PP2A inhibition (Simon and Vernoux 1994). Pan et al. demonstrated that OA, DTX-1 and DTX-2 are more abundant in the culture medium than in the cells (Pan et al., 1999). The large scale isolation of okadaic acid from filtered seawater was recently described by Rundberget et al. (2007). We therefore reasoned that testing of the culture medium would be a simple and efficient way to perform preliminary screens for OA production. C18 extracts of strains 6–9, 25 and 40 (Prorocentrum hoffmanianum) were positive for PP2A inhibition. All fifty-one of the remaining field strains, including six strains (21, 22, 23, 27, 54, 55) whose D1D2 sequence matched most closely with other Prorocentrum species (micans, minimum, donghaiense and dentatum) (Scorzetti et al., 2009) tested negative for PP2A inhibition.
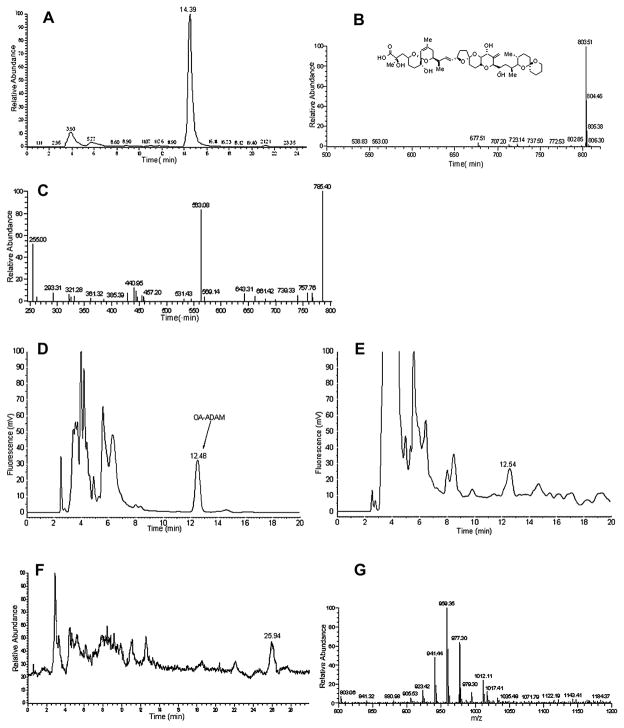
To confirm the presence of OA by LC-MS, two hundred liters of culture medium from P. rhathymum, strain 8 was extracted. LC-MS of the crude extract was performed in negative ion mode with the full scan range of m/z 500–820. Both the OA standard and the crude extract of P. rhathymum revealed a [M-1] peak of m/z 803.5 at a retention time of 14.4 min (Fig. 2A and B). Only a single peak, with an m/z of 803.5 was observed, nor did we observe a peak with m/z 817, suggesting that only OA was present and not DTX-1 or DTX-2. LC-MS/MS analyses of standard OA and P. rhathymum extract were performed in negative ion mode for the fragmentation of m/z 803.5. The MS/MS spectrum shows four major product ions at m/z 785, 563 and 255 and two minor product ions at m/z 767, 785 and 321 (Fig. 2C) identical to those produced from OA (Torgersen et al., 2008).
Fig. 2.
A. The chromatogram of P. rhathymum extract monitored for m/z 803.5. B. MS spectrum of 14.39 min. peak from Fig. 2A. C. The LC-MS/MSof the m/z 803.5 ion as the precursor. D. HPLC-FLD analysis for reaction of “in situ” ADAM reagent system with 8.65 ng of an okadaic acid standard. E. HPLC-FLD analysis for reaction of “in situ” ADAM reagent system with P. rhathymum supernatant extract. F. Total ion chromatogram (TIC) of the “in situ” ADAM reaction of P. rhathymum supernatant extract. G. MS spectrum of 25.94 min. peak from Fig. 2F.
Standard OA and P. rhathymum extract were derivatized with ADAM. Fig. 2D shows the HPLC-FLD analysis of the OA-ADAM reaction. The peak at a retention time of 12.49 min was absent from the blank and corresponds to ADAM esterified OA. Fig. 2E is the HPLC-FLD analysis of culture extract derivatized with ADAM. A peak was observed with the retention time of 12.54 minutes. The same sample that gave the chromatogram in Fig. 2E was subsequently analyzed by LC-MS and the results are presented in Fig. 2F. The total ion current chromatogram (TIC) showed one major peak, whose relative retention time was 26 min. The mass spectrum recorded in Fig. 2F confirmed this to be the ADAM esterified OA. An [M+NH4]+ ion was observed at m/z 1012.11 and fragment ions due to sequential losses of water were observed at m/z 977.6 and 959.6. Thus the unambiguous identification of OA from P. rhathymum was made based on the retention time, molecular mass and fragment ion information. Quantitative analysis of the OA-ADAM derivative indicated that the concentration of OA in the culture medium of P. rhathymum was 0.153 μg/L.
We demonstrated the production of okadaic acid by P. rhathymum isolated from Florida Bay. While the presence of OA was confirmed by LC-MS or HPLC-FLD for strain 6 only, strains 7–9 and 25 were positive for PP2A inhibition. As we know of no other PP2A inhibitor produced by a dinoflagellate, we consider this indicative of OA production as well. Recently, OA has been identified in another strain of P. rhathymum from Malaysia, further supporting our findings (Caillaud et al., 2009). In our work, okadaic acid was isolated from the culture medium and not, as is traditionally done, from the cells. We are not aware of any other report of screening for OA in this way. The concentration of OA in the medium of our cultures of P. rhathymum was as much as two to three orders of magnitude lower (0.153 μg/L) than other known OA producers such as P. lima or P. hoffmanianum when the culture medium was analyzed in the same way (Perez et al., 2008). However, OA and DTX-1/2 are stored in the dinoflagellate cells as esters, while the free acids are released into the medium. This suggests that the total OA in the cultures could be much higher than our analysis indicates. At the time of sampling, strains 6–9 were the most abundant protists and may impact seafood safety in Florida Bay. In addition, future research should consider the pectenotoxins, which are often associated with OA producing organisms.
Supplementary Material
Acknowledgments
This work was supported by the NSF-NIEHS Oceans and Human Health Center Program (National Science Foundation grant 0432368 and NIEHS grant P50 ES12736-01). Grant support was also provided by the National Institute of Environmental Health Sciences (NIEHS) Grant S11 ES11181.
Appendix. Supplementary information
Supplementary data associated with this article including experimental details may be found at XXXXXXX
Footnotes
Conflict of interest: The authors declare that there are no conflicts of interest.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. Journal of Molecular Biology. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Caillaud A, de la Iglesia P, Campas M, Elandaloussi L, Fernandez M, Mohammad-Noor N, Andree K, Diogene J. Evidence of okadaic acid production in a cultured strain of the dinoflagellate Prorocentrum rhathymum from Malaysia. Toxicon. 2009 doi: 10.1016/j.toxicon.2009.07.016. In Press, Corrected Proof, Available online 23 July 2009. [DOI] [PubMed] [Google Scholar]
- Cortes-Altamirano R, Sierra-Bltran AP. Morphology and taxonomy of Prorocentrum mexicanum and reinstatement of Prorocentrum rhathymum (Dinophyceae) Journal of Phycology. 2003;38:221–225. [Google Scholar]
- Daugbjerg N, Hansen G, Larsen J, Moestrup Ø. Phylogeny of some of the major genera of dinoflagellates based on ultrastructure and partial LSU rDNA sequence data, including the erection of three new genera of unarmored dinoflagellates. Phycologia. 2000;39:302–317. [Google Scholar]
- Dickey RW, Bobzin SC, Faulkner DJ, Bencsath FA, Andrzejewski D. Identification of okadaic acid from a caribbean dinoflagellate, Prorocentrum concavum. Toxicon. 1990;28:371–377. doi: 10.1016/0041-0101(90)90074-h. [DOI] [PubMed] [Google Scholar]
- D’Onofrio G, Marino D, Bianco L, Busico E, Montresor M. Toward an assessment on the taxonomy of dinoflagellates that produce calcarerous cysts (Calciodinelloideaea, Dinophyceaae): a morphological and molecular approach. Journal of Phycology. 1999;35:1063–1078. [Google Scholar]
- Dounay AB, Forsyth CJ. Okadaic acid: The archetypal serine/threonine protein phosphatase inhibitor. Current Medicinal Chemistry. 2002;9:1939–1980. doi: 10.2174/0929867023368791. [DOI] [PubMed] [Google Scholar]
- Draisci R, Giannetti L, Lucentini L, Marchiafava C, James KJ, Bishop AG, Healy BM, Kelly SS. Isolation of a new okadaic acid analogue from phytoplankton implicated in diarrhetic shellfish poisoning. Journal of Chromatography A. 1998;798:137–145. doi: 10.1016/s0021-9673(97)01200-4. [DOI] [PubMed] [Google Scholar]
- Faust MA. Morphologic details of six benthic species of Prorocentrum (Pyrrophyta) from a mangrove island, Twin Cays, Belize, including two new species. Journal of Phycology. 1990;26:548–558. doi: 10.1111/j.1529-8817.2007.00450.x. [DOI] [PubMed] [Google Scholar]
- Gestal-Otero JJ. Nonneurotoxic toxins. Food Science and Technology (New York, NY, United States) 2000;103 (Seafood and Freshwater Toxins):45–64. [Google Scholar]
- Hu T, deFreitas ASW, Doyle J, Jackson D, Marr J, Nixon ES, Quilliam MA, Walter JA, Wright JLC. New DSP toxin derivatives isolated from toxic mussels and the dinoflagellates, Prorocentrum lima and Prorocentrum concavum. In: Smayda TJ, Shimizu Y, editors. Developments in Marine Biology. Vol. 3. Elsevier; New York: 1993. pp. 507–512. Toxic Phytoplankton Blooms in the Sea. [Google Scholar]
- Montresor M, Gross S, Procaccini G, Koistra WHCE. Intraspecific diversity in Scrippsiella trochoidea (Dinophyceae): evidence for criptic species. Phycologia. 2003;42:56–70. [Google Scholar]
- Morton SL, Moeller PD, Young KA, Lanoue B. Okadaic acid production from the marine dinoflagellate Prorocentrum belizeanum Faust isolated from the Belizean coral reef ecosystem. Toxicon. 1998;36:201–206. doi: 10.1016/s0041-0101(97)00054-8. [DOI] [PubMed] [Google Scholar]
- Morton SL. Morphology and toxicology of Prorocentrum fausitae sp. nov., a toxic species of non-planktonic dinoflagellate from Heron Island. Botanica Marina. 1998;41:565–569. [Google Scholar]
- Murakami Y, Oshima Y, Yasumoto T. Identification of okadaic acid as a toxic component of a marine dinoflagellate Prorocentrum lima. Nippon Suisan Gakkaishi. 1982;48:69–72. [Google Scholar]
- Murata M, Shimatani M, Sugitani H, Oshima Y, Yasumoto T. Isolation and structural elucidation of the causative toxin of the diarrhetic shellfish poisoning. Nippon Suisan Gakkaishi. 1982;48:549–552. [Google Scholar]
- Pan Y, Cembella AD, Quilliam MA. Cell cycle and toxin production in the benthic dinoflagellate Prorocentrum lima. Marine Biology. 1999;134:541–549. [Google Scholar]
- Perez R, Liu L, Lopez J, An T, Rein KS. Diverse Bacterial PKS Sequences Derived From Okadaic Acid-Producing Dinoflagellates. Marine Drugs. 2008;6:164–179. doi: 10.3390/md20080009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quilliam MA, Gago-Martinez A, Rodriguez-Vazquez J. Improved method for preparation and use of 9-anthryldiazomethane for derivatization of hydroxycarboxylic acids: application to diarrhetic shellfish poisoning toxins. Journal of Chromatography A. 1998;807:229–239. doi: 10.1016/s0021-9673(98)00069-7. [DOI] [PubMed] [Google Scholar]
- Rundberget T, Sandvik M, Larsen, Pizarro GM, Reguera B, Castberg T, Gustad E, Loader JI, Rise F, Wilkins AL, Miles CO. Extraction of microalgal toxins by large - scale pumping of seawater in Spain and Norway, and isolation of okadaic acid and dinophysistoxin-2. Toxicon. 2007;50:960–970. doi: 10.1016/j.toxicon.2007.07.003. [DOI] [PubMed] [Google Scholar]
- Scorzetti G, Brand LE, Hitchcock GL, Rein KS, Sinigalliano CD, Fell JW. Multiple simultaneous detection of Harmful Algal Blooms (HABs) through a high throughput bead array technology, with potential use in phytoplankton community analysis. Harmful Algae. 2009;8:196–211. doi: 10.1016/j.hal.2008.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scorzetti G, Fell JW, Fonseca A, Statzell-Tallman A. Systematics of basidiomycetous yeasts: a comparison of large sub-unit D1D2 and internal transcribed spacer rDNA regions. FEMS Yeast Research. 2002;2:495–517. doi: 10.1111/j.1567-1364.2002.tb00117.x. [DOI] [PubMed] [Google Scholar]
- Simon JF, Vernoux JP. Highly sensitive assay of okadaic acid using protein phosphatase and paranitrophenyl phosphate. Natural Toxins. 1994;2:293–301. doi: 10.1002/nt.2620020508. [DOI] [PubMed] [Google Scholar]
- Sinigalliano CD, Winshell J, Guerrero MA, Scorzetti G, Fell JW, Eaton RW, Rein KS. Viable cell sorting of dinoflagellates by multi-parametric flow cytometry. Phycologia. 2009;44:249–257. doi: 10.2216/08-51.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steidinger KA. A re-evaluation of the toxic dinoflagellate biology and ecology. Progress in Phycological Research. 1983;2:147–188. [Google Scholar]
- Ten-Hage L, Delaunay N, Pichon V, Coute A, Puiseux-Dao S, Turquet J. Okadaic acid production from the marine benthic dinoflagellate Prorocentrum arenarium Faust (dinophyceae) isolated from Europa Island coral reef ecosystem (SW Indian Ocean) Toxicon. 2000;38:1043–1054. doi: 10.1016/s0041-0101(99)00216-0. [DOI] [PubMed] [Google Scholar]
- Torgersen T, Wilkins AL, Rundberget T, Miles CO. Characterization of fatty acid esters of okadaic acid and related toxins in blue mussels (Mytilus edulis) from Norway. Rapid Communications in Mass Spectrometry. 2008;22:1127–1136. doi: 10.1002/rcm.3490. [DOI] [PubMed] [Google Scholar]
- Yasumoto T, Murata M, Oshima Y, Sano M, Matsumoto GK, Clardy J. Diarrhetic shellfish toxins. Tetrahedron. 1985;41:1019–25. [Google Scholar]
- Yasumoto T, Seino N, Murakami Y, Murata M. Toxins produced by benthic dinoflagellates. Biological Bulletin (Woods Hole, MA, United States) 1987;172:128–131. [Google Scholar]
- Zhou J, Fritz L. Ultrastructure of two toxic marine dinoflagellates, Prorocentrum lima and Prorocentrum maculosum. Phycologia. 1993;32:444–450. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.