Abstract
Guanidinylated bioreducible polymer (GBP) was developed for gene delivery systems utilizing cellular penetrating ability of guanidine groups. GBP could retard pDNA from a weight ratio of 5 completely in agarose gel electrophoresis but pDNA was released from GBP polyplexes in reducing condition (2.5 mM DTT) due to their biodegradation. GBP also could construct 200 nm-sized and positively charged (~30 mV) polyplex nanoparticles with pDNA. The cytotoxicity of GBP was found to be minimal and GBP showed about 8 folds improved transfection efficiency than a scaffold polymer, poly(cystaminebisacrylamide-diaminohexane) (poly(CBA-DAH)) and even higher transfection efficiency than PEI25k in mammalian cell lines. Its high cellular uptake efficiency (96.1 %) and strong nuclear localization ability for pDNA delivery due to the structural advantage of bioreducible polymer and guanidine groups were also identified, suggesting GBP is a promising candidate for efficient gene delivery systems.
Keywords: bioreducible polymer, gene transfer, guanidinylation, cytotoxicity, nuclear localization ability
1. Introduction
For non-viral gene delivery systems, lots of cationic polymers have been developed and used to form nanoparticles with genetic materials such as plasmid DNA (pDNA) or siRNA via electrostatic interaction. These polymeric gene delivery carriers were known to have large gene delivery capacities, to be non-immunogenic and easy to manufacture, appearing as promising alternatives of viral gene delivery carriers [1,2]. Moreover, polymers possess multi-functional groups modifiable with biofunctional moieties such as endosome buffering moieties, targeting ligands or intracellular trafficking signals [3,4]. Therefore, these polyplex nanoparticles comprising polymers and nucleic acids can mimic viral vectors with efficient transduction ability. However, several drawbacks such as cytotoxicity and relatively low transfection efficiency limited their applications and there have been many researches developing novel polymeric gene delivery carriers to overcome these defects [5].
Introduction of biodegradability such as ester [6–9], phosphoester [10], or acetal [11] bonds to polymeric vectors has been proved to be effective for this goal by decreasing the cytotoxicity and controlling the release of genetic materials from polyplexes with response to the external stimulus such as acidification of endosome after endocytosis of polyplexes. End group-modified hydrolytically degradable poly(β-amino ester)s were reported to facilitate gene delivery to even human embryonic stem cells without causing side effects or nonspecific differentiation because of their minimal toxicity and high transfection efficiency [12]. Meanwhile, various bioreducible polymers containing disulfide bonds have been developed for gene delivery systems because they are also known to be degraded by intracellular reducing agents such as glutathione and possess more convenience for handling due to their low susceptibility to humidity in comparison with hydrolytic polymers [13–16]. Recently, another bioreducible polymers have been synthesized by Michael reaction of commercial monomers such as N,N′-cystaminebisacrylamide for gene delivery systems [17–19]. Besides simplicity and high yield of synthesis, their low cytotoxicity due to biodegradability and high transfection efficiency in mammalian cells showed a potential for efficient gene delivery carriers.
Another one of the strategies for enhancing transfection ability of polymeric vectors is to modify the primary amine groups of polymers with guanidine groups. The guanidine groups are known to function importantly in cellular penetrating peptides (CPPs) [20]. These peptides such as Tat sequence or Antennapedia homeodomain have received intense interest in drug and gene delivery field due to their efficient translocation of non-permeant molecules into cells [21–23]. Several works reported that guanidinylation of polymeric vectors could improve the transfection efficiency greatly, meaning that not only CPP conjugation to the polymers but also simple chemical modification such as guanidinylation also can transport polyplex nanoparticles into cells efficiently [24–26].
Therefore, combining the unique properties of bioreducible polymers with the guanidine function, it is expected that this newly synthesized guanidinylated bioreducible polymer (GBP) would construct polyplexes with pDNA showing high transfection efficiency and sustaining its low cytotoxicity.
Here, we report the first attempt to modify bioreducible polymer with gianidinylation and characterize polyplexes consisted of this novel polymer and pDNA, which includes physiochemical properties, cytotoxicity, transfection efficiency. Additionally, we examine the cellular uptake profile and intracellular trafficking of GBP polyplexes and demonstrate their high nuclear localization ability for pDNA delivery due to guanidinylation for the first time.
2. Materials and methods
2.1. Materials
Hyperbranched poly(ethylenimine) (bPEI, 25 kDa), tert-Butyl-N-(6-aminohexyl)carbamate (N-Boc-1,6-diaminohexane, N-Boc-DAH), 1H-pyrazole-1-carboxamidine hydrochloride, trifluoroacetic acid (TFA), triisobutylsilane, dithiothreitol (DTT), N,N-diisopropylethylamine (DIPEA), and 3-[4,5-dimethylthiazol-2-yl]-2,5-diphenyltetrazolium bromide (MTT) were purchased from Sigma-Aldrich (St. Louis, MO). N,N′-cystaminebisacrylamide (CBA) was purchased from PolySciences, Inc. (Warrington, PA). The plasmid pCMV-Luc, containing a firefly luciferase reporter gene was amplified in E.coli DH5α and isolated by standard Maxiprep kit (Invitrogen, Carlsbad, CA). Luciferase assay system and reporter lysis buffer were purchased from Promega (Madison, WI). SYBR safe DNA gel stain (10,000X in DMSO), fetal bovine serum (FBS), Dulbecco’s phosphate buffered saline (DPBS), and Dulbecco’s modified Eagle’s medium (DMEM) were purchased from Invitrogen (Carlsbad, Ca). BCA™ protein assay kit was purchased from PIERCE (Rocford, Il). YOYO-1 iodide (1 mM solution in DMSO) was purchased from Molecular Probes (Eugene, OR). All other chemicals were purchased and used without any further purification.
2.2. Synthesis of GBP
The bioreducible scaffold polymer, poly(CBA-DAH) was synthesized by Michael reaction of equivalent moles of N-Boc-DAH and CBA in MeOH/H2O solution (9:1, v/v). Polymerization reaction was conducted in the absence of light under nitrogen atmosphere at 60 °C for 5 days. Then, 10 % mole of N-Boc-DAH was added to the reaction mixture to mask unreacted acrylamide groups of polymer products with amine monomers and the reaction was further maintained for 2 days. After precipitation of reaction mixture with excess of diethyl ether, the Boc groups of product was removed by the reagent solution (TFA: triisobutylsilane: H2O = 95: 2.5: 2.5, v/v) at ice bath for 30 min. After additional precipitation with diethyl ether, the polymer product was dialyzed against ultra-pure water with dialysis membrane (MWCO = 1000, Spectrum Laboratories, Inc., Rancho Dominguez, CA), followed by lyophilization to leave poly(CBA-DAH) as sticky solid.
Then, the primary amines of poly(CBA-DAH) was guanidinylated in water with equivalent moles of 1H-pyrazole-1-carboxamidine hydrochloride and DIPEA. After 1 day of reaction at room temperature, the reaction mixture was dialyzed against ultra-pure water overnight and lyophilized before use for analysis and assay. The syntheses of poly(CBA-DAH) and guanidinylated bioreducible polymer (abbreviated to GBP) were confirmed by 1H NMR (400 MHz, D2O).
2.3. Molecular weight measurement of GBP
The molecular weight of GBP was measured by size-exclusion chromatography (SEC) (Superose 12 column, calibrated with standard poly[N-(2-hydroxypropyl)methacrylamide] (pHPMA)) using AKTA FPLC system. The polymer was dissolved at a concentration of 3 mg/mL. 0.1 M acetate buffer (30 % Acetonitrile, v/v, pH 6.5) was used as an eluent. Flow rate was 0.4 mL/min.
2.4. Agarose gel electrophoresis
pDNA condensing ability of GBP was examined by agarose gel electrophoresis. Polyplexes were prepared in Hepes buffered saline (10 mM Hepes, 1 mM NaCl, pH 7.4) at various weight ratios ranging from 0.5 to 20. Agarose gel (0.7 %, w/v) containing SYBR safe DNA gel stain solution was prepared in TAE (Tris-Acetate-EDTA) buffer. After 30 min of incubation at room temperature, the polyplex solutions were electrophoresed at 100 V for 30 min. In addition, the identical polyplexes were incubated in the presence of 2.5 mM DTT for 30 min at room temperature and electrophoresed in order to investigate the effect of biodegradation on pDNA condensing profile of GBP in reducing environment. Non-bioreducible PEI25k polyplex (weight ratio = 1) was used as a control. The location of pDNA bands was visualized by UV illuminator (Gel Documentation Systems, Bio-Rad, Hercules, CA).
2.5. Zeta-potential values and average sizes measurements of GBP polyplexes
Zeta-potential values and average sizes of GBP polyplexes were examined by using Zetasizer 3000 (Malvern Instruments, USA) with a He–Ne Laser beam (633 nm, fixed scattering angle of 90°) at 25 °C. 0.5 mL of polyplex solutions (10 μg pDNA) were prepared in Hepes buffered saline at various weight ratios ranging from 0.5 to 40. After 30 min incubation, polyplex solutions were diluted to final volume of 4 mL before measurement. Measured Zeta-potential values and average sizes were presented as the average values of 5 runs.
2.6. TEM observation
5 μL of GBP polyplex solutions (0.5 μg pDNA) are prepared and deposited on TEM copper grid plates. The identical polyplexe solutions were prepared for TEM after incubation in the presence of 2.5 mM DTT for 30 min at room temperature. The samples were then stained with filtered phosphotungstenic acid (PTA) for 30 s. Images were visualized using a Technai T12 scope (EFM) with an accelerating voltage of 80kV.
2.7. Cell culture
C2C12 mouse myoblast cells, HEK293 human embryonic kidney cells and HepG2 human hepatocellular carcinoma cells were obtained from American Type Culture Collection (Manassas, VA). They were maintained in DMEM medium supplemented with 10 % FBS in a humidified atmosphere containing 5 % CO2 at 37 °C.
2.8. Cytotoxicity
The cytotoxicity of the polymers was measured by MTT assay. C2C12, HEK293 and HepG2 cells were seeded in a 96-well tissue culture plate at 1 × 104 cells per well in 90 μL DMEM medium containing 10% FBS. Cells achieving 70~80 % confluence after 24 h were exposed to 10 μL of the polymer solutions having various concentration for 4 h in serum-free DMEM medium. After exchange of medium for fresh DMEM medium containing 10% FBS, the cells were further maintained for 24 h. Then, 25 μL of stock solution of MTT (2 mg/ml in PBS) were added to each well. After 2 h of incubation at 37 °C, each medium was removed carefully and 150 μL of DMSO was added to each well to dissolve the formazan crystal formed by proliferating cells. Absorbance was measured at 570 nm using a microplate reader (Model 680, Bio-Rad Lab, Hercules, CA) and recorded as a percentage relative to untreated control cells. All experiments were performed in quadruplicate.
2.9. Transfection experiments in vitro
C2C12, HEK293 and HepG2 cells were seeded at a density of 5 × 104 cells/well in a 24-well plate in DMEM medium containing 10% FBS and grown to reach 70~80 % confluence prior to transfection. Before transfection, medium of each well was exchanged for fresh serum-free. The cells were treated with polyplex solution (0.5 μg pMV-Luc) at different weight ratios for 4 h at 37 °C. After exchange with fresh medium containing 10 % FBS, cells were further incubated for 2 days before assay. Then, the growth medium was removed and the cells were rinsed with DPBS and shaken for 30 min at room temperature in 100 μL of Reporter Lysis Buffer. Luciferase activity was measured by a luminescence assay and a protein quantification assay was performed using a BCA™ Protein Assay Reagent Kit. The luciferase activity of 25 μL cell lysate was measured by using 100 μL of luciferase assay reagent on a luminometer (Dynex Technologies Inc., Chantilly, CA). The final results were reported in terms of RLU/mg cellular protein. All experiments were performed in triplicate.
2.10. Flow cytometry
C2C12 cells were seeded at a density of 1 × 05 cells/well in a 12-well plate in DMEM medium containing 10 % FBS and grown to reach 70~80% confluence prior to transfection. Before transfection, medium of each well was exchanged for fresh serum-free medium. pDNA was labeled with YOYO-1 iodide (1 molecule of the dye per 50 base pairs of the nucleotide). The cells were treated with polyplex solutions (1 μg of pDNA) at different weight ratios for 4 h at 37 °C. Then, medium was aspirated off from the wells and the cells were washed two times with ice-cold DPBS. After trypsinization, the cells were suspended in 1 mL DPBS.
The cellular uptake of fluorescence-labeled polyplexes was examined by using the BD FACScan analyzer (Becton Dickinson, San Jose, CA) at a minimum of 1 × 104 cells gated per sample. Analysis was performed by using Becton Dickinson CellQuest software. Data were processed by using Windows Multiple Document interface software (WinMDI).
2.11. Confocal microscopy
For confocal microscopy, C2C12 cells were seeded in confocal imaging dishes (Glass Bottom microwells, MatTek Corp., Ashland, MA) at a density of 5 × 104 cells per dish. The polyplexes containing 1 μg of YOYO-1 iodide labeled pDNA (1 molecule of the dye per 50 base pairs of the nucleotide) were prepared and treated in cells in the absence of serum (PEI25k: weight ratio=1, poly(CBA-DAH) and GBP: weight ratio=40). After 4 h of incubation at 37 °C, cells were washed with cold DPBS and further incubated for 3 h in DMEM medium (10 % FBS). The localization of YOYO-1 iodide labeled pDNA within the cells was observed by a confocal laser scanning microscope (FV1000-XY confocal Olympus IX81 microscope, Melville, NY) with a 60× oil immersion objective lens using an argon laser (Ex = 488 nm).
3. Results and discussion
3.1. Synthesis and characterization of GBP
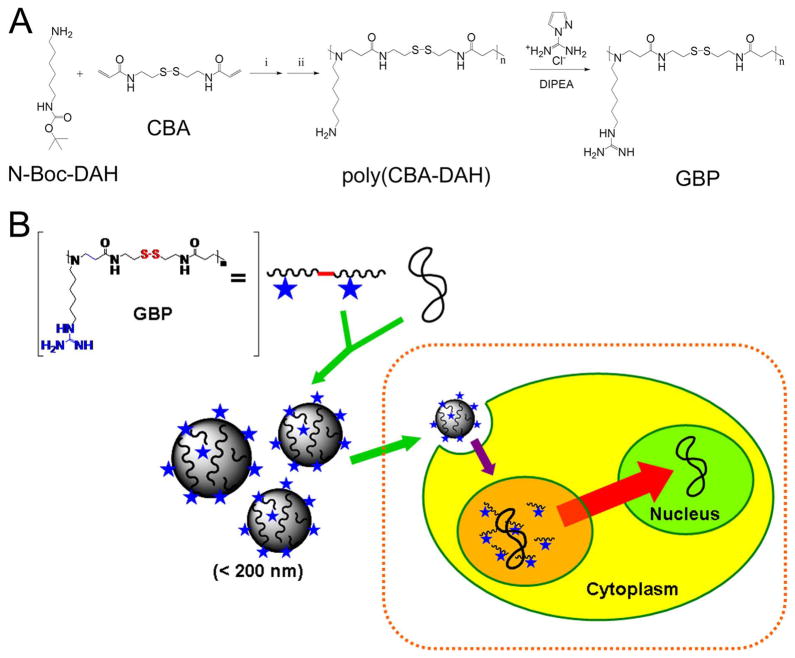
The bioreducible poly(CBA-DAH) polymer was selected as a scaffold polymer based on the previous report showing its high transfection efficiency and minimal cytotoxicity [27]. Moreover, it possesses modifiable primary amines. Michael reaction of N,N′-cystaminebisacrylamide (CBA) and N-Boc-1,6-diaminohexane (N-Boc-DAH) were conducted at 1:1 molar ratio to synthesize poly(CBA-DAH). After removal of Boc groups, exposed primary amines of poly(CBA-DAH) were then modified into guanidine groups with 1H-pyrazole-1-carboxamidine hydrochloride and DIPEA. Figure 1 shows the synthetic scheme of polymers and the schematic diagram of gene delivery based on GBP polyplexes. The guanidinylation of polymer was confirmed by 1H NMR as followings. G means guanidine groups.
Figure 1.
(A) synthetic scheme of guanidinylated bioreducible polymer (GBP). i) MeOH: H2O = 9:1, v/v, 60 °C, 5 days, ii) TFA: triisobutylsilane: H2O = 95: 2.5: 2.5, v/v, 0 °C, 30 min. (B) schematic diagram of gene delivery based on GBP polyplexes.
GBP; 1H NMR(D2O): δ (G-NCH2CH2CH2CH2CH2CH2N) = 1.22, δ (G-NCH2CH2CH2CH2CH2CH2N) = 1.36~1.54, δ (NCH2CH2CONHCH2CH2SS) = 2.32, δ (CH2SSCH2) = 2.73, δ (protons next to tertiary amines) = 2.75~2.88, δ (G-NCH2CH2CH2CH2CH2CH2N) = 3.05, δ (NCH2CH2CONHCH2CH2) = 3.40
The modification yield of guanidinylation was estimated to be 85 % by comparing the integration values of proton peak next to guanidine (3.05 ppm) and proton peak of DAH methylene protons (1.22 ppm).
The molecular weight of polymer was measured by size-exclusion chromatography (SEC) combined with AKTA FPLC system. Poly[N-(2-hydroxypropyl)methacrylamide] (pHPMA) was used as a standard. Mw of polymer was evaluated to be 5.35 × 103 Da and its PDI value was 1.11.
3.2. Agarose gel electrophoresis
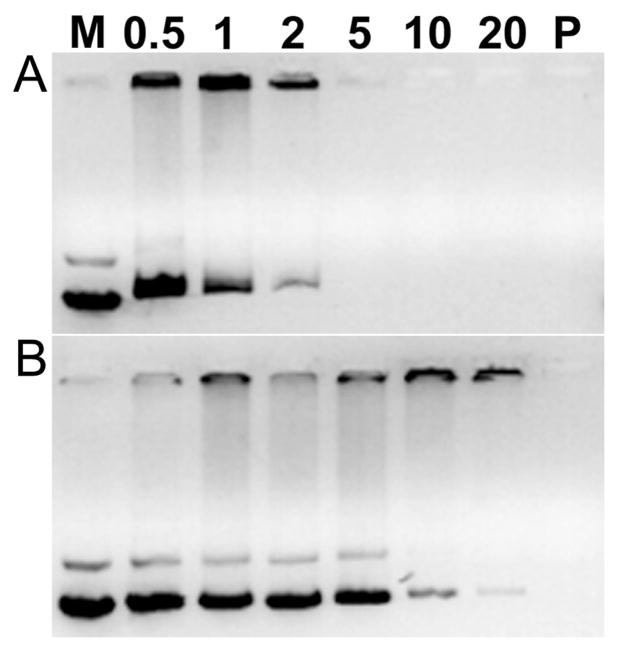
Agarose gel electrophoresis was performed in order to examine the pDNA condensing ability of GBP for polyplexes formation. It is well-known that cationic polymers can retard negatively charged pDNA by forming complexes via electrostatic interaction. After 30 min of polyplex formation, it was found that GBP completely retarded pDNA from a weight ratio of 5 in the absence of DTT (Figure 2A), displaying that it could condense pDNA succsessfully. However, pDNA released from GBP polyplexes was still observed even at a weight ratio of 20 in the presence of 2.5 mM DTT, a well-known reducing agent (Figure 2B). Non-bioreducible PEI25k (P in Figure 2) was observed to retard pDNA at a weight ratio of 1 irrespective of DTT condition as expected. It means that GBP polyplexes can be degraded and release pDNA under reducing environment because of reducible cleavages of disulfide bonds within the GBP backbone. Therefore, it is also suggested that GBP polyplexes can be biodegraded in the cytoplasm containing 0.1~10 mM glutathione after cellular uptake. Considering the fact that the biodegradation of polymer into small molecules can decrease the cytotoxicity and lead to efficient release of pDNA from polyplex [28], this finding deduces that polymer also may have these benefits for gene delivery.
Figure 2.
Agarose gel electrophoresis results of GBP polyplexes (A) without and (B) with 2.5 mM DTT. M: pDNA marker. Numbers mean weight ratios of polyplexes. P: PEI25k polyplexes (weight ratio = 1).
3.3. Zeta-potential values and average sizes of GBP polyplexes
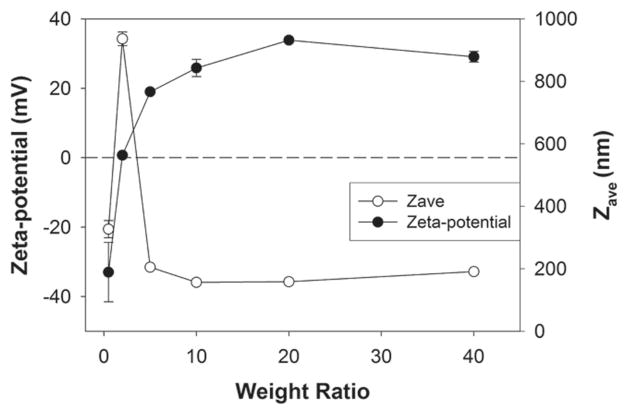
We surveyed Zeta-potential values and average sizes of GBP polyplexes. It was reported that the positive charges of polyplexes can improve their adhesion onto cellular membrane, increasing the chance of cellular uptake [29] and the proper size (< 200 nm) of polyplexes may be required for efficient cellular uptake of polyplex [30]. At a weight ratio of 0.5, Zeta-potential value and average size of GBP polyplexes were measured to be about −33.0 mV and 327.0 nm, respectively, indicating that negatively charged pDNA is not condensed with cationic polymers completely and on the contrary pDNA wraps polyplex particles at that ratio (Figure 3). Interestingly, when Zeta-potential value was almost zero (0.7 mV) at a weight ratio of 2, average size of GBP polyplexes was found to be 936.1 nm. It means that electrically neutral polyplex particles construct large aggregates via hydrophobic interactions in aqueous solution. From a weight ratio of 5 to 40, GBP polyplex particles showed positive Zeta-potential values converging 30 mV and almost 200 nm-size, suggesting that GBP condenses pDNA completely and form positively charged and stable polyplex nanoparticles with pDNA. This result exactly corresponds with agarose gel electrophoresis result. Therefore, it was confirmed that GBP can form positively charged and 200 nm-sized polyplex particles with pDNA, satisfying the requirements mentioned above.
Figure 3.
Average size and Zeta-potential value measurements of GBP polyplexes.
3.4. TEM observation of GBP polyplexes
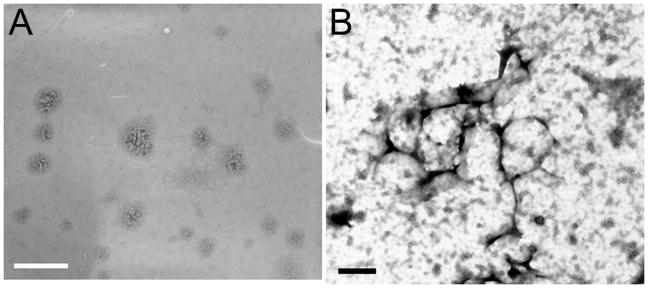
Morphology of GBP polyplex nanoparticles were also examined by TEM. In Figure 4A, GBP polyplex nanoparticles appeared as spheres with sizes less than 250 nm. It is believed that pDNA (white rope-like material) was wound with GBP (gray material) like the thread of bobbin in these polyplex nanostructures. Contrary to the former observation, after treatment of 2.5 mM DTT for 30 min to the same sample, TEM could detect the scattered pDNA (black rope-like material) and some spherical structures (black circle) regarded as non-degraded polyplex particles (Figure 4B). This result strongly supports the presumption that pDNA would be released from GBP polyplex nanoparticles due to the degradation by DTT reduction. Theses observation agrees well with previous agarose gel electrophoresis and size measurement experiment results.
Figure 4.
TEM images of GBP polyplexes (A) without and (B) with 2.5 mM DTT. Scale bars (A: white, B: black) are 500 nm.
3.5. Cytotoxicity measurement by MTT assay
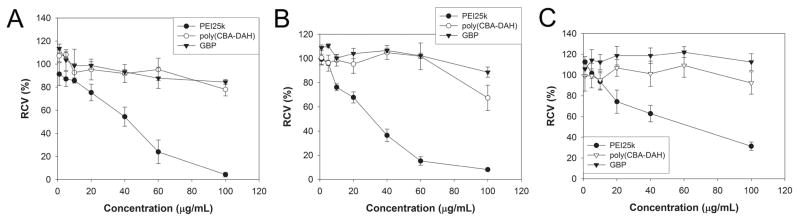
We conducted MTT assay in order to examine the cytotoxicity of polymer. C2C12 mouse myoblast cells (Figure 5A), HEK293 human embryonic kidney cells (Figure 5B) and HepG2 human hepatocellular carcinoma cells (Figure 5C) were used. PEI25k and poly(CBA-DAH) were used as controls. RCV (relative cell viability) of PEI25k was abruptly decreased in three cell lines as the concentration was increased, showing its severe cytotoxicity. In contrast with PEI25k, GBP displayed high RCV (above 85 %) even at 100 μg/mL concentration in all cell lines. HepG2 cells seem to have the more tolerance to the cytotoxicity of cationic polymers than other cells examined. Backbone polymer, poly(CBA-DAH) was also found to have little cytotoxicity due to its biodegradation in cytoplasm, which was reported in our previous work [27]. From this result, it was investigated that GBP has low cytotoxicity despite guanidinylation.
Figure 5.
MTT assay results in (A) C2C12 cells, (B) HEK 293 cells, and (C) HepG2 cells.
3.6. Transfection experiments in vitro
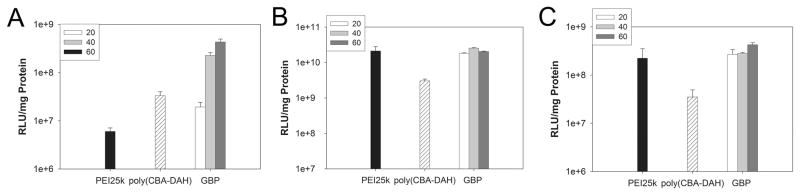
Transfection experiments were performed in same three cell lines. PEI25k polyplex and poly(CBA-DAH) polyplex were prepared at a weight ratio of 1 and 40, respectively as controls. In C2C12 cells, the transfection efficiency of GBP was almost 40 times higher than that of PEI25k (Figure 6A). In HEK293 cells (Figure 6B) and HepG2 cells (Figure 6C), GBP showed high transfection efficiency similar to or higher than PEI25k which is known to be a one of the most highly efficient transfection reagent. In comparison with poly(CBA-DAH), GBP displayed almost 8 times increased transfection efficiency in all cell lines. Therefore, it was identified that polymer has highly improved transfection efficiency due to guanidinylation and possesses great potential to apply for gene delivery systems.
Figure 6.
Transfection experiment results in (A) C2C12 cells, (B) HEK 293 cells, and (C) HepG2 cells. Numbers in small boxes mean weight ratios of GBP polyplexes. PEI25k polyplexes and poly(CBA-DAH) polyplexes were prepared at a weight ratio of 1 and 40, respectively.
3.7. Cellular uptake of GBP polyplexes by flow cytometry
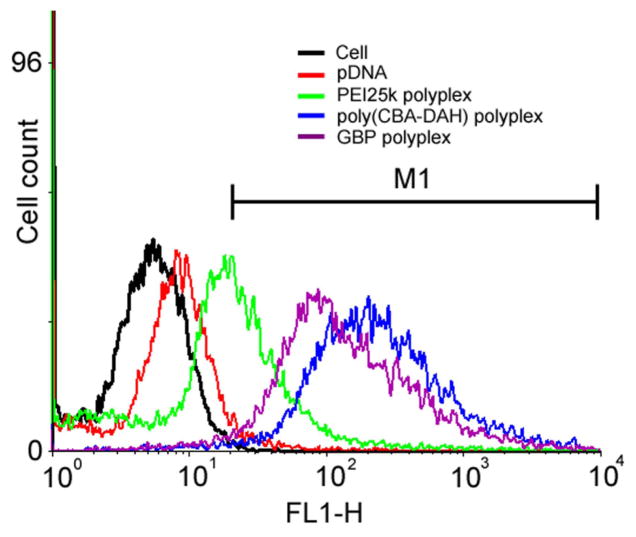
In order to investigate the factors inducing the high transfection efficiency of GBP, cellular uptake of polyplexes was investigated by flow cytometry in C2C12 cells. Polyplexes were prepared with YOYO-1 iodide-labeled pDNA. Each cellular uptake value (%) was calculated by setting up M1 region as a gate. As shown in Figure 7, the cellular uptake value of PEI25k polyplexes was found to be 44.8 % when those of untreated cells and only pDNA were 0.64 % and 4.92 %, respectively. However, both poly(CBA-DAH) and GBP polyplexes displayed 2 times or higher cellular uptake profile (97.2 % and 96.1 %, respectively) than PEI25k polyplexes. Interestingly, this result shows that the poly(CBA-DAH) polyplexes originally possess good cellular penetrating ability despite the lack of cellular penetrating moieties such as guanidine. Regarding the fact that cellular uptake of polyplexes is one of barriers which polyplexes must overcome for transfection, this result partially explains their higher transfection efficiency in comparison with that of PEI25k in C2C12 cells but it still could not illustrate the increased transfection efficiency of GBP than that of poly(CBA-DAH). Previously, we reported that cellular uptake of arginine-grafted poly(CBA-DAH) (ABP) was almost similar with that of poly(CBA-DAH) regardless of transfection efficiency difference between them, hypothesizing that greatly improved transfection efficiency of ABP may be caused by other factors such as nuclear localization ability of arginine moieties [31].
Figure 7.
Flow cytometry results of polyplexes in C2C12 cells. M1 region was set up as a gate for the estimation of cellular uptake values.
3.8. Intracellular trafficking of polyplexes by confocal microscopy
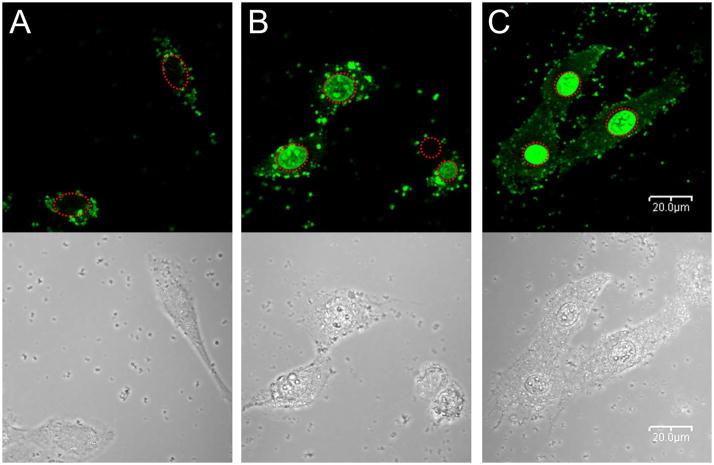
So, we observed the intracellular trafficking of polyplexes by confocal microscopy in C2C12 cells. As shown in Figure 8A, almost all PEI25k polyplexes were found to be accumulated in cytoplasm as weak green dots (fluorescence from YOYO-1 iodide-labeled pDNA), not in cell nuclei at this time. They have particle shapes, meaning that pDNA was still condensed with PEI25k. However, for the case of poly(CBA-DAH) polyplex, some particles and widely distributed fluorescence were present in both cytoplasm and cell nuclei (Figure 8B). Therefore, first it is confirmed that bioreducible polymers can be degraded after cellular uptake due to the reductive cleavages of internal disulfide bonds by intracellular glutathione. On the other hand, very strong fluorescence of YOYO-1 labeled pDNA was observed to be mainly located in cell nuclei for the case of GBP polyplexes (Figure 8C). Considering the structural differences between poly(CBA-DAH) and GBP, this intense accumulation of pDNA in cell nuclei is surely induced by nuclear localization ability of guanidine groups as well as fast release of pDNA from destabilized polyplexes by GBP biodegradation. This nuclear localization ability is strongly believed to be able to induce greatly enhanced transfection of GBP in comparison with poly(CBA-DAH), because the translocation of pDNA across the nuclear membrane is another barrier for gene delivery. There is a good possibility that the degraded GBP molecules may enter cell nuclei, still binding to pDNA, based on some works showing molecular transporters containing guanidine groups can target and permeate cell nuclei successfully [32,33], although the mechanism is not clearly known. Some studies reported the chemical conjugation of nuclear localization signal (NLS) [34,35] or the insertion of nuclear factor κB (NF κB) binding sites to DNA [36] could improve the nuclear entry. However, to our knowledge, this transporting pDNA preferentially into cell nuclei without any modifications by employing guanidinylated bioreducible polymeric nanoparticles is reported for the first time.
Figure 8.
Intracellular trafficking observation by confocal microscopy in C2C12 cells. YOYO-1 iodide-labeled pDNA was used. (A) PEI25k polyplexes-treated cells, (B) poly(CBA-DAH) polyplexes-treated cells, and (C) GBP polyplexes-treated cells. Red dashed circles indicate cell nuclei.
Interestingly, it was also informed by using confocal microscopy that arginine-grafted bioreducible polymer (ABP), which is structurally similar with GBP, can efficiently localize siRNA only in cytoplasm, excluding cell nuclei [37]. This obvious discordance between the delivering location of pDNA and siRNA by similar cellular penetrating polymers shows the evident difference of intracellular fate between pDNA and siRNA after cellular uptake.
4. Conclusions
We synthesized guanidinylated bioreducible polymer for gene delivery systems in order to take the advantages for combining the biodegradability of reducible disulfide bonds and the cell penetrating ability of guanidine groups. It was found that GBP could form polyplex nanoparticles with about 200 nm-size and 30 mV Zeta-potential value. However, in reducing environment (2.5 mM DTT), GBP could not condense pDNA successfully due to the biodegradation of GBP polyplexes. Its cytotoxicity still remained minimal like scaffold polymer, poly(CBA-DAH). GBP polyplexes displayed high transfection efficiency even higher than PEI25k or poly(CBA-DAH) polyplexes. The high cellular uptake profile and strong pDNA nuclear localization ability of GBP polyplexes were discovered and it is thought to be due to the structural benefit from scaffold bioreducible polymer and the guanidine groups. Therefore, it can be concluded that the strategy for the synthesis of GBP by introduction of biodegradation moieties and guainidine groups is very successful for the development of efficient gene delivery systems.
Acknowledgments
This work was supported by NIH grant (DK 077703) and Research fund (WCU 200900000000024) of the Ministry of Education, Science and Technology, Korea. We thank Monika Sima for FPLC-SEC experiments and thank Prof. You Han Bae for generous permission of Zeta-sizer usage and also thank Dr. James Yockman for helpful discussion.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Contributor Information
Tae-il Kim, Email: Taeil.Kim@utah.edu, Center for Controlled Chemical Delivery, Department of Pharmaceutics and Pharmaceutical Chemistry, University of Utah, Salt Lake City, UT 84112, USA, Phone: 801-585-3707, Fax: 801-581-7848
Minhyung Lee, Email: minhyung@hanyang.ac.kr, Department of Bioengineering, College of Engineering, Hanyang University, Seoul, Korea, Phone: 82-2-2220-0424, Fax: 82-2-2220-1998
Sung Wan Kim, Email: SW.Kim@pharm.utah.edu, Center for Controlled Chemical Delivery, Department of Pharmaceutics and Pharmaceutical Chemistry, University of Utah, Salt Lake City, UT 84112, USA, Phone: 801-581-6654, Fax: 801-581-7848
References
- 1.Luo D, Saltzman WM. Synthetic DNA delivery systems. Nat Biotechnol. 2000;18:33–37. doi: 10.1038/71889. [DOI] [PubMed] [Google Scholar]
- 2.Liu F, Huang L. Development of non-viral vectors for systemic gene delivery. J Control Release. 2002;78:259–266. doi: 10.1016/s0168-3659(01)00494-1. [DOI] [PubMed] [Google Scholar]
- 3.Park TG, Jeong JH, Kim SW. Current status of polymeric gene delivery systems. Adv Drug Deliv Rev. 2006;58:467–486. doi: 10.1016/j.addr.2006.03.007. [DOI] [PubMed] [Google Scholar]
- 4.Merdan T, Kopec ek J, Kissel T. Prospects for cationic polymers in gene and oligonucleotide therapy against cancer. Adv Drug Deliv Rev. 2002;54:715–758. doi: 10.1016/s0169-409x(02)00046-7. [DOI] [PubMed] [Google Scholar]
- 5.Wong SY, Pelet JM, Putnam D. Polymer systems for gene delivery-Past, present, and future. Prog Polym Sci. 2007;32:799–837. [Google Scholar]
- 6.Lynn DM, Langer R. Degradable poly(β-amino esters): synthesis, characterization, and self-assembly with plasmid DNA. J Am Chem Soc. 2000;122:10761–10768. [Google Scholar]
- 7.Tinsley-Brown AM, Fretwell R, Dowsett AB, Davis SL, Farrar GH. Formulation of poly(D, L-lactic-co-glycolic acid) microparticles for rapid plasmid DNA delivery. J Control Release. 2000;66:229–241. doi: 10.1016/s0168-3659(99)00275-8. [DOI] [PubMed] [Google Scholar]
- 8.Lim Y-b, Kim C-h, Kim K, Kim SW, Park J-s. Development of a safe gene delivery system using biodegradable polymer, poly[α-(4-aminobutyl)-L-glycolic acid] J Am Chem Soc. 2002;122:6524–6525. [Google Scholar]
- 9.Kim T-i, Seo HJ, Choi JS, Yoon JK, Baek J-u, Kim K, et al. Synthesis of biodegradable cross-linked poly(β-amino ester) for gene delivery and its modification, inducing enhanced transfection efficiency and stepwise degradation. Bioconjugate Chem. 2005;16:1140–1148. doi: 10.1021/bc0497012. [DOI] [PubMed] [Google Scholar]
- 10.Wang J, Mao H, Leong KW. A novel biodegradable gene carrier based on polyphosphoester. J Am Chem Soc. 2001;123:9480–9481. doi: 10.1021/ja016062m. [DOI] [PubMed] [Google Scholar]
- 11.Goh SL, Murthy N, Xu M, Fréchet JMJ. Cross-linked microparticles as carriers for the delivery of plasmid DNA for vaccine development. Bioconjugate Chem. 2004;15:467–474. doi: 10.1021/bc034159n. [DOI] [PubMed] [Google Scholar]
- 12.Green JJ, Zhou BY, Mitalipova MM, Beard C, Langer R, Jaenisch R, et al. Nanoparticles for gene transfer to human embryonic stem cell colonies. Nano Lett. 2008;8:3126–3130. doi: 10.1021/nl8012665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gosselin MA, Guo W, Lee RJ. Efficient gene transfer using reversibly cross-linked low molecular weight polyethylenimine. Bioconjugate Chem. 2001;12:989–994. doi: 10.1021/bc0100455. [DOI] [PubMed] [Google Scholar]
- 14.Ahn C-H, Chae SY, Bae YH, Kim SW. Biodegradable poly(ethylenimine) for plasmid DNA delivery. J Control Release. 2002;80:273–282. doi: 10.1016/s0168-3659(01)00547-8. [DOI] [PubMed] [Google Scholar]
- 15.Miyata K, Kakizawa Y, Nishiyama N, Harada A, Yamasaki Y, Koyama H, et al. Block catiomer polyplexes with regulated densities of charge and disulfide crosslinking directed to enhance gene expression. J Am Chem Soc. 2004;126:2355–2361. doi: 10.1021/ja0379666. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Mo H, Koo H, Park J-Y, Cho MY, Jin G-w, et al. Visualization of the degradation of a disulfide polymer, linear poly(ethylenimine sulfide), for gene delivery. Bioconjugate Chem. 2007;18:13–18. doi: 10.1021/bc060113t. [DOI] [PubMed] [Google Scholar]
- 17.Christensen LV, Chang CW, Kim WJ, Kim SW, Zhong Z, Lin C, et al. Reducible poly(amido ethylenimine)s designed for triggered intracellular gene delivery. Bioconjugate Chem. 2006;17:1233–1240. doi: 10.1021/bc0602026. [DOI] [PubMed] [Google Scholar]
- 18.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, et al. Linear poly(amido amine)s with secondary and tertiary amino groups and variable amounts of disulfide linkages: synthesis and in vitro gene transfer properties. J Control Release. 2006;116:130–137. doi: 10.1016/j.jconrel.2006.09.009. [DOI] [PubMed] [Google Scholar]
- 19.Lin C, Zhong Z, Lok MC, Jiang X, Hennink WE, Feijen J, et al. Novel bioreducible poly(amido amine)s for highly efficient gene delivery. Bioconjugate Chem. 2007;18:138–145. doi: 10.1021/bc060200l. [DOI] [PubMed] [Google Scholar]
- 20.Wender PA, Galliher WC, Goun EA, Jones LR, Pillow TH. The design of guanidinium-rich transporters and their internalization mechanisms. Adv Drug Deliv Rev. 2008;60:452–472. doi: 10.1016/j.addr.2007.10.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brooks H, Lebleu B, Vivès E. Tat peptide-mediated cellular delivery: back to basics. Adv Drug Delivery Rev. 2005;57:559–577. doi: 10.1016/j.addr.2004.12.001. [DOI] [PubMed] [Google Scholar]
- 22.Zhao M, Weissleder R. Intracellular cargo delivery using Tat peptide and derivatives. Med Res Rev. 2004;24:1–12. doi: 10.1002/med.10056. [DOI] [PubMed] [Google Scholar]
- 23.Zorko M, Langel Ü. Cell-penetrating peptides: mechanism and kinetics of cargo delivery. Adv Drug Deliv Rev. 2005;57:529–545. doi: 10.1016/j.addr.2004.10.010. [DOI] [PubMed] [Google Scholar]
- 24.Funhoff AM, van Nostrum CF, Lok MC, Fretz MM, Crommelin DJA, Hennink WE. Poly(3-guanidinopropyl methacrylate): a novel cationic polymer for gene delivery. Bioconjugate Chem. 2004;15:1212–1220. doi: 10.1021/bc049864q. [DOI] [PubMed] [Google Scholar]
- 25.Tziveleka LA, Psarra AM, Tsiourvas D, Paleos CM. Synthesis and characterization of guanidinylated poly(propylene imine) dendrimers as gene transfection agents. J Control Release. 2007;117:137–146. doi: 10.1016/j.jconrel.2006.10.019. [DOI] [PubMed] [Google Scholar]
- 26.Bromberg L, Raduyk S, Hatton TA, Concheiro A, Rodriguez-Valencia C, Silva M, et al. Guanidinylated Polyethyleneimine–Polyoxypropylene–Polyoxyethylene Conjugates as Gene Transfection Agents. Bioconjugate Chem. 2009;20:1044–1053. doi: 10.1021/bc900119t. [DOI] [PubMed] [Google Scholar]
- 27.Ou M, Wang XL, Xu R, Chang CW, Bull DA, Kim SW. Novel biodegradable poly(disulfide amine)s for gene delivery with high efficiency and low cytotoxicity. Bioconjugate Chem. 2008;19:626–633. doi: 10.1021/bc700397x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luten J, van Nostrum CF, De Smedt SC, Hennink WE. Biodegradable polymers as non-viral carriers for plasmid DNA delivery. J Control Release. 2008;126:97–110. doi: 10.1016/j.jconrel.2007.10.028. [DOI] [PubMed] [Google Scholar]
- 29.Kopatz I, Remy J-S, Behr JP. A model for non-viral gene delivery: through syndecan adhesion molecules and powered by actin. J Gene Med. 2004;6:769–776. doi: 10.1002/jgm.558. [DOI] [PubMed] [Google Scholar]
- 30.Zauner W, Ogris M, Wagner E. Polylysine-based transfection systems utilizing receptor-mediated delivery. Adv Drug Del Rev. 1998;30:97–113. doi: 10.1016/s0169-409x(97)00110-5. [DOI] [PubMed] [Google Scholar]
- 31.Kim T-i, Ou M, Lee M, Kim SW. Arginine-grafted bioreducible poly(disulfide amine) for gene delivery systems. Biomaterials. 2009;30:658–664. doi: 10.1016/j.biomaterials.2008.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huang K, Voss B, Kumar D, Hamm HE, Harth E. Dendritic molecular transporters provide control of delivery to intracellular compartments. Bioconjugate Chem. 2007;18:403–409. doi: 10.1021/bc060287a. [DOI] [PubMed] [Google Scholar]
- 33.Schröder T, Niemeier N, Afonin S, Ulrich AS, Krug HF, Bräse S. Peptoidic amino- and guanidinium-carrier systems: targeted drug delivery into the cell cytosol or the nucleus. J Med Chem. 2008;51:376–379. doi: 10.1021/jm070603m. [DOI] [PubMed] [Google Scholar]
- 34.Sebestyén MG, Ludtke JJ, Bassik MC, Zhang G, Budker V, Lukhtanov EA, et al. DNA vector chemistry: the covalent attachment of signal peptides to plasmid DNA. Nat Biotechnol. 1998;16:80–85. doi: 10.1038/nbt0198-80. [DOI] [PubMed] [Google Scholar]
- 35.Zanta MA, Belguise-Valladier P, Behr JP. Gene delivery: a single nuclear localization signal peptide is sufficient to carry DNA to the cell nucleus. Proc Natl Acad Sci USA. 1999;96:91–96. doi: 10.1073/pnas.96.1.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mesika A, Grigoreva I, Zohar M, Reich Z. A regulated, NFkappaB-assisted import of plasmid DNA into mammalian cell nuclei. Mol Ther. 2001;3:653–657. doi: 10.1006/mthe.2001.0312. [DOI] [PubMed] [Google Scholar]
- 37.Kim SH, Jeong JH, Kim T-i, Kim SW, Bull DA. VEGF siRNA delivery system using arginine-grafted bioreducible poly(disulfide amine) Mol Pharm. 2009;6:718–726. doi: 10.1021/mp800161e. [DOI] [PMC free article] [PubMed] [Google Scholar]