Abstract
There have been many attempts to increase concentrations of the nutritionally essential sulphur amino acids by modifying their biosynthetic pathway in leaves of transgenic plants. This report describes the first modification of cysteine biosyntheis in developing seeds; those of the grain legume, narrow leaf lupin (Lupinus angustifolius, L.). Expression in developing lupin embryos of a serine acetyltransferase (SAT) from Arabidopsis thaliana (AtSAT1 or AtSerat 2;1) was associated with increases of up to 5-fold in the concentrations of O-acetylserine (OAS), the immediate product of SAT, and up to 26-fold in free cysteine, resulting in some of the highest in vivo concentrations of these metabolites yet reported. Despite the dramatic changes in free cysteine in developing embryos of SAT overexpressers, concentrations of free methionine in developing embryos, and the total cysteine and methionine concentrations in mature seeds were not significantly altered. Pooled F2 seeds segregating for the SAT transgene and for a transgene encoding a methionine- and cysteine-rich sunflower seed storage protein also had increased OAS and free cysteine, but not free methionine, during development, and no increase in mature seed total sulphur amino acids compared with controls lacking SAT overexpression. The data support the view that the cysteine biosynthetic pathway is active in developing seeds, and indicate that SAT activity limits cysteine biosynthesis, but that cysteine supply is not limiting for methionine biosynthesis or for storage protein synthesis in maturing lupin embryos in conditions of adequate sulphur nutrition. OAS and free methionine, but not free cysteine, were implicated as signalling metabolites controlling expression of a gene for a cysteine-rich seed storage protein.
Keywords: Aspartate, methionine, nutritive value, OAS, plant, SAT, seed storage protein, SerAT, sulphur, sulphur amino acid
Introduction
Sulphur is channelled into a wide range of essential metabolites via the reductive assimilation pathway that culminates in the formation of cysteine, the first stable, reduced sulphur metabolite in the cell (Saito, 2004; Hawkesford and De Kok, 2006). Cysteine is a precursor of methionine, which is generated in a series of three further reactions involving the transfer of the reduced sulphur moiety from cysteine to an amino acid backbone derived from homoserine, a product of the aspartate amino acid biosynthetic pathway (Fig. 1; Ravanel et al., 1998, 2004). Methionine and other aspartate-derived amino acids are nutritionally essential because of the lack of this pathway in animals (Azevedo et al., 2006). Similarly, animals cannot perform reductive sulphur assimilation or cysteine biosynthesis de novo, but they can derive cysteine from catabolism of methionine; hence dietary cysteine and methionine are often considered together (Tabe and Higgins, 1998).
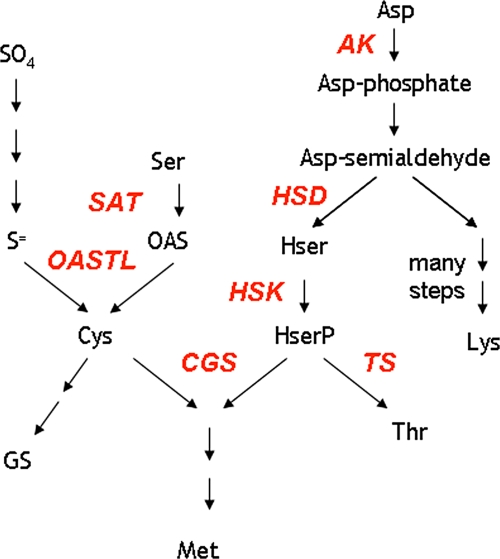
Fig. 1.
Schematic pathway of sulphur amino acid biosynthesis. SAT, serine acetyltransferase; OASTL, OAS (thiol) lyase; AK, aspartate kinase; HSD, homoserine dehydrogenase; HSK, homoserine kinase; CGS, cystathionine γ-synthase; TS, threonine synthase; Asp, aspartic acid; Asp-phosphate, aspartate-4-phosphate; Asp-semialdehyde, aspartate-4-semialdehyde; Hser, homoserine; HserP, O-phosphohomoserine; Lys, lysine; SO4, sulphate; S=, sulphide; Ser, serine; OAS, O-acetylserine; Cys, cysteine; GS, glutathione; Thr, threonine; Met, methionine. (This figure is available in colour at JXB online.)
Substantial effort has been expended over many years to enrich the plant parts used as food and feed in under-represented essential amino acids, most notably lysine in cereal grains, and cysteine and methionine in legume seeds (Tabe and Higgins, 1998; Shewry, 2007). Two distinct strategies have been used: manipulation of the pathways of amino acid biosynthesis, and creation of a storage sink by expression of a protein rich in the relevant amino acid (Amir and Tabe, 2006; Shewry, 2007). In the case of the sulphur amino acids, many different steps of the pathways of reductive sulphur assimilation and sulphur amino acid biosynthesis have been manipulated in a range of plant species. A critical control point occurs at the synthesis of cysteine by the cysteine synthase complex, which is a hetero-oligomer of the enzymes, serine acetyltransferase (SAT) and O-acetylserine-(thiol)-lyase (OASTL) (Wirtz and Hell, 2006). In plant tissues, OASTL is present in large excess over SAT, making SAT the limiting activity (Wirtz and Hell, 2006; Haas et al., 2008). A number of authors have reported the effects of overexpressing SAT in transgenic plants using constitutive promoters. The effects of these manipulations were assayed mainly in vegetative plant parts, and usually included several-fold increases in steady-state pools of free cysteine (Sirko et al., 2004). In contrast, plant modification with genes encoding heterologous, ‘sulphur-sink’ proteins mainly involved transgene expression in developing seeds, with the aim of accumulating high levels of the foreign protein stored stably in the mature seeds (Tabe and Higgins, 1998; Amir and Tabe, 2006).
Transgenic chickpeas and narrow leaf lupins were previously enriched with methionine by seed-specific expression of a naturally cysteine- and methionine-rich protein from sunflower. The sunflower seed albumin (SSA), containing 8% cysteine residues and 16% methionine residues, accumulated to ∼4–5% of total seed protein in some transgenic lines. The concentration of total methionine was doubled in mature seeds of the transgenic lupins compared with non-transgenic controls. Surprisingly, seed cysteine concentrations were not increased, meaning that total seed sulphur amino acids were only increased by ∼20% in the transgenic lupins compared with controls (Molvig et al., 1997). Although the enrichment in seed methionine was enough to increase significantly the nutritive value of the transgenic lupin seeds for chickens (Ravindran et al., 2002) and for ruminant (White et al., 2001) and non-ruminant animals (Molvig et al., 1997), further increases in total seed sulphur amino acids are needed before the transgenic lupins could satisfy the full dietary requirements of animals for these essential nutrients. Developing seeds of transgenic lupins and chickpeas expressing SSA had decreased pools of free cysteine and methionine, indicating that the supply of these amino acids for storage protein synthesis may become limiting for seed sulphur amino acid accumulation in these transgenic plants grown with adequate sulphur supply (Tabe and Droux, 2002; Chiaiese et al., 2004). Developing embryos of chickpea and narrow leaf lupin have the capacity to assimilate sulphur supplied ex vivo in the form of sulphate. Furthermore, these grain legume embryos have significant amounts of several of the enzymes of cysteine and methionine biosynthesis, indicating that the sulphur amino acid biosynthetic pathway is active in the developing seeds (Tabe and Droux, 2001; Chiaiese, 2002).
The aim of the current work was to increase further the sulphur amino acid content of narrow leaf lupin seeds by combining two approaches. First, a seed-expressed gene for a cysteine feedback-insensitive, plastid-localized SAT from Arabidopsis thaliana (AtSAT1 or AtSERAT 2.1, At1g55920) was transferred to lupin, producing transgenic events with greatly increased SAT activity in developing embryos. Secondly, this modification was combined with expression of the SSA sulphur storage sink by crossing transgenic lines strongly expressing each transgene separately. Overexpression of the SAT enzyme resulted in dramatic increases in the concentrations of O-acetylserine (OAS), free cysteine, and, to a lesser extent, glutathione in developing embryos of transgenic lupins. The concentrations of free cysteine were the highest yet reported in planta, providing proof of the functionality of the rest of the cysteine biosynthetic pathway in the developing, transgenic lupin seeds. However, concentrations of free methionine were unaffected, and de-regulation of SAT throughout seed development did not detectably increase total sulphur amino acid concentrations in the mature seeds. Similarly, overexpression of AtSAT1 did not consistently augment the increase in mature seed sulphur amino acid concentrations achieved by expression of SSA alone in transgenic lupins. The data indicate roles for free methionine and OAS, but not free cysteine, in controlling expression of the cysteine-rich seed storage protein, conglutin δ.
Materials and methods
Gene constructs and plant transformation
A full-length cDNA encoding the SAT1 enzyme from A. thaliana (Ruffet et al., 1995) was cloned into the pBI121 binary vector, replacing the Escherichia coli β-glucuronidase protein-coding region. The cauliflower mosaic virus 35S promoter in this construct was then replaced with a 1.5 kb PCR fragment containing the promoter and 5′-untranslated region of a bean phaseolin gene (Slightom et al., 1983). All sequences were confirmed by DNA sequencing. This phaseolin promoter has been reported by several authors to direct seed-specific gene expression in transgenic dicot host plants (Sengupta-Gopalan et al., 1985; Bustos et al., 1989). A study in which the promoter was used to drive expression of the cell-autonomous, highly toxic diphtheria toxin A demonstrated that the phaseolin promoter was expressed in embryo and endosperm, put not seed coat of developing transgenic tobacco seeds. This extremely sensitive assay also revealed low level expression of the promoter in pollen mother cells of the transgenic tobacco (van der Geest et al., 1995).
The gene cassette, containing the promoter and 5′-untranslated regions of bean phaseolin, the complete protein-coding region of AtSAT1, and the 3′ region of the nos gene from Agrobacterium tumefaciens, was excised with PstI from the pBI121 vector backbone and ligated into pRM66, a twin T-DNA binary vector containing a cauliflower mosaic virus 35S-bar gene for plant selection, and tetracycline resistance for bacterial selection. Plasmid pRM66 was constructed according to the method of Matthews et al. (2001). A DNA fragment containing a pair of right and left T-DNA border regions was created by PCR amplification of the ‘twinning insert’ of Matthews et al., using these primers: GAATTCTAGAACTAGTGCGGTGTCATCTATGT and GAATTCTGCAGAAGCTTCGCGTCATCGGCGGG. This DNA fragment was digested with EcoRI and cloned into the EcoRI site of pKSB10, which was the binary vector pTAB10 (Tabe et al., 1995), with the XbaI and PstI sites removed. The final AtSAT1 binary plasmid was transferred to A. tumefaciens strain Agl0 (Lazo et al., 1991) by triparental mating.
Narrow leaf lupins (Lupinus angustifolius, L.) of cultivar (cv) Kalya were transformed with the AtSAT1 gene construct using the method of Pigeaire et al. (1997), except that insoluble polyvinylpyrrolidone was added to all regeneration and selection media at 3 g l−1. Regenerated shoots surviving phosphinothricin selection were induced to form roots by dipping for 20 s in 1 mg ml−1 indole-3-butyric acid followed by culture in B5 medium (Gamborg et al., 1968) containing 1 mg l−1 indole-3-butyric acid and 150 mg l−1 Timentin (SmithKline Beecham, Australia). Following induction of roots on transformed shoots, plantlets were transferred to soil.
Plant material
The ‘SSA Tg’ lupin was a homozygous, transgenic line of lupins (from cv Warrah), which expressed a chimeric SSA gene under control of the seed-specific promoter from a pea vicilin gene (Higgins et al., 1988). The pea vicilin and bean phaseolin genes both encode 7S seed storage proteins, and are thought to have evolved from a common ancestral gene (Borroto and Dure, 1987). The SSA Tg line accumulated SSA at ∼4–5% of total protein in mature seed. The presence of SSA was demonstrated in the protein bodies of cotyledons of the embryo, indicating faithful expression of the seed-specific promoter in the transgenic legume host (Molvig et al., 1997). Northern blotting demonstrated high level expression of the chimeric vicilin–SSA gene from at least 21 days after anthesis (DAA) in developing transgenic lupin embryos. (Tabe and Droux, 2002). Thus, the vicilin promoter controlling the SSA transgene was active in embryos at the stage when altered SAT activity and free cysteine were detected in the same tissues of the SAT transgenic lupins (see Results section).
Lupins were grown in soil containing 0.6 g l−1 of slow-release fertilizer [‘Aboska’, containing 15.2% (w/w) nitrogen, 6.9% (w/w) phosphorus, 5.2% (w/w) potassium sulphate] in 25 cm pots (one plant per pot) in a controlled-temperature greenhouse (23 °C 12 h day, 18 °C night). Pots were randomized over three greenhouse benches, and were watered from the surface as required. Lupin flowers were tagged on the day of anthesis, and developing pods were collected from upper lateral branches between 11 am and noon at 25 DAA. This developmental stage corresponded to approximately half of the maximum fresh weight. The pods were transferred to the laboratory in a humid container where seeds were removed and immediately dissected to separate the developing embryos (cotyledons and embryonic axis) and the seed coat. The embryo is the storage tissue of legume seeds, which have a transient, liquid endosperm at an early stage of development. At 25 DAA, the endosperm had been absorbed and the embryo and seed coat each contributed ∼50% of the total fresh weight of the narrow leaf lupin seed (see Results section). Between four and eight individual embryos were pooled and frozen in liquid nitrogen for each sample. Mature seed samples consisted of pools of at least 50 individual whole seeds (∼7–10 g). In mature seeds, the embryo contributes ∼75% and the seed coat 25% of total seed weight (data not shown). Pooled seed or embryo samples from three individual plants (three biological replicates) were analysed for each genotype, except in the case of the initial screening of T2 embryos from individual T1 plants.
Protein extraction and enzyme assays
Protein extraction from the frozen, pooled samples and assays for SAT (EC 2.3.1.30) and OASTL (EC 2.5.1.47) were performed as previously described, except that the frozen (undried) embryos were homogenized in 1.5 vols of the extraction buffer. Enzyme assays were performed as previously described (Tabe and Droux, 2001).
Analysis of amino acids and thiols
Samples of 4–8 pooled, frozen embryos were powdered in liquid nitrogen and extracted with 0.1 N HCl. Thiols and amino acids were quantified after derivatization with monobromobimane (Calbiochem) and AccQ-Tag (Waters), respectively. The derivatization procedure and separation of derivatives were performed as described in Wirtz et al. (2004) using the same HPLC system. The total amino acid composition of pooled, mature seed samples was determined using an amino acid analyser after complete hydrolysis of finely ground flour by an accredited commercial laboratory (Werribee Centre, Primary Industries Research, Victoria, Australia).
RNA expression analysis
Total RNA was extracted from pools of 4–8 frozen lupin embryos with phenol/chloroform, followed by precipitation with lithium chloride as previously described (Tabe and Droux, 2001). Samples of RNA were DNase treated using a ‘DNA-Free’ kit from Ambion. DNase-treated RNA was reverse transcribed with Invitrogen Superscript II using the method recommended by the manufacturer. Quantitative, real-time reverse transcriptase PCR (qRT-PCR) was carried out in a RotorGene 3000 thermal cycler (Corbett Research) using Hotstar Taq polymerase (Qiagen) as described by the manufacturer, with the following program: 15 min at 95 °C, followed by 40 cycles of 30 s at 94 °C, 30 s at 50 °C, 30 s at 72 °C; followed by melt analysis ramping from 50 °C to 99 °C with 5 s intervals. Probe sequences were derived as follows. A small cDNA library was made from RNA from 25 DAA developing L. angustifolius embryos using a cDNA synthesis kit from Pharmacia. Randomly selected clones were sequenced to identify those encoding either the cysteine- and methionine-poor conglutin β, or a control for normalizing gene expression across samples. A sequence with 91% identity to that of a putative 60S acidic ribosomal protein P1 from Lupinus albus (GenBank accession CA410528) was used as a control gene for normalizing expression of the target conglutins. A sequence with 86% identity to conglutin β from L. albus (TIGR transcript assembly TA306_3870, Childs et al., 2007) and 88% and 83% identity with two cDNA sequences for putative members of the conglutin β family from L. angustifolius (GenBank accessions EF455724 and EU352876, respectively) was used to design primers to quantify L. angustifolius conglutin β transcripts. Primers for conglutin δ were designed from the published sequence (Gayler et al., 1990). The primer sequences were as follows: 60S ribosomal acidic protein P1, ACCACTTGCAGCAGCAACTG and TCGCTTGGTGAAACTGCTTCC; conglutin β, TCCTCAAATCTCCGCTTGCTTG and TGACCCCTTCTTCCACGTCTAC; conglutin δ, AGGCACTGCGAGAACCACATAG and CCCTCACATTGCTCGCTTTGAC. The sequences of the L. angustifolius cDNA clones are available on request.
Results
Generation and characterization of SAT transgenic events
Narrow leaf lupin, cv Kalya, was transformed via A. tumefaciens, with a seed-expressed gene encoding a SAT isoform from A. thaliana. The SAT sequence used was originally denoted SAT-1, and was initially thought to encode a cytosol-localized protein (Murillo et al., 1995; Ruffet et al., 1995). The same gene was subsequently named SAT-p (Noji et al., 1998) and AtSerat 2;1 (Kawashima et al., 2005), and corresponds to locus At1g55920. The name AtSAT1 will be used in this report. In Arabidopsis, the AtSAT1 enzyme was shown to be plastid localized and cysteine feedback insensitive (Noji et al., 1998). Expression in transgenic lupin of the AtSAT1 transgene was controlled by the seed-specific promoter from a bean phaseolin gene (Slightom et al., 1983).
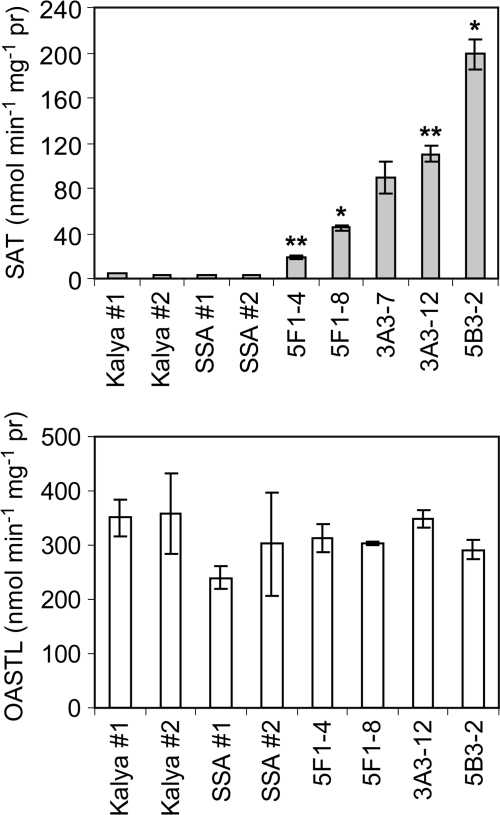
Nine independent T0 transgenic events were confirmed by PCR to contain the SAT transgene. Because of the limited number of seed on T0 plants, analysis was conducted in developing seed of the T2 generation. Seeds were sampled for analysis at 25 DAA, when they had attained approximately half their maximum fresh weight. This stage corresponds to the maturation phase of seed development, which is characterized by expression of seed storage proteins and accumulation of seed reserves. Protein extracts from pools of developing T2 embryos from individual T1 plants were screened for SAT enzyme activity. Of the nine confirmed transgenic events analysed, three events showed significantly increased SAT activity in T2 embryos. SAT activity was increased over the wild-type or SSA Tg controls in event 5F1 by 5- to 12-fold, in event 3A3 by 24- to 31-fold, and in event 5B3, for which a single T1 plant survived, by 49-fold (Fig. 2). There were no significant differences in the OASTL activities of any of the transgenic events compared with controls. Further characterization was performed on progeny of the following T1 plants: 5F1-4, 3A3-12, and 5B3-2. These genetic lineages are subsequently referred to as 5F1, 3A3, and 5B3.
Fig. 2.
SAT and OASTL enzyme activities in developing embryos of T2 transgenic narrow leaf lupins and controls. Each bar represents the mean ±SE for T2 embryos from one T1 transgenic plant. Between two and five technical replicates were performed, with extracts from 1–5 independent subsamples of embryos for each T1 plant. Symbols indicate the significance of the differences between each mean value and that of the wild-type control, cv Kalya (#P <0.1; *P <0.05; **P <0.01). Kalya #1 and #2 and SSA #1 and # 2 represent measurements for pooled embryo extracts for two separate plants of either the non-transgenic control Kalya or a transgenic lupin line homozygous for the SSA transgene.
Analysis of SAT activity and metabolite concentrations in developing embryos of transgenic events with increased SAT expression
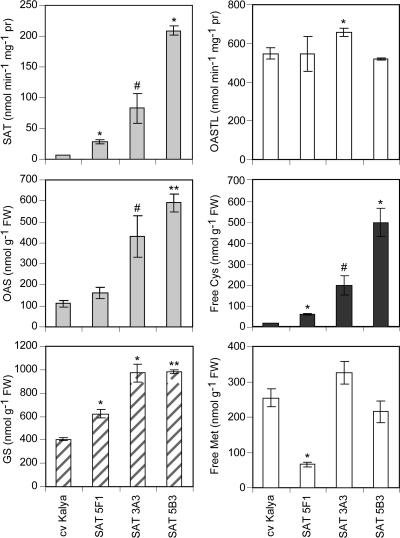
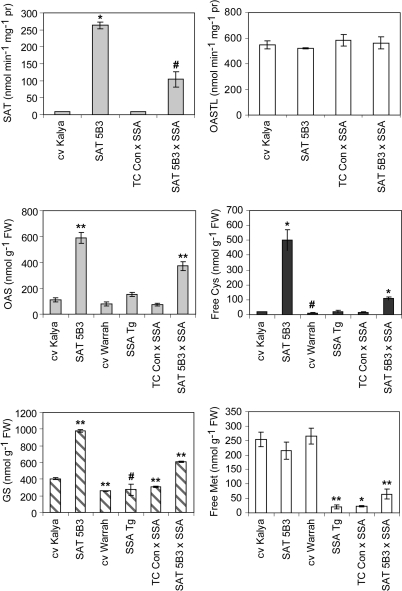
SAT PCR-positive T3 plants of the three selected transgenic genotypes, plus control plants, were grown in matched soil and glasshouse conditions, to produce T4 seed. SAT and OASTL activities, as well as the levels of several sulphur metabolites were measured in pools of developing T4 embryos from each of three individual T3 plants for each genotype (i.e. three PCR-confirmed biological replicates per genotype). In vitro SAT activity was significantly increased by 4-, 12-, and 30-fold in embryos of genotypes 5F1, 3A3, and 5B3, respectively, relative to the non-transgenic control, cv Kalya. There were no consistent differences in OASTL activity between the transgenic genotypes and the non-transgenic controls (Fig. 3). Between-plant variation was quite large, presumably partially due to segregation of transgenes in the developing seeds. Low variance in the OASTL activities for line 3A3 resulted in a statistically significantly higher OASTL activity in this line compared with the non-transgenic control. However, the OASTL activities of the other two lines with increased SAT, including 5B3 in which SAT was 30-fold higher than the wild type, had unchanged OASTL activities compared with the control. Therefore, despite the statistical significance, the 20% higher OASTL activity in line 3A3 was probably not biologically meaningful.
Fig. 3.
SAT and OASTL enzyme activities, and concentrations of sulphur metabolites in developing T4 embryos of three transgenic lupin events with increased SAT enzyme activity. Each bar represents the mean of three biological replicates ±SE; symbols indicate the significance of the differences between each mean value and that of the wild-type control, cv Kalya (#P <0.1; *P <0.05; **P <0.01). Free Cys, acid-soluble cysteine; GS, glutathione; free Met, acid-soluble methionine.
The levels of several key intermediates of the sulphur amino acid biosynthetic pathway were clearly altered in developing embryos of the SAT-overexpressing transgenic genotypes. Steady-state concentrations of OAS were increased by 1.5-, 3.9-, and 5.4-fold in genotypes 5F1, 3A3, and 5B3, respectively, relative to the non-transgenic control (Fig. 3). The corresponding increases in free cysteine concentrations were 3.3-, 10.5-, and 26.4-fold. Glutathione increased significantly by 1.5-, 2.4-, and 2.4-fold, respectively, in the same material; however, there were no consistent differences in the concentrations of free methionine in the transgenic embryos compared with the non-transgenic control. The free methionine level in embryos of line 5F1 was significantly less than that of the non-transgenic control, but the other SAT-overexpressing genotypes showed no significant difference in free methionine relative to controls. Therefore, as in the case of the OASTL activity in line 3A3, the low free methionine concentration in embryos of line 5F1 probably reflects biological variation rather than a consistent effect of altered SAT activity.
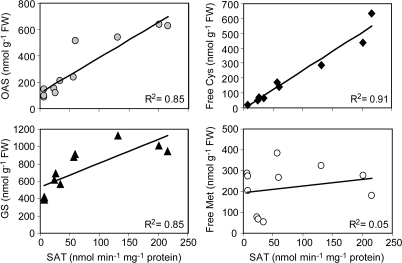
A direct, causal relationship between increased SAT activity and increased OAS and free cysteine concentrations was supported by the high correlations seen when SAT activities in individual developing T4 embryo samples for all genotypes were plotted against their corresponding OAS and free cysteine concentrations (Fig. 4). A clear correlation was also evident between total SAT activities and glutathione concentrations, although the graph suggested a possible threshold effect in this relationship. In contrast, there was no detectable correlation between SAT activities and concentrations of free methionine in individual samples (Fig. 4). The specificity of the observed effects was investigated further by determining the concentrations of sulphur metabolites in the embryos and seed coats separately of developing lupin seeds. At 25 DAA, the embryos constituted a mean of 53±2% of total seed weight for the four samples for which embryo and seed coat were analysed separately (see Table 1). The phaseolin promoter used to drive the SAT transgene is known to be expressed in filial, developing embryonic tissues where seed storage proteins accumulate, rather than in the maternal seed coat of developing dicotyledonous seeds (Bustos et al., 1989; van der Geest et al., 1995). As expected, it was found that the large increases in OAS and free cysteine concentrations observed in line 5B3 were specific to the embryo of the developing seeds (Table 1). Little or no difference was seen in the concentrations of these metabolites in seed coats of the 5B3 transgenic genotype compared with non-transgenic controls. Similarly, glutathione concentrations were increased specifically in the developing embryos rather than the seed coats of the strongest expressing SAT transgenic line. No differences in free methionine concentrations between genotypes were seen in either tissue. Sulphate was 10-fold more abundant in the embryos compared with the seeds coats, but the concentrations did not differ between the control and SAT 5B3 genotypes.
Fig. 4.
Correlations between SAT activities, and concentrations of sulphur metabolites in three individual biological replicates of developing T4 embryos of three transgenic lupin events with increased SAT enzyme activity, and the cv Kalya control. Free Cys, acid-soluble cysteine; GS, glutathione; free Met, acid-soluble methionine.
Table 1.
Concentrations of sulphur metabolites in embryos and seed coats from pooled samples of developing transgenic lupin seeds overexpressing SAT
| Sample | Metabolite (nmol g−1 FW) | ||||
| OAS | Free Cys | GS | Free Met | Sulphate | |
| cv Kalya embryo #1 | 157 | 17 | 367 | 298 | 6164 |
| cv Kalya embryo #2 | 132 | 19 | 406 | 248 | 5345 |
| SAT 5B3 embryo #1 | 628 | 637 | 949 | 180 | 4137 |
| SAT 5B3 embryo #2 | 506 | 427 | 984 | 187 | 5771 |
| cv Kalya seed coat #1 | 33 | 6 | 111 | 54 | 451 |
| cv Kalya seed coat #2 | 106 | 11 | 117 | 41 | 411 |
| SAT 5B3 seed coat #1 | 48 | 19 | 105 | 74 | 521 |
| SAT 5B3 seed coat #2 | 54 | 18 | 225 | 45 | 448 |
Samples labelled #1 and #2 were pooled embryo and corresponding seed coat samples (4–8 seeds per sample) from two separate plants for line 5B3, and for two separate seed samples from a single plant for cv Kalya.
OAS, O-acetylserine; free Cys, acid-soluble cysteine; GS, glutathione; free Met, acid-soluble methionine.
Analysis of mature seed composition and lupin seed storage protein gene expression in transgenic events with increased SAT expression
The composition of mature seeds represents the final result of gene expression throughout seed development. Mature seed nitrogen is mainly in the form of acid-insoluble amino acids incorporated in seed storage proteins. In order to assess the effects of SAT overexpression during seed development on lupin seed composition at the end of maturation, total amino acids (i.e. soluble amino acids plus protein-incorporated amino acids) were determined in hydrolysed flour from pooled mature seed samples of two plants of the control, cv Kalya, and the strongest SAT overexpresser, line 5B3. Because of the limited number of values (necessitated by the cost of the analyses), statistical tests were not appropriate. However, comparisons of the values in Table 2 reveal that total sulphur amino acid concentrations were not obviously different in mature seed flour of the SAT transgenic line compared with the non-transgenic control.
Table 2.
Concentrations of total cysteine and methionine in mature seeds of transgenic lupins overexpressing SAT
| Sample | Sulphur amino acid (μmol g−1 FW) |
Sulphur amino acid (% of total amino acids) |
||
| Cys | Met | Cys | Met | |
| cv Kalya #1 | 30.8 | 14.7 | 1.03 | 0.61 |
| cv Kalya #2 | 32.5 | 16.0 | 1.14 | 0.70 |
| SAT 5B3 #1 | 31.7 | 14.7 | 0.97 | 0.56 |
| SAT 5B3 #2 | 31.7 | 15.3 | 0.98 | 0.59 |
Values represent determinations on hydrolysed flour from pools of at least 7 g of whole mature seed from each of two plants of the non-transgenic control, cv Kalya and the SAT-overexpressing genotype 5B3. Flour samples contained ∼12% moisture (i.e. dry weight ∼88% of FW).
To facilitate comparison of developing and mature seeds, Table 3 shows the levels of free cysteine and glutathione per seed at both developmental stages. In the metabolically active developing seeds, the pool of free cysteine was ∼17-fold higher in the SAT-overexpressing line 5B3 than in the control. In the quiescent, mature seeds, the pool of free cysteine was 2-fold higher in the SAT overexpresser than in the control. Glutathione pools were 2-fold higher in seeds of line 5B3 than in cv Kalya at both stages, and were higher in mature seeds than in developing seeds. Consequently, in the mature seeds of line 5B3, glutathione was increased to 13% of total seed cysteine compared with an average of 6% in the cv Kalya control seeds. In contrast, the levels of protein-incorporated cysteine were slightly lower in seeds of line 5B3. The net result of these differences in individual pools of cysteine was a slight increase (mean 5.3%) in the levels of total cysteine per mature seed in the SAT overexpresser (Table 3).
Table 3.
Free cysteine and glutathione in mature versus developing seeds of transgenic lupins overexpressing SAT
| Sample | Free Cys | GS | Free Cys | GS | Protein Cys | Total Cys |
| nmol developing seed−1 | nmol mature seed−1 | |||||
| cv Kalya #1 | 4.4 | 92.3 | 2.9 | 292 | 5474 | 5782 |
| cv Kalya #2 | 5.0 | 88.2 | 3.0 | 431 | 5020 | 5472 |
| SAT 5B3 #1 | 84.5 | 135.2 | 5.7 | 705 | 5182 | 5921 |
| SAT 5B3 #2 | 78.3 | 212.7 | 8.4 | 909 | 4980 | 5931 |
For developing seeds, values were calculated from the concentrations of the metabolites in the embryo and seed coat samples shown in Table 1. For mature seeds, free Cys (acid-soluble cysteine) and GS (glutathione) were measured in hydrolysed flour from pools of at least 7 g of whole mature seed from each of two plants of the non-transgenic control, cv Kalya, and SAT-overexpressing genotype 5B3. Protein Cys was calculated by subtracting the sum of free Cys, GS, γ-glutamylcysteine and cysteinyl-glycine from total Cys.
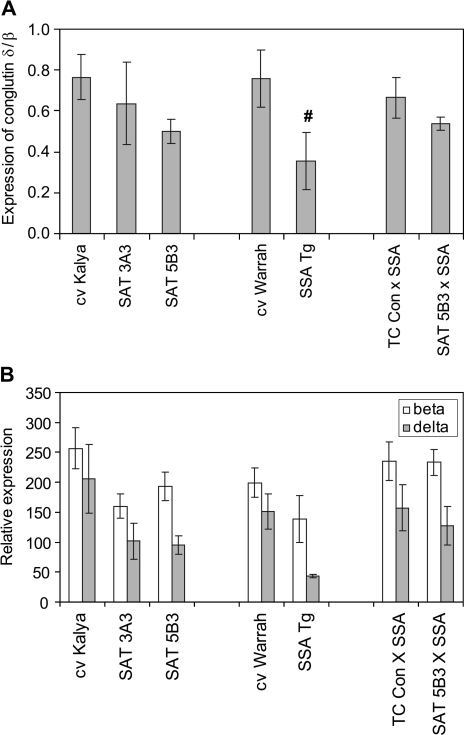
Seed protein composition can change in response to plant nutrition. In the case of grain legumes in particular, seed composition is modulated in response to plant sulphur nutrition. Metabolites of the sulphur amino acid biosynthetic pathway have been reported to influence expression of specific sulphur-rich or sulphur-poor seed storage proteins in soybean (Tabe et al., 2002), thereby maximizing protein accumulation for the amount of sulphur available. In order to assess the potential effects of altered sulphur metabolite concentrations on expression of genes encoding lupin seed storage proteins, qRT-PCR was used to measure the relative abundance of transcripts for the sulphur-rich conglutin δ class of storage proteins and the sulphur-poor conglutin β class of storage proteins in developing lupin embryos overexpressing SAT. Expression of conglutin δ was quantified relative to conglutin β, and expression of both conglutins was quantified relative to a gene for a putative ribosomal protein as a normalizing control for RNA amount. The data in Fig. 5A (first three genotypes) showed a possible trend towards decreased abundance of conglutin δ relative to conglutin β in SAT-overexpressing lines 3A3 and 5B3 compared with the cv Kalya control, but these differences were not statistically significant due to high biological variation within genotypes.
Fig. 5.
Relative abundance determined by qRT-PCR of transcripts for sulphur-rich conglutin δ and sulphur-poor conglutin β in developing transgenic lupin embryos expressing SAT and/or SSA. (A) Ratios of the relative abundance of conglutin δ/conglutin β. (B) Relative abundance of each conglutin alone (relative to the normalizing gene for a putative 60S ribosomal acid protein P1). Each bar represents the mean of three biological replicates ±SE; the symbol indicates the significance of the differences between each mean value and that of the wild-type control, cv Kalya (#P <0.1).
Analysis of F2 progeny of a cross between transgenic genotype 5B3 with 30-fold overexpression of SAT and a transgenic event with strong expression of the sulphur-rich storage protein, SSA
Crosses were made between T2 plants of SAT transgenic line 5B3 in which the presence of the transgene was confirmed by PCR (female parent) and a homozygous transgenic lupin event expressing a transgene for the sulphur-rich seed storage protein, SSA (SSA Tg, male parent; Molvig et al., 1997). The SAT 5B3 transgenic event was in a cv Kalya genetic background, while the SSA Tg transgenic event was in a cv Warrah genetic background. Individual F1 plants were screened by PCR, and three F1 plants confirmed to contain both transgenes were grown in matched soil and glasshouse conditions along with three F1 plants from a control cross. Because there were no negative segregants from line 5B3 itself, the control cross was made between a negative segregant from the transgenic genotype 5F1 [tissue culture control (TC Con), female parent] and the SSA Tg line (male parent). The cv Warrah genotype was also grown as an additional control.
SAT and OASTL activities and sulphur metabolite concentrations were measured in pools of F2 developing embryos. SAT activity was significantly higher in F2 embryos of the SAT 5B3×SSA Tg cross than in either the cv Kalya control or the TC Con×SSA Tg F2 embryo population (Fig. 6). There were no differences in OASTL activities among the embryo samples. A pattern similar to that of SAT activity was seen for OAS and free cysteine, with both metabolites showing significantly higher concentrations in the SAT 5B3×SSA Tg F2 embryos than in the controls, cv Kalya and cv Warrah, or the TC Con×SSA Tg F2 population. Glutathione was also significantly higher in the SAT 5B3×SSA Tg embryos than in controls. Glutathione concentrations were slightly lower in cv Warrah, and in the crosses involving the SSA Tg line in the Warrah genetic background, than in the cv Kalya control. Concentrations of free methionine were significantly lower than the Kalya control in all embryo samples expressing SSA. This is in agreement with our previous report that free methionine was strongly reduced in developing embryos of the SSA Tg event compared with cv Warrah control embryos, indicating a limitation in supply of this amino acid for protein synthesis in the transgenic SSA lupin seeds (Tabe and Droux, 2002). The concentration of free methionine in the F2 embryos segregating for both SAT and SSA genes was not significantly different from those in F2 embryos of the control cross that were segregating for SSA alone (Fig. 6).
Fig. 6.
SAT and OASTL enzyme activities, and concentrations of sulphur metabolites in developing lupin embryos. Data for cv Kalya and SAT 5B3 are as shown in Fig. 3, reproduced here for easy comparison. Data for the crosses were from pooled developing F2 embryos. The SSA Tg line was homozygous for the SSA transgene. Each bar represents the mean of three biological replicates ±SE; symbols indicate the significance of the differences between each mean value and that of the wild-type control, cv Kalya (#P <0.1; *P <0.05; **P <0.01). Free Cys, acid-soluble cysteine; GS, glutathione; free Met, acid-soluble methionine.
Total sulphur amino acid composition of pools of the F2 mature seeds was determined for two plants of each of the two crosses, and for the cv Kalya control. The data in Table 4 show that, as expected, total seed methionine was higher in both segregating seed samples expressing SSA than in the cv Kalya control. However, the total seed methionine in the SAT 5B3×SSA Tg sample was not higher than that in the TC Con×SSA Tg sample by more than the error of the two readings for each genotype (8%), indicating that overexpression of SAT, along with SSA, during seed development did not result in increased flow of sulphur amino acids into seed protein over and above that achieved by expression of SSA alone. Investigation of the expression of endogenous seed storage protein genes in these genotypes showed that the expression of conglutin δ relative to conglutin β was significantly decreased in the homozygous SSA transgenic parent line compared with the control cv Kalya. This effect was less (and not significant) in the segregating TC Con×SSA Tg sample. This diminution was not significantly affected by the presence of the SAT transgene in the SAT 5B3×SSA Tg sample (Fig. 5A).
Table 4.
Concentrations of total cysteine and methionine in mature F2 seeds of crosses between a transgenic lupin expressing SSA and either a strong SAT-overexpressing transgenic lupin (SAT 5B3) or a control (TC Con, a negative segregant from transgenic event 5F1)
| Sample | Sulphur amino acid (μmol g−1 FW) |
Sulphur amino acid (% of total amino acid) |
||
| Total Cys | Total Met | Total Cys | Total Met | |
| cv Kalya | 31.7 | 15.3 | 1.08 | 0.66 |
| TC Con×SSA | 32.9 | 23.3 | 1.06 | 0.98 |
| SAT 5B3×SSA | 32.5 | 24.0 | 1.10 | 0.97 |
Values are the mean of determinations on hydrolysed flour from pools of at least 7 g of mature F2 seed for each of two plants per genotype. The SDs of the measurements were ≤8%. Flour samples contained ∼12% moisture (i.e. dry weight ∼88% of FW).
Cys, cysteine; Met, methionine.
Discussion
Activity of cysteine biosynthesis in developing lupin seeds
Expression of a transgene encoding the plastid-targeted, cysteine feedback-insensitive AtSAT1 enzyme (At1g55920), driven by a seed storage protein gene promoter, gave large increases in total extractable SAT activity and large increases in the concentrations of OAS and free cysteine in developing lupin embryos. The concentrations of free cysteine measured in embryos of the best expresser of the introduced SAT gene were the highest of which we are aware (up to 500 nmol g−1 FW). These findings indicate that the other enzyme of the cysteine synthase complex (OASTL) is active in developing lupin embryos. Since cysteine is the first stable, reduced sulphur compound in the cell, the results also imply that the pathway of sulphur reduction to sulphide is active in the developing lupin embryos. In agreement with this, sulphate, the substrate for sulphur reduction, was found to be abundant in the developing embryos (Table 1).
These data support the findings of a previous report showing that maturing pods of lupins are supplied with sulphur predominantly in the form of sulphate via the phloem, and that the sulphur amino acids for storage protein synthesis may be largely synthesized in the developing embryos themselves (Tabe and Droux, 2001). The data from lupin, along with reports of significant activities of enzymes of sulphur reduction and cysteine biosynthesis in developing soybean seeds and chickpea seeds, suggest that this may be a general feature of grain legume seeds (Sexton and Shibles, 1999; Chiaiese, 2002; Chronis and Krishnan, 2003; Phartiyal et al., 2008). The situation may be different for oilseeds and cereals, in which it appears that sulphur is transported in the phloem predominantly in reduced forms (Lappartient and Touraine, 1996; Bourgis et al., 1999; Kuzuhara et al., 2000). However, it is noteworthy that recent characterization of knockout mutants of Arabidopsis suggested that putative segregants lacking all five SAT genes in the Arabidopsis genome were arrested at the torpedo stage during seed development. The results indicated that complete lack of SAT activity in the embryo was lethal, implying that SAT is essential for seed development, and that cysteine or other reduced forms of sulphur supplied through maternal tissues are not sufficient to allow embryo maturation (Watanabe et al., 2008).
Overexpression of a plastid-localized, cysteine feedback-insensitive SAT had large effects on cysteine accumulation in developing lupin seeds
A number of reports describe constitutive SAT overexpression in transgenic plants. Expression of plastid-targeted, cysteine feedback-sensitive or insensitive forms of a SAT from E. coli in transgenic tobacco or potato resulted in 2- to 3-fold increases in steady-state concentrations of free cysteine and glutathione in leaves (Harms et al., 2000; Blaszczyk et al., 2002). The relative importance of subcellular localization and cysteine feedback sensitivity was subsequently examined by expression of four different forms of a watermelon SAT in transgenic Arabidopsis. Increases of up to 7-fold and 10-fold in leaf free cysteine and glutathione, respectively, were achieved. The authors concluded that cysteine feedback insensitivity of the heterologous SAT enzyme was more important than its subcellular targeting to the plastid versus the cytosol for increasing cysteine accumulation in leaves of the transgenic plants (Noji and Saito, 2002). The highest levels of free cysteine and glutathione reported before the current work were achieved by overexpressing modified forms of the cysteine feedback-insensitive AtSAT3 enzyme from Arabidopsis (At3g13110) in transgenic tobacco. Concentrations of free cysteine achieved in that work reached 220 nmol g−1 FW in leaves (Wirtz and Hell, 2003, 2007). In the context of the previous work, the main reasons for the success of the current strategy in lupin were probably the cysteine feedback insensitivity of the SAT enzyme, along with the strong seed-specific expression of the transgene. Further work would be needed to determine the importance of the subcellular targeting of the heterologous SAT enzyme to the plastid.
Contrary to earlier beliefs, recent studies have demonstrated that although plastids are the unique subcellular site of sulphur reduction to sulphide, mitochondria, which normally contain the bulk of the cellular SAT activity, are the main site of OAS synthesis, while the cytoplasm is the main site of cysteine biosynthesis by OASTL. Consequently, sulphide, OAS, and cysteine must equilibrate freely across intracellular membranes (Haas et al., 2008; Heeg et al., 2008; Watanabe et al., 2008; Krueger et al., 2009). In the present study, correct targeting of the AtSAT1 transgene product to the plastid would mean that the plastid compartment contained the bulk of cellular SAT activity in the transgenic lupin embryos. Thus, the results indicate that either OAS and sulphide formed in the plastid were exchanged freely with other compartments, allowing increased cysteine synthesis in the cytosol, or that the increased cysteine synthesis occurred in the plastid itself in the SAT-overexpressing embryos. Furthermore, the data suggest that in developing, wild-type lupin embryos grown with adequate sulphur nutrition, the activity of SAT rather than the supply of sulphide was limiting for cysteine biosynthesis.
Overexpression of SAT in lupins did not augment free methionine pools in developing seeds
Despite the increased concentrations in free cysteine observed, there was no increase in free methionine concentrations in transgenic developing lupin embryos overexpressing SAT. The results imply that supplies of free cysteine were not limiting for methionine synthesis in developing lupin seeds. The data are consistent with the suggestion of others that flux to free methionine, at least in vegetative seedlings of Arabidopsis, is controlled ultimately by the supply of homoserine through the aspartate amino acid biosynthetic pathway (Lee et al., 2005). The enzymes cystathionine γ-synthase and threonine synthase regulate the partitioning of their common substrate, O-phosphohomoserine, between synthesis of methionine and threonine (see Fig. 1 and Amir and Tabe, 2006). Recent work with transgenic tobacco expressing de-regulated forms of both cystathionine γ-synthase and a bacterial aspartate kinase in their leaves confirms the importance of this branch of the biosynthetic pathway for controlling methionine and threonine levels in plants (Hacham et al., 2008).
Overexpression of SAT in lupins increased storage of cysteine in glutathione but not in protein in mature seeds
Overexpression of SAT in embryos inflated pools of free cysteine by up to 17-fold during the maturation phase of seed development. This resulted in a number of subtle changes in the sizes of cysteine pools in mature seeds. Free cysteine made up the smallest pool of mature seed cysteine, and was 2-fold higher in the strongest SAT overexpresser, 5B3 (Table 3). Free amino acid pools tend to be larger in metabolically active, developing seeds than in quiescent, mature seeds, due to incorporation of the free amino acids into more stable storage products during maturation, and to turnover of excess free amino acid, as reported, for example, for lysine in both wild-type tobacco and transgenic tobacco overexpressing enzymes of lysine biosynthesis (Karchi et al., 1994). The next largest pool of cysteine in mature lupin seeds was glutathione, which accumulated to a 2-fold greater extent in seeds of the SAT overexpresser, where it constituted 13% of total seed cysteine, in contrast to 6% in the control mature seeds. However, the largest pool of cysteine, that which was incorporated into protein, was slightly lower in seeds of line 5B3 (Table 3). From these results it can be concluded that increasing the supply of free cysteine during the phase of seed storage protein synthesis did not ‘push’ more cysteine into protein, although it did result in a doubling of stored glutathione in the mature seeds.
In contrast, previous reports have shown that increasing demand for sulphur amino acids by expressing high levels of the cysteine- and methionine-rich storage protein, SSA, was able to ‘pull’ 20% more total cysteine plus methionine into the mature seed protein. Unexpectedly, this was due to a doubling of seed total methionine concentration, with no increase in seed cysteine (Molvig et al., 1997). From the measured amount of SSA present, it could be calculated that all the extra methionine was in the SSA protein. SSA contains a high concentration of cysteine as well as methionine. The sequestration of cysteine in SSA was apparently made possible by decreased synthesis of endogenous, cysteine-rich seed storage proteins, as evidenced by strongly decreased levels of transcripts for the cysteine-rich conglutin δ in the SSA transgenic lupin seeds (Tabe and Droux, 2002). Thus, increasing demand for both cysteine and methionine was apparently met by increased flux through the biosynthetic pathway, as well as by re-allocation of cysteine via changed expression of seed storage protein genes in response to altered levels of signalling sulphur metabolites (Tabe et al., 2002).
Free methionine and OAS have been reported to act as opposed signals that modulate expression of sulphur-rich and sulphur-poor soybean seed storage proteins in response to plant sulphur nutrition (Holowach et al., 1984a, b, 1986; Kim et al., 1999). Thus, transcription of the gene for the sulphur-poor β-subunit of soybean conglycinin is reported to be up-regulated by OAS and down-regulated by methionine (Hirai et al., 2002). In the same study, the converse was observed for the sulphur-rich glycinin protein in soybean cotyledons treated with OAS and/or methionine. It was previously hypothesized that synthesis of SSA in transgenic lupin and chickpea seeds triggered signals (such as decreased free methionine and increased OAS) that mimicked the effects of sulphur nutritional deficiency on expression of endogenous seed storage proteins (Tabe and Droux, 2002; Tabe et al., 2002; Chiaiese et al., 2004).
In the current work, levels of both these signalling metabolites were affected by SAT or SSA expression during seed development. OAS was strongly increased in developing embryos of the two highest SAT-overexpressing transgenic lupin lines (Fig. 3). This correlated with a trend towards lower transcripts for the sulphur-rich conglutin δ relative to the sulphur-poor conglutin β in embryos of these transgenic events (Fig. 5A). In agreement with this, measurement of cysteine pools in mature seeds showed that protein-incorporated cysteine was in fact slightly lower in mature seeds of the SAT transgenic line 5B3 compared with controls (Table 3). In the SSA transgenic lupin embryos, transcripts of conglutin δ were significantly lower than in either the cv Kalya or cv Warrah controls (Fig. 5A, P <0.1 for both). These differences correlated with a markedly lower concentration of free methionine in the homozygous, transgenic SSA embryos compared with the non-transgenic controls.
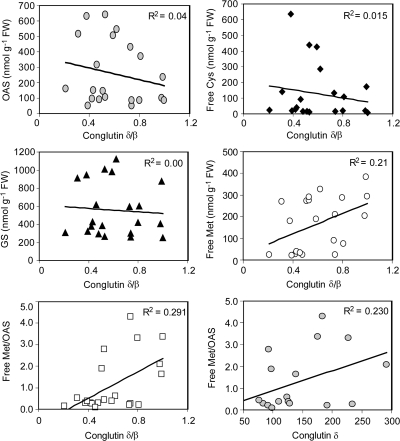
When the transcripts of conglutins β and δ were considered separately, it was clear that the main reasons for differences in the ratios of the two conglutins were decreases in the conglutin δ transcripts rather than increases in conglutin β (both quantified relative to the normalizing control gene encoding a putative ribosomal protein, Fig. 5B). The high degree of variation between biological replicates within a genotype meant that there were no statistically significant differences between genotypes in either conglutin β or δ transcripts (relative to the normalizing control gene). Despite the lack of statistical significance, the mean relative abundances of transcripts for conglutin δ were obviously lower in the SAT transgenic lines compared with the cv Kalya control (by 50%), and decreased by 70% in the SSA Tg compared with the cv Warrah control. These differences correlated with increased levels of OAS in the former case and decreased levels of free methionine in the latter case. Transcripts for the sulphur-poor conglutin β were also decreased (by 25–37%) in the above comparisons (Fig. 5B), indicating that this gene was not regulated by sulphur metabolites in the same way as reported for the sulphur-poor soybean conglycinin β. Together, these results indicate that free methionine and, to a lesser extent, OAS modulated transcripts of the endogenous, sulphur-rich conglutin δ. Consistent with this, comparison of the concentrations of all the sulphur metabolites with the ratios and individual levels of conglutin β and δ transcripts across all the genotypes showed that the best correlations were between the concentrations of free methionine (or the ratios of free methionine over OAS) and the relative abundance of conglutin δ transcripts (or the ratios of conglutin δ over β; Fig. 7, and results not shown). However, the relatively weak correlations indicate that other factors also influenced regulation of seed storage protein gene expression. The lack of any detectable correlations between the conglutin transcripts and either free cysteine or glutathione suggests little signalling role for these metabolites, despite the fact that conglutin δ is rich in cysteine, but contains no methionine (Gayler et al., 1990).
Fig. 7.
Correlations between concentrations of sulphur metabolites in developing embryos and relative abundance of conglutins in mature lupin seeds. The data are for each of three individual biological replicates of the following genotypes: cv Kalya, SAT 5B3 (T4 seed), cv Warrah, SSA Tg, and F2 progeny of the crosses TC Con×SSA and SAT 5B3×SSA. Free Cys, acid-soluble cysteine; GS, glutathione; free Met, acid-soluble methionine.
In summary, overexpression of SAT dramatically increased pools of free cysteine and OAS, but not free methionine, in developing lupin embryos during the phase of storage protein accumulation. Increased OAS, and, in SSA-containing genotypes, decreased free methionine probably acted as signals that slightly down-regulated expression of an endogenous cysteine-rich seed storage protein. The end results of these processes were mature seeds that contained elevated levels of free cysteine and glutathionine, but slightly less protein-incorporated cysteine. Because protein cysteine is by far the largest pool of cysteine in mature seeds, the net result was little change in total cysteine levels in mature seeds. Thus, increasing the supply of free cysteine during the phase of seed storage protein gene expression was not able to increase incorporation of cysteine into storage proteins. Crossing with the strongest SAT overexpresser could not correct the apparent limitation to free sulphur amino acid supply in a transgenic lupin with increased demand from synthesis of SSA, indicating that the inferred limits to supply did not lie in the cysteine biosynthetic pathway. Further characterization of progeny lines homozygous for both SAT and SSA transgenes will be needed to determine whether there were any subtle effects of SAT overexpression on storage of cysteine and methionine in SSA-expressing transgenic lupins. The present results indicate that it will be necessary to increase free methionine availability in order to stimulate accumulation of both the foreign SSA protein and endogenous cysteine-rich seed proteins. Alternative strategies focused on manipulation of other branches of the methionine biosynthetic pathway are being pursued.
Acknowledgments
This work was partly supported by funding from the Grains Research and Development Corporation. We gratefully acknowledge the competent technical assistance of Joerg Kittelmann during a 6-month traineeship at CSIRO Plant Industry.
Glossary
Abbreviations
- AtSAT1.
serine acetyltransferase encoded at Arabidopsis thaliana locus At1g55920
- cv
cultivar
- DAA
days after anthesis
- FW
fresh weight
- OAS
O-acetylserine
- OASTL
O-acetylserine (thiol) lyase
- qRT-PCR
quantitative real-time reverse transcriptase PCR
- SAT
serine acetyltransferase
- SSA
sunflower seed albumin
References
- Amir R, Tabe L. Molecular approaches to improving plant methionine content. In: Pawan KJ, Rana PS, editors. Plant genetic engineering. Metabolic engineering and molecular farming II. Vol. 8. Houston, TX: Studium Press LLC; 2006. pp. 1–26. [Google Scholar]
- Azevedo RA, Lancien M, Lea PJ. The aspartic acid metabolic pathway, an exciting and essential pathway in plants. Amino Acids. 2006;30:143–162. doi: 10.1007/s00726-005-0245-2. [DOI] [PubMed] [Google Scholar]
- Blaszczyk A, Sirko L, Hawkesford MJ, Sirko A. Biochemical analysis of transgenic tobacco lines producing bacterial serine acetyltransferase. Plant Science. 2002;162:589–597. [Google Scholar]
- Borroto K, Dure L. The globulin seed storage proteins of flowering plants are derived from two ancestral genes. Plant Molecular Biology. 1987;8:113–131. doi: 10.1007/BF00025323. [DOI] [PubMed] [Google Scholar]
- Bourgis F, Roje S, Nuccio ML, et al. S-Methylmethionine plays a major role in phloem sulfur transport and is synthesized by a novel type of methyltransferase. The Plant Cell. 1999;11:1485–1498. doi: 10.1105/tpc.11.8.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bustos MM, Guiltinan MJ, Jordano J, Begum D, Kalkan FA, Hall TC. Regulation of β-glucuronidase expression in transgenic tobacco plants by an A/T-rich, cis-acting sequence found upstream of a French bean β-phaseolin gene. The Plant Cell. 1989;1:839–853. doi: 10.1105/tpc.1.9.839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaiese P. Modifying chickpea seed composition by manipulating sulfur supply and demand. 2002 PhD Thesis, Australian National University, Canberra, Australia. [Google Scholar]
- Chiaiese P, Ohkama-Ohtsu N, Molvig L, Godfree R, Dove H, Hocart C, Fujiwara T, Higgins TJV, Tabe LM. Sulphur and nitrogen nutrition influence the response of chickpea seeds to an added, transgenic sink for organic sulphur. Journal of Experimental Botany. 2004;55:1889–1901. doi: 10.1093/jxb/erh198. [DOI] [PubMed] [Google Scholar]
- Childs KL, Hamilton JP, Zhu W, Ly E, Cheung F, Wu H, Rabinowicz PD, Town CD, Buell CR, Chan AP. The TIGR plant transcript assemblies database. Nucleic Acids Research. 2007;35:D846–D851. doi: 10.1093/nar/gkl785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chronis D, Krishnan HB. Sulfur assimilation in soybean: molecular cloning and characterization of O-acetylserine (thiol) lyase (cysteine synthase) Crop Science. 2003;43:1819–1827. [Google Scholar]
- Gamborg OL, Miller RA, Ojima K. Nutrient requirements of suspension cultures of soybean root cells. Experimental Cell Research. 1968;50:151–158. doi: 10.1016/0014-4827(68)90403-5. [DOI] [PubMed] [Google Scholar]
- Gayler KR, Kolivas S, Macfarlane AJ, Lilley GG, Baldi M, Blagrove RJ, Johnson ED. Biosynthesis, cDNA and amino-acid-sequences of a precursor of conglutin-delta, a sulfur-rich protein from Lupinus angustifolius. Plant Molecular Biology. 1990;15:879–893. doi: 10.1007/BF00039427. [DOI] [PubMed] [Google Scholar]
- Haas FH, Heeg C, Queiroz R, Bauer A, Wirtz M, Hell R. Mitochondrial serine acetyltransferase functions as a pacemaker of cysteine synthesis in plant cells. Plant Physiology. 2008;148:1055–1067. doi: 10.1104/pp.108.125237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacham Y, Matityahu I, Schuster G, Amir R. Over-expression of mutated forms of aspartate kinase and cystathionine γ-synthase in tobacco leaves resulted in the high accumulation of methionine and threonine. The Plant Journal. 2008;54:260–271. doi: 10.1111/j.1365-313X.2008.03415.x. [DOI] [PubMed] [Google Scholar]
- Harms K, Von Ballmoos P, Brunold C, Hofgen R, Hesse H. Expression of a bacterial serine acetyltransferase in transgenic potato plants leads to increased levels of cysteine and glutathione. The Plant Journal. 2000;22:335–343. doi: 10.1046/j.1365-313x.2000.00743.x. [DOI] [PubMed] [Google Scholar]
- Hawkesford MJ, De Kok LJ. Managing sulphur metabolism in plants. Plant, Cell and Environment. 2006;29:382–395. doi: 10.1111/j.1365-3040.2005.01470.x. [DOI] [PubMed] [Google Scholar]
- Heeg C, Kruse C, Jost R, Gutensohn M, Ruppert T, Wirtz M, Hell R. Analysis of the Arabidopsis O-acetylserine(thiol)lyase gene family demonstrates compartment-specific differences in the regulation of cysteine synthesis. The Plant Cell. 2008;20:168–185. doi: 10.1105/tpc.107.056747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Higgins TJV, Newbigin EJ, Spencer D, Llewellyn DJ, Craig S. The sequence of a pea vicilin gene and its expression in transgenic tobacco plants. Plant Molecular Biology. 1988;11:683–695. doi: 10.1007/BF00017468. [DOI] [PubMed] [Google Scholar]
- Hirai MY, Kim H, Hayashi H, Chino M, Naito S, Fujiwara T. Independent roles of methionine and O-acetyl-L-serine in the regulation of the β-subunit gene of β-conglycinin. Soil Science and Plant Nutrition. 2002;48:87–94. [Google Scholar]
- Holowach LP, Madison JT, Thompson JF. Studies on the mechanism of regulation of the mRNA level for a soybean storage protein subunit by exogenous L-methionine. Plant Physiology. 1986;80:561–567. doi: 10.1104/pp.80.2.561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowach LP, Thompson JF, Madison JT. Storage protein composition of soybean cotyledons grown in vitro in media of various sulfate concentrations in the presence and absence of exogenous L-methionine. Plant Physiology. 1984a;74:584–589. doi: 10.1104/pp.74.3.584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holowach LP, Thompson JF, Madison JT. Effects of exogenous methionine on storage protein composition of soybean cotyledons cultured in vitro. Plant Physiology. 1984b;74:576–583. doi: 10.1104/pp.74.3.576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karchi H, Shaul O, Galili G. Lysine synthesis and catabolism are coordinately regulated during tobacco seed development. Proceedings of the National Academy of Sciences, USA. 1994;91:2577–2581. doi: 10.1073/pnas.91.7.2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawashima CG, Berkowitz O, Hell R, Noji M, Saito K. Characterization and expression analysis of a serine acetyltransferase gene family involved in a key step of the sulfur assimilation pathway in Arabidopsis. Plant Physiology. 2005;137:220–230. doi: 10.1104/pp.104.045377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H, Hirai MY, Hayashi H, Chino M, Naito S, Fujiwara T. Role of O-acetyl-L-serine in the coordinated regulation of the expression of a soybean seed storage-protein gene by sulfur and nitrogen nutrition. Planta. 1999;209:282–289. doi: 10.1007/s004250050634. [DOI] [PubMed] [Google Scholar]
- Krueger S, Niehl A, Martin MC, Steinhauser D, Donath A, Hildebrandt T, Romero LC, Hoefgen R, Gotor C, Hesse H. Analysis of cytosolic and plastidic serine acetyltransferase mutants and subcellular metabolite distributions suggests interplay of the cellular compartments for cysteine biosynthesis in Arabidopsis. Plant, Cell and Environment. 2009;32:349–367. doi: 10.1111/j.1365-3040.2008.01928.x. [DOI] [PubMed] [Google Scholar]
- Kuzuhara Y, Isobe A, Awazuhara M, Fujiwara T, Hayashi H. Glutathione levels in phloem sap of rice plants under sulfur deficient conditions. Soil Science and Plant Nutrition. 2000;46:265–270. [Google Scholar]
- Lappartient AG, Touraine B. Demand-driven control of root ATP sulfurylase activity and SO42–uptake in intact canola. The role of phloem-translocated glutathione. Plant Physiology. 1996;111:147–157. doi: 10.1104/pp.111.1.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lazo GR, Stein PA, Ludwig RA. A DNA transformation-competent Arabidopsis genomic library in Agrobacterium. Bio-Technology. 1991;9:963–967. doi: 10.1038/nbt1091-963. [DOI] [PubMed] [Google Scholar]
- Lee M, Martin MN, Hudson AO, Lee J, Muhitch MJ, Leustek T. Methionine and threonine synthesis are limited by homoserine availability and not the activity of homoserine kinase in Arabidopsis thaliana. The Plant Journal. 2005;41:685–696. doi: 10.1111/j.1365-313X.2004.02329.x. [DOI] [PubMed] [Google Scholar]
- Matthews PR, Wang MB, Waterhouse PM, Thornton S, Fieg SJ, Gubler F, Jacobsen JV. Marker gene elimination from transgenic barley, using co-transformation with adjacent ‘twin T-DNAs’ on a standard Agrobacterium transformation vector. Molecular Breeding. 2001;7:195–202. [Google Scholar]
- Molvig L, Tabe LM, Eggum BO, Moore AE, Craig S, Spencer D, Higgins TJV. Enhanced methionine levels and increased nutritive value of seeds of transgenic lupins (Lupinus angustifolius L.) expressing a sunflower seed albumin gene. Proceedings of the National Academy of Sciences, USA. 1997;94:8393–8398. doi: 10.1073/pnas.94.16.8393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murillo M, Foglia R, Diller A, Lee S, Leustek T. Serine acetyltransferase from Arabidopsis thaliana can functionally complement the cysteine requirement of a cysE mutant strain of Escherichia coli. Cellular and Molecular Biology Research. 1995;41:425–433. [PubMed] [Google Scholar]
- Noji M, Inoue K, Kimura N, Gouda A, Saito K. Isoform-dependent differences in feedback regulation and subcellular localization of serine acetyltransferase involved in cysteine biosynthesis from Arabidopsis thaliana. Journal of Biological Chemistry. 1998;273:32739–32745. doi: 10.1074/jbc.273.49.32739. [DOI] [PubMed] [Google Scholar]
- Noji M, Saito K. Molecular and biochemical analysis of serine acetyltransferase and cysteine synthase: towards sulfur metabolic engineering in plants. Amino Acids. 2002;22:231–243. doi: 10.1007/s007260200011. [DOI] [PubMed] [Google Scholar]
- Phartiyal P, Kim WS, Cahoon RE, Jez JM, Krishnan HB. The role of 5′-adenylylsulfate reductase in the sulfur assimilation pathway of soybean: molecular cloning, kinetic characterization, and gene expression. Phytochemistry. 2008;69:356–364. doi: 10.1016/j.phytochem.2007.07.013. [DOI] [PubMed] [Google Scholar]
- Pigeaire A, Abernethy D, Smith PM, Simpson K, Fletcher N, Lu CY, Atkins CA, Cornish E. Transformation of a grain legume (Lupinus angustifolius L.) via Agrobacterium tumefaciens-mediated gene transfer to shoot apices. Molecular Breeding. 1997;3:341–349. [Google Scholar]
- Ravanel S, Block MA, Rippert P, Jabrin S, Curien G, Rebeille F, Douce R. Methionine metabolism in plants—chloroplasts are autonomous for de novo methionine synthesis and can import S-adenosylmethionine from the cytosol. Journal of Biological Chemistry. 2004;279:22548–22557. doi: 10.1074/jbc.M313250200. [DOI] [PubMed] [Google Scholar]
- Ravanel S, Gakiere B, Job D, Douce R. The specific features of methionine biosynthesis and metabolism in plants. Proceedings of the National Academy of Sciences, USA. 1998;95:7805–7812. doi: 10.1073/pnas.95.13.7805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ravindran V, Tabe LM, Molvig L, Higgins TJV, Bryden WL. Nutritional evaluation of transgenic high-methionine lupins (Lupinus angustifolius L) with broiler chickens. Journal of the Science of Food and Agriculture. 2002;82:280–285. [Google Scholar]
- Ruffet ML, Lebrun M, Droux M, Douce R. Subcellular distribution of serine acetyltransferase from Pisum sativum and characterization of an Arabidopsis thaliana putative cytosolic isoform. European Journal of Biochemistry. 1995;227:500–509. doi: 10.1111/j.1432-1033.1995.tb20416.x. [DOI] [PubMed] [Google Scholar]
- Saito K. Sulfur assimilatory metabolism. The long and smelling road. Plant Physiology. 2004;136:2443–2450. doi: 10.1104/pp.104.046755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengupta-Gopalan C, Reichert NA, Barker RF, Hall TC, Kemp JD. Developmentally regulated expression of the bean beta-phaseolin gene in tobacco seed. Proceedings of the National Academy of Sciences, USA. 1985;82:3320–3324. doi: 10.1073/pnas.82.10.3320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sexton PJ, Shibles RM. Activity of ATP sulfurylase in reproductive soybean. Crop Science. 1999;39:131–135. [Google Scholar]
- Shewry PR. Improving the protein content and composition of cereal grain. Journal of Cereal Science. 2007;46:239–250. [Google Scholar]
- Sirko A, Blaszczyk A, Liszewska F. Overproduction of SAT and/or OASTL in transgenic plants: a survey of effects. Journal of Experimental Botany. 2004;55:1881–1888. doi: 10.1093/jxb/erh151. [DOI] [PubMed] [Google Scholar]
- Slightom JL, Sun SM, Hall TC. Complete nucleotide sequence of a French bean storage protein gene: phaseolin. Proceedings of the National Academy of Sciences, USA. 1983;80:1897–1901. doi: 10.1073/pnas.80.7.1897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe LM, Droux M. Sulfur assimilation in developing lupin cotyledons could contribute significantly to the accumulation of organic sulphur reserves in the seed. Plant Physiology. 2001;126:176–187. doi: 10.1104/pp.126.1.176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe LM, Droux M. Limits to sulfur accumulation in transgenic lupin seeds expressing a foreign sulphur-rich protein. Plant Physiology. 2002;128:1137–1148. doi: 10.1104/pp.010935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabe L, Hagan N, Higgins T. Plasticity of seed protein composition in response to nitrogen and sulfur availability. Current Opinion in Plant Biology. 2002;5:212–217. doi: 10.1016/s1369-5266(02)00252-2. [DOI] [PubMed] [Google Scholar]
- Tabe L, Higgins TJV. Engineering plant protein composition for improved nutrition. Trends in Plant Science. 1998;3:282–286. [Google Scholar]
- Tabe LM, Wardley-Richardson T, Ceriotti A, Aryan A, McNabb W, Moore A, Higgins TJV. A biotechnological approach to improving the nutritive value of alfalfa. Journal of Animal Science. 1995;73:2752–2759. doi: 10.2527/1995.7392752x. [DOI] [PubMed] [Google Scholar]
- van der Geest AHM, Frisch DA, Kemp JD, Hall TC. Cell ablation reveals that expression from the phaseolin promoter is confined to embryogenesis and microsporogenesis. Plant Physiology. 1995;109:1151–1158. doi: 10.1104/pp.109.4.1151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watanabe M, Mochida K, Kato T, Tabata S, Yoshimoto N, Noji M, Saito K. Comparative genomics and reverse genetics analysis reveal indispensable functions of the serine acetyltransferase gene family in Arabidopsis. The Plant Cell. 2008;20:2484–2496. doi: 10.1105/tpc.108.060335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White CL, Tabe LM, Dove H, Hamblin J, Young P, Phillips N, Taylor R, Gulati S, Ashes J, Higgins TJV. Increased efficiency of wool growth and live weight gain in Merino sheep fed transgenic lupin seed containing sunflower albumin. Journal of the Science of Food and Agriculture. 2001;81:147–154. [Google Scholar]
- Wirtz M, Droux M, Hell R. O-Acetylserine (thiol) lyase: an enigmatic enzyme of plant cysteine biosynthesis revisited in Arabidopsis thaliana. Journal of Experimental Botany. 2004;55:1785–1798. doi: 10.1093/jxb/erh201. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R. Production of cysteine for bacterial and plant biotechnology: application of cysteine feedback-insensitive isoforms of serine acetyltransferase. Amino Acids. 2003;24:195–203. doi: 10.1007/s00726-002-0313-9. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R. Functional analysis of the cysteine synthase protein complex from plants: structural, biochemical and regulatory properties. Journal of Plant Physiology. 2006;163:273–286. doi: 10.1016/j.jplph.2005.11.013. [DOI] [PubMed] [Google Scholar]
- Wirtz M, Hell R. Dominant-negative modification reveals the regulatory function of the multimeric cysteine synthase protein complex in transgenic tobacco. The Plant Cell. 2007;19:625–639. doi: 10.1105/tpc.106.043125. [DOI] [PMC free article] [PubMed] [Google Scholar]