Abstract
Current approaches to reaction discovery focus on one particular transformation. Typically, researchers choose substrates based on their predicted ability to serve as precursors for the target structure, then evaluate reaction conditions1–6 for their ability to effect product formation. This approach is ideal for addressing specific reactivity problems, but its focused nature might leave many areas of chemical reactivity unexplored. Here we report a reaction discovery approach that uses DNA-templated organic synthesis7–10 and in vitro selection to simultaneously evaluate many combinations of different substrates for bond-forming reactions in a single solution. Watson–Crick base pairing controls the effective molarities of substrates tethered to DNA strands; bond-forming substrate combinations are then revealed using in vitro selection for bond formation, PCR amplification and DNA microarray analysis. Using this approach, we discovered an efficient and mild carbon–carbon bond-forming reaction that generates an enone from an alkyne and alkene using an inorganic palladium catalyst. Although this approach is restricted to conditions and catalysts that are at least partially compatible with DNA, we expect that its versatility and efficiency will enable the discovery of additional reactions between a wide range of substrates.
A reaction discovery system capable of simultaneously evaluating many combinations of substrates for bond-forming reactivity in a single solution must meet several requirements. First, the system must organize complex substrate mixtures into discrete pairs that can react (or not react) without affecting the reactivity of the other substrate pairs. Second, the system requires a general method for separating reactive substrate pairs from unreactive pairs. Last, the reactive substrate pairs must be identified efficiently.
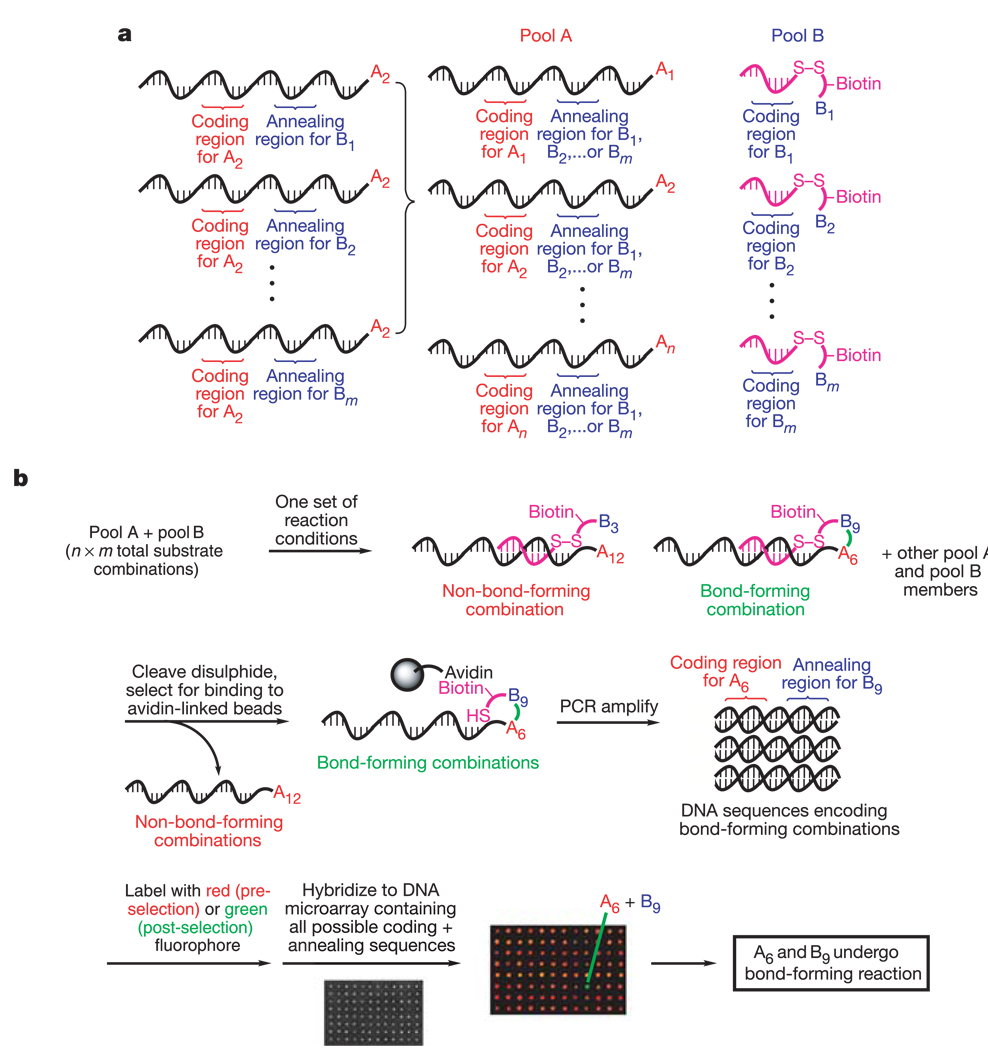
Recent developments in DNA-templated organic synthesis7–10 indicate that DNA annealing can organize many substrates in a single solution into DNA sequence-programmed pairs. To this end, we prepared two pools of DNA-linked substrates, with n substrates in pool A and m substrates in pool B. Each substrate in pool A is covalently linked to the 5′ end of a set of DNA oligonucleotides containing one ‘coding region’ (uniquely identifying that substrate) and one of m different ‘annealing regions’ (Fig. 1a). Each of the m substrates in pool B is attached to the 3′ end of an oligonucleotide containing a coding region that uniquely identifies the substrate and complements one of the m annealing regions in pool A.
Figure 1.
Key elements of a new approach to reaction discovery. a, Two pools of DNA-linked organic functional groups that associate each of n × m substrate combinations with a unique DNA sequence. b, A general one-pot selection and analysis method for the detection of bond-forming reactions between DNA-linked substrates.
When pools A and B are combined in a single aqueous solution at nanomolar concentrations, Watson–Crick base pairing organizes the mixture into n × m discrete pairs of substrates attached to complementary sequences. Only substrates linked to complementary oligonucleotides experience effective molarities in the millimolar range8; substrates linked to non-complementary oligonucleotides experience nanomolar solution concentrations and hence do not react with each other at a significant rate. This effective molarity-based design enables reactions that might otherwise be suppressed by the preferential dimerization of one or both substrates. The possibility of interference by the structure of DNA during reactions between substrates is minimized by using long and flexible substrate–DNA linkers11.
The separation of reactive pairs of substrates is based on selection concepts used in the directed evolution of catalytic RNA and DNA12,13 (Fig. 1b). We covalently linked each pool B substrate to its corresponding oligonucleotide by a linker containing a biotin group and a disulphide bond (Fig. 1b and Supplementary Information). After incubation under a set of chosen reaction conditions, followed by cleavage of the disulphide bonds, only pool A sequences encoding bond formation between a pool A and pool B substrate remain covalently linked to biotin. Streptavidin affinity selection of the resulting solution separates biotinylated from non-biotinylated sequences. In contrast to many existing reaction discovery screens1,5 focused on a particular reaction type, our selection-based approach does not depend on any specific substrate or product property but instead can identify all substrate pairs capable of forming a covalent bond under the reaction conditions.
Reactive substrate pairs are identified by amplification of the selected sequences by polymerase chain reaction (PCR) followed by DNA microarray analysis (see below). Because PCR amplification is extremely sensitive, femtomole quantities of substrates are sufficient for the entire reaction discovery process.
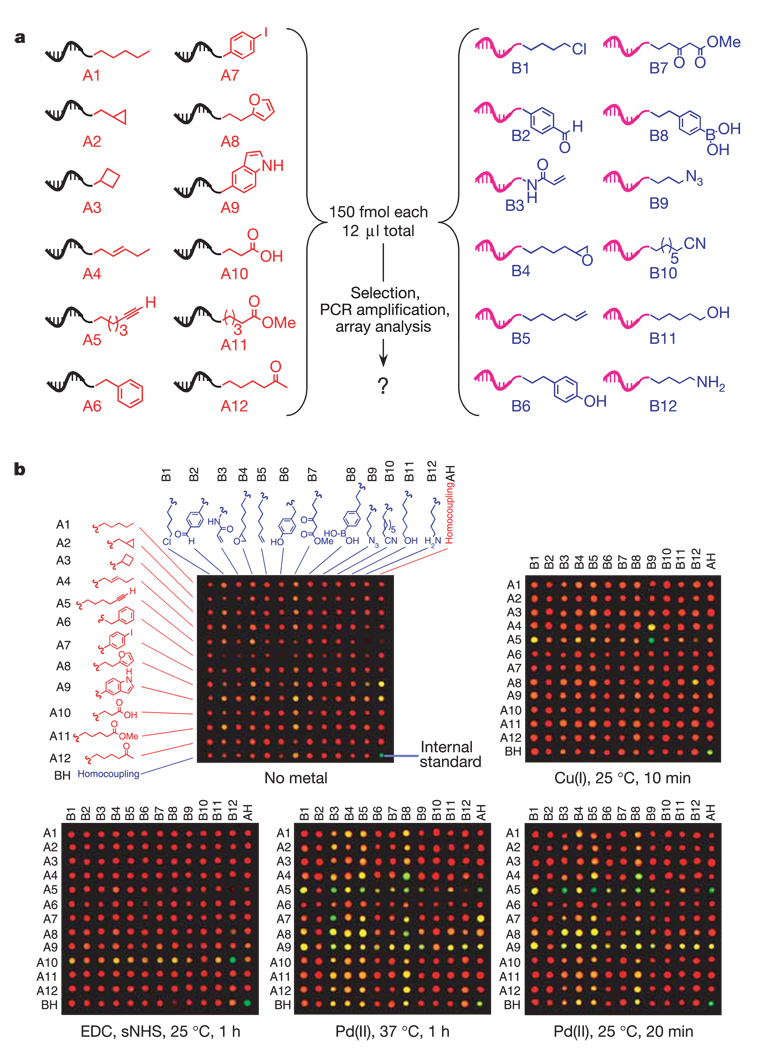
We prepared pool A and pool B containing 12 substrates each (Fig. 2a), representing 144 heterocoupling combinations (see Supplementary Methods and Supplementary Fig. S1 for details). We also prepared DNA-linked substrates to enable the detection of homocoupling of any of the 24 different substrates, bringing the total number of unique substrate combinations to 168 (Fig. 2a). The 24 substrates in Fig. 2a were chosen to represent simple functional groups commonly encountered in organic molecules. New reactions forming bonds between simple functionalities are of special interest because they might provide more accessible alternatives to coupling reactions that require more complex substrates. Although our approach requires the preparation of DNA-linked substrates, a single nanomole-scale preparation of each pool provided sufficient material for more than 1,000 reaction discovery experiments that could collectively evaluate more than 168,000 combinations of substrates and reaction conditions.
Figure 2.
Results from reaction discovery selections and analysis. a, Pool A and pool B substrates used in this work. b, Qualitative results of reaction discovery selections after exposure to the reaction conditions listed below each array image. Spots that are significantly green suggest bond formation between the corresponding substrates (quantitative fluorescence ratios in Fig. 3 and the Supplementary Data are used for actual interpretations). The 840 reaction possibilities in these five experiments were evaluated by one researcher in two days. See Supplementary Methods for detailed reaction conditions.
To test the ability of our system to detect a single reactive combination of substrates out of 168 possibilities, we combined pool A and pool B in the presence of Cu(I), conditions known to promote a cycloaddition between a terminal alkyne (A5 in Fig. 2a) and an azide (B9 in Fig. 2a)14. Pool A members (2 pmol total) were combined with 2 pmol total of pool B members (12 fmol of material encoding each possible substrate combination) in the presence of 500 µM Cu(I) in a total volume of 12 µl. After 10 min at 25 °C, the salts were removed and disulphide linkages were cleaved with 0.1 M tris-carboxyethylphosphine hydrochloride. A control experiment lacking Cu(I) but identical in all other respects was also performed.
Sequences encoding bond-forming substrate pairs were captured with streptavidin-linked magnetic particles and amplified by PCR with a DNA primer labelled with the cyanine fluorophore Cy3. For comparison, an aliquot of the pool A sequences before selection was amplified by PCR with a Cy5-labelled primer. The Cy3-labelled and Cy5-labelled PCR products were combined and hybridized to a DNA microarray containing all 168 possible reaction-encoding sequences. The ratios of Cy3 (green) to Cy5 (red) fluorescence for all array locations were calculated and ordered by rank, and spots with green/red fluorescence ratios significantly higher than the majority of spots (in the experiments below, ratios above 1.5) were considered to be positive. A pre-quantified internal standard (bottom right corner of the array images in Fig. 2b) corresponding to a moderate level of reactivity was used as a positive control and as a reference for comparing different arrays.
For the experiment performed in the presence of Cu(I), the array spot corresponding to the combination of the alkyne (A5) and the azide (B9) was the sole spot that had a significant green/red fluorescence ratio (A5 + B9 = 8.5, standard = 2.8; see Fig. 2b and Supplementary Data). In contrast, the experiment performed in the absence of any added metal yielded no green array spots other than the standard (Fig. 2b). These results show that the above method can detect a single bond-forming substrate combination from a complex mixture of 168 combinations, and confirm the high chemoselectivity characteristic of the Sharpless-modified Huisgen cycloaddition reaction15.
Cu(I) damages DNA by promoting radical-mediated processes16. Indeed, we observed 48% degradation of a DNA oligonucleotide exposed for 10 min to the Cu(I)-containing conditions (Supplementary Data). The successful detection of the alkyne–azide cycloaddition under conditions that degrade DNA shows that reaction conditions do not need to be fully DNA compatible, because the extreme sensitivity of PCR amplification17 enables bond formation to be revealed even if a significant fraction of the total DNA in an experiment is destroyed.
To validate this reaction discovery system further, we performed a selection for bond formation after exposure to organic reagents (1-(3-(dimethylamino)propyl)-3-ethylcarbodiimide hydrochloride and N-hydroxysuccinimide at pH 6.0) known to promote the coupling of amines with carboxylic acids18. Microarray analysis revealed one positive; this spot indeed corresponded to the combination of the carboxylic acid from pool A (A10) and the amine from pool B (B12) (A10 + B12 = 6.9, standard = 15.6; see Fig. 2b and Supplementary Data).
After successfully ‘rediscovering’ known bond-forming reactions, we examined the reactivity of a simple Pd(II) salt. Inorganic Pd salts and complexes containing Pd are known to mediate diverse coupling reactions19, indicating that Pd can activate a variety of organic functional groups. Selection for bond formation in the presence of 500 µM Na2PdCl4 for 1 h at 37 °C resulted in five strong positives (A7 + B3, A5 + B3, A4 + B8, A5 + B5 and A5 homocoupling) with significant green/red fluorescence ratios (3.6 to 2.6; standard = 2.9). In addition, we observed five weaker positives (A9 + B3, A8 + B3, A8 + B8, A5 + B8 and A5 + B9) with lower, but possibly significant, green/red fluorescence ratios ranging from 1.9 to 1.6 (Fig. 2b and Supplementary Data). One positive representing the combination of aryl iodide (A7) and acrylamide (B3) is consistent with the well-known Heck reaction20,21 (assuming the formation of some Pd(0) under the reaction conditions). Other positives could be rationalized with mechanistic steps precedented in known Pd-mediated chemistries. For example, the spot indicating bond formation between an olefin (A4) and an aryl boronic acid (B8) is consistent with transmetallation of Pd(II) by the aryl boronic acid22 followed by the insertion of an olefin into the Pd–aryl bond and subsequent β-hydride elimination23.
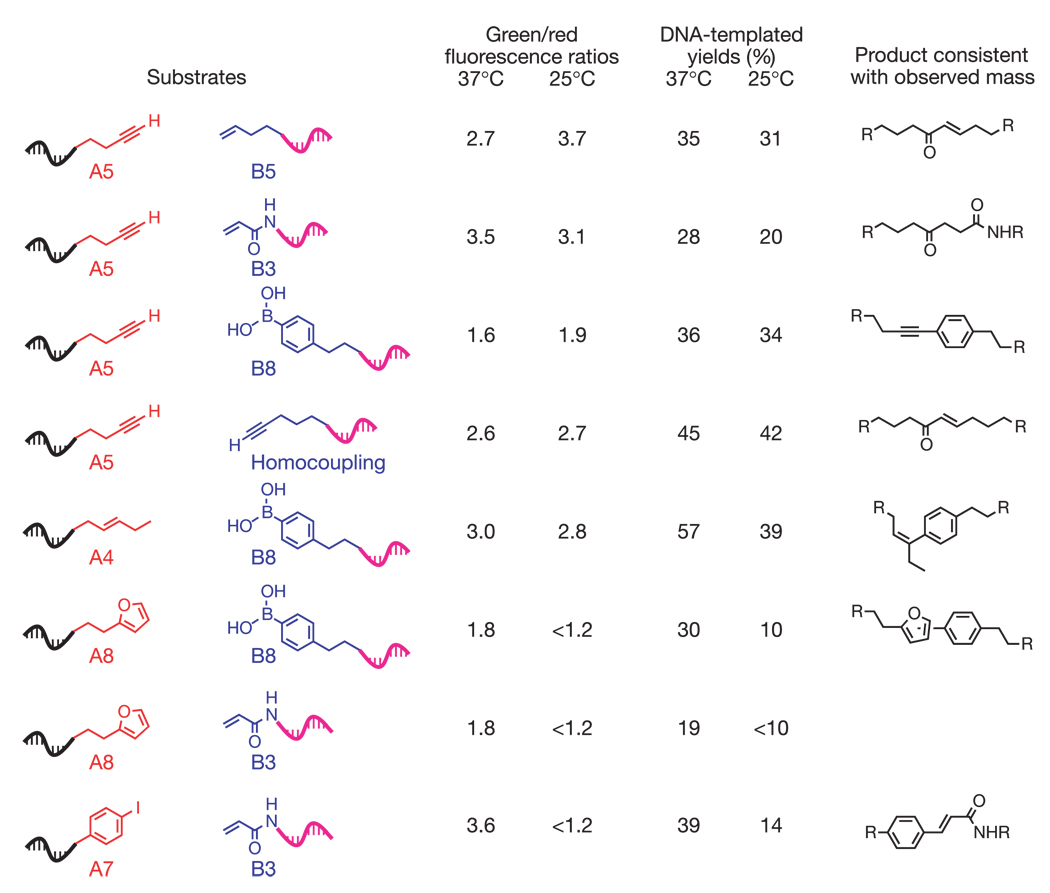
To determine whether these array results reflect genuine bond-forming events, we examined the putative reactions corresponding to the above ten spots in separate DNA-templated reactions. Denaturing polyacrylamide gel electrophoresis (PAGE) analysis indicated that all five strong positives and three of the five weak positives corresponded to authentic DNA-templated reactions, whereas two weak positives (A9 + B3 and A5 + B9) showed little or no product formation (Fig. 3 and Supplementary Fig. S2). We observed no product formation in control reactions in which Na2PdCl4 was omitted, or in which either of the reactive substrates was replaced with an unreactive alkane group. These findings are consistent with the detection of Pd-dependent reactions that couple substrate groups and do not couple the functionality present in DNA.
Figure 3.
Characterization in a DNA-templated format of array positives resulting from exposure to 500 µM Na2PdCl4 at 37 °C for 1 h, or at 25 °C for 20 min. Putative reactions were screened by PAGE and MALDI–TOF mass spectrometric analysis (Supplementary Information). Template architectures and reaction conditions were chosen to match those used in the selection rather than to maximize product yields. Product structures other than those proposed are possible. The green/red fluorescence ratio for the internal standard at 37 °C and 25 °C is 2.9 and 3.5, respectively. For A8 + B3, reactivity was not sufficient to obtain reliable product mass characterization.
We further characterized the PAGE-validated Pd-mediated DNA-templated reactions using matrix assisted laser desorption ionization–time-of-flight (MALDI–TOF) mass spectrometry (Fig. 3). The observed product masses are consistent with covalent bond formation. We suggest possible structures consistent with the observed masses for each of the reaction products but note that other products or mixtures of products are also consistent with the data. Together with the PAGE analysis, these results show that this reaction discovery system reliably and efficiently reveals bond-forming events even under conditions that cause multiple combinations of substrates to react.
Increasing the stringency of the reaction conditions by decreasing the temperature to 25 °C and quenching the reaction after 20 min decreased the number of strong positives to four (A5 + B5, A5 + B3, A4 + B8 and A5 homocoupling; green/red fluorescence ratios = 3.7 to 2.7; standard = 3.5) and the number of weak positives to two (A5+B8 and A5+B9; fluorescence ratios = 1.9 and 1.7; see Fig. 2b and Supplementary Data). Consistent with these results, the five authentic bond-forming reactions among this set (all four strong positives and weak positive A5+B8) showed substantial product formation by PAGE analysis when performed under the 25 °C conditions, whereas the three additional reactions listed in Fig. 3 but not appearing as positives in the 25 °C experiment (A8+B8, A8+B3 and A7+B3) showed significantly weaker product formation (Fig. 3). Varying the stringency of the reaction conditions before selection can therefore efficiently distinguish substrate combinations on the basis of their level of reactivity.
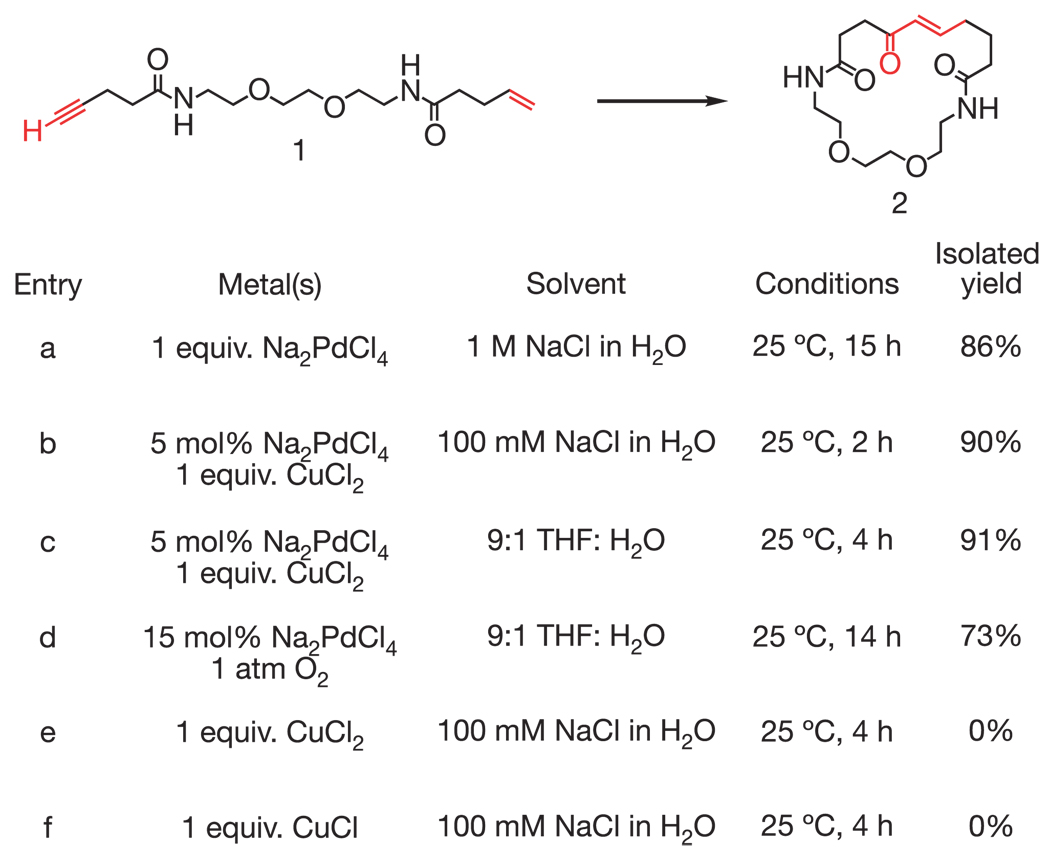
The above results suggested that the Pd(II)-mediated carbon–carbon bond formation between a simple terminal alkyne and terminal alkene (A5+B5) proceeds efficiently to generate a possible enone product (Fig. 2b and Fig. 3), prompting a detailed investigation of this reaction. We synthesized a small-molecule substrate (1) to study a non-DNA-templated, intramolecular version of this alkyne–alkene coupling reaction on a multi-milligram scale (Fig. 4). The addition of 1 to one equivalent of Pd(II) in 1 M aqueous NaCl over 15 h followed by purification by reverse-phase high-performance liquid chromatography provided the 20-membered macrocyclic trans-enone 2 as a single olefin stereoisomer in 86% isolated yield (Fig. 4, entry a). The structure of 2 was confirmed by 1H-NMR, 13C-NMR, homonuclear correlated spectroscopy (COSY) and high-resolution mass spectrometry (Supplementary Data). To our knowledge, this is the first example of macrocyclic enone formation from a simple alkyne–alkene precursor. The finding also shows that a reaction discovered by a selection for DNA-templated covalent bond formation can operate in a non-DNA-templated format on a much larger (109-fold) scale.
Figure 4.
Characterization of a new alkyne–alkene macrocyclization reaction in a non-DNA-templated format. The macrocyclic enone product (2) was characterized by 1H-NMR, 13C-NMR, COSY, UV–visible spectrometry and high-resolution mass electrospray (Supplementary Information). We speculate that product formation proceeds through the following sequence: soft deprotonation of the alkyne to form a Pd(II)–alkynyl intermediate; insertion of the alkene into the Pd-alkyne bond; β-hydride elimination to form a conjugated enyne; Pd(II)-catalysed hydration of the alkyne to form an enol π-allyl Pd complex; enol tautomerization and π-allyl Pd protonation to generate the trans-enone.
Because the formation of an enone from an alkyne and an alkene represents an oxidative coupling, we proposed that the reaction could be performed with catalytic quantities of Pd by introducing an oxidant such as CuCl2 + air to reoxidize Pd(0) to Pd(II) (ref. 24). Indeed, the addition of 1 to 5 mol% Pd and 1 equivalent of CuCl2 over 2 h in water at 25 °C provided enone 2 as the sole observed product in 90% isolated yield (Fig. 4, entry b). Significantly, this reaction maintains its high efficiency in solvent containing 9:1 tetrahydrofuran:water (Fig. 4, entry c). Product was formed in similar yields, although at a slower rate, using 1 atm O2 instead of CuCl2 to reoxidize Pd(0) (Fig. 4, entry d). Control reactions with CuCl2 or CuCl alone yielded no observed product formation (Fig. 4, entries e and f).
The discovery of this alkyne–alkene coupling reaction suggests the value of searching a large number of substrate combinations for unexpected reactions. While aqueous Pd(II) has been known for more than 40 years to oxidize alkenes rapidly to ketones24,25, which are unreactive towards alkynes under these conditions (Fig. 2b and Supplementary Data below Supplementary Fig. S4), our approach to reaction discovery revealed that carbon–carbon bond formation between alkynes and alkenes can outcompete alkene oxidation. Although other enone-forming coupling reactions such as the Horner–Wadsworth–Emmons reaction26 or aldol condensation are known, the mild reaction conditions, simple hydrocarbon starting materials and high efficiency of the transformation discovered here might render it an attractive alternative for addressing some macrocyclization problems in organic synthesis. In addition, the compatibility of this reaction with both organic and aqueous solvents might facilitate the synthesis of highly functionalized macrocyclic enones, of which there are numerous known biologically active examples27.
Six of the eight observed heterocoupling reactions in Fig. 3 involve a substrate that couples with itself under the reaction conditions (reactions involving either the alkyne or the aryl boronic acid; see Supplementary Data below Supplementary Fig. S4). In a DNA-templated format, heterocoupling without competitive homocoupling is possible because a substrate experiences its sequence-programmed heterocoupling partner at a much higher effective concentration than the concentration of another identical substrate molecule.
Biological macromolecules have previously been used to address specific problems in chemical reactivity28–30. In contrast, our approach uses the ability of nucleic acids to direct effective molarities and undergo in vitro selection and amplification to reveal bond-forming reactivity in a general manner. Once DNA-linked substrate pools are prepared, a single researcher can evaluate thousands of combinations of substrates and reaction conditions in a two-day experiment. Our approach requires water-soluble catalysts and aqueous reaction conditions that are at least partially compatible with DNA; however, as illustrated by the alkyne–alkene coupling reaction, some of the discovered reactions will proceed in a non-DNA-templated format in organic solvents. We expect that using this approach in a broad examination of reaction conditions including transition-metal complexes, Lewis acids, mild oxidants or reductants, and organic reagents will lead to the discovery of additional bond-forming reactions between simple and relatively unreactive functional groups.
Supplementary Material
Acknowledgements
We thank P. Kehayova for the initial analysis of early reaction discovery selections, and C. Bailey (Bauer Center for Genomics Research) for assistance with DNA microarray preparation and analysis. We thank DNA Software for assistance with screening coding and annealing sequences. This work was supported by the National Institutes of Health, the Office of Naval Research, and the Arnold and Mabel Beckman Foundation. M.W.K. and T.M.S. are NSF Graduate Research Fellows. M.M.R. is an NDSEG Graduate Research Fellow. K.S. is a Helen Hay Whitney Postdoctoral Research Fellow.
Footnotes
Supplementary Information accompanies the paper on www.nature.com/nature.
Competing interests statement The authors declare competing financial interests: details accompany the paper on www.nature.com/nature.
References
- 1.Stambuli JP, Hartwig JF. Recent advances in the discovery of organometallic catalysts using high-throughput screening assays. Curr. Opin. Chem. Biol. 2003;7:420–426. doi: 10.1016/s1367-5931(03)00056-5. [DOI] [PubMed] [Google Scholar]
- 2.Reetz MT. Combinatorial and evolution-based methods in the creation of enantioselective catalysts. Angew. Chem. Int. Edn Engl. 2001;40:284–310. [PubMed] [Google Scholar]
- 3.Stambuli JP, Stauffer SR, Shaughnessy KH, Hartwig JF. Screening of homogeneous catalysts by fluorescence resonance energy transfer. Identification of catalysts for room-temperature Heck reactions. J. Am. Chem. Soc. 2001;123:2677–2678. doi: 10.1021/ja0058435. [DOI] [PubMed] [Google Scholar]
- 4.Taylor SJ, Morken JP. Thermographic selection of effective catalysts from an encoded polymer-bound library. Science. 1998;280:267–270. doi: 10.1126/science.280.5361.267. [DOI] [PubMed] [Google Scholar]
- 5.Lober O, Kawatsura M, Hartwig JF. Palladium-catalyzed hydroamination of 1,3-dienes: a colorimetric assay and enantioselective additions. J. Am. Chem. Soc. 2001;123:4366–4367. doi: 10.1021/ja005881o. [DOI] [PubMed] [Google Scholar]
- 6.Evans CA, Miller SJ. Proton-activated fluorescence as a tool for simultaneous screening of combinatorial chemical reactions. Curr. Opin. Chem. Biol. 2002;6:333–338. doi: 10.1016/s1367-5931(02)00324-1. [DOI] [PubMed] [Google Scholar]
- 7.Calderone CT, Puckett JW, Gartner ZJ, Liu DR. Directing otherwise incompatible reactions in a single solution by using DNA-templated organic synthesis. Angew. Chem. Int. Edn Engl. 2002;41:4104–4108. doi: 10.1002/1521-3773(20021104)41:21<4104::AID-ANIE4104>3.0.CO;2-O. [DOI] [PubMed] [Google Scholar]
- 8.Gartner ZJ, Liu DR. The generality of DNA-templated synthesis as a basis for evolving non-natural small molecules. J. Am. Chem. Soc. 2001;123:6961–6963. doi: 10.1021/ja015873n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gartner ZJ, Kanan MW, Liu DR. Expanding the reaction scope of DNA-templated synthesis. Angew. Chem. Int. Edn Engl. 2002;41:1796–1800. doi: 10.1002/1521-3773(20020517)41:10<1796::aid-anie1796>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 10.Rosenbaum DM, Liu DR. Efficient and sequence-specific DNA-templated polymerization of peptide nucleic acid aldehydes. J. Am. Chem. Soc. 2003;125:13924–13925. doi: 10.1021/ja038058b. [DOI] [PubMed] [Google Scholar]
- 11.Li X, Liu DR. Stereoselectivity in DNA-templated organic synthesis and its origins. J. Am. Chem. Soc. 2003;125:10188–10189. doi: 10.1021/ja035379e. [DOI] [PubMed] [Google Scholar]
- 12.Wilson DS, Szostak JW. In vitro selection of functional nucleic acids. Annu. Rev. Biochem. 1999;68:611–647. doi: 10.1146/annurev.biochem.68.1.611. [DOI] [PubMed] [Google Scholar]
- 13.Joyce GF. Directed evolution of nucleic acid enzymes. Annu. Rev. Biochem. 2004;73:791–836. doi: 10.1146/annurev.biochem.73.011303.073717. [DOI] [PubMed] [Google Scholar]
- 14.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective ‘ligation’ of azides and terminal alkynes. Angew. Chem. Int. Edn Engl. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, et al. Bioconjugation by copper(I)-catalyzed azide-alkyne [3+2] cycloaddition. J. Am. Chem. Soc. 2003;125:3192–3193. doi: 10.1021/ja021381e. [DOI] [PubMed] [Google Scholar]
- 16.Burrows CJ, Muller JG. Oxidative nucleobase modifications leading to strand scission. Chem. Rev. 1998;98:1109–1152. doi: 10.1021/cr960421s. [DOI] [PubMed] [Google Scholar]
- 17.Kramer MF, Coen DM. In: Current Protocols in Molecular Biology. Ausubel FM, et al., editors. Vol. 2. New York: Wiley; 2001. pp. 15.1.1–15.1.3. [Google Scholar]
- 18.Bailey PD, Collier ID, Morgan KM. In: Comprehensive Organic Functional Group Transformations. Katritzky AR, Meth-Cohn O, Rees CW, editors. Vol. 5. New York: Pergamon; 1995. pp. 257–307. [Google Scholar]
- 19.Tsuji J. Palladium Reagents and Catalysts. New York: Wiley; 1995. [Google Scholar]
- 20.Heck RF. Palladium-catalyzed vinylation of organic halides. Org. React. 1982;27:345–390. [Google Scholar]
- 21.Li C-J, Chan T-H. Organic Reactions in Aqueous Media. New York: Wiley; 1997. [Google Scholar]
- 22.Miyaura N, Suzuki A. Palladium-catalyzed cross-coupling reactions of organoboron compounds. Chem. Rev. 1995;95:2457–2483. [Google Scholar]
- 23.Crabtree RH. The Organometallic Chemistry of the Transition Metals. New York: Wiley; 2001. [Google Scholar]
- 24.Smidt J, et al. Olefinoxydationmit palladiumchlorid-katalysatoren. Angew. Chem. 1962;74:93–102. [Google Scholar]
- 25.Smidt J, et al. Katalytische umsetzungen von olefinen an platinmetall-verbindungen: das consortium-verfahren zur herstellung von acetaldehyd. Angew. Chem. 1959;71:176–182. [Google Scholar]
- 26.Wadsworth WSJ, Emmons WD. Utility of phosphonate carbanions in olefin synthesis. J. Am. Chem. Soc. 1961;83:1733–1738. [Google Scholar]
- 27.Staunton J, Weissman KJ. Polyketide biosynthesis: a millennium review. Nat. Prod. Rep. 2001;18:380–416. doi: 10.1039/a909079g. [DOI] [PubMed] [Google Scholar]
- 28.Kohli RM, Walsh CT, Burkart MD. Biomimetic synthesis and optimization of cyclic peptide antibiotics. Nature. 2002;418:658–661. doi: 10.1038/nature00907. [DOI] [PubMed] [Google Scholar]
- 29.Breslow R. Biomimetic chemistry and artificial enzymes: catalysis by design. Acc. Chem. Res. 1995;28:146–153. [Google Scholar]
- 30.Schultz PG, Lerner RA. Completing the circle. Nature. 2002;418:485. doi: 10.1038/418485a. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.