Abstract
The translation of nucleic acid libraries into corresponding synthetic compounds would enable selection and amplification principles to be applied to man-made molecules. We used multistep DNA-templated organic synthesis to translate libraries of DNA sequences, each containing three “codons,” into libraries of sequence-programmed synthetic small-molecule macrocycles. The resulting DNA-macrocycle conjugates were subjected to in vitro selections for protein affinity. The identity of a single macrocycle possessing known target protein affinity was inferred through the sequence of the amplified DNA template surviving the selection. This work represents the translation, selection, and amplification of libraries of nucleic acids encoding synthetic small molecules rather than biological macromolecules.
Nature generates functional biological molecules by subjecting libraries of nucleic acids to iterated cycles of translation, selection, amplification, and diversification (1–4). Compared with analogous synthesis and screening methods currently used to discover synthetic molecules with desired properties, these evolution-based approaches are attractive because of the much larger numbers of molecules that can be simultaneously evaluated, the minute quantities of material needed, and the relatively modest infrastructure requirements for library synthesis and processing (1–4).
Despite these attractions, evolution-based approaches can only be applied to molecules that can be translated from amplifiable information carriers. We previously described the generality of DNA-templated organic synthesis (DTS) and explored its potential for translating DNA sequences into corresponding synthetic products by using DNA hybridization to modulate the effective molarity of DNA-linked reactants (5). DTS can generate products unrelated in structure to the DNA backbone in a sequence-specific manner (5, 6), does not require functional group adjacency to proceed efficiently (5, 7), can mediate sequence-programmed multistep small-molecule synthesis (8), and can enable reaction pathways that are difficult or impossible to realize with the use of conventional synthetic strategies (9).
These features of DTS raise the possibility of translating single-solution libraries of DNA sequences into corresponding libraries of synthetic small molecules conjugated to their respective templates. Because each member of a DNA-templated synthetic library is linked to an encoding nucleic acid, these libraries are suitable for in vitro selection (10), polymerase chain reaction (PCR) amplification, and DNA sequence characterization to reveal the identity of synthetic library members possessing functional properties (Fig. 1). Below, we describe the integration of these concepts into the DNA-templated synthesis of a library of macrocycles (Fig. 2A), the selection of this pilot library for affinity to a target protein, and the identification of a functional library member through the amplification and characterization of DNA sequences surviving the selection.
Fig. 1.
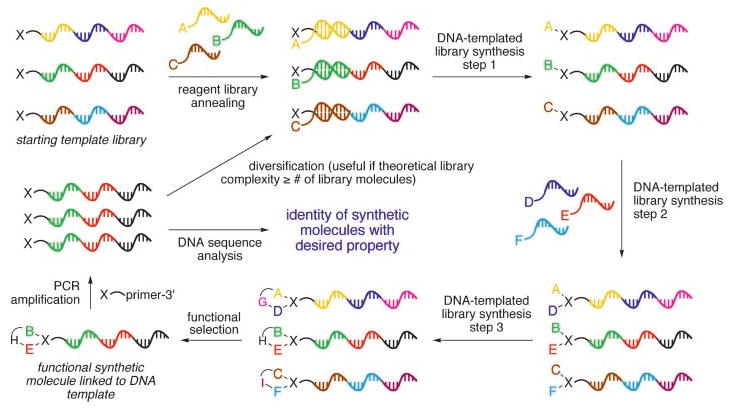
Scheme for the translation, selection, and amplification of libraries of DNA templates encoding synthetic small molecules. When the number of different possible library structures approaches or exceeds the number generated, template diversification after selection can be added to evolve the pool of synthetic molecules toward structures possessing desired properties. X is a starting material common to all library members.
Fig. 2.
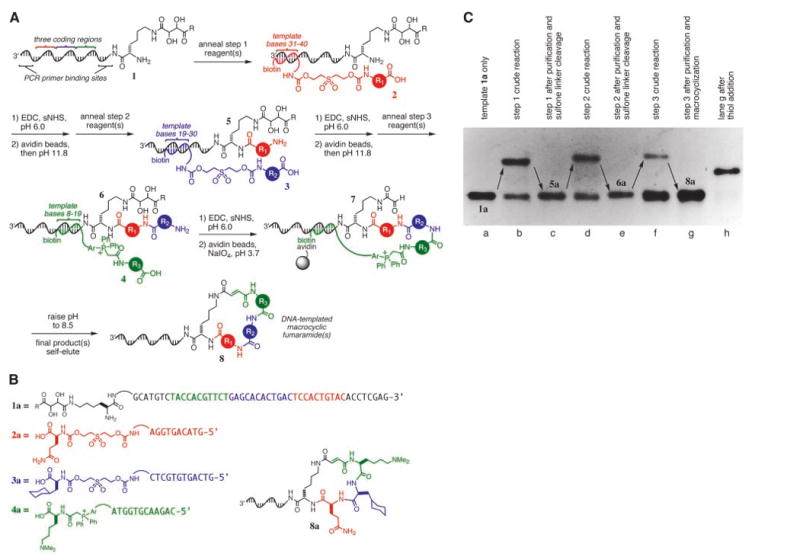
(A) DNA-templated macrocycle library synthesis scheme. R is NHCH3 where R is NHCH3 or tryptamine; Ar, –(p-C6H4)–. The macrocyclization reaction is confirmed to give predominantly trans alkene stereochemistry for one library member (fig. S4) (13) but may give other outcomes for different macrocyclic structures. (B) Template and reagents used in the DNA-templated synthesis of 8a. (C) Denaturing PAGE of each step in the DNA-templated synthesis of 8a. Lane h is the product of a DNA-templated thiol addition to the product shown in lane g, confirming the formation of the fumaramide group during macrocyclization.
Although macrocycles can be challenging targets for conventional synthesis (11), the compatibility of DTS with nM reactant concentrations, aqueous solvents (12), and purification methods not available to conventional synthesis (8) suggested that macrocycle synthesis might proceed efficiently in this format. We subjected a 48-base DNA-linked lysine derivative (1a, the “template,” analogous to an mRNA during protein biosynthesis) to three successive DNA-templated amine acylation reactions (6) with building blocks conjugated to DNA 10-mer or 12-mer oligonucleotides (2a, 3a, or 4a, the “reagents,” analogous to tRNAs) (Figs. 2B and 3). Each reagent oligonucleotide complemented one of three unique coding regions in the template sequence. The reagent oligonucleotides were biotinylated to allow products from each DNA-templated step (5a, 6a, and 7a) to be purified by capture with, and release from, streptavidin-linked magnetic beads (8) (Fig. 2, A and B). This direct selection for bond formation facilitates multistep synthesis by enabling the one-pot purification of products independent of their structure.
Fig. 3.
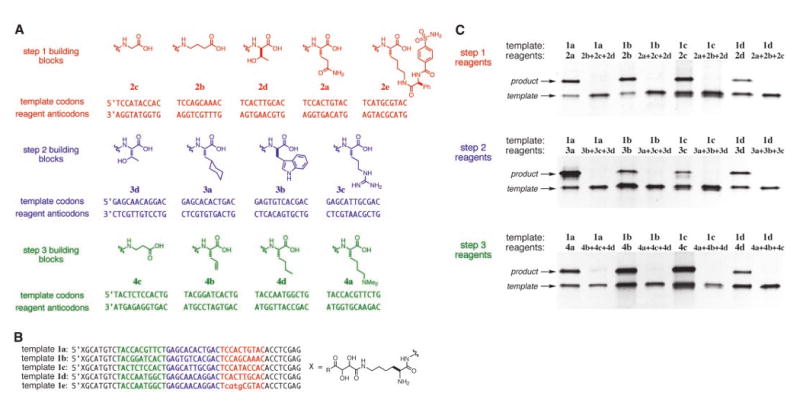
(A) Building blocks and anticodons used in reagents for DNA-templated macrocycle library synthesis. The variable regions within each anticodon are underlined. The NlaIII cleavage site within template 1e is shown in lower case. (B) Representative DNA templates used in macrocycle library synthesis. Templates 1a to 1e (R is NHCH3 or tryptamine) collectively call for each of the reagents in (A). (C) Denaturing PAGE analysis confirming the sequence specificity of each template-reagent combination used in the macrocycle library synthesis. The listed template and reagent(s) were combined under the conditions shown in Fig. 2A, and the reactions were analyzed before reagent-linker cleavage. Products appear as bands of higher molecular weight above templates.
The diol group in the captured product of the third DNA-templated reaction (7a) was oxidatively cleaved with NaIO4 to reveal an aldehyde. The phosphonium group was then deprotonated by elevating the pH of the buffer to 8.5, which induced Wittig olefination and macrocyclization. Because macrocyclization results in the cleavage of the reagent oligonucleotide-product bond, the desired macrocyclic fumaramide (8a) self-eluted in pure form from the streptavidin-linked magnetic beads (Fig. 2, A and B). The generality of this reaction was examined by assaying the macrocyclization of 11 molecules related to 7a (fig. S1) (13). Macrocyclization was efficient for a wide variety of precyclized structures (60 to 90% yields with no contaminating uncyclized material). Control reactions lacking NaIO4 confirmed that the cyclization reaction required the presence of an aldehyde group (fig. S1) (13).
The progress of each DNA-templated step during the transformation of 1a to 8a was followed by denaturing polyacrylamide gel electrophoresis (PAGE) (Fig. 2C) and by matrix-assisted laser desorption/ionization-time-of-flight (MALDI-TOF) mass spectrometry (Fig. 4A) after endonuclease-catalyzed removal of all but seven nucleotides at the 5′ end of the template. The presence of the electrophilic fumaramide group in the final product was confirmed by mass spectrometry and by its ability to accept a thiol nucleophile during a DNA-templated conjugate addition (Figs. 2C and 4A). Macrocycle 8a (expected mass is 2908.8 daltons; observed mass is 2910.4 ± 6 daltons) was synthesized with high purity in 1 to 5% overall yield for three DNA-templated steps, the macrocyclization, and all associated purifications. To verify that the reaction conditions do not induce epimerization of the amino acid–derived chiral centers, we performed large-scale amine acylation reactions of analogous non–DNA-linked substrates under conditions that mimicked steps 1 to 3. The stereochemical integrity of the resulting products was confirmed by comparison to authentic diastereomeric standards (figs. S2 and S3) (13). In addition, exposure of a non–DNA-linked version of 7 to the above macrocyclization conditions provided the corresponding macrocycle (a non–DNA-linked version of 8) containing a predominantly trans alkene (14) by nuclear magnetic resonance analysis (figs. S4 and S5) (13).
Fig. 4.
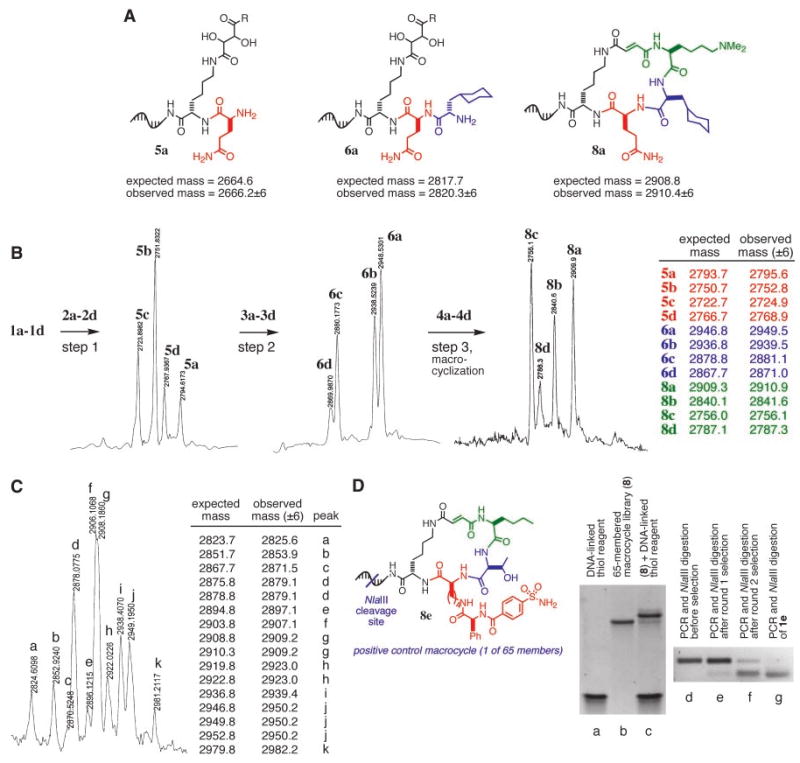
MALDI-TOF mass spectrometric analyses (M–H negative ion mode) of (A) the multistep DNA-templated synthesis of 8a starting from 1a (R is NHCH3), (B) the translation of four templates (1a to 1d; R is tryptamine) into four corresponding macro-cycles (8a to 8d), and (C) step two of the translation of 65 templates (the 64 templates containing all possible combinations of codons complementing 2a to 2d, 3a to 3d, 4a to 4d, plus template 1e) into 65 corresponding macrocycles. (D) Analysis of the DNA-templated 65-member macrocyclic fumaramide library by denaturing PAGE (lanes a to c) and agarose gel electrophoresis (lanes d to g). Lane a, DNA-linked thiol reagent complementing the constant 5′ region of all macro-cycle template sequences; lane b, the 65-member library of template-linked macro-cycles (8); lane c, the library after DNA-templated thiol addition, confirming the presence of the fumaramide group formed during macrocyclization; lane d, NlaIII digestion of PCR-amplified templates from the 65-member macrocycle library before selection; lanes e and f, NlaIII digestion of PCR-amplified templates from the 65-member macrocycle library after one and two rounds of selection for carbonic anhydrase affinity, respectively; lane g, NlaIII digestion of authentic PCR-amplified template 1e that directs the synthesis of 8e.
In addition to the multistep DTS of one synthetic small molecule, implementation of the scheme in Fig. 1 requires that DTS proceed in a sequence-specific manner in a library format in which multiple templates and multiple reagents are present in the same solution. Although DNA-templated reactions have been shown to be sequence-specific (5, 9), library-format DTS to generate multistep small-molecule products has not been previously achieved.
We chose unique template “codons” to encode four or five reactants for each of the three DNA-templated steps in the macrocycle synthesis (13 codons total) (Fig. 3A). Each of the 13 codons was assigned to encode a different building block. The building blocks were chosen to include diverse functionalities, stereochemistries, and backbone lengths (Fig. 3A). Four different templates (1a to 1d) (Fig. 3B), each containing three codons, were prepared such that the maximum number of 12 different codons (complementing reagents 2a to 2d, 3a to 3d, and 4a to 4d) were represented within the four templates. The corresponding reagents, each consisting of an amino acid building block conjugated through the linkers shown in Fig. 2A to a decoding DNA “anticodon,” were also prepared (Fig. 3A).
We tested the sequence specificity of DTS in the presence of multiple reagents by exposing template 1a, 1b, 1c, or 1d separately to a mixture of all step 1 reagents except the reagent complementing the step 1 codon present in each template. As a positive control, each of the four templates was also separately reacted with its complementary step 1 reagent. These two DNA-templated reactions were repeated for each of the four step 2 codons and for each of the four step 3 codons. In contrast with the positive control, the reaction lacking the complementary reagent in all 12 cases did not generate significant product (Fig. 3C). These results indicate that templates do not react with mismatched reagents under the conditions in Fig. 2, even in the absence of complementary reagents.
To examine the sequence specificity of true library-format DNA-templated synthesis involving multiple templates and multiple reagents in a single solution, we reacted templates 1a to 1d in one solution with the four step 1 reagents (2a to 2d). After reagent-linker cleavage (Fig. 2, A and C), the solution containing the step 1 products was reacted with the four step 2 reagents (3a to 3d), and the resulting purified products were then reacted with the four step 3 reagents (4a to 4d) before undergoing Wittig macrocyclization.
In all cases, the major products observed by MALDI-TOF mass spectrometry after each step consisted of all four of the sequence-programmed products (Fig. 4B). If the 12 reagents used to synthesize 8a to 8d reacted with templates randomly, rather than in a sequence-programmed manner, up to 64 different macrocycles would have been synthesized rather than the exclusive formation of the four observed products. The faithful translation of four DNA templates (1a to 1d) into four sequence-programmed macrocyclic fumaramides (8a to 8d) indicates a one-to-one correspondence between the DNA sequence that enters the above process and the structure of the resulting macrocycle.
After developing a robust multistep DNA-templated macrocycle synthesis and establishing the sequence specificity of library-format DNA-templated reactions, we prepared a library of 65 templates (1) that contained all 64 possible combinations of the four codons at each coding region and a 65th template (1e) that uniquely contained a step 1 codon encoding a phenyl sulfonamide building block (Fig. 3A). Because carbonic anhydrase is known to bind phenyl sulfonamides with high affinity [dissociation constant Kd = ∼1 nM (15)], the macrocycle encoded by the 65th template serves as a positive control to evaluate the ability of a DNA-templated small molecule library to be selected in vitro for target protein affinity (see below).
The single-solution library of 65 equimolar DNA templates was translated into 65 corresponding macrocyclic fumaramides through the scheme shown in Fig. 2A. Each of the three coupling steps was executed in a single solution containing all 65 templates (typically 500 pmol total) and all five (step 1) or four (steps 2 and 3) reagents as described above. Denaturing PAGE analysis of each library synthesis step indicated yields similar to those of the single-template and four-template cases. MALDI-TOF mass spectrometry was used to observe the formation of the four major step 1 small-molecule products (all but the product encoded by 1e, which is expected to be 16-fold less abundant than the four major step 1 products). After step 2, mass spectrum peaks consistent with the presence of all mass-resolvable step 2 small-molecule products were also observed (Fig. 4C).
After step 3 and macrocyclization, exposure of the completed 65-member library to a DNA-linked thiol reagent efficiently (84% yield) converted the library to higher molecular weight species. This result is consistent with the formation of the fumaramide group during the macrocyclization (Fig. 4D). Beginning with 0.1 to 3 nmol of starting template (1), the multistep DNA-templated library synthesis described above provided sufficient final product to undergo many in vitro selections (10) for library members with protein binding properties. Taken together, these results represent the translation of a library of DNA templates into a library of corresponding synthetic molecules.
Each member of a DNA-templated synthetic library of the type described in Fig. 1 is associated with an amplifiable DNA strand that not only encodes but has actually directed that molecule's synthesis. DNA-templated libraries, like libraries made by DNA display (16), are conceptually analogous to genetically encoded protein libraries (2, 4) except that the structures generated are not limited to those that can be biosynthesized by the ribosome. Similar to nucleic acid–templated protein libraries, DNA-templated synthetic libraries, in principle, can be selected for desired properties such as target binding affinity or specificity (10).
To test this possibility, we subjected a minute quantity (100 fmol) of the 65-member DNA-templated macrocyclic fumaramide library to an in vitro selection for binding carbonic anhydrase, a well-studied protein (17). Carbonic anhydrase was immobilized by reaction with N-hydroxysuccinimide ester–linked agarose beads, combined with 100 fmol of the 65-member macrocyclic fumaramide library, and washed with buffer. Bound molecules were eluted and subjected to a second iteration of the selection. The DNA templates encoding macrocycles surviving each round of selection were amplified by PCR and digested with the restriction endonuclease NlaIII. The 65th template (1e) uniquely contains a 5′-CATG-3′ sequence in the coding region of the phenyl sulfonamide building block that is cleaved by NlaIII (Fig. 3B).
Before selection for binding to carbonic anhydrase, NlaIII digestion reveals that the templates from the macrocycle library do not contain a noticeable representation of template 1e, as expected because the library consists predominantly of other templates. Each selection for binding to carbonic anhydrase successively enriches the template pool for the sequence in 1e, such that after two selections the pool predominantly contains the sequence encoding the phenyl sulfonamide– containing macrocycle (8e) (Fig. 4D). We therefore conclude that a single member of the 65-member DNA-templated macrocycle library was efficiently selected for carbonic anhydrase binding activity.
Macrocycles of the general structure 8 are promising compounds for perturbing the activity of biologically important proteases (18) because of their partial peptidic and conformationally constrained nature. In addition, the electrophilic fumaramide group can capture proximal nucleophiles (Figs. 2C and 4D) such as those present in protease active sites. On the basis of the above findings, efforts to generate and select DNA-templated synthetic libraries of high complexity and structural diversity are under way in our laboratory.
Supplementary Material
Acknowledgments
Supported by NIH (National Institute of General Medical Sciences R01GM065865), the Office of Naval Research (N00014-03-1-0749), the Arnold and Mabel Beckman Foundation, the Searle Scholars Foundation (00-C-101), and the Alfred P. Sloan Foundation (BR-4141). Z.J.G. is a Bristol-Myers Squibb Graduate Research Fellow. B.N.T. and T.M.S. are NSF Graduate Research Fellows. J.B.D. is a National Defense Science and Engineering Graduate Research Fellow. We are grateful to the Bauer Center for Genomics Research for MALDI-TOF mass spectrometric analyses, G. Verdine for LC-MSinstrument access, and DNA Software for assistance with codon screening. The rights to commercial development of DNA-templated synthesis have been licensed to Ensemble Discovery, a company for which D.R.L. is a consultant and shareholder.
Footnotes
References and Notes
- 1.Wilson DS, Szostak JW. Annu Rev Biochem. 1999;68:611. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 2.Lin H, Cornish VW. Angew Chem Int Ed Engl. 2002;41:4402. doi: 10.1002/1521-3773(20021202)41:23<4402::AID-ANIE4402>3.0.CO;2-H. [DOI] [PubMed] [Google Scholar]
- 3.Joyce GF. Annu Rev Biochem. 2004;73:791. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 4.Taylor SV, Kast P, Hilvert D. Angew Chem Int Ed Engl. 2001;40:3310. doi: 10.1002/1521-3773(20010917)40:18<3310::aid-anie3310>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 5.Gartner ZJ, Liu DR. J Am Chem Soc. 2001;123:6961. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gartner ZJ, Kanan MW, Liu DR. Angew Chem Int Ed Engl. 2002;41:1796. doi: 10.1002/1521-3773(20020517)41:10<1796::aid-anie1796>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 7.Gartner ZJ, Grubina R, Calderone CT, Liu DR. Angew Chem Int Ed Engl. 2003;42:1370. doi: 10.1002/anie.200390351. [DOI] [PubMed] [Google Scholar]
- 8.Gartner ZJ, Kanan MW, Liu DR. J Am Chem Soc. 2002;124:10304. doi: 10.1021/ja027307d. [DOI] [PubMed] [Google Scholar]
- 9.Calderone CT, Puckett JW, Gartner ZJ, Liu DR. Angew Chem Int Ed Engl. 2002;41:4104. doi: 10.1002/1521-3773(20021104)41:21<4104::AID-ANIE4104>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 10.Doyon JB, Snyder TM, Liu DR. J Am Chem Soc. 2003;125:12372. doi: 10.1021/ja036065u. [DOI] [PubMed] [Google Scholar]
- 11.Woodward RB, et al. J Am Chem Soc. 1981;103:3210. [Google Scholar]
- 12.Li CJ, Chan TH. Organic Reactions in Aqueous Media. Wiley; New York: 1997. [Google Scholar]
- 13.Synthesis and characterization details are available on Science Online.
- 14.Maryanoff BE, Reitz AB. Chem Rev. 1989;89:863. [Google Scholar]
- 15.Jain A, Whitesides GM, Alexander RS, Christianson DW. J Med Chem. 1994;37:2100. doi: 10.1021/jm00039a023. [DOI] [PubMed] [Google Scholar]
- 16.Halpin DR, Harbury PB. PLoS Biol. 2004;2:E174. doi: 10.1371/journal.pbio.0020174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tripp BC, Smith K, Ferry JG. J Biol Chem. 2001;276:48615. doi: 10.1074/jbc.R100045200. [DOI] [PubMed] [Google Scholar]
- 18.Lamarre D, et al. Nature. 2003;426:186. doi: 10.1038/nature02099. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


