Abstract
Paramyxovirus matrix (M) proteins organize virus assembly, functioning as adapters that link together viral ribonucleoprotein complexes and viral glycoproteins at infected cell plasma membranes. M proteins may also function to recruit and manipulate host factors to assist virus budding, similar to retroviral Gag proteins. By yeast two-hybrid screening, angiomotin-like 1 (AmotL1) was identified as a host factor that interacts with the M protein of parainfluenza virus 5 (PIV5). AmotL1-M protein interaction was observed in yeast, in transfected mammalian cells, and in virus-infected cells. Binding was mapped to a 83 amino acid region derived from the C-terminal portion of AmotL1. Overexpression of M-binding AmotL1-derived polypeptides potently inhibited production of PIV5 VLPs and impaired virus budding. Expression of these polypeptides moderately inhibited production of mumps VLPs, but had no effect on production of Nipah VLPs. siRNA-mediated depletion of AmotL1 protein reduced PIV5 budding, suggesting that this interaction is beneficial to paramyxovirus infection.
Keywords: virus budding, virus assembly, matrix protein, paramyxovirus, parainfluenza virus 5, PIV5, mumps virus, Nipah virus, angiomotin-like 1, AmotL1
Introduction
Paramyxovirus particles, like those of other enveloped viruses, are formed by budding from cellular membranes. Budding occurs after viral structural components, including viral glycoproteins and viral ribonucleoprotein complexes (RNPs), have assembled together on infected cell plasma membranes (reviewed in (Schmitt and Lamb, 2004; Takimoto and Portner, 2004). Paramyxovirus assembly is coordinated by viral matrix (M) proteins, which occupy a position in virions that is between the glycoproteins and the RNPs. M proteins interact strongly with cellular membranes, and are known to self-oligomerize, forming dense layers along the inner surfaces of plasma membranes. During virus assembly, M proteins are thought to interact directly with the cytoplasmic tails of viral glycoproteins, and also with the nucleocapsid (NP) protein components of RNPs, effectively bridging these elements and concentrating them at locations from which virus particles will bud (reviewed in (Schmitt and Lamb, 2004; Takimoto and Portner, 2004).
For many paramyxoviruses, the processes of particle formation and release have been recapitulated in cells transfected to produce one or more viral proteins. The resulting virus-like particles (VLPs) are morphologically similar to authentic virions and have proven to be useful tools for dissecting steps involved in virus exit. Not surprisingly, paramyxovirus VLP production is critically dependent on the presence of the viral M proteins (Schmitt and Lamb, 2004). Nonetheless, substantial differences have been identified among paramyxoviruses in the requirements for efficient VLP production. For example, optimal production of Sendai VLPs requires expression of the viral C protein in addition to the M protein and viral glycoproteins (Sugahara et al., 2004). For the rubulaviruses, which do not encode C proteins, efficient VLP production requires co-expression of M proteins together with NP proteins and viral glycoproteins (fusion (F) protein for mumps virus; F protein or hemagglutinin-neuraminidase (HN) protein for parainfluenza virus 5 (PIV5)) (Li et al., 2009; Schmitt et al., 2002). Other paramyxoviruses, including Newcastle disease virus and Nipah virus (NiV), produce VLPs very efficiently even when M protein is expressed by itself (Ciancanelli and Basler, 2006; Pantua et al., 2006; Patch et al., 2008). Here, VLP production does not appear to increase upon co-expression of other viral proteins, although the co-expressed proteins are efficiently incorporated into the budding particles.
PIV5 (formerly SV5) is a prototype paramyxovirus. Like other paramyxoviruses, it possesses an RNA genome of negative-sense polarity that is encapsidated by the viral NP protein. The encapsidated genome is associated with the viral RNA-dependent RNA polymerase complex, composed of large (L) protein and phospho (P) protein subunits. Together, these components comprise the viral RNP core. The core is packaged into a viral envelope, which is obtained from the host cell plasma membrane during particle budding. The viral envelope contains densely-packed integral membrane spike proteins. The HN protein facilitates virion attachment to target cells by binding to sialic acid receptors and, in addition, possesses a sialidase activity that allows newly-formed virions to separate from infected cells. The F protein mediates fusion between viral and host cell membranes during virus entry. The small hydrophobic (SH) protein is a low-abundance membrane component that functions to block apoptosis of infected cells (He et al., 2001). The viral envelope is linked to the RNP core by the highly-abundant M protein.
Enveloped viruses typically do not encode all of the machinery necessary for proper transport, assembly, and budding of virus particles. Instead, host machinery is manipulated to allow efficient virus exit. For example, many retroviruses employ protein-protein interaction sequences (late domains) within their Gag polypeptides to recruit host factors to virus assembly sites (reviewed in (Bieniasz, 2005; Calistri et al., 2009; Chen and Lamb, 2008; Freed, 2002). Preventing this host factor recruitment significantly impairs retrovirus budding in many cases (Bieniasz, 2005; Freed, 2003; Fujii, Hurley, and Freed, 2007). The same late domains found in retroviruses are also present in the matrix proteins of some negative-strand RNA viruses such as Ebola virus (Harty et al., 2000) and Lassa fever virus (Perez, Craven, and de la Torre, 2003), suggesting that the overall strategy of host protein manipulation to achieve efficient virus budding is highly conserved. Consequently, it is thought that paramyxovirus M proteins are also likely to bind host factors to facilitate virus exit. These M proteins lack the prototypical PPxY, P[T/S]AP, and YP(x)nL late domains found in retroviral Gag proteins, suggesting that the details of host factor recruitment must be different for the paramyxoviruses. Alternative sequences have been identified in several paramyxovirus M proteins that have been proposed to direct host factor recruitment. These include the sequence FPIV within the PIV5 M protein (Schmitt et al., 2005), the sequence YLDL within the Sendai virus M protein (Irie et al., 2006), and the sequences YMYL (Ciancanelli and Basler, 2006) and YPLGVG (Patch et al., 2008) within the NiV M protein.
In this study, yeast two-hybrid screening was conducted in an attempt to identify host proteins that interact with the PIV5 M protein. Among the candidates identified was human angiomotin-like 1 (AmotL1), a tight junction-localized protein that contains PPxY motifs as well as a PDZ-binding motif. AmotL1 is thought to function in cells to allow normal endothelial migration and has been shown to colocalize with F-actin (Gagne et al., 2009; Zheng et al., 2009). Here, AmotL1 is shown to interact with PIV5 M protein in yeast, in transfected mammalian cells, and in virus-infected cells. Binding was mapped to a C-terminal region of the AmotL1 protein, and occurred independent of the FPIV sequence within PIV5 M protein. Overexpression of the C-terminal region of AmotL1 inhibited the budding of PIV5-like particles, and siRNA depletion of AmotL1 reduced the budding efficiency of PIV5. These results provide the first demonstration of physical and functional interaction between a host factor and a rubulavirus M protein.
Results
Identification of PIV5 M-interacting host proteins by yeast two-hybrid screening
To define cellular proteins that interact with PIV5 M protein, a yeast two-hybrid screening strategy was employed. To this end, two bait expression plasmids were constructed: one encodes full-length PIV5 M protein with LexA DNA binding domain fused to its N-terminus, and the other encodes full-length PIV5 M protein with LexA DNA binding domain fused to its C-terminus. Each of these could be expressed efficiently in yeast cells as judged by immunoblotting, and neither self-activated transcription in yeast (not shown). A premade HeLa cell-derived cDNA library containing 9.6 × 106 primary clones was screened using each of the two PIV5 M protein baits. The screens were performed in yeast strain L40, in which transcription of LexA-driven reporters results in the production of beta-galactosidase, and also allows growth of yeast cells on media lacking histidine. Approximately 5 × 107 clones were screened with each of the two baits. Library-derived cDNAs were PCR-amplified from His+, beta-galactosidase+ yeast colonies, and analyzed by DNA sequencing. Each of the two screens identified several candidate PIV5 M-interacting proteins, but only one of these candidates, angiomotin-like 1 (AmotL1), was identified in both screens.
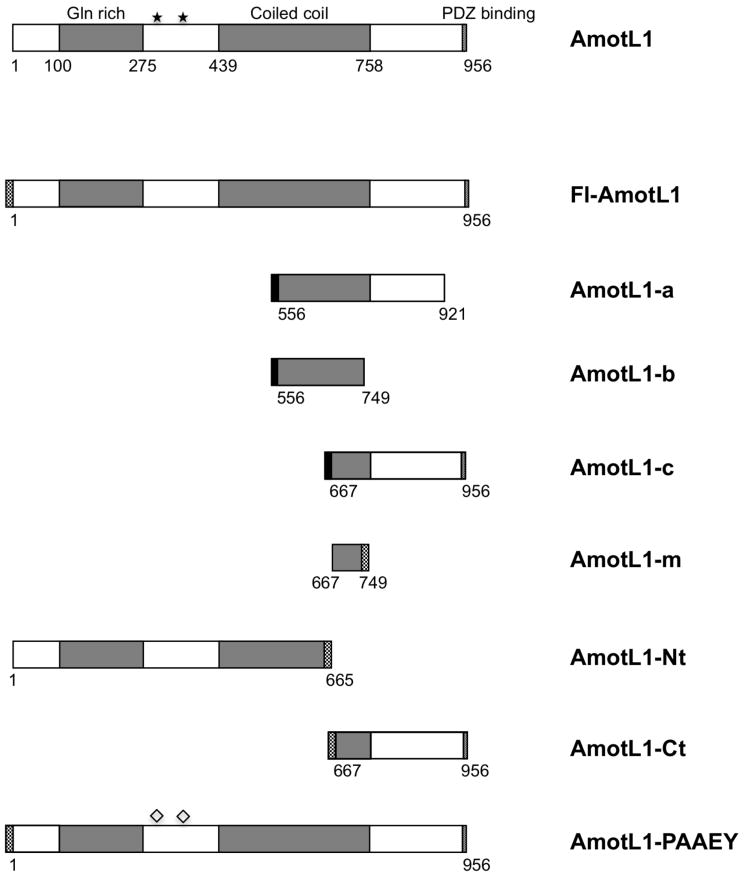
Human AmotL1 (GenBank NM_130847; largest potential isoform) is a 956 amino acid long protein. AmotL1 (also known as JEAP) belongs to a family of proteins called the motins, together with angiomotin (Amot) and angiomotin-like 2 (AmotL2) (Bratt et al., 2002). These proteins are known to have cellular functions related to endothelial cell migration, angiogenesis, embryonic cell movements, and maintenance of cell polarity (Bratt et al., 2005; Gagne et al., 2009; Huang et al., 2007; Levchenko et al., 2003; Shimono and Behringer, 2003; Troyanovsky et al., 2001; Zheng et al., 2009). The N-terminal region of AmotL1 protein contains a glutamine-rich domain, and two PPPEY sequences. The central region of the protein contains a coiled-coil domain, and at the C-terminal end of the AmotL1 protein is a PDZ-binding motif (Fig. 1). None of the AmotL1 prey sequences identified by yeast two-hybrid screening corresponded to the full-length protein. Rather, all of these sequences corresponded to one of three AmotL1 subfragments designated here as AmotL1-a, AmotL1-b, and AmotL1-c (Fig. 1). Each of the three prey fragments was identified multiple times and with both baits. A short region of overlap is common to all three fragments. The overlap region spans amino acid residues 667-749 of the full-length protein, and corresponds to the C-terminal portion of the coiled-coil region.
Figure 1. Schematic illustration of AmotL1 protein and AmotL1-derived polypeptides.
Numbers indicate the span of amino acid residues derived from AmotL1 that is contained within each construct. Checkered regions denote Flag tags, and solid black regions denote HA tags. Stars mark the positions of the two PPPEY sequences, and diamonds are used to indicate replacement of PPPEY with PAAEY.
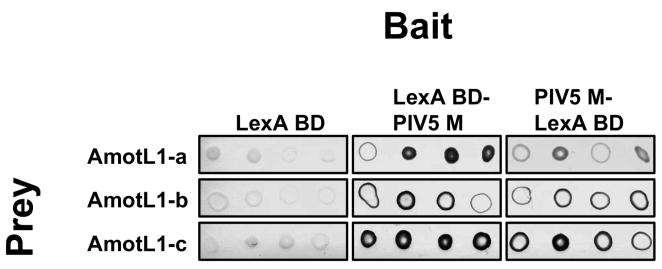
Pairwise yeast two-hybrid assays were used to confirm the screening results. Prey plasmid DNAs were isolated from His+, beta-galactosidase+ yeast clones. The prey plasmids were re-introduced into fresh L40 yeast together with bait plasmids. Beta-galactosidase activities of the transformants were measured using a colony filter assay. As shown in Fig. 2, each of the three AmotL1-derived preys tested positive for interaction with both of the PIV5 M baits, but tested negative for interaction with unfused LexA DNA binding domain.
Figure 2. Interaction between PIV5 M protein and AmotL1 detected with a pairwise yeast two-hybrid assay.
Yeast cells (strain L40) were transformed with plasmids corresponding to the indicated baits, and then transformed a second time with plasmids corresponding to the indicated preys. Growing yeast colonies were selected, suspended in water, and spotted onto a nitrocellulose membrane in replicates of four. Yeast cells on the membrane were disrupted with freeze-thaw cycles, and beta-galactosidase activity was detected using the colorimetric substrate X-gal.
AmotL1 interacts with PIV5 M protein in mammalian cells
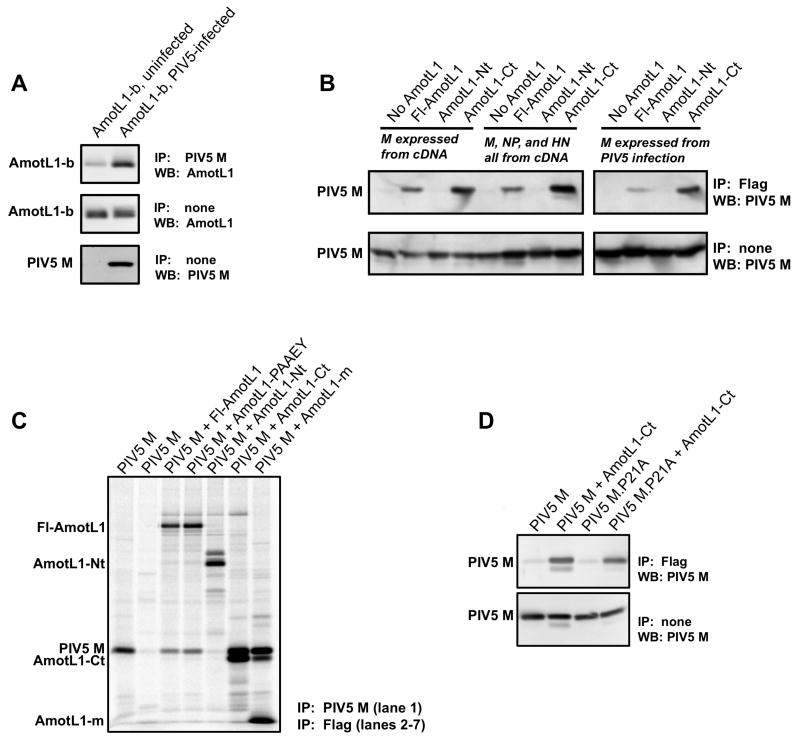
To test whether AmotL1 can bind to M protein in mammalian cells capable of supporting paramyxovirus infection, co-immunoprecipitation experiments were performed. In the first experiment, PIV5-infected or mock-infected 293T cells were transfected to produce AmotL1-b, which corresponds to the smallest prey fragment identified by yeast two-hybrid screening. Immunoprecipitation of PIV5 M protein resulted in the co-precipitation of AmotL1-b, as judged by immunoblotting with AmotL1-specific antibody (Fig. 3A). To confirm and further explore this interaction, additional experiments were conducted using the reverse procedure (immunoprecipitation of AmotL1 followed by immunodetection of M protein; Fig. 3B). For these experiments, a series of Flag-tagged AmotL1 protein derivatives were constructed as illustrated in Fig. 1. These include full-length AmotL1 protein (Fl-AmotL1), the N-terminal 70% of AmotL1 protein (AmotL1-Nt) and the C-terminal 30% of AmotL1 protein (AmotL1-Ct). Note that the region of overlap among the three AmotL1 prey fragments recovered in the yeast two-hybrid screen – the region that is anticipated to direct binding to M protein – is contained within AmotL1-Ct and is absent from AmotL1-Nt. The Flag-tagged AmotL1 constructs were expressed in 293T cells together with PIV5 M protein, by transient transfection. Precipitation of Fl-AmotL1 with Flag tag-specific antibody resulted in co-precipitation of PIV5 M protein, judged by immunoblotting with M protein-specific antibody (Fig. 3B, left panel). Likewise, precipitation of AmotL1-Ct led to co-precipitation of M protein. Precipitation of AmotL1-Nt, on the other hand, led to no detectable co-precipitation of M protein. Similar results were obtained in a related experiment in which cells were metabolically labeled and the immunoprecipitated proteins detected directly using a phosphorimager (Fig. 3C). Again, PIV5 M protein co-precipitated with Fl-AmotL1 and AmotL1-Ct, but not with AmotL1-Nt. These results suggested that elements within the N-terminal region of AmotL1 protein, including the two PPPEY sequences, are not important for the PIV5 M protein interaction. To further rule out involvement of the PPPEY sequences, a mutated version of full-length AmotL1 was generated in which both copies of PPPEY were changed to PAAEY. PIV5 M protein was co-precipitated with AmotL1-PAAEY similar to the co-precipitation with Fl-AmotL1, confirming that the PPPEY sequences do not contribute to this interaction (Fig. 3C). An additional AmotL1 derivative, AmotL1-m, comprising only the 83 amino acid long overlap region within the C-terminal part of AmotL1 found in the three yeast two-hybrid preys, was also examined. PIV5 M protein was co-precipitated efficiently with this small AmotL1-m polypeptide (Fig. 3C). These results are consistent with those obtained by yeast two-hybrid screening and define a region comprising amino acid residues 667 to 749 of AmotL1 that can direct binding to PIV5 M protein in transfected mammalian cells.
Figure 3. Interaction between PIV5 M protein and AmotL1 in mammalian cells detected by co-immunoprecipitation.
(A) 293T cells were infected with PIV5 or were mock-infected. At 6 h p.i., cells were transfected with a pCAGGS plasmid to produce AmotL1-b. Cell lysates were subjected to immunoprecipitation with mouse anti-PIV5 M monoclonal antibody. A portion of each cell lysate was reserved and not subjected to immunoprecipitation. Immunoprecipitated proteins, as well as the reserved total cell lysate fractions, were fractionated by SDS-PAGE and analyzed by immunoblotting with either a rabbit polyclonal anti-AmotL1 antibody or a rabbit polyclonal anti-PIV5 M antibody. (B) 293T cells were transfected with pCAGGS plasmids to produce the indicated Flag-tagged AmotL1-derived polypeptides together with PIV5 M protein, or together with PIV5 M, NP, and HN proteins. Alternatively, cells were first infected with PIV5, and at 6 h. p.i. were transfected to produce AmotL1-derived polypeptides. Cell lysates were processed for immunoprecipitation with mouse anti-Flag monoclonal antibody. Immunoprecipitated proteins were fractionated by SDS-PAGE and analyzed by immunoblotting with a rabbit polyclonal anti-PIV5 M antibody. (C) 293T cells were transfected to produce PIV5 M protein together with the indicated Flag-tagged AmotL1-derived polypeptides. Proteins synthesized in the transfected cells were metabolically labeled using 35S amino acids. Cell lysates were processed for immunoprecipitation with either a mouse monoclonal anti-PIV5 M antibody (left-most lane) or a mouse monoclonal anti-Flag antibody (all other lanes). Immunoprecipitated proteins were fractionated by SDS-PAGE and detected using a phosphorimager. (D) 293T cells were transfected with pCAGGS plasmids to produce AmotL1-Ct together with either wt PIV5 M protein or M.P21A protein in which the sequence 20-FPIV-23 is disrupted, as indicated. Cell lysates were processed for immunoprecipitation with mouse anti-Flag monoclonal antibody. Immunoprecipitated proteins were fractionated by SDS-PAGE and analyzed by immunoblotting with a rabbit polyclonal anti-PIV5 M antibody. Each result shown is a representative example of at least three independent experiments.
Additional AmotL1 co-precipitation experiments were performed in cells expressing the PIV5 M, NP, and HN proteins together. Under this circumstance, the viral M protein is known to be actively engaged in the budding process, evidenced by the efficient production of VLPs (Schmitt et al., 2002). Here, M protein was co-precipitated together with Fl-AmotL1 and with AmotL1-Ct (but not with AmotL1-Nt), similar to the results obtained in the absence of NP and HN protein coexpression (Fig. 3B, left panel). Additional experiments were conducted to confirm these results in virus-infected cells, and once again binding was observed with Fl-AmotL1 and AmotL1-Ct, but not with AmotL1-Nt (Fig. 3B, right panel). Hence, this interaction can be detected in virus infected cells, in VLP-producing cells, and in transfected cells expressing PIV5 M protein in the absence of any other viral proteins. To test whether the FPIV sequence within PIV5 M protein, proposed to function as a protein-protein interaction domain (Schmitt et al., 2005), is important for the AmotL1-M protein interaction, co-immunoprecipitation experiments were carried out in cells transfected to produce AmotL1-Ct together with PIV5 M.P21A, in which the FPIV sequence has been disrupted (Schmitt et al., 2005). Co-precipitation of the mutant M protein was still observed in this case, indicating that this interaction is not FPIV-dependent (Fig. 3D).
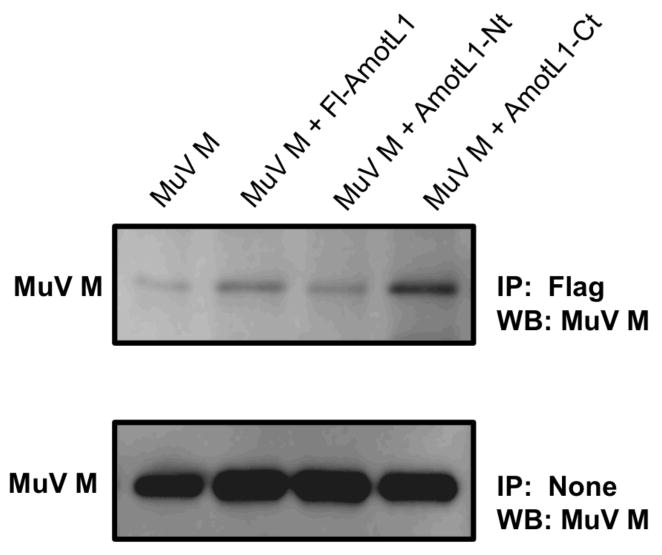
To explore the possibility that AmotL1 might interact with other viral matrix proteins in addition to the PIV5 M protein, co-immunoprecipitation experiments were carried out using the MuV and NiV M proteins. Co-precipitation of the MuV M protein with Fl-AmotL1 and with AmotL1-Ct could be detected (Fig. 4), but these signals were relatively faint, suggesting a weak interaction approaching the limits of detection with this assay system. We did not detect co-precipitation of NiV M protein with any of the AmotL1-derived fragments (not shown). These results indicate that AmotL1 protein interacts with only a subset of viral matrix proteins, and suggest that the binding to PIV5 M protein is more efficient than the binding to MuV M protein.
Figure 4. Co-immunoprecipitation of mumps virus M protein with AmotL1.
293T cells were transfected with pCAGGS plasmids to produce the indicated Flag-tagged AmotL1-derived polypeptides together with MuV M protein. Cell lysates were processed for immunoprecipitation with mouse anti-Flag monoclonal antibody. Immunoprecipitated proteins were fractionated by SDS-PAGE and analyzed by immunoblotting with a rabbit polyclonal anti-MuV M antibody. The data shown is representative of two independent experiments.
Overexpression of M-binding AmotL1 fragments blocks production of PIV5 VLPs
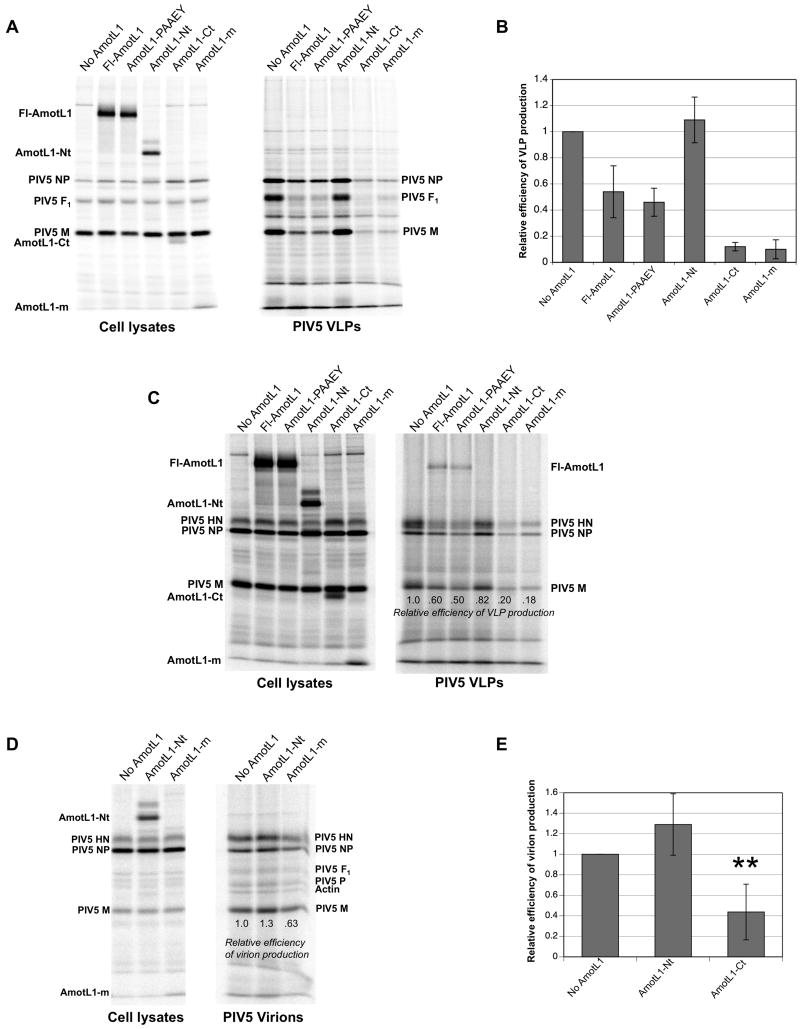
Short fragments of host proteins that bind to viral Gag or M proteins are often potent inhibitors of virus budding, likely because they either act as competitive inhibitors and prevent binding by the endogenous host proteins, or otherwise disrupt viral protein function on binding (Chen et al., 2005; Demirov et al., 2002; Freed, 2003; Fujii, Hurley, and Freed, 2007; Martin-Serrano et al., 2003; Munshi et al., 2006). To test the possibility that expression of M-interacting AmotL1 fragments might affect the production of PIV5 particles, 293T cells were transfected with various AmotL1-derived fragments together with the PIV5 M, NP, and F proteins for VLP production. After metabolic labeling of transfected cells, VLPs were collected from the culture medium, purified by centrifugation through sucrose cushions, further purified by flotation on sucrose gradients, and loaded directly onto SDS gels. As expected, VLP production was efficient in cells lacking exogenous AmotL1 expression, as well as in cells expressing AmotL1-Nt that fails to bind M protein (Fig. 5A). In contrast, VLP production was markedly inhibited in cells expressing either of the two M-binding AmotL1 fragments, AmotL1-Ct and AmotL1-m. Overexpression of full-length AmotL1 protein also reduced VLP production, although in this case the reduction (to about 55% of the normal level on average) was more variable and not as severe as that observed upon expression of the smaller M-binding AmotL1 fragments. VLP production was quantified on the basis of M protein content, normalized to M protein expression levels in cell lysate fractions, and results from three independent experiments were analyzed (Fig. 5B). Expression of AmotL1-Ct or AmotL1-m reduced VLP production to levels that were approximately 10% of those obtained in the absence of AmotL1 overexpression (Fig. 5B). A similar inhibitory effect was observed on the production of VLPs harboring HN glycoprotein in place of F protein (Fig. 5C), consistent with the notion that this inhibition is mediated through binding to the viral M protein, and not through targeting viral glycoproteins (which are also critical for VLP production). Together, these results define a short, 83 amino acid long AmotL1-derived polypeptide that potently blocks PIV5-like particle production when expressed in transfected cells.
Figure 5. Overexpression of M-binding AmotL1 fragments blocks production of PIV5 VLPs.
(A) 293T cells were transfected to express the indicated Flag-tagged AmotL1-derived polypeptides, together with the PIV5 M, NP, and F proteins for VLP production. Radiolabeled proteins from cell lysates were collected by immunoprecipitation using a mixture of antibodies specific to the PIV5 M, NP, and F proteins together with anti-Flag antibody. VLPs from culture supernatants were purified by centrifugation through sucrose cushions and flotation on sucrose gradients. Viral proteins from both cell lysates and purified VLPs were fractionated by SDS-PAGE and detected using a phosphorimager. (B) Efficiency of VLP production was calculated as the amount of M protein detected in VLPs divided by the amount of M protein detected in the corresponding cell lysate, and was normalized relative to the value obtained in the absence of AmotL1 expression. Relative VLP production efficiency values were calculated from four independent experiments performed as in (A), with standard deviations indicated by error bars. (C) The effect of AmotL1-derived polypeptide expression on VLP production was measured as in (A) except that VLPs resulted from the co-expression of PIV5 M, NP, and HN proteins, instead of PIV5 M, NP, and F proteins, and HN-specific antibody was used for immunoprecipitation in place of F-specific antibody. The result shown is representative of three independent experiments. (D) 293T cells were infected with PIV5, followed by transfection with the indicated AmotL1-derived polypeptides. Radiolabeled proteins from cell lysates were collected by immunoprecipitation using a mixture of antibodies specific to the PIV5 M, NP, and HN proteins together with anti-Flag antibody. Virions from culture supernatants were purified by centrifugation through sucrose cushions and flotation on sucrose gradients. Viral proteins from both cell lysates and purified virions were fractionated by SDS-PAGE and detected using a phosphorimager. The result shown is representative of two independent experiments. (E) The effect of AmotL1-derived polypeptide expression on virion production was measured as in (D), except that AmotL1-Ct was expressed in place of AmotL1-m. Relative efficiency of virion production was calculated from four independent experiments. Differences from control values (no AmotL1 expression) were assessed for statistical significance using Student's t test, with P values < 0.01 denoted by **.
We considered the possibility that the two PPPEY sequences within AmotL1 protein might function to indirectly recruit to PIV5 assembly sites the same WW domain-containing ubiquitin ligases that are directly recruited by the PPxY late domains important for the budding of Rous sarcoma virus, Ebola virus, and other enveloped viruses (reviewed in (Bieniasz, 2005; Calistri et al., 2009; Freed, 2002). In this case, we reasoned that overexpression of AmotL1-PAAEY, in which both PPxY sequences are disrupted, might have a potent inhibitory effect on VLP production, as the nonfunctioning AmotL1-PAAEY would act as a competitive inhibitor and prevent interaction with endogenous AmotL1. However, overexpression of AmotL1-PAAEY led only to a moderate effect on VLP production that was indistinguishable from the effect of Fl-AmotL1 overexpression (Figs. 5A-C). While this result does not rule out the possibility that AmotL1 PPPEY domains contribute to PIV5 budding, no positive evidence in support of this hypothesis has yet been obtained.
To further explore the potential of M-binding AmotL1-derived polypeptides as inhibitors of virus particle production, cells were infected with PIV5, followed by transient transfection to produce these polypeptides. This experiment is somewhat limited due to incomplete overlap of AmotL1-transfected and PIV5-infected cell populations, a general limitation that has been characterized previously for PIV5 infections (He et al., 2002). Nonetheless, a moderate reduction in virion production was observed in cells expressing AmotL1-m, to a level that was approximately 65% of that observed in the absence of AmotL1-m expression (Fig. 5D). A more significant effect on PIV5 particle production was observed upon expression of AmotL1-Ct (Fig. 5E), as particle production in this case was reduced to levels that were 44% of control levels, on average. Thus, expression of M-binding AmotL1-derived polypeptides negatively affects PIV5 virion production from infected cells.
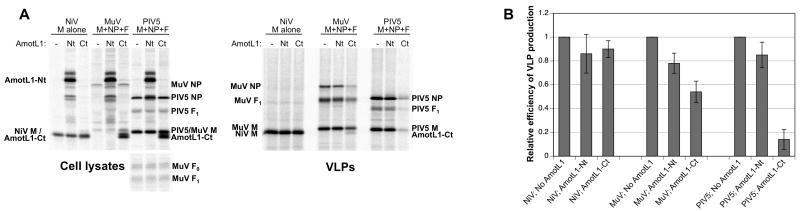
To determine if AmotL1-derived polypeptides can more generally affect the assembly and budding of paramyxovirus particles, AmotL1-Ct was expressed in 293T cells via transient transfection together with the NiV M protein for production of Nipah VLPs, or with the MuV M, NP, and F proteins for production of mumps VLPs (Figs. 6). Expression of AmotL1-Ct had no effect on Nipah M-alone VLP production. However, a moderate effect was observed on mumps VLP production, with particle release falling to levels that were on average about 55% of those obtained in the absence of AmotL1-Ct expression. These results are consistent with co-immunoprecipitation results, as there was a correlation between the ability of paramyxovirus M proteins to bind AmotL1-derived polypeptides and the susceptibility of the corresponding VLPs to AmotL1 fragment-mediated inhibition.
Figure 6. Overexpression of AmotL1 fragments affects the production of mumps VLPs, but not the production of Nipah VLPs.
(A) 293T cells were transfected to express the indicated Flag-tagged AmotL1-derived polypeptides, together with the NiV M protein for production of Nipah VLPs, the MuV M, NP, and F proteins for the production of mumps VLPs, or the PIV5 M, NP, and F proteins for the production of PIV5 VLPs. Radiolabeled proteins from cell lysates were collected by immunoprecipitation using antibodies specific to each of the viral proteins expressed, together with anti-Flag antibody. VLPs from culture supernatants were purified by centrifugation through sucrose cushions and flotation on sucrose gradients. Viral proteins from both cell lysates and purified VLPs were fractionated by SDS-PAGE and detected using a phosphorimager. (B) Relative VLP production efficiency values were calculated from three independent experiments performed as in (A), with standard deviations indicated by error bars.
Depletion of AmotL1 from cells reduces PIV5 virion production
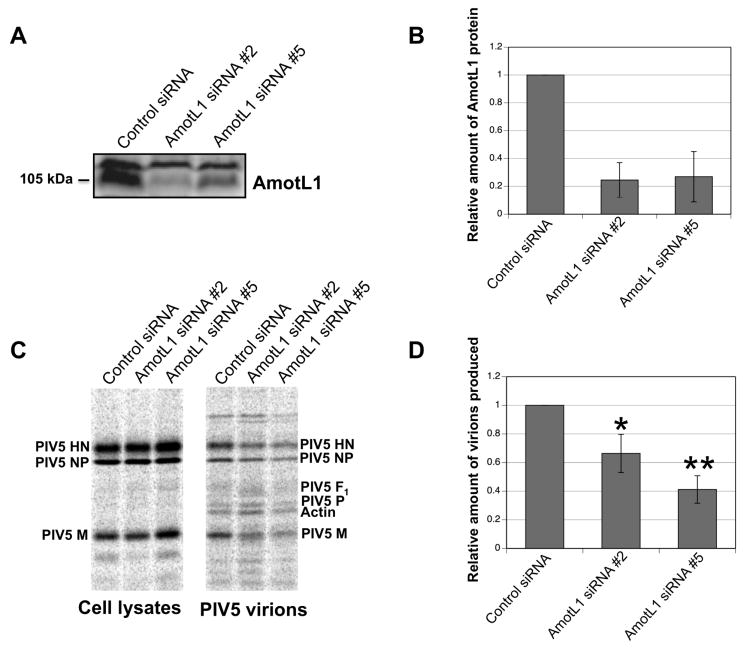
To assess the importance of endogenous AmotL1 protein for PIV5 particle production, a series of AmotL1-specific siRNAs were obtained. Of five siRNAs tested, two were partially effective at reducing endogenous full-length AmotL1 protein accumulation in 293T cells, judged by immunoblotting (Figs. 7A-B), with protein levels reduced to approximately 25% of control levels. To measure the effect of AmotL1 depletion on virion formation and release, cells transfected with AmotL1-specific siRNAs or control siRNA were infected with PIV5. siRNA-mediated AmotL1 depletion led to moderate but statistically significant reductions in the production of PIV5 virions (Figs. 7C-D). Depletion mediated by siRNA #2 led to PIV5 virion production that was 66% of control levels on average, while depletion mediated by siRNA #5 led to levels that were 41% of control levels (Fig. 7D). For these experiments, virion production was measured as the amount of M protein detected in particles, without normalizing to M protein expression in cell lysates. This is because somewhat elevated levels of M protein were consistently detected in cell lysates transfected with siRNA #5, compared with those transfected with siRNA #2 and control siRNA. This had the effect of potentially overestimating the effect of siRNA #5 on virion production (when normalized to cell lysate M protein content, budding efficiency after siRNA #5 transfection was 35% of control levels, on average). Overall, these results are consistent with the possibility that efficient PIV5 particle production is, in part, dependent on the presence of physiological levels of AmotL1 protein within host cells.
Figure 7. Depletion of AmotL1 reduced PIV5 budding.
(A) 293T cells were transfected with siRNAs specific to AmotL1, or with an unrelated control siRNA. At 72 h p.t., cell lysates were prepared and AmotL1 protein levels were measured by immunoblotting using a rabbit polyclonal AmotL1-specific antibody. (B) AmotL1 protein levels were quantified from three independent experiments performed as in (A), using a Storm 860 imaging system. Standard deviations are indicated by error bars. (C) 293T cells were transfected with the indicated siRNAs. At 24 h. p.t., cells were infected with PIV5. Radiolabeled viral proteins from infected cell lysates were collected by immunoprecipitation using antibodies specific to the PIV5 M, NP, and HN proteins. Virions from culture supernatants were purified by centrifugation through sucrose cushions and flotation on sucrose gradients. Viral proteins from both cell lysates and purified virions were fractionated by SDS-PAGE and detected using a phosphorimager. (D) Virion production was calculated as the amount of M protein detected in virions, and was normalized relative to the value obtained in cells transfected with control siRNA. Relative virion production values were calculated from three independent experiments, with standard deviations indicated by error bars. Differences from control values were assessed for statistical significance using Student's t test, with P values < 0.05 denoted by * and P values < 0.01 denoted by **.
Discussion
Host factors are recruited and manipulated to assist assembly and budding of a wide variety of enveloped viruses including retroviruses such as HIV-1 and Rous sarcoma virus, as well as negative-strand RNA viruses such as Ebola virus and Lassa fever virus. In many cases, these host factors are recruited by viral late domains, and are thought to facilitate the pinching off of virions from infected cells (reviewed in (Bieniasz, 2005; Calistri et al., 2009). Relatively little is known about the contributions of host proteins to paramyxovirus budding, however, and prior to this study M-interacting host proteins recruited during infections by many paramyxoviruses, such as PIV5 and mumps virus, had not been identified. Here, host protein AmotL1 was identified as a PIV5 M-interacting factor, and M-binding ability was mapped to a region of AmotL1 protein that spans amino acid residues 667 to 749. Interaction between PIV5 M protein and AmotL1 was observed in yeast, in transfected mammalian cells, and in virus-infected cells. siRNA-mediated depletion of AmotL1 protein reduced PIV5 budding efficiency. Remarkably, M-binding AmotL1 fragments acted as potent inhibitors of PIV5-like particle production, and also inhibited the production of virions from virus-infected cells. Thus, the binding interface between the AmotL1 and PIV5 M proteins represents a possible target for the inhibition of virus replication.
Motin proteins have been characterized mainly as factors that regulate migration of endothelial cells which line the surfaces of capillaries and larger blood vessels (Bratt et al., 2005; Troyanovsky et al., 2001; Zheng et al., 2009). The best-studied of the three motins is Amot, which was initially identified in a yeast two-hybrid screen as an angiostatin-binding protein (Troyanovsky et al., 2001), and which can function to promote tumor angiogenesis (Holmgren et al., 2006; Levchenko et al., 2004; Levchenko et al., 2008). The other motin proteins, AmotL1 and AmotL2, share glutamine rich domains, conserved coiled coil regions, and C-terminal PDZ-binding domains with Amot, but apparently lack the angiostatin-binding domain (Bratt et al., 2002). Amot and AmotL1 localize together at endothelial tight junctions and regulate the maintenance of endothelial cell polarity and junction stability (Bratt et al., 2005; Zheng et al., 2009). In addition to controlling migration and polarity of endothelial cells, motin proteins also function in epithelial cells, the primary targets of infection by many respiratory viruses including PIV5. AmotL1 was first identified as a tight junction-associated protein in epithelial cells (Nishimura et al., 2002). Endogenous levels of all three motin proteins can be detected in epithelial cells, where they are found to localize to tight junctions as well as to apical membranes and vesicular compartments within the cytoplasm (Sugihara-Mizuno et al., 2007). All three motin proteins interact via their PDZ-binding domains with Patj, a critical regulator of epithelial cell tight junction formation (Ernkvist et al., 2008; Michel et al., 2005; Shin, Straight, and Margolis, 2005; Sugihara-Mizuno et al., 2007; Wells et al., 2006), and Amot has been shown to be a functional component of Patj-containing polarity complexes, necessary for long-term stability of epithelial cell tight junctions (Wells et al., 2006). PIV5, like many other paramyxoviruses as well as influenza A virus, buds preferentially from apical surfaces of polarized epithelial cells (Blau and Compans, 1995; Boulan and Sabatini, 1978). It is possible that the trafficking of PIV5 components to apical membranes in preparation for polarized budding could be facilitated by AmotL1-M protein interaction. Efforts are currently underway to assess the importance of AmotL1 and other motin proteins in polarized budding of PIV5 by mapping the AmotL1-binding region within PIV5 M protein, which will potentially allow the generation of binding-defective recombinant viruses.
Two distinct isoforms of Amot have been characterized. In addition to the p80 isoform which promotes endothelial cell migration, a p130 Amot splice isoform has been identified in cells and shown to interact with actin fibers, influencing cell shape (Ernkvist et al., 2006). Recent evidence also links AmotL1 to the actin cytoskeleton, as AmotL1 co-localizes with F-actin, and expression of exogenous full-length AmotL1 protein modifies the architecture of the actin cytoskeleton, causing F-actin to adopt a punctate-like appearance (Gagne et al., 2009). Paramyxovirus assembly and budding has long been thought to involve the actin cytoskeleton. Large quantities of actin are contained within the virions of Sendai virus, mumps virus, and other paramyxoviruses (Lamb, Mahy, and Choppin, 1976; Orvell, 1978; Takimoto and Portner, 2004; Ulloa et al., 1998). Recruitment of actin to paramyxovirus assembly sites may occur via interactions with the viral M proteins (Giuffre et al., 1982; Takimoto et al., 2001). Treatment of infected cells with cytochalsin D or nocodazole to disrupt the actin cytoskeleton inhibits release of paramyxoviruses such as measles virus and respiratory syncytial virus (Burke et al., 1998; Stallcup, Raine, and Fields, 1983). Here, overexpression of full-length AmotL1 protein resulted in a moderate decrease in PIV5-like particle production (Figs. 5A-C). It is plausible that this decrease, to about half of the normal particle release efficiency, could have been caused by modifications to the actin cytoskeleton as a result of AmotL1 overexpression in the transfected cells. It is unlikely, however, that modification of the actin cytoskeleton can account for the much larger decreases in PIV5-like particle production that were observed upon overexpression of AmotL1-Ct or AmotL1-m, as these two AmotL1 fragments are both derived from the C-terminal half of the protein, which lacks the ability to colocalize with F-actin (Gagne et al., 2009). We did consider the possibility that overexpression of these short M-binding fragments of AmotL1 might prevent endogenous AmotL1 protein from binding with M protein, and thereby affect potential M protein interplay with cellular actin through AmotL1. However, if the main contribution of AmotL1 to PIV5 assembly was to facilitate M protein interaction with actin, then one might have expected incorporation of actin into PIV5 virions to have been affected by AmotL1-m overexpression or by siRNA-mediated AmotL1 depletion, and this was not observed (Figs. 5D and 7C).
PPxY motifs function as late domains necessary for the efficient budding of many enveloped viruses, including Rous sarcoma virus (Parent et al., 1995), murine leukemia virus (Yuan, Li, and Goff, 1999), Ebola virus (Harty et al., 2000), vesicular stomatitis virus (Craven et al., 1999), and Lassa fever virus (Perez, Craven, and de la Torre, 2003). Recruitment of WW domain-containing Nedd4-like ubiquitin ligases to virus assembly sites is thought to facilitate the pinching off of virus particles, although the precise mechanism by which these host factors influence virus budding is unclear. Ubiquitin is thought to have an important role in virus budding (reviewed in (Martin-Serrano, 2007), and monoubiquitination of viral and/or cellular factors may be facilitated through PPxY-mediated host factor recruitment. It is also possible that Nedd4-like proteins function mainly as adapters, allowing the further recruitment of other host factors to virus budding sites in a way that is independent of ubiquitin ligase activity (Martin-Serrano et al., 2004). The experiments described here present the interesting possibility that PIV5 might recruit WW domain-containing host proteins indirectly to virus assembly sites via AmotL1, which harbors two PPxY motifs in its N-terminal region. If this is the case, we reasoned that overexpression of PPxY-disrupted AmotL1 protein should result in competitive inhibition and prevent such recruitment via endogenous AmotL1. However, overexpression of PPxY-disrupted AmotL1 protein did not affect particle production to any greater extent than overexpression of wt AmotL1. Thus, no positive evidence in support of a role for AmotL1 PPxY motifs in PIV5 budding has been obtained. Nonetheless, further investigation is warranted to determine if WW domain-containing Nedd4-like proteins are indeed associated with PIV5 M protein through AmotL1.
In addition to PPxY, enveloped viruses are known to recruit host factors for budding via a number of other protein interaction motifs including P[T/S]AP and YP(x)nL (reviewed in (Bieniasz, 2005; Calistri et al., 2009; Freed, 2002). Although these late domain sequences direct interactions to separate host factors, they can function interchangeably in some cases to allow virus budding (Accola, Strack, and Göttlinger, 2000; Craven et al., 1999; Li et al., 2002; Yuan et al., 2000). Indeed, an additional late domain sequence, FPIV, derived from the N-terminal region of PIV5 M protein and critical for PIV5 budding, was identified on the basis of its ability to restore budding function to PTAP-disrupted HIV-1 Gag protein (Schmitt et al., 2005). Mumps virus contains a similar sequence, FPVI, within its N-terminal region that is necessary for efficient budding of mumps VLPs (Li et al., 2009). FPIV-like sequences presumably function to recruit host factors to virus assembly sites, similar to other viral late domains. Hence, identification of specific host proteins that bind to paramyxovirus M proteins via FPIV-like sequences is an important goal towards understanding paramyxovirus budding mechanisms. Here, we describe for the first time a specific interaction between a host protein and PIV5 M protein. This AmotL1-M protein interaction, readily detected by co-immunoprecipitation in transfected as well as virus-infected cells, was not substantially affected when wt M protein was replaced with mutant M protein in which the critical proline residue within FPIV was changed to alanine (Fig. 3D). Thus, AmotL1 does not appear to be recruited to PIV5 assembly sites via FPIV. However, it remains possible that PIV5 M protein interacts, either simultaneously or sequentially, with multiple host proteins during its intracellular transport, assembly, and release from cells.
M-binding AmotL1-derived polypeptides were potent inhibitors of PIV5-like particle production, and also negatively affected the production of mumps VLPs and the production of PIV5 virions from infected cells. The shortest of these M-binding fragments, AmotL1-m, is just 83 amino acid residues in length yet caused ten-fold reduction of PIV5 VLP production efficiency. These results parallel earlier findings made with retroviral Gag-binding host factors Tsg101 and Alix. Expression of a Gag-binding fragment of Tsg101 (TSG-5′) caused a 60% reduction in HIV-1 particle production (Demirov et al., 2002; Freed, 2003). Similarly, expression of Gag-binding fragments of Aip1/Alix reduced HIV-1 and equine infectious anemia virus particle production by 5- to 10-fold (Chen et al., 2005; Fujii, Hurley, and Freed, 2007; Martin-Serrano et al., 2003; Munshi et al., 2006). Even expression of full-length Aip1/Alix was found to reduce virus particle production (Chen et al., 2005), providing a further similarity to results described here in which expression of full-length AmotL1 partially inhibited PIV5-like particle production. Together, these studies reinforce the general notion that binding interfaces between host proteins and viral Gag or M proteins represent potentially attractive targets for antiviral drug design (Harty, 2009). In the case of HIV-1, the structure of Tsg101 in complex with a PTAP-containing peptide has been determined, opening new doors for rational design of antiviral drugs that disrupt this critical interaction (Freed, 2003; Pornillos et al., 2002). Our findings suggest that generally similar approaches may also be effective towards combating paramyxovirus pathogens.
Materials and methods
Plasmids
Plasmids pHybLex-PIV5 M-Zeo and pHyb-PIV5 M-LexZeo were constructed by subcloning full-length PIV5 M protein cDNA into the yeast two-hybrid bait plasmid pHybLexZeo (Invitrogen, Carlsbad, CA) to generate hybrid proteins with LexA DNA binding domain appended to the N-terminus or the C-terminus of PIV5 M protein, respectively. Cloning details will be provided upon request.
cDNA corresponding to full-length human AmotL1 (GenBank accession no. NM_130847) was obtained from OriGene Technologies (Rockville, MD). This sequence was amplified and modified by PCR to incorporate an N-terminal Flag tag (amino sequence DYKDDDDK), and the product was subcloned into the eukaryotic expression plasmid pCAGGS (Niwa, Yamamura, and Miyazaki, 1991), to generate plasmid pCAGGS-Fl-AmotL1. Subfragments of AmotL1 were amplified by PCR using full-length AmotL1 cDNA as template, with boundaries and positioning of Flag tags and HA tags as illustrated in Fig. 1. The products were subcloned into pCAGGS to generate pCAGGS-AmotL1-a, pCAGGS AmotL1-b, pCAGGS-AmotL1-c, pCAGGS-AmotL1-m, pCAGGS-AmotL1-Nt, and pCAGGS-AmotL1-Ct. pCAGGS-AmotL1-PAAEY was generated by PCR mutagenesis of pCAGGS-Fl-AmotL1, first changing the PPPEY sequence spanning amino acid residues 309-313 to PAAEY, and then using this product as template for a second round of PCR mutagenesis changing the PPPEY sequence spanning amino acid residues 366-370 to PAAEY. Nucleotide sequences of all AmotL1-derived plasmids were verified by DNA sequencing of the entire AmotL1 coding regions (Macrogen Inc., South Korea).
The plasmids pCAGGS-PIV5 M, pCAGGS-PIV5 NP, pCAGGS-PIV5 HN, and pCAGGS-PIV5 F were kind gifts of Robert Lamb (Northwestern University, Evanston, IL) (Schmitt et al., 2002). The plasmid pCAGGS-PIV5M.P21A in which the sequence 20-FPIV-23 within PIV5 M protein is changed to 20-FAIV-23 has been described before (Schmitt et al., 2005). The plasmids pCAGGS-MuV M, pCAGGS-MuV NP, and pCAGGS-MuV F that express the M, NP, and F proteins, respectively, of mumps virus strain 88-1961, have been described previously (Li et al., 2009). Plasmid pCAGGS-NiV M has also been described previously (Li et al., 2009).
Antibodies
Monoclonal antibodies M-h, HN1b, and NPa, specific to the PIV5 M, HN, and NP proteins, respectively, were kind gifts of Richard Randall (St. Andrews University, St. Andrews, Scotland, United Kingdom) (Randall et al., 1987). Polyclonal antibodies specific to the MuV M, NP, and F proteins, as well as the NiV M protein, were generated as described previously (Li et al., 2009). Polyclonal antibodies Fsol, specific to the PIV5 F protein, and Dudet, specific to the PIV5 M protein, were kind gifts of Robert Lamb (Northwestern University, Evanston, IL). To generate antibody specific to AmotL1 protein, full-length AmotL1 cDNA was subcloned into plasmid pRSETB (Invitrogen, Carlsbad, CA) and the protein was expressed in Escherichia coli by autoinduction (Studier, 2005). The resulting His-tagged recombinant protein was purified by metal affinity chromatography and sent to Harlan Bioproducts for Science (Indianapolis, IN) for rabbit immunization and polyclonal antibody production. Monoclonal antibody specific to Flag tag (clone M2) was purchased from Stratagene (La Jolla, CA).
Yeast two-hybrid library screening and pairwise assays
Yeast two-hybrid screening and assays were performed according to the manufacturer's instructions for the Hybrid Hunter yeast two-hybrid system (Invitrogen, Carlsbad, CA) using yeast strain L40 in which LexA-driven transcription results in expression of both histidine and beta-galactosidase reporters. Briefly, yeast cells were transformed with either of the two bait plasmids pHybLex-PIV5 M-Zeo or pHyb-PIV5 M-LexZeo, and selection was maintained with Zeocin. Yeast cells harboring the bait plasmids were transformed with a pre-made HeLa cell-derived yeast two-hybrid cDNA library constructed in plasmid pJG4-5 in which preys are fused with B42 transcription activation domain (Invitrogen, Carlsbad, CA). For screening, transformants were plated on synthetic medium lacking tryptophan, uracil, lysine, and histidine and containing 300 μg/ml Zeocin. After incubation for 3 days at 30°C, growing colonies were selected as primary positives and further tested using a beta-galactosidase colony filter assay, as instructed by the manufacturer. Yeast clones testing positive were subjected to DNA sequence analysis using prey plasmid-specific primer sets. For pairwise assays to confirm binding in yeast, prey plasmids were isolated from His+, beta-galactosidase+ yeast clones. Prey plasmids were re-transformed into L40 cells harboring pHybLex-PIV5 M-Zeo, pHyb-PIV5 M-LexZeo, or the empty bait plasmid pHybLexZeo. Beta-galactosidase activity was measured in replicates of four using a colony filter assay.
Co-immunoprecipitation
293T cells in 10-cm-diameter dishes (70 to 80% confluent) grown in Dulbecco's modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum were transfected with pCAGGS plasmids encoding PIV5 M protein (0.8 μg of plasmid DNA per dish), MuV M protein (0.4 μg of plasmid DNA per dish), or NiV M protein (0.8 μg of plasmid DNA per dish) together with plasmids encoding AmotL1-derived polypeptides (1.5 μg of plasmid DNA per dish). Transfections were supplemented with a pCAGGS plasmid lacking an insert when necessary to equalize total plasmid DNA quantities. For co-immunoprecipitation from virus-infected cells, 293T cells were infected with PIV5 at a multiplicity of infection (MOI) of 1 plaque forming unit (PFU) / cell. At six h post-infection (p.i.), the cells were transfected with plasmids encoding AmotL1-derived polypeptides (1.5 μg of plasmid DNA per dish). Transfection experiments were performed in Opti-MEM using Lipofectamine-Plus reagents (Invitrogen, Carlsbad, CA). At 16 h post-transfection (p.t.), cells were washed with ice-cold phosphate-buffered saline (PBS), suspended in whole cell extract buffer (20 mM Tris-HCl, pH 7.5; 280 mM NaCl; 10% glycerol; 0.5% Nonidet P-40; 2 mM EGTA; 0.2 mM EDTA; 100 mM phenylmethylsulphonyl fluoride), and incubated on ice for 30 min. After centrifugation for 10 min. at 14,000 × g, lysate supernatants were precleared with protein A-Sepharose (Invitrogen, Carlsbad, CA) for 30 min at 4 °C, followed by incubation for 3 h at 4°C with Flag tag-specific antibody, or with mouse monoclonal antibody specific to PIV5 M protein. Immune complexes were incubated with protein A-Sepharose for 30 min. at 4°C, and the resin was then washed twice with whole cell extract buffer. Immunoprecipitated proteins were eluted from the resin with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer containing 2.5% (wt/vol) dithiothreitol. Samples were fractionated by SDS-PAGE using 10% gels and transferred to PVDF membranes. Immunodetection was carried out using rabbit polyclonal antibodies specific to AmotL1 protein, PIV5 M protein, or MuV M protein, followed by alkaline phosphatase-conjugated goat anti-rabbit secondary antibody (Jackson ImmunoResearch Laboratories, Inc., West Grove, PA). A Storm 860 laser scanner was used for detection (GE Healthcare, Piscataway, NJ).
Alternatively, for co-precipitation of radiolabeled proteins, 293T cells were transfected as described above. At 16 h p.t., culture media was replaced with DMEM lacking methionine and cysteine. After 30 min., this media was replace with fresh DMEM lacking methionine and cysteine, supplemented with 40 μCi of [35S]Promix/ml (Perkin Elmer, Waltham, MA). After an additional 2 h, the cells were harvested, lysed, and subjected to immunoprecipitation analysis as described above. Samples were fractionated by SDS-PAGE using 10% gels and protein bands were detected using a Storm 860 laser scanner.
Measurements of VLP production and virion production
For VLP production, 293T cells in 10-cm-diameter dishes were transfected with plasmids encoding viral proteins or AmotL1-derived polypeptides as described above. Plasmid quantities per dish were as follows: pCAGGS-PIV5 M, 800 ng; pCAGGS-PIV5 NP, 300 ng; pCAGSS-PIV5 F, 3.0 μg; pCAGGS-PIV5 HN, 3.0 μg; pCAGGS-MuV M, 800 ng; pCAGGS-MuV NP, 250 ng; pCAGGS-MuV F, 1.5 μg; pCAGGS-NiV M, 1.6 μg; pCAGGS-Fl-AmotL1 and derivatives, 1.5 μg. Transfections were supplemented with a pCAGGS plasmid lacking an insert when necessary to equalize total plasmid DNA quantities. For virion production, 293T cells were infected with PIV5 at a MOI of 1 PFU / cell. At 6 h p.i., the cells were transfected with plasmids encoding AmotL1-derived polypeptides.
At 16 h p.t., the culture medium was replaced with DMEM containing 1/10 the normal amounts of methionine and cysteine and 40 μCi of [35S]Promix/ml. After an additional 24 h, the cells and media were harvested for protein expression and VLP/virion production analysis.
To analyze the VLPs or virions released from cells, culture media were centrifuged at 7,500 × g for 2 min to remove cell debris and then layered onto 20% sucrose cushions (10 ml in NTE [0.1 M NaCl; 0.01 M Tris-HCl, pH 7.4; 0.001 M EDTA]). Samples were centrifuged at 140,000 × g for 1.5 h. Pellets were resuspended in 0.5 ml of NTE and mixed with 1.3 ml of 80% sucrose in NTE. Layers containing 50% sucrose (1.8 ml) and 10% sucrose (0.6 ml) in NTE were applied to the tops of the samples, which were then centrifuged at 110,000 × g for 3 h. A 2.1-ml volume was collected from the top of each gradient, and VLPs/virions contained within this floated fraction were pelleted by centrifugation at 140,000 × g for 1.5 h. VLP/virion pellets were resuspended in SDS-PAGE loading buffer containing 2.5% (wt/vol) dithiothreitol.
Cell lysate preparation and immunoprecipitation of proteins was performed as described previously (Schmitt et al., 2002). Immunoprecipitated proteins from cell lysates as well as purified VLPs from culture media were analyzed by SDS-PAGE using 10% gels. Detection and quantification were performed using either a Storm 860 laser scanner or a Fuji FLA-7000 laser scanner (FujiFilm Medical Systems, Stamford, CT). Budding efficiency was calculated as the M-protein-specific counts in culture media divided by the M-protein-specific counts in the corresponding cell lysates and was normalized relative to values obtained in control experiments.
RNA interference
siRNAs targeting AmotL1 as well as control siRNA GL3 targeting luciferase were purchased from Sigma (Saint Louis, MO). AmotL1 siRNA #2 targets the sequence 5′CCAACAUGCCGGAAUACAA3′. AmotL1 siRNA #5 targets the sequence 5′GAAUGAUUUGAACUGAUAA3′. 293T cells in 6-cm-diameter dishes (50-60% confluent) were transfected with 30 pmol/dish of siRNA using Lipofectamine and Plus reagents. At 24 h p.t., the cells were infected with PIV5 at a MOI of 1 PFU / cell. At 24 h p.i., the culture medium was replaced with DMEM containing 1/10 the normal amounts of methionine and cysteine and 20 μCi of [35S]Promix/ml. After an additional 24 h, the cells and media were harvested for protein expression and virion production analysis as described above.
Acknowledgments
We thank Bob Lamb for PIV5 cDNAs, and polyclonal antibodies to PIV5 F and M proteins. We thank Steve Rubin and Biao He for mumps virus cDNAs. We are grateful to Phuong Schmitt for assistance with co-immunoprecipitation experiments, and to Megan Harrison for critical reading of the manuscript.
This work was supported in part by the Middle Atlantic Regional Center of Excellence (MARCE) for Biodefense and Emerging Infectious Disease Research NIH grant AI057168, and research grant AI070925 from the National Institute of Allergy and Infectious Diseases to A.P.S. This project is funded, in part, under a grant with the Pennsylvania Department of Health using Tobacco Settlement funds to A.P.S. The Department specifically disclaims responsibility for any analyses, interpretations or conclusions.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Accola MA, Strack B, Göttlinger HG. Efficient particle production by minimal Gag constructs which retain the carboxy-terminal domain of human immunodeficiency virus type 1 capsid-p2 and a late assembly domain. J Virol. 2000;74(12):5395–5402. doi: 10.1128/jvi.74.12.5395-5402.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bieniasz PD. Late budding domains and host proteins in enveloped virus release. Virology. 2005;344(1):55–63. doi: 10.1016/j.virol.2005.09.044. [DOI] [PubMed] [Google Scholar]
- Blau DM, Compans RW. Entry and release of measles virus are polarized in epithelial cells. Virology. 1995;210(1):91–99. doi: 10.1006/viro.1995.1320. [DOI] [PubMed] [Google Scholar]
- Boulan ER, Sabatini DD. Asymmetric budding of viruses in epithelial monlayers: a model system for study of epithelial polarity. Proc Natl Acad Sci USA. 1978;75(10):5071–5075. doi: 10.1073/pnas.75.10.5071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bratt A, Birot O, Sinha I, Veitonmäki N, Aase K, Ernkvist M, Holmgren L. Angiomotin regulates endothelial cell-cell junctions and cell motility. J Biol Chem. 2005;280(41):34859–34869. doi: 10.1074/jbc.M503915200. [DOI] [PubMed] [Google Scholar]
- Bratt A, Wilson WJ, Troyanovsky B, Aase K, Kessler R, Van Meir EG, Holmgren L, Meir EG. Angiomotin belongs to a novel protein family with conserved coiled-coil and PDZ binding domains. Gene. 2002;298(1):69–77. doi: 10.1016/s0378-1119(02)00928-9. [DOI] [PubMed] [Google Scholar]
- Burke E, Dupuy L, Wall C, Barik S. Role of cellular actin in the gene expression and morphogenesis of human respiratory syncytial virus. Virology. 1998;252(1):137–148. doi: 10.1006/viro.1998.9471. [DOI] [PubMed] [Google Scholar]
- Calistri A, Salata C, Parolin C, Palu G. Role of multivesicular bodies and their components in the egress of enveloped RNA viruses. Rev Med Virol. 2009;19(1):31–45. doi: 10.1002/rmv.588. [DOI] [PubMed] [Google Scholar]
- Chen BJ, Lamb RA. Mechanisms for enveloped virus budding: can some viruses do without an ESCRT? Virology. 2008;372(2):221–232. doi: 10.1016/j.virol.2007.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen C, Vincent O, Jin J, Weisz OA, Montelaro RC. Functions of early (AP-2) and late (AIP1/ALIX) endocytic proteins in equine infectious anemia virus budding. J Biol Chem. 2005;280(49):40474–40480. doi: 10.1074/jbc.M509317200. [DOI] [PubMed] [Google Scholar]
- Ciancanelli MJ, Basler CF. Mutation of YMYL in the Nipah virus matrix protein abrogates budding and alters subcellular localization. J Virol. 2006;80(24):12070–12078. doi: 10.1128/JVI.01743-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craven RC, Harty RN, Paragas J, Palese P, Wills JW. Late domain function identified in the vesicular stomatitis virus M protein by use of rhabdovirus-retrovirus chimeras. J Virol. 1999;73(4):3359–3365. doi: 10.1128/jvi.73.4.3359-3365.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demirov DG, Ono A, Orenstein JM, Freed EO. Overexpression of the N-terminal domain of TSG101 inhibits HIV-1 budding by blocking late domain function. Proc Natl Acad Sci USA. 2002;99(2):955–960. doi: 10.1073/pnas.032511899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ernkvist M, Aase K, Ukomadu C, Wohlschlegel J, Blackman R, Veitonmäki N, Bratt A, Dutta A, Holmgren L. p130-angiomotin associates to actin and controls endothelial cell shape. FEBS J. 2006;273(9):2000–2011. doi: 10.1111/j.1742-4658.2006.05216.x. [DOI] [PubMed] [Google Scholar]
- Ernkvist M, Birot O, Sinha I, Veitonmaki N, Nyström S, Aase K, Holmgren L. Differential roles of p80- and p130-angiomotin in the switch between migration and stabilization of endothelial cells. Biochim Biophys Acta. 2008;1783(3):429–437. doi: 10.1016/j.bbamcr.2007.11.018. [DOI] [PubMed] [Google Scholar]
- Freed EO. Viral late domains. J Virol. 2002;76(10):4679–4687. doi: 10.1128/JVI.76.10.4679-4687.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freed EO. The HIV-TSG101 interface: recent advances in a budding field. Trends Microbiol. 2003;11(2):56–59. doi: 10.1016/s0966-842x(02)00013-6. [DOI] [PubMed] [Google Scholar]
- Fujii K, Hurley JH, Freed EO. Beyond Tsg101: the role of Alix in ‘ESCRTing’ HIV-1. Nat Rev Microbiol. 2007;5(12):912–916. doi: 10.1038/nrmicro1790. [DOI] [PubMed] [Google Scholar]
- Gagne V, Moreau J, Plourde M, Lapointe M, Lord M, Gagnon E, Fernandes MJ. Human angiomotin-like 1 associates with an angiomotin protein complex through its coiled-coil domain and induces the remodeling of the actin cytoskeleton. Cell Motil Cytoskeleton. 2009;66(9):754–768. doi: 10.1002/cm.20405. [DOI] [PubMed] [Google Scholar]
- Giuffre RM, Tovell DR, Kay CM, Tyrrell DL. Evidence for an interaction between the membrane protein of a paramyxovirus and actin. J Virol. 1982;42(3):963–968. doi: 10.1128/jvi.42.3.963-968.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN. No exit: targeting the budding process to inhibit filovirus replication. Antiviral Res. 2009;81(3):189–197. doi: 10.1016/j.antiviral.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harty RN, Brown ME, Wang G, Huibregtse JM, Hayes FP. A PPxY motif within the VP40 protein of Ebola virus interacts physically and functionally with a ubiquitin ligase: implications for filovirus budding. Proc Natl Acad Sci USA. 2000;97(25):13871–13876. doi: 10.1073/pnas.250277297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Lin GY, Durbin JE, Durbin RK, Lamb RA. The SH integral membrane protein of the paramyxovirus simian virus 5 is required to block apoptosis in MDBK cells. J Virol. 2001;75(9):4068–4079. doi: 10.1128/JVI.75.9.4068-4079.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He B, Paterson RG, Stock N, Durbin JE, Durbin RK, Goodbourn S, Randall RE, Lamb RA. Recovery of paramyxovirus simian virus 5 with a V protein lacking the conserved cysteine-rich domain: the multifunctional V protein blocks both interferon-beta induction and interferon signaling. Virology. 2002;303(1):15–32. doi: 10.1006/viro.2002.1738. [DOI] [PubMed] [Google Scholar]
- Holmgren L, Ambrosino E, Birot O, Tullus C, Veitonmäki N, Levchenko T, Carlson LM, Musiani P, Iezzi M, Curcio C, Forni G, Cavallo F, Kiessling R. A DNA vaccine targeting angiomotin inhibits angiogenesis and suppresses tumor growth. Proc Natl Acad Sci USA. 2006;103(24):9208–9213. doi: 10.1073/pnas.0603110103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Lu FI, Jia S, Meng S, Cao Y, Wang Y, Ma W, Yin K, Wen Z, Peng J, Thisse C, Thisse B, Meng A. Amotl2 is essential for cell movements in zebrafish embryo and regulates c-Src translocation. Development. 2007;134(5):979–988. doi: 10.1242/dev.02782. [DOI] [PubMed] [Google Scholar]
- Irie T, Shimazu Y, Yoshida T, Sakaguchi T. The YLDL sequence within sendai virus M protein is critical for budding of virus-like particles and interacts with Alix/AIP1 independently of C protein. J Virol. 2006;81(5):2263–2273. doi: 10.1128/JVI.02218-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb RA, Mahy BW, Choppin PW. The synthesis of sendai virus polypeptides in infected cells. Virology. 1976;69(1):116–131. doi: 10.1016/0042-6822(76)90199-9. [DOI] [PubMed] [Google Scholar]
- Levchenko T, Aase K, Troyanovsky B, Bratt A, Holmgren L. Loss of responsiveness to chemotactic factors by deletion of the C-terminal protein interaction site of angiomotin. J Cell Sci. 2003;116(Pt 18):3803–3810. doi: 10.1242/jcs.00694. [DOI] [PubMed] [Google Scholar]
- Levchenko T, Bratt A, Arbiser JL, Holmgren L. Angiomotin expression promotes hemangioendothelioma invasion. Oncogene. 2004;23(7):1469–1473. doi: 10.1038/sj.onc.1207264. [DOI] [PubMed] [Google Scholar]
- Levchenko T, Veitonmäki N, Lundkvist A, Gerhardt H, Ming Y, Berggren K, Kvanta A, Carlsson R, Holmgren L. Therapeutic antibodies targeting angiomotin inhibit angiogenesis in vivo. FASEB J. 2008;22(3):880–889. doi: 10.1096/fj.07-9509com. [DOI] [PubMed] [Google Scholar]
- Li F, Chen C, Puffer BA, Montelaro RC. Functional replacement and positional dependence of homologous and heterologous L domains in equine infectious anemia virus replication. J Virol. 2002;76(4):1569–1577. doi: 10.1128/JVI.76.4.1569-1577.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M, Schmitt PT, Li Z, McCrory TS, He B, Schmitt AP. Mumps virus matrix, fusion, and nucleocapsid proteins cooperate for efficient production of virus-like particles. J Virol. 2009;83(14):7261–7272. doi: 10.1128/JVI.00421-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J. The Role of Ubiquitin in Retroviral Egress. Traffic. 2007;8(10):1297–1303. doi: 10.1111/j.1600-0854.2007.00609.x. [DOI] [PubMed] [Google Scholar]
- Martin-Serrano J, Eastman SW, Chung W, Bieniasz PD. HECT ubiquitin ligases link viral and cellular PPXY motifs to the vacuolar protein-sorting pathway. J Cell Biol. 2004;168(1):89–101. doi: 10.1083/jcb.200408155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin-Serrano J, Yarovoy A, Perez-Caballero D, Bieniasz PD, Yaravoy A. Divergent retroviral late-budding domains recruit vacuolar protein sorting factors by using alternative adaptor proteins. Proc Natl Acad Sci USA. 2003;100(21):12414–12419. doi: 10.1073/pnas.2133846100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel D, Arsanto JP, Massey-Harroche D, Béclin C, Wijnholds J, Le Bivic A. PATJ connects and stabilizes apical and lateral components of tight junctions in human intestinal cells. J Cell Sci. 2005;118(Pt 17):4049–4057. doi: 10.1242/jcs.02528. [DOI] [PubMed] [Google Scholar]
- Munshi UM, Kim J, Nagashima K, Hurley JH, Freed EO. An Alix fragment potently inhibits HIV-1 budding: Characterization of binding to retroviral YPXL late domains. J Biol Chem. 2006;282(6):3847–3855. doi: 10.1074/jbc.M607489200. [DOI] [PubMed] [Google Scholar]
- Nishimura M, Kakizaki M, Ono Y, Morimoto K, Takeuchi M, Inoue Y, Imai T, Takai Y. JEAP, a novel component of tight junctions in exocrine cells. J Biol Chem. 2002;277(7):5583–5587. doi: 10.1074/jbc.M110154200. [DOI] [PubMed] [Google Scholar]
- Niwa H, Yamamura K, Miyazaki J. Efficient selection for high-expression transfectants with a novel eukaryotic vector. Gene. 1991;108(2):193–199. doi: 10.1016/0378-1119(91)90434-d. [DOI] [PubMed] [Google Scholar]
- Orvell C. Structural polypeptides of mumps virus. J Gen Virol. 1978;41(3):527–539. doi: 10.1099/0022-1317-41-3-527. [DOI] [PubMed] [Google Scholar]
- Pantua HD, McGinnes LW, Peeples ME, Morrison TG. Requirements for the assembly and release of Newcastle disease virus-like particles. J Virol. 2006;80(22):11062–11073. doi: 10.1128/JVI.00726-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parent LJ, Bennett RP, Craven RC, Nelle TD, Krishna NK, Bowzard JB, Wilson CB, Puffer BA, Montelaro RC, Wills JW. Positionally independent and exchangeable late budding functions of the Rous sarcoma virus and human immunodeficiency virus Gag proteins. J Virol. 1995;69(9):5455–5460. doi: 10.1128/jvi.69.9.5455-5460.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patch JR, Han Z, McCarthy SE, Yan L, Wang LF, Harty RN, Broder CC. The YPLGVG sequence of the Nipah virus matrix protein is required for budding. Virol J. 2008;5(1):137. doi: 10.1186/1743-422X-5-137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez M, Craven RC, de la Torre JC. The small RING finger protein Z drives arenavirus budding: implications for antiviral strategies. Proc Natl Acad Sci USA. 2003;100(22):12978–12983. doi: 10.1073/pnas.2133782100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pornillos OW, Alam SL, Davis DR, Sundquist WI. Structure of the Tsg101 UEV domain in complex with the PTAP motif of the HIV-1 p6 protein. Nat Struct Biol. 2002;9(11):812–817. doi: 10.1038/nsb856. [DOI] [PubMed] [Google Scholar]
- Randall RE, Young DF, Goswami KK, Russell WC. Isolation and characterization of monoclonal antibodies to simian virus 5 and their use in revealing antigenic differences between human, canine and simian isolates. J Gen Virol. 1987;68(Pt 11):2769–2780. doi: 10.1099/0022-1317-68-11-2769. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Lamb RA. Escaping from the cell: assembly and budding of negative-strand RNA viruses. Curr Top Microbiol Immunol. 2004;283:145–196. doi: 10.1007/978-3-662-06099-5_5. [DOI] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Morita E, Sundquist WI, Lamb RA. Evidence for a new viral late-domain core sequence, FPIV, necessary for budding of a paramyxovirus. J Virol. 2005;79(5):2988–2997. doi: 10.1128/JVI.79.5.2988-2997.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt AP, Leser GP, Waning DL, Lamb RA. Requirements for budding of paramyxovirus simian virus 5 virus-like particles. J Virol. 2002;76(8):3952–3964. doi: 10.1128/JVI.76.8.3952-3964.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimono A, Behringer RR. Angiomotin regulates visceral endoderm movements during mouse embryogenesis. Curr Biol. 2003;13(7):613–617. doi: 10.1016/s0960-9822(03)00204-5. [DOI] [PubMed] [Google Scholar]
- Shin K, Straight S, Margolis B. PATJ regulates tight junction formation and polarity in mammalian epithelial cells. J Cell Biol. 2005;168(5):705–711. doi: 10.1083/jcb.200408064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stallcup KC, Raine CS, Fields BN. Cytochalasin B inhibits the maturation of measles virus. Virology. 1983;124(1):59–74. doi: 10.1016/0042-6822(83)90290-8. [DOI] [PubMed] [Google Scholar]
- Studier FW. Protein production by auto-induction in high density shaking cultures. Protein Expr Purif. 2005;41(1):207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- Sugahara F, Uchiyama T, Watanabe H, Shimazu Y, Kuwayama M, Fujii Y, Kiyotani K, Adachi A, Kohno N, Yoshida T, Sakaguchi T. Paramyxovirus Sendai virus-like particle formation by expression of multiple viral proteins and acceleration of its release by C protein. Virology. 2004;325(1):1–10. doi: 10.1016/j.virol.2004.04.019. [DOI] [PubMed] [Google Scholar]
- Sugihara-Mizuno Y, Adachi M, Kobayashi Y, Hamazaki Y, Nishimura M, Imai T, Furuse M, Tsukita S. Molecular characterization of angiomotin/JEAP family proteins: interaction with MUPP1/Patj and their endogenous properties. Genes Cells. 2007;12(4):473–486. doi: 10.1111/j.1365-2443.2007.01066.x. [DOI] [PubMed] [Google Scholar]
- Takimoto T, Murti KG, Bousse T, Scroggs RA, Portner A. Role of matrix and fusion proteins in budding of Sendai virus. J Virol. 2001;75(23):11384–11391. doi: 10.1128/JVI.75.23.11384-11391.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takimoto T, Portner A. Molecular mechanism of paramyxovirus budding. Virus Res. 2004;106(2):133–145. doi: 10.1016/j.virusres.2004.08.010. [DOI] [PubMed] [Google Scholar]
- Troyanovsky B, Levchenko T, Månsson G, Matvijenko O, Holmgren L. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J Cell Biol. 2001;152(6):1247–1254. doi: 10.1083/jcb.152.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulloa L, Serra R, Asenjo A, Villanueva N. Interactions between cellular actin and human respiratory syncytial virus (HRSV) Virus Res. 1998;53(1):13–25. doi: 10.1016/s0168-1702(97)00121-4. [DOI] [PubMed] [Google Scholar]
- Wells CD, Fawcett JP, Traweger A, Yamanaka Y, Goudreault M, Elder K, Kulkarni S, Gish G, Virag C, Lim C, Colwill K, Starostine A, Metalnikov P, Pawson T. A Rich1/Amot complex regulates the Cdc42 GTPase and apical-polarity proteins in epithelial cells. Cell. 2006;125(3):535–548. doi: 10.1016/j.cell.2006.02.045. [DOI] [PubMed] [Google Scholar]
- Yuan B, Campbell S, Bacharach E, Rein A, Goff SP. Infectivity of Moloney murine leukemia virus defective in late assembly events is restored by late assembly domains of other retroviruses. J Virol. 2000;74(16):7250–7260. doi: 10.1128/jvi.74.16.7250-7260.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan B, Li X, Goff SP. Mutations altering the moloney murine leukemia virus p12 Gag protein affect virion production and early events of the virus life cycle. EMBO J. 1999;18(17):4700–4710. doi: 10.1093/emboj/18.17.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng Y, Vertuani S, Nystrom S, Audebert S, Meijer I, Tegnebratt T, Borg JP, Uhlen P, Majumdar A, Holmgren L. Angiomotin-like protein 1 controls endothelial polarity and junction stability during sprouting angiogenesis. Circ Res. 2009;105(3):260–270. doi: 10.1161/CIRCRESAHA.109.195156. [DOI] [PubMed] [Google Scholar]