Abstract
The differentiation of human embryonic stem cells (hESCs) into cardiomyocytes (CMs) using embryoid bodies (EBs) is relatively inefficient and highly variable. Formation of EBs using standard enzymatic disaggregation techniques results in a wide range of sizes and geometries of EBs. Use of a 3-D cuboidal microwell system to culture hESCs in colonies of defined dimensions, 100 to 500 μm in lateral dimensions and 120 μm in depth, enabled formation of more uniform sized EBs. The 300 μm microwells produced highest percentage of contracting EBs, but flow cytometry for myosin light chain 2A (MLC2a) expressing cells revealed a similar percentage (~3%) of cardiomyocytes formed in EBs from 100 μm and 300 μm microwells. These data, and immunolabeling with anti-MF20 and MLC2a, suggest that the smaller EBs are less likely to form contracting EBs, but those contracting EBs are relatively enriched in cardiomyocytes compared to larger EB sizes where CMs make up a proportionately smaller fraction of the total cells. We conclude that microwell-engineered EB size regulates cardiogenesis and can be used for more efficient and reproducible formation of hESC-CMs needed for research and therapeutic applications.
Keywords: human embryonic stem cells, microwells, embryoid body, cardiomyocytes, differentiation
1. Introduction
Efficiently guiding the differentiation of human embryonic stem cells (hESCs) has proven to be a key roadblock in the development of therapeutic applications and further scientific discovery using hESCs. In the absence of controlling factors, spontaneous hESC differentiation is highly variable, dictated by factors such as hESC colony characteristics including size and cell density [1,2], as well as the cellular microenvironment [3,4]. Multiple methods have been employed to induce hESC differentiation, including embryoid body (EB) formation, inductive co-culture with specific cell lines, and directed differentiation using particular growth factors [1,5–12]. However, for most cell lineages, the protocols are inefficient and poorly reproducible.
Differentiation of ESCs into cardiomyocytes has been extensively studied since the initial report of cardiogenesis from mouse embryonic stem cells (mESCs) in EBs in 1985 [13]. Research utilizing mESCs identified several variables which influence the extent of cardiogenesis in EBs including the particular cell line, the starting number of cells used to form each EB, medium and growth factors, and the duration of suspension culture prior to EB plating [14]. Early studies using enzymatic digestion of the mESC colonies resulted in a large range of EB sizes which differed in their ability to undergo cardiac differentiation [14–16]. This source of variability was overcome by using the hanging drop method of EB formation in which a defined number of enzymatically isolated mESCs could be added to a drop where they aggregate [17,18].
In the case of differentiation of hESCs to cardiac cell lineages, the differentiation protocols have been relatively inefficient and are still undergoing rapid development. Formation of EBs is the most common approach to hESC cardiac differentiation, but substantial variability exists with this technique, likely in part due to the inconsistency in starting aggregate size. Unfortunately extrapolating the hanging drop technique directly to hESCs has not been possible in most hands because enzymatically isolated single hESCs fail to aggregate and form an EB [19,20]. A variety of techniques have recently been explored to promote formation of uniform-sized aggregates for reproducible differentiation of hESCs. Enzymatically isolated hESCs have been subjected to forced aggregation using centrifugation to promote EB-mediated hematopoietic and cardiac differentiation [19,21]. Micro-contact printing has been used to patterned 2-D aggregates of isolated hESCs to promote uniform EB size [22,23]. Another approach involves use of microtextured surfaces composed of square-pyramidal pits in a silicon wafer in which ES cells could be seeded [24]. Each approach has a variety of advantages and disadvantages. For example, the enzymatic isolation to single cells and forced-aggregation could be stressful to hESCs and disrupt the cell-cell signaling required for hESC growth, survival, and differentiation. Some approaches are labor intensive and not readily scalable. Overall, these techniques have provided advances, but the goal of highly reproducible, efficient, scalable cardiac differentiation has not been obtained.
Our recent study and others have demonstrated that hESC colonies can be efficiently grown and maintained in engineered 3-D microwells [25,26]. The microwells, which are surrounded by a cell and protein repellant self-assembled monolayer (SAM), promote hESC self-renewal far longer than standard cultures, while still allowing differentiation to derivatives of each of the three primary germ layers upon removal from microwells. By constraining colony growth within microwells we are able to maintain cell-cell signaling and colony characteristics necessary for hESC survival and proliferation better than unconstrained standard culture techniques. Thus we postulated that microwells could be used to impose a uniform 3-D size of hESC colonies used for formation of EBs and allow the use of uniform populations of undifferentiated hESCs for EB formation. The present study examines the impact of varying microwell dimensions on cardiogenesis from hESCs in EBs.
2. Materials and methods
2.1 Microwell formation and functionalization
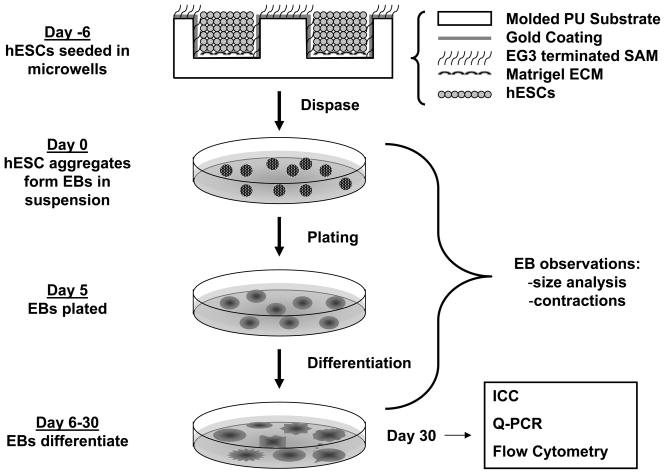
The detailed microwell manufacturing and functionalization protocol are described elsewhere [25]. Briefly, silicon masters were manufactured through photolithography and plasma etching. Masters were passivated under a fluorinated silane vapor to facilitate detachment of PDMS molds. PDMS stamps were attached to glass slides and Norland optical adhesive 61 (Norland Products Inc., Cranbury, NJ, USA) polyurethane prepolymer was distributed between stamp and glass slide via capillary action. After crosslinking, stamps were removed, yielding microwells (Figure 1).
Figure 1. Schematic of microwell controlled EB formation and differentiation.
Cuboidal microwells were molded using polyurethane as described in the Methods, and hESCs seeded to the Matrigel coated bottom of the wells. hESC colonies from the microwells were used to form EBs which were cultured and characterized as schematically indicated.
Using e-beam evaporation, microwells were coated with a thin layer of gold at oblique angles (>45°) to coat the sides of wells and areas between wells. Samples were then sterilized prior to self-assembled monolayer (SAM) formation and cell seeding by immersion in 100% ethanol and 1 hour UV sterilization. Microwells were functionalized with a tri-ethylene glycol terminated alkanethiol (EG3) (Prochimia, Poland) by immersion in a 2 mM ethanolic EG3 solution for 2 hours. Microwell slides were then washed in 100% ethanol and air dried prior to cell seeding.
2.2 hESC culture
hESC line H9 was used throughout all experiments. hESCs were cultured on either 6-well tissue culture polystyrene culture dishes (unconstrained), or in microwells. Both were coated by growth factor reduced Matrigel (Beckton-Dickinson Bioscience, Medford, MA, USA). Culture medium was conditioned on MEFs for 24 hours and supplemented with 4 ng/ml bFGF. MEF medium is composed of DMEM (Invitrogen), fetal bovine serum (10%, heat inactivated, Invitrogen), and MEM non-essential amino acid solution (1%, Invitrogen). The hESC medium without bFGF (UMF-) consists of DMEM/F12 (Invitrogen), Knockout Serum Replacer (KOSR, 20%, Invitrogen), L-glutamine (2 mmol/L, Invitrogen), and MEM non-essential amino acid solution (1%, Invitrogen). The MEF conditioned hESC medium with bFGF (CMF+) is supplemented with 4 ng/ml bFGF. hESCs were passaged from unconstrained dishes to microwells using pre-warmed 1 mg/ml dispase in DMEM/F12. Plates were incubated at 37°C until colonies were easily washed out of wells or off the plate.
2.3 Microwell cell seeding
Prior to seeding cells in microwells, each microwell slide was placed in a well of a 6-well plate and was immersed in 100% ethanol to wet the surface and prevent air bubble entrapment in microwells. Microwell slides were washed in cold DMEM/F12 to remove all traces of ethanol. Matrigel solution was prepared by resuspending 2 mg growth factor reduced Matrigel in 24 ml cold DMEM/F12. Microwells were coated with Matrigel by adding 2 ml of cold Matrigel solution to each well of a 6-well plate and incubated for one hour at 37°C. Following the Matrigel incubation, microwell slides were washed in 2 ml PBS and then transferred to a fresh 6-well plate. hESCs were passaged from 1 well of a 6-well plate to one microwell slide. hESCs were incubated up to 5 minutes in 1 mg/ml dispase solution until colony edges began lifting off plate. Cells were gently scraped off the plate and washed once in UMF-. Cells were then filtered by 70 μm cell strainer to remove large clumps to facilitate seeding in microwell slides. Cells were washed twice in UMF- and pelleted. The pellet was then resuspended in 300 μl CMF+. In order to maximize cell seeding efficiency, the cell suspension was aliquoted to the top of the microwell slide, taking care to maintain the entire cell suspension on the slide. Slides were held at room temperature at least 30 minutes to allow cells to settle into wells and then 2 ml/well CMF+ was added carefully to each well of the 6-well plate to prevent cells being washed out of microwells.
2.4 EB culture and differentiation
The general strategy for using microwells to form EBs and for the culture of the resulting EBs is shown schematically in Fig. 1. In brief, EBs were formed from hESC colonies removed from microwells or standard 6-well plates as described above and then cultured in suspension for 5 days in 4 ml/well medium in Corning 3471 low attachment plates. EBs were suspended 1 day in UMF-, followed by 4 days in an EB medium consisting of DMEM/F12 (Invitrogen), fetal bovine serum (20%, cardiac differentiation qualified, Hyclone, Catalog No. SH3007003, Lot No. ARH 27209), L- glutamine (2 mmol/L, Invitrogen), and MEM non-essential amino acid solution (1%, Invitrogen). EBs were plated on 0.1% gelatin-coated 6-well plates or coverslips in 24-well plates and cultured in EB medium for the duration of the experiment. After 10 days of differentiation, FBS concentration in EB medium was reduced to 2%. At day 7, the total attached EBs were counted under the microscope. EBs were observed daily, and contracting EBs were counted every three days beginning at day 9. The percentage of contracting EBs was obtained by dividing the number of contracting EBs by the total number of EBs for each well of 6-well plate and for each independent experiment.
2.5 QPCR analysis
Total RNA was isolated using Trizol Reagent (Invitrogen, Carlsbad, CA) from one well of EB culture in 6-well plate according to the manufacturer’s instruction. 2 μg of total RNA was used per RT reaction using High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). Each PCR was performed in triplicate with TaqMan® Universal PCR Kit (Applied Biosystems, Foster City, CA) and TaqMan® Gene Expression Assays. TaqMan Primers for GAPDH (Hs99999905_m1), NKX2-5 (Hs00231763_m1), TNNT2 (Hs00165960_m1), MYL7 (Hs00221909_ml) and PLN (Hs00160179_m1) were used. Fold change was calculated using the comparative Ct method (ΔΔCT method) [27].
2.6 Flow cytometry
Differentiated cells were detached from culture dishes for flow cytometry analysis by first washing twice in 2 ml/well PBS for 5 minutes at 37°C. Cells were then incubated in 1 ml/well 0.25% trypsin-EDTA (Invitrogen), 2% chick serum (Sigma) for 10 minutes at 37°C, followed by neutralization using 4 ml/well FACS buffer (PBS without Ca/Mg2+, 2% FBS, 0.1% NaN3). Remaining cell aggregates were disrupted by gently pipetting. Cells were centrifuged 5 minutes at 1000 rpm, supernatant discarded, and the pellet was resuspended in 1 ml PBS with 60 μl 16% paraformaldehyde for fixation. Samples were incubated 10 minutes in 37°C water bath and then chilled on ice 1 minute. Samples were centrifuged, pellet resuspended in 1 ml ice-cold 90% methanol, and incubated on ice for 30 minutes to permeabilize the cells. Cells were washed once in FACS buffer plus 0.1% Triton, centrifuged, and supernatant discarded leaving about 50 μl. Primary antibody was diluted in 50 μl/sample FACS buffer plus 0.1% Triton and aliquoted to each sample for total sample volume of 100 μl. Samples were incubated overnight at 4°C. MF20 and MLC2a primary antibodies and their dilutions were the same as described in immunolabeling. Cells were washed twice in 3 ml FACS buffer plus 0.1% Triton, centrifuged, and supernatant discarded leaving ~ 50 μl. Secondary antibody, Alexa Fluor 633 goat anti-mouse IgG2b (Invitrogen, Carlsbad, CA), was diluted 1:1000 in 50 μl/sample FACS buffer plus Triton and aliquoted to each sample (final sample volume 100 μl). Samples were incubated 30 to 45 min in the dark at room temperature, then washed twice in FACS buffer. Samples were centrifuged, resuspended in 300 μl FACS buffer, transferred to flow cytometry tubes, and stored on ice until analysis. Data were collected on a FACSCaliber flow cytometer (Beckton Dickinson) and analysis performed using CellQuest (Beckton Dickinson) software. All cells were gated according to light scatter and fluorescence.
2.7 Immunolabeling
EBs plated on coverslips were fixed in 4% Paraformaldehyde by diluting 16% Paraformaldehyde (EMS Catalog No. 15710-S) in 1X PBS (Invitrogen, Catalog No. 14190–144) and incubating for 15 minutes at room temperature. Cells were rinsed twice in PBS after fixation, followed by permeabilization in 0.2% Triton X-100 in PBS solution (Sigma Catalog No. T-9284) for 1 hour at room temperature. Samples were blocked by preparing fresh blocking solution, 5% non-fat dry milk in 0.2% Triton X-100 solution, and incubated for two hours at room temperature on a rotator, or at 4°C overnight covered with Parafilm. Samples were washed twice with PBS, 5 minutes per wash. Primary antibodies, MF20 (Developmental Studies Hybridoma Bank, Iowa City, IA) and anti-MLC2a (Synaptic Systems, Germany, Cat. No. 311011) were diluted 1:20 or 1:400, respectively, in 0.1% Triton X-100, 1% BSA in PBS solution and incubated overnight at 4oC. Cells were washed with 0.2% Tween 20 in PBS three times, 5 minutes each, and 1X PBS once. Secondary antibody, Alexa Fluor 594 goat anti-mouse IgG2b (Invitrogen, Carlsbad, CA), was diluted in the same solution as the primary antibody and incubated at room temperature for 1.5 hours in the dark. Three washes in 0.2% Tween 20 in PBS, 5 minutes each, were followed by one 1X PBS wash. One drop of antifade reagent with DAPI (Invitrogen, Catalog No. P36935) was placed on each slide, and coverslips were applied with cell surface down.
2.8 Statistics
Data are presented as mean ± standard error of the mean (SEM). Statistical significance was determined by one-way ANOVA or blocked one-way ANOVA where appropriate with post hoc testing using Tukey method using Microcal Origin, v7.5 and R. P < 0.05 was considered statistically significant. Data histograms were fit to Gaussian distributions using nonlinear least squares regression with Microcal Origin, version 7.5.
3. Results
3.1 Size distribution of EBs from microwells
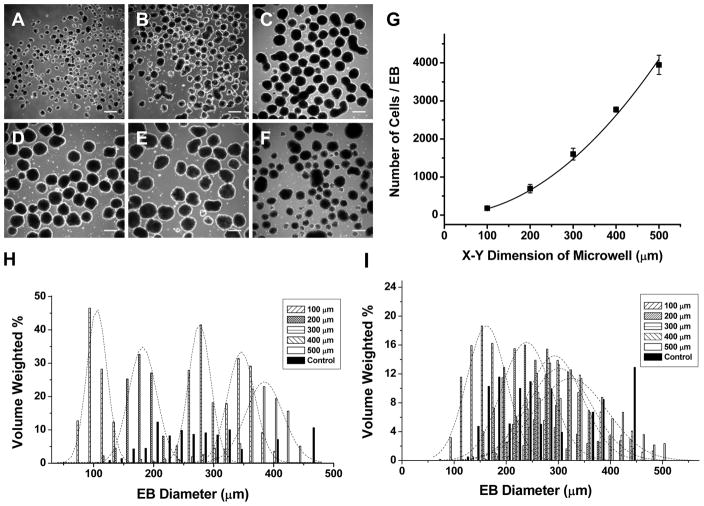
hESCs were seeded into Matrigel-coated cuboidal microwells, 100 to 500 μm in lateral dimensions and 120 μm in depth. After 6 days of growth in CMF+, hESC colonies were enzymatically removed from microwells and cultured in EB medium in suspension to initiate differentiation. Upon removal from microwells and suspension culture for one day, spherical EBs formed and exhibited relatively homogeneous sizes (Fig 2A-E) directly related to the initial microwell dimensions in contrast to the more heterogeneous size distribution of EBs formed from standard hESC culture (Fig 2F). EB size distributions were quantified using image analysis by measuring the diameter of each EB and calculating the volume with the assumption of spherical EBs. The volume of each size of EB was then normalized to the total EB volume in culture for each microwell size. Fig 2H shows the volume-weighted size distribution based on EB diameter, and the data for each microwell size could be well described by a Gaussian distribution. The average EB diameters (±SD) at one day of culture for the different microwell sizes were 88±16 μm (100), 162±24 μm (200), 256±28 μm (300), 322±36 μm (400), 350±51 μm (500), and 196±68 μm (control). Thus the microwell constrained hESC culture system led to predictable, uniform-sized EBs, particularly for the smaller sized microwells, in contrast to more heterogeneous EB sizes resulting from unconstrained, conventional hESC culture on Matrigel.
Figure 2. hESC EBs formed from microwells yield homogeneous size distributions with predictable number of cells per EB.
EBs formed following one day of suspension culture from hESCs cultured in microwells of 100 μm (A), 200 μm (B), 300 μm (C), 400 μm (D) and 500 μm (E) lateral dimensions and 120 μm depth compared to EBs from standard hESC culture on Matrigel (F). Between 68 and 185 EBs were evaluated for each sample set done in parallel in each experiment. Scale bar is 300 μm. (G) EBs derived from microwells 120 μm deep x 100–500 μm lateral dimensions were isolated and trypsinized after 1 day suspension culture, and the average number of cells per EB was counted for a total of 3 independent experiments. Assuming the volume of the starting microwell determined cell number, the data were fit to a simple polynomial equation, cell number=A(X-Y dimension)2 where A is a constant. The data were well fit to the model (R2= 0.99) with A=0.0163±0.0004. (H) Histogram of the volume-weighted distribution of EB size relative to EB diameter for each starting microwell size. The microwell EBs sizes could well be described by Gaussian distributions (dashed curves) centered at 105 μm (100), 181 μm (200), 276 μm (300), 345 μm (400), 384 μm (500). In contrast, the EBs derived from unconstrained culture were more broadly size distributed and not well described by a simple Gaussian distribution. (I) Histogram of EB size distribution after 5 days in suspension culture showed differential growth of EBs from different sized microwells. The microwell EB sizes could well be described by Gaussian distributions centered at 162 μm (100), 237 μm (200), 291 μm (300), 301 μm (400), and 323 μm (500). A range of 189 to 536 EBs were counted for each sample set.
If microwell size directly regulates EB size, then we postulated that the number of cells present in EBs should be directly proportional to the microwell volume. Microwell EBs were placed in suspension culture for 1 day, and then subjected to enzymatic separation using trypsin to isolate individual cells which were counted using a hemacytometer. The average number of cells per EB for each microwell size is plotted in Fig. 2G, and the data were fit to a simple polynomial function based on the square of the X-Y dimension given that the depth of the microwells was kept constant at 120 μm. Thus the microwell system allowed us to systematically vary the number of cells in an EB after one day of culture over a relatively wide range, from ~140 cells/EB to ~3900 cells/EB.
We next assessed the effect of microwell size on EB dimensions following five days of suspension culture. The growth of EBs in culture reflects a complex combination of factors including cellular proliferation, differentiation, apoptosis, and possible disaggregation. EB size distributions were quantified using image analysis by measuring the diameter of each EB as described for day 1 EBs. The volume-weighted percent size distributions of EBs from the different microwells could all be well described by Gaussian distributions, in contrast to more heterogeneous EB size from the control EBs, as shown in Fig 2I. We observed that the 100 μm and 200 μm microwells resulted in distinct sized EBs with mean diameters (±SD) of 135±37 μm and 206±44 μm, respectively; however, the large sized microwell EBs (300–500 μm dimensions) all tended to form similar sized EBs; mean EB diameters were 258±53 μm, 264±58 μm and 285±61 μm, respectively. Thus the smaller sized microwell EBs (100 and 200 μm) tended to grow in size during five days of suspension culture, but the EBs from larger sized microwells (300,400, and 500 μm) either did not increase in size or slightly decreased. Thus starting EB size can impact the growth properties of similarly maintained EBs.
3.2 Impact of microwell size on cardiogenesis
Once predictable EB size distributions had been obtained via microwell culture, we tested the effect of EB size on cardiogenesis. To induce differentiation, hESCs were removed from microwells 120 μm deep x 100–500 μm lateral dimensions, after six days of culture in CMF+. The aggregates were cultured in suspension for one day in UMF which minimized aggregate to aggregate adhesion which otherwise occurred in the presence of serum containing medium. After the first day, the medium was changed to EB medium and after four days in suspension, the EBs were attached to gelatin-coated plates and cultured in EB medium for a total of 30 days.
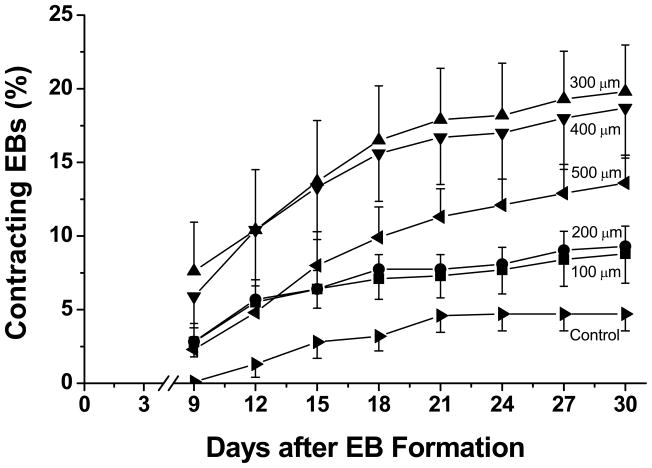
We first measured the fraction of EBs that formed spontaneously contracting areas as a function of microwell dimensions and time in culture. Contracting EBs were initially observed within three days of plating on gelatin coated plates, and the fraction of contracting EBs increased through time to reach a plateau at approximately 21 days (Fig 3). The 300 and 400 μm microwells produced the most spontaneously contracting EBs, achieving ~18–20% at 30 days compared to 100 and 200 μm microwells which produced ~8–10% contracting EBs. The 500 μm microwells were intermediate with 14% contracting EBs at 30 days. In contrast to the relatively high percentages of contracting EBs afforded through microwell culture, unconstrained controls yielded only 5% contracting EBs, significantly less than 300 and 400 μm microwell EBs. The time over which contracting EBs developed was comparable for all microwell sizes and the control EBs.
Figure 3. Time course of development of spontaneously contracting EBs comparing microwell and control EBs.
EBs formed from microwells of 100, 200, 300, 400 and 500 μm lateral dimensions and from unconstrained standard culture were plated and observed for development of spontaneous contractions. The number of EBs contracting was scored every three days starting at day 9 after EB formation until day 30 as a percentage of attached EBs. Average contracting percentages were obtained by pooling EB counts from three independent experiments done in triplicate using at least 474 total EBs for each sample set. Error bars represent SEM. Data were compared using one-way ANOVA and Tukey post tests. At day 9, 12 and 15, there were no significant differences between any of the microwell EBs and control EBs. On day 18, the 300 and 400 μm EBs were significantly different from the control EBs (P < 0.05), and from day 21 to day 30, the 300 and 400 μm EBs were significantly different from the control EBs (P < 0.05), and the 300 μm EBs were also significantly different from the 100 μm EBs (P < 0.05).
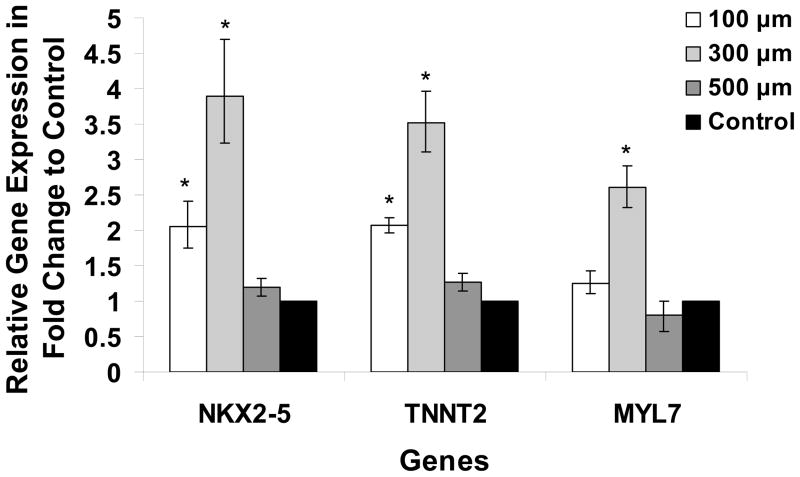
To provide another assessment of cardiogenesis from the microwell EBs compared to standard unconstrained culture for formation of EBs, we performed real-time quantitative PCR examining the expression of cardiac genes after 30 days of culture. TaqMan gene expression assays for cardiac markers included a transcription factor critical for cardiac development, Nkx2.5 (human genome NKX2-5), a cardiac specific myofilament protein, cardiac troponin T (human genome TNNT2), and another cardiac myofilament protein, myosin light chain 2A (human genome MYL7). We compared expression of these three genes in EBs generated from three distinct microwell sizes to control EBs and found that like the measurements of percentage of contracting EBs, the greatest expression of NKX2-5, TNNT2, and MYL7 was in the 300 μm microwells with a 2–4 fold increase and were significantly different from the gene expression of the control EBs (Fig. 4). The 100 μm microwell EBs also resulted in a significant increase of NKX2-5 and TNNT2 expression over the control EBs. The 500 μm microwells showed no significant difference in expression of the genes under study compared to control EBs. Overall, the quantitative PCR data were consistent with the measurements of percentage of contracting EBs, with the 300 μm microwells demonstrating the greatest expression of cardiac markers.
Figure 4. Expression of cardiac genes NKX2-5, TNNT2, and MYL7 in microwell and control EBs.
Total RNA was isolated from each size of microwell EB culture and unconstrained control EBs. Real-time RT-PCR was performed using Taqman gene expression assays, and the fold change in expression for each gene is plotted for the tested microwell size relative to the control. Error bars indicate fold change range calculated by the comparative Ct method for triplicates from three independent experiments. Data were compared using one-way ANOVA and Tukey post tests with * indicating the gene expression in microwell EBs is significantly different from that of the control EBs, P < 0.05.
Flow cytometry provided another quantitative assessment of cardiogenesis in the EBs by measuring the fraction of cells expressing cardiac-specific proteins. EBs were subjected to enzymatic separation to single cells, which were labeled with an antibody recognizing MLC2a, a ubiquitous myofilament protein expressed in all early cardiomyocytes types [28]. Data were obtained from samples composed of a collection of EBs from a single well of a six well plate containing 50-200 EBs for each experiment, so both contracting and noncontracting EBs were included in the sample. In this collection of cells, the number of MLC2a positive cells from control EBs was ~1.2%. Evaluation of EBs from the different sized microwells revealed that 500 μm microwells resulted in a comparable number of MLC2a positive cells (~2.0%) compared to control EBs; however, the 100 and 300 μm microwells produced on average approximately three fold more cardiomyocytes (~3.3%) in comparison to the control cells (Fig. 5). Interestingly, the 100 μm microwell EBs formed as many or more cardiomyocytes than the 300 μm microwell EBs in contrast to the finding that the 300 μm microwell EBs formed about twice as many contracting EBs compared to 100 μm EBs. This apparent discrepancy could be explained by differences in the size of contracting areas in EBs, the presence of MLC2a in noncontracting EBs, or technical limitations in the flow cytometry assay. To evaluate for limitations related to the chosen antibody against MLC2a, we also performed flow cytometry using the antibody MF20 which recognizes sarcomeric myosin present in cardiomyocytes, and found quantitatively similar results (data not shown).
Figure 5. Flow cytometric analysis of cells expressing MLC2a, a marker for cardiomyocytes, in EBs from microwell and control EBs.
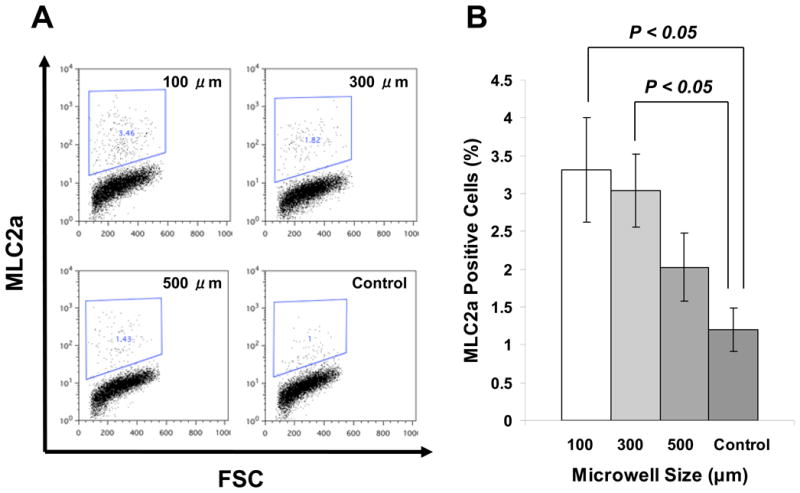
A) Flow cytometric analysis of cells expressing MLC2a in the microwell EBs and the control EBs. Cells were harvested from microwell EBs of 100, 300, and 500 μm lateral dimensions compared to EBs from standard unconstrained culture following 30 days of culture. B) Average number MLC2a positive cells comparing different sized microwell EBs to control EBs from 3 independent experiments, each experiment had 3 wells of EB culture in 6-well plate for flow cytometry. Error bars indicate SEM. Data were compared using blocked one-way ANOVA and Tukey post tests.
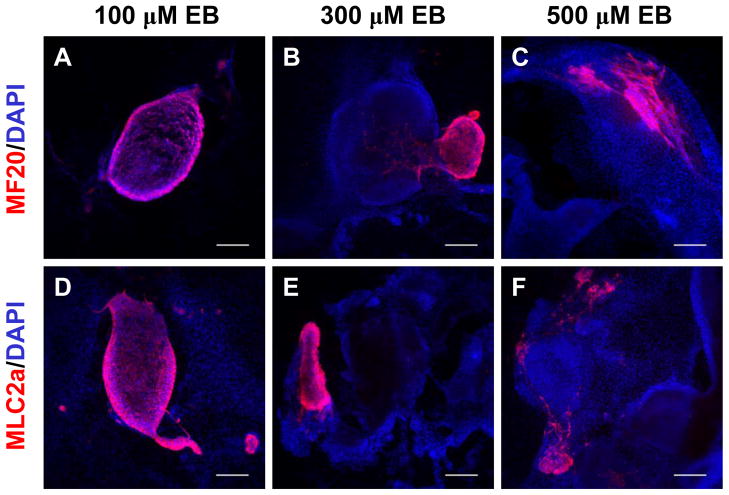
To understand the distribution of cardiomyocytes in the differentiating EBs, we next performed immunolabeling for cardiac proteins in EBs. The EBs were fixed at day 30 after plating on gelatin coated coverslips and immunolabeling was performed to identify all hESC-derived cardiomyocytes using antibodies to sarcomeric myosin (MF20) and MLC2a. Only 5–20% of the EBs exhibited any immunolabeling for the cardiac proteins, and this fraction was qualitatively similar to the fraction of contracting EBs. In the EBs immunolabeled with anti-MF20 or anti-MLC2a, positive cells were tightly clustered, consistent with observations of localized regions of contraction in EBs. Typically, only a single outgrowth of cardiomyocytes was observed for an EB with the exception of some large EBs that contained multiple regions. Fig. 6 compares representative immunolabeled EBs from 100 μm, 300 μm, and 500 μm microwells. These images demonstrate that smaller EBs typically formed from the 100 μm microwell cultures, but the density of cardiomyocytes in the EBs was high, often composing the majority of the cells in the EB. In contrast, the presence of cardiomyocytes in larger EBs such as those from 300 μm and 500 μm microwell EBs was more restricted and limited to a relatively small fraction of the EB. These data demonstrate that the size of microwells used to form EBs can strongly impact the efficiency of cardiac differentiation in EBs with 100 μm microwells leading to EBs which are greatly enriched in cardiomyocytes relative to other cell types.
Figure 6. Immunolocalization of cardiomyocytes in EBs formed from different sized microwells.
Panels A – C show immunolabeling with MF20 antibody (red) which recognizes sarcomeric myosin present in cardiomyocytes and nuclei are stained with DAPI (blue), in 30 days old EBs formed from 100 μm, 300 μm and 500 μm microwells , respectively. Panels D - F show immunolabeling with anti-MLC2a antibody (red), which recognizes a myofilament protein expressed in all early embryonic cardiomyocytes, and nuclei are stained with DAPI (blue), in 30 day old EBs formed from 100 μm, 300 μm and 500 μm microwells, respectively. Scale bar is 300 μm.
4. Discussion
Cardiac differentiation of hESCs in EBs has demonstrated a wide range of efficiency ranging 0% to 70% of EBs containing contracting cardiomyocytes in published studies [5,16,19,29,30]. Some of the variability may be related to the use of different hESC lines with distinct predispositions for cardiogenesis [19,31]. But even within a laboratory and using the same hESC line, there can be substantial variability. In recent years, microscale engineering approaches have been utilized to improve reproducibility, efficiency and scalability of stem cell differentiation. The present study used engineered microwells to produce hESC colonies of defined 3-dimensional sizes and to generate specific-sized aggregates of hESCs for EB formation. By testing a range of microwell sizes, we demonstrated improved efficiency and reproducibility of cardiac differentiation using microwells compared to standard enzymatic methods to form EBs.
The described microwell approach has several potential advantages over standard EB formation as well as over some micro-engineered techniques. First, the use of feeder free culture of hESCs on Matrigel removes the potential confounding factors related to feeder cells which can inhibit cardiogenesis [32]. Secondly, the microwells provide a confined 3 dimensional structure that permits the hESC colonies to grow to uniform sizes and shapes that show less variability compared to standard culture of hESCs [25]. Uniformity in starting hESC colony properties is important for reproducibility of differentiation, and 2-dimensional patterning of hESC culture using micro-contact printing could not eliminate the variability in the starting hESC gene expression patterns [33]. Finally, controlled 3-dimensional aggregate size used in EB formation is advantageous as we demonstrated that 300 μm wells were most efficient at generating contracting EBs, comparable to the 250–350 μm range found in the forced aggregation method for forming human EBs as well as similar work with mouse ESCs (~300 μm) [19,34]. Another microwell approach has been recently described which generates different sized inverted cuboidal pyramid microwells using similar silicon micropatterning [24]. However, seeding of the inverted cuboidal pyramid microwells requires enzymatically isolated single cells and exposes the cells to stresses from centrifugation or chemical means to promote aggregation in contrast to the present work where the ESCs are grown in the microwells. An advantage of the pyramidal microwells is that they can be seeded with various cell numbers to readily generate different sized aggregates which is not readily possible with our rectangular cuboidal microwells which require formation of different sized microwells for such variation in aggregate size. Whether the shape of the microwell and thus the initial aggregate shape impacts the differentiation process is not yet known.
The present study revealed that controlling EB size regulates cardiogenesis from hESCs not only by impacting the likelihood an EB will generate some cardiomyocytes, but also the density of cardiomyocytes relative to other cell types in the differentiating EB. By measuring both the percentage of EBs contracting as well as the number of cardiomyocytes by flow cytometry, we discovered that the likelihood of a smaller 100 μm microwell EB undergoing cardiogenesis was lower than 300 μm microwell EB, but if the 100 μm microwell EB developed cardiomyocytes, a much greater density of cardiomyocytes was found in the EB. This observation has not been previously described in part because most prior studies only quantify contracting EBs and not absolute cardiomyocytes number. Although the mechanism of this size effect remains to be determined, this provides a potential tool to get the highest density of cardiomyocytes from preparations if conditions can be optimized to initiate cardiogenesis in small wells.
The importance of EB size in regulating cardiac differentiation likely is related to diffusion of critical substrates as well as the role of various chemical cues such as growth factors. Observations of the patterns of expansion of the EBs revealed that the smaller sized EBs (100 and 200 μm) showed the greatest growth in five days and larger EBs either did not expand or contracted in size during this time period. Whether these distinct growth properties are related to a maximal size for EBs that can be supported by passive diffusion of substrates or due to other regulatory mechanisms is not known. Spatial gradients of a variety of signaling molecules are essential for normal cardiac embryological development. For example, factors secreted from neighboring anterior lateral endoderm such as BMPs promote cardiogenesis while canonical Wnt signals from adjacent neuroectoderm inhibit cardiogenesis from mesoderm [35–39]. In EB differentiation, previous studies have suggested that at least a fraction of EBs exhibit radial multilaminar organization with an outer layer of extraembryonic endoderm that may be critical for the inner mass of cells to undergo cardiogenesis [24,40] [41–43]. Simple geometrical considerations suggest that changes in EB diameter will greatly impact the relative ratio of this outer layer of extraembryonic endoderm and related signaling molecules to the inner mass of cells. Future studies will be needed to test how microwell technology can regulate such signaling.
Progress in using signaling molecules to direct differentiation of hESCs to cardiomyocytes has been made using Activin A, BMP4, bFGF, and Wnt signaling inhibitor [6,12]. Most of these techniques involve the formation of aggregates of ESCs which potentially could be standardized by microwell technology. Ultimately combining microwell technology to regulate the size of hESC colonies with controlled growth factor delivery in defined microenvironments may provide a powerful next step in the reproducible differentiation of cardiomyocytes from ESCs as well as the more recently described induced pluripotent stem cells [44–46].
5. Conclusion
Our results demonstrate that microwell technology can be used to culture hESCs and form EBs of defined sizes and cell numbers in contrast to more highly variable EBs formed using standard techniques based on enzymatic or mechanical procedures. Furthermore, systematically varying the microwell size demonstrated that EBs from intermediate sized (300 μm) microwells generated the highest percentage of contracting EBs. Evaluation of the EBs demonstrated that microwell size also influenced the density of CMs in EBs with the smallest microwells (100 μm) showing the greatest percentage of cells that were CMs in a contracting EB. Therefore, the process of cardiac differentiation of hESCs can be modulated in multiple ways by confined culture conditions using microwells to grow hESCs and form EBs to improve the efficiency of cardiogenesis. Decreasing the heterogeneity inherent in traditional EB methods will be invaluable in optimizing the process for guiding differentiation to CMs and likely many other lineages. Microwell technology can advance both basic research applications using hESCs as well as contribute to the processes necessary to generate highly reproducible and abundant cell populations for clinical applications using human pluripotent stem cells.
Acknowledgments
NIH/NIBIB R01 EB007534, NIH/NHLBI R01 HL08846150, NSF EFRI-0735903, and the WiCell Research Institute provided support for the study. The authors express appreciation for the assistance in manuscript preparation provided by Thankful Sanftleben. The authors confirm that there are no known conflicts of interest associated with this publication, and there has been no significant financial support for this work that could have influenced its outcome.
Abbreviations
- hESC
human embryonic stem cell
- CM
cardiomyocytes
- EB
embryoid body
Footnotes
A preliminary report was presented at the 6th annual meeting of the International Society for Stem Cell Research.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Odorico JS, Kaufman DS, Thomson JA. Multilineage differentiation from human embryonic stem cell lines. Stem Cells. 2001;19:193–204. doi: 10.1634/stemcells.19-3-193. [DOI] [PubMed] [Google Scholar]
- 2.Watt FM, Hogan BL. Out of Eden: stem cells and their niches. Science. 2000;287:1427–30. doi: 10.1126/science.287.5457.1427. [DOI] [PubMed] [Google Scholar]
- 3.Spradling A, Drummond-Barbosa D, Kai T. Stem cells find their niche. Nature. 2001;414:98–104. doi: 10.1038/35102160. [DOI] [PubMed] [Google Scholar]
- 4.Streuli C. Extracellular matrix remodelling and cellular differentiation. Curr Opin Cell Biol. 1999;11:634–40. doi: 10.1016/s0955-0674(99)00026-5. [DOI] [PubMed] [Google Scholar]
- 5.Mummery C, Ward-van Oostwaard D, Doevendans P, Spijker R, van den BS, Hassink R, et al. Differentiation of human embryonic stem cells to cardiomyocytes: role of coculture with visceral endoderm-like cells. Circulation. 2003;107:2733–40. doi: 10.1161/01.CIR.0000068356.38592.68. [DOI] [PubMed] [Google Scholar]
- 6.Laflamme MA, Chen KY, Naumova AV, Muskheli V, Fugate JA, Dupras SK, et al. Cardiomyocytes derived from human embryonic stem cells in pro-survival factors enhance function of infarcted rat hearts. Nat Biotechnol. 2007;25:1015–24. doi: 10.1038/nbt1327. [DOI] [PubMed] [Google Scholar]
- 7.Schuldiner M, Yanuka O, Itskovitz-Eldor J, Melton DA, Benvenisty N. From the cover: effects of eight growth factors on the differentiation of cells derived from human embryonic stem cells. Proc Natl Acad Sci U S A. 2000;97:11307–12. doi: 10.1073/pnas.97.21.11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Itskovitz-Eldor J, Schuldiner M, Karsenti D, Eden A, Yanuka O, Amit M, et al. Differentiation of human embryonic stem cells into embryoid bodies compromising the three embryonic germ layers. Mol Med. 2000;6:88–95. [PMC free article] [PubMed] [Google Scholar]
- 9.Dvash T, Benvenisty N. Human embryonic stem cells as a model for early human development. Best Pract Res Clin Obstet Gynaecol. 2004;18:929–40. doi: 10.1016/j.bpobgyn.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 10.Tian X, Morris JK, Linehan JL, Kaufman DS. Cytokine requirements differ for stroma and embryoid body-mediated hematopoiesis from human embryonic stem cells. Exp Hematol. 2004;32:1000–9. doi: 10.1016/j.exphem.2004.06.013. [DOI] [PubMed] [Google Scholar]
- 11.Wang L, Li L, Shojaei F, Levac K, Cerdan C, Menendez P, et al. Endothelial and hematopoietic cell fate of human embryonic stem cells originates from primitive endothelium with hemangioblastic properties. Immunity. 2004;21:31–41. doi: 10.1016/j.immuni.2004.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Yang L, Soonpaa MH, Adler ED, Roepke TK, Kattman SJ, Kennedy M, et al. Human cardiovascular progenitor cells develop from a KDR+ embryonic-stem-cell-derived population. Nature. 2008;453:524–8. doi: 10.1038/nature06894. [DOI] [PubMed] [Google Scholar]
- 13.Doetschman TC, Eistetter H, Katz M, Schmidt W, Kemler R. The in vitro development of blastocyst-derived embryonic stem cell lines: formation of visceral yolk sac, blood islands and myocardium. J Embryol Exp Morphol. 1985;87:27–45. [PubMed] [Google Scholar]
- 14.Boheler KR, Czyz J, Tweedie D, Yang HT, Anisimov SV, Wobus AM. Differentiation of pluripotent embryonic stem cells into cardiomyocytes. Circ Res. 2002;91:189–201. doi: 10.1161/01.res.0000027865.61704.32. [DOI] [PubMed] [Google Scholar]
- 15.Singla DK, Jayaraman S, Zhang J, Kamp TJ. Cardiomyocyte Derivation from Human Embryonic Stem Cells. In: Master JR, Palsson BO, Thomson JA, editors. Human Cell Culture 6: Embryonic Stem Cells. New York: Springer-Verlag; 2007. pp. 211–34. [Google Scholar]
- 16.Xu C, Police S, Rao N, Carpenter MK. Characterization and enrichment of cardiomyocytes derived from human embryonic stem cells. Circ Res. 2002;91:501–8. doi: 10.1161/01.res.0000035254.80718.91. [DOI] [PubMed] [Google Scholar]
- 17.He J-Q, January CT, Thomson J, Kamp TJ. Human embryonic stem cell-derived cardiomyocytes: Drug discovery and safety pharmacology. Expert Opinion on Drug Discovery. 2007;2:39–753. doi: 10.1517/17460441.2.5.739. [DOI] [PubMed] [Google Scholar]
- 18.Segev H, Kenyagin-Karsenti D, Fishman B, Gerecht-Nir S, Ziskind A, Amit M, et al. Molecular analysis of cardiomyocytes derived from human embryonic stem cells. Dev Growth Differ. 2005;47:295–306. doi: 10.1111/j.1440-169X.2005.00803.x. [DOI] [PubMed] [Google Scholar]
- 19.Burridge PW, Anderson D, Priddle H, Barbadillo M, Chamberlain S, Allegrucci C, et al. Improved human embryonic stem cell embryoid body homogeneity and cardiomyocyte differentiation from a novel V-96 plate aggregation system highlights interline variability. Stem Cells. 2007;25:929–38. doi: 10.1634/stemcells.2006-0598. [DOI] [PubMed] [Google Scholar]
- 20.Reubinoff BE, Pera MF, Fong CY, Trounson A, Bongso A. Embryonic stem cell lines from human blastocysts: somatic differentiation in vitro. Nat Biotechnol. 2000;18:399–404. doi: 10.1038/74447. [DOI] [PubMed] [Google Scholar]
- 21.Ng ES, Davis RP, Azzola L, Stanley EG, Elefanty AG. Forced aggregation of defined numbers of human embryonic stem cells into embryoid bodies fosters robust, reproducible hematopoietic differentiation. Blood. 2005;106:1601–3. doi: 10.1182/blood-2005-03-0987. [DOI] [PubMed] [Google Scholar]
- 22.Bauwnes CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, et al. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells Express. 2008;26:2300–10. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 23.Peerani R, Rao BM, Bauwens C, Yin T, Wood GA, Nagy A, et al. Niche-mediated control of human embryonic stem cell self-renewal and differentiation. The EMBO Jorunal. 2007;26:4744–55. doi: 10.1038/sj.emboj.7601896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ungrin MD, Joshi C, Nica A, Bauwens C, Zandstra PW. Reproducible, ultra high- throughput formation of multicellular organization from single cell suspension-derived human embryonic stem cell aggregates. PLoS ONE. 2008;3:e1565. doi: 10.1371/journal.pone.0001565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mohr JC, de Pablo JJ, Palecek SP. 3-D microwell culture of human embryonic stem cells. Biomaterials. 2006;27:6032–42. doi: 10.1016/j.biomaterials.2006.07.012. [DOI] [PubMed] [Google Scholar]
- 26.Khademhosseini A, Ferreira L, Blumling J, III, Yeh J, Karp JM, Fukuda J, et al. Co-culture of human embryonic stem cells with murine embryonic fibroblasts on microwell-patterned substrates. Biomaterials. 2006;27:5968–77. doi: 10.1016/j.biomaterials.2006.06.035. [DOI] [PubMed] [Google Scholar]
- 27.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 28.Kubalak SW, Miller-Hance WC, O'Brien TX, Dyson E, Chien KR. Chamber specification of atrial myosin light chain-2 expression precedes septation during murine cardiogenesis. J Biol Chem. 1994;269:16961–70. [PubMed] [Google Scholar]
- 29.Kehat I, Kenyagin-Karsenti D, Snir M, Segev H, Amit M, Gepstein A, et al. Human embryonic stem cells can differentiate into myocytes with structural and functional properties of cardiomyocytes. J Clin Invest. 2001;108:407–14. doi: 10.1172/JCI12131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.He JQ, Ma Y, Lee Y, Thomson JA, Kamp TJ. Human embryonic stem cells develop into multiple types of cardiac myocytes: action potential characterization. Circ Res. 2003;93:32–9. doi: 10.1161/01.RES.0000080317.92718.99. [DOI] [PubMed] [Google Scholar]
- 31.Osafune K, Caron L, Borowiak M, Martinez RJ, Fitz-Gerald CS, Sato Y, et al. Marked differences in differentiation propensity among human embryonic stem cell lines. Nat Biotechnol. 2008;26:313–5. doi: 10.1038/nbt1383. [DOI] [PubMed] [Google Scholar]
- 32.Tomescot A, Leschik J, Bellamy V, Dubois G, Messas E, Bruneval P, et al. Differentiation in vivo of cardiac committed human embryonic stem cells in postmyocardial infarcted rats. Stem Cells. 2007;25:2200–5. doi: 10.1634/stemcells.2007-0133. [DOI] [PubMed] [Google Scholar]
- 33.Bauwens CL, Peerani R, Niebruegge S, Woodhouse KA, Kumacheva E, Husain M, et al. Control of human embryonic stem cell colony and aggregate size heterogeneity influences differentiation trajectories. Stem Cells. 2008;26:2300–10. doi: 10.1634/stemcells.2008-0183. [DOI] [PubMed] [Google Scholar]
- 34.Rudy-Reil D, Lough J. Avian precardiac endoderm/mesoderm induces cardiac myocyte differentiation in murine embryonic stem cells. Circ Res. 2004;94:e107–16. doi: 10.1161/01.RES.0000134852.12783.6e. [DOI] [PubMed] [Google Scholar]
- 35.Schultheiss TM, Burch JB, Lassar AB. A role for bone morphogenetic proteins in the induction of cardiac myogenesis. Genes Dev. 1997;11:451–62. doi: 10.1101/gad.11.4.451. [DOI] [PubMed] [Google Scholar]
- 36.Sugi Y, Lough J. Anterior endoderm is a specific effector of terminal cardiac myocyte differentiation of cells from the embryonic heart forming region. Dev Dyn. 1994;200:155–62. doi: 10.1002/aja.1002000207. [DOI] [PubMed] [Google Scholar]
- 37.Climent S, Sarasa M, Villar JM, Murillo-Ferrol NL. Neurogenic cells inhibit the differentiation of cardiogenic cells. Dev Biol. 1995;171:130–48. doi: 10.1006/dbio.1995.1266. [DOI] [PubMed] [Google Scholar]
- 38.Cohen ED, Tian Y, Morrisey EE. Wnt signaling: an essential regulator of cardiovascular differentiation, morphogenesis and progenitor self-renewal. Development. 2008;135:789–98. doi: 10.1242/dev.016865. [DOI] [PubMed] [Google Scholar]
- 39.Wagner M, Siddiqui MA. Signal transduction in early heart development (I): cardiogenic induction and heart tube formation. Exp Biol Med (Maywood ) 2007;232:852–65. [PubMed] [Google Scholar]
- 40.Conley BJ, Trounson AO, Mollard R. Human embryonic stem cells form embryoid bodies containing visceral endoderm-like derivatives. Fetal Diagn Ther. 2004;19:218–23. doi: 10.1159/000076701. [DOI] [PubMed] [Google Scholar]
- 41.Coucouvanis E, Martin GR. Signals for death and survival: a two-step mechanism for cavitation in the vertebrate embryo. Cell. 1995;83:279–87. doi: 10.1016/0092-8674(95)90169-8. [DOI] [PubMed] [Google Scholar]
- 42.Li X, Chen Y, Scheele S, Arman E, Haffner-Krausz R, Ekblom P, et al. Fibroblast growth factor signaling and basement membrane assembly are connected during epithelial morphogenesis of the embryoid body. J Cell Biol. 2001;153:811–22. doi: 10.1083/jcb.153.4.811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Conley BJ, Ellis S, Gulluyan L, Mollard R. BMPs regulate differentiation of a putative visceral endoderm layer within human embryonic stem-cell-derived embryoid bodies. Biochem Cell Biol. 2007;85:121–32. doi: 10.1139/o06-145. [DOI] [PubMed] [Google Scholar]
- 44.Yu J, Vodyanik MA, Smuga-Otto K, Antosiewicz-Bourget J, Frane JL, Tian S, et al. Induced pluripotent stem cell lines derived from human somatic cells. Science. 2007;318:1917–20. doi: 10.1126/science.1151526. [DOI] [PubMed] [Google Scholar]
- 45.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, et al. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–72. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 46.Zhang J, Wilson GF, Soerens AG, Koonce CH, Yu J, Palecek SP, et al. Functional cardiomyocytes derived from human induced pluripotent stem cells. Circ Res. 2009;104:e30–41. doi: 10.1161/CIRCRESAHA.108.192237. [DOI] [PMC free article] [PubMed] [Google Scholar]