Abstract
The self-incompatibility mechanism that reduces inbreeding in many plants of the Rosaceae is attributed to a multi-allelic S locus which, in the Prunoideae and Maloideae subfamilies, comprises two complementary genes, a stylar-expressed S-RNase and a pollen-expressed SFB. To elucidate incompatibility in the subfamily Rosoideae, stylar-specific RNases and self-(in)compatibility status were analysed in various diploid strawberries, especially Fragaria nubicola and F. viridis, both self-incompatible, and F. vesca, self-compatible, and in various progenies derived from them. Unexpectedly, two unlinked RNase loci, S and T, were found, encoding peptides distinct from Prunoideae and Maloideae S-RNases; the presence of a single active allele at either is sufficient to confer self-incompatibility. By contrast, in diploid Maloideae and Prunoideae a single locus encodes S-RNases that share several conserved regions and two active alleles are required for self-incompatibility. Our evidence implicates the S locus in unilateral inter-specific incompatibility and shows that S and T RNases can, remarkably, confer not only allele-specific rejection of cognate pollen but also unspecific rejection of Sn Tn pollen, where n indicates a null allele, consistent with the the presence of the pollen component, SFB, activating the cognitive function of these RNases. Comparison of relevant linkage groups between Fragaria and Prunus suggests that Prunus S-RNases, unique in having two introns, may have resulted from gene conversion in an ancestor of Prunus. In addition, it is shown that there is a non-S locus that is essential for self-incompatibility in diploid Fragaria.
Keywords: Fragaria, Rosaceae, Rosoideae, self-incompatibility, S/T RNases, unilateral incompatibility
Introduction
Self-incompatibility prevents fertile hermaphrodite plants from self-fertilizing, and promotes heterozygosity and the long-term adaptability of populations in the wild (De Nettancourt, 1977). In various members of the Rosaceae, especially economically important fruit crops, self-incompatibility has been attributed to the gametophytic multi-allelic locus S (Crane and Lawrence, 1929; Kobel et al., 1939), as in the Solanaceae (East and Mangelsdorf, 1925). The high polymorphism of the S locus is a consequence of balancing selection favouring pollinations by pollen carrying rarer S-alleles (Wright, 1939). Studies in rosaceous species belonging to the subfamilies Prunoideae, such as Prunus avium L., sweet cherry, and Maloideae, such as Malus pumila Mill., apple, have shown that there are at least two genes at the rosaceous S-locus, one encoding a stylar glycoprotein with ribonuclease activity (S-RNase) (Sassa et al., 1994; Bošković and Tobutt, 1996) which seems to be essential for pollen rejection in related species (Sassa et al., 1997; Bošković et al., 1999), and the other encoding a pollen-specific F-Box protein (SFB or SLF) (Ushijima et al., 2003; Cheng et al., 2006). These components complement each other and the interaction of cognate alleles prevents successful pollen growth in the style and self-fertilization. Lack of expression of either component confers self-compatibility, at least in Prunus (Bošković et al., 1999; Sonneveld et al., 2005). Incompatibility in Solanaceae is also RNase-based (McClure et al., 1989; Lee et al., 1994) and, in addition to the two-part S locus, various modifier loci have been proposed to explain some examples of self-compatibility (McClure and Franklin-Tong, 2006). Rosaceous and solanaceous S-RNases share several structural features including five conserved regions (Ushijima et al., 1998) although S-RNases of Prunus differ from those of the Maloideae and the Solanaceae in having two introns rather than one (Igic and Kohn, 2001). RNase-based self-incompatibility is regarded as the ancestral state in the majority of eudicots (Igic and Kohn, 2001), implying that self-compatibility is a derived character resulting from a loss of function. Phylogenetic analysis shows that S-RNases of the Prunoideae and Maloideae form two separate clades within which are trans-specific rather than species-specific clusters, a consequence of balancing selection and the longevity of alleles (Ushijima et al., 1998; Igic and Kohn, 2001). In contrast to Maloideae and Solanaceae, the S-RNase genealogies of Prunoideae show very little phylogenetic structure, consistent with reduced diversity, and perhaps indicating an increased level of intragenic recombination (Kohn, 2008).
Fragaria, the strawberries, belong to another subfamily, the Rosoideae. It was established through selfing and intercrossing of several diploid species (2n=2x=14) that F. daltoniana J. Gay, F. nilgerrensis Schltdl. ex J. Gay and F. vesca L. are self-compatible (SC), whereas F. nipponica Makino, F. nubicola (Hook. f.) Lindl. ex Lacaita, F. pentaphylla Losinsk., F. sp. nova 301, and F. viridis Weston are self-incompatible (SI) (Evans and Jones, 1967; Staudt, 1989; Sargent et al., 2004). Moreover, the SC species failed to pollinate the SI species, indicating that SC pollen fails on SI styles, whereas reciprocal crosses resulted in viable seed (Evans and Jones, 1967), an example of unilateral interspecific incompatibility (Lewis and Crowe, 1958). The F1 hybrids from SC by SI crosses were SI and rejected the pollen of SC, but not SI, species (Evans and Jones, 1967). Involvement of S-RNases in mediation of SI in the Rosoideae has not been demonstrated.
The S-locus in Prunus maps to linkage group PG6 (Ballester et al., 1998) and in Malus to MG17 (Maliepaard et al., 1998). However, the S-bearing section of MG17 is not syntenic to PG6, but to PG3 (Dirlewanger et al., 2004). PG6 showed synteny with FG1 and FG6 of diploid Fragaria (Vilanova et al., 2008).
To test whether S-RNases mediate incompatibility in Rosoideae and to clarify genetic control, stylar-specific RNases and self-(in)compatibility status were first analysed in accessions of SC and SI diploid strawberry species and then several available interspecific F1, F2 or BC (back-cross) progenies raised from SC and SI species were analysed and various test pollinations performed. In addition, the genes encoding the Fragaria S-RNases were mapped and their protein products were sequenced. Preliminary genetic evidence for a non-S locus that is essential for mediation of SI in diploid Fragaria is also presented.
Materials and methods
To investigate the association between stylar ribonucleases and (in)compatibility status in diploid Fragaria, stylar ribonucleases were analysed in accessions of eight species maintained at East Malling Research: F. daltoniana, F. nilgerrensis, F. vesca, SC, and F. nipponica, F. nubicola, F. pentaphylla, F. sp. nova 301, and F. viridis, SI. To elucidate the genetic control of stylar ribonucleases and their relationship with (in)compatibility status, stylar ribonucleases and (in)compatibility status were analysed in four available interspecific progenies (Table 1). These were: the F1 of F. vesca 801 by F. nubicola 601 (FV×FN); the F2 of F. vesca 815 by F. nubicola 601 (FV×FN)2 (Sargent et al., 2006); the F1 of F. vesca 815 by F. viridis 903 (FVe×FVi); and the back-cross of F. vesca 815 by FVe×FVi F1 seedling 4 [FVe×(FVe×FVi)] (Nier et al., 2006). Chi2 tests were used to compare single locus segregations to Mendelian ratios and appropriate annotations are provided for the observed ratios differing from expectation at the probabilities 0.001 (***), 0.01 (**) or 0.05 (*).
Table 1.
The four diploid Fragaria progenies used to investigate the genetic control of incompatibility RNases together with parental and seedling phenotypes, segregations predicted if control is by two loci, labelled S and T, and observed segregations
| Progeny code and size | Parentage | Parental RNase phenotypes | Segregation of progeny RNase phenotypes | Interpretation according to two loci, S and T | Observed segregations at loci S and T | Chi2a |
| FV×FN | F1 of F. vesca 801×F. nubicola 601 | N×AB | 11A:6B:4N:9AB | SnSn×SaSn = 1SaSn:1SnSn | 20:10 | 3.72 |
| 30 seedlings | TnTn×TbTn = 1TbTn:1TnTn | 15:15 | 0.0 | |||
| (FV×FN)2 | F2 of F. vesca 815×F. nubicola 601 | AB×AB | 17A:14B:12N:19AB | SaSn×SaSn = 3Sa−:1SnSn | 36:26 | 11.63*** |
| 62 seedlings | TbTn×TbTn = 3Tb−:1TnTn | 33:29 | 16.51*** | |||
| FVe×FVi | F1 of F. vesca 815×F. viridis 903 | N×CDE | 5C:3CE:3D:0DE | SnSn×SeSn = 1SeSn:1SnSn | 3.8 | 0.98 |
| 11 seedlings | TnTn×TcTd = 1TcTn:1TdTn | 8.3 | 0.98 | |||
| FVe×(FVe×FVi) | BC of F. vesca 815×F. viridis 903 | N×C | 23C:32N | SnSn×SnSn = all SnSn | ||
| 55 seedlings | to F. vesca 815 | TnTn×TcTn = 1TcTn:1TnTn | 23:32 | 1.19 |
***Observed ratios significantly different from expected at P=0.001.
For protein extraction for stylar ribonuclease analysis of the species accessions and progenies, whole receptacles (4–8 per plant) with carpels from newly opened flowers were snap-frozen in liquid nitrogen in a 2 ml microcentrifuge tube, ground to a fine powder, and washed with 1.8 ml acetone containing 0.07% (v/v) β-mercaptoethanol until the samples were colourless. The acetone was removed and the samples were dried overnight. Resultant powders were resuspended in 1 ml of extraction buffer containing 10% (v/v) dimethyl sulphoxide, 3% (w/v) sucrose, 0.1% (w/v) sodium metabisulphite, and 0.2% (v/v) Pharmalyte 3-10 and incubated on ice for 2 h. After centrifugation, supernatants containing the stylar native protein extract were stored at −80 °C until required. Isoelectric focusing (IEF) of the proteins and staining for ribonuclease activity were conducted in accordance with published methods (Bošković and Tobutt, 1996).
To determine the self-(in)compatibility status of seedlings of the four progenies, potted plants grown in an insect-proof greenhouse were self-pollinated with a fine brush or a finger and monitored for fruit set.
For mapping the loci in the (FV×FN)2 mapping population, anodal and cathodal RNases were scored as present or absent in 62 seedlings and assigned to loci S and T, respectively. The data were integrated with available marker data (Vilanova et al., 2008) and a map was constructed with JOINMAP software (Sargent et al., 2007).
To clarify the patterns of cross-compatibility relationships and to check fertility, test-crosses among various SI hybrids of progeny FV×FN (detailed later) were effected by using flowers of the male parents as brushes to dust pollen directly onto the styles of the female parents.
As consensus primers developed from conserved regions of rosaceous S-RNases failed to amplify RNases from Fragaria genomic and stylar cDNA (data not shown) samples of ribonucleases from F. nubicola and F. viridis were prepared for tandem mass spectrometric (MS–MS) sequencing. The acetone powders described above were resuspended in 1% Pharmalyte 3-10 and 5 mM DTT. After 2 h, proteins were precipitated using trichloracetic acid and deoxycholate with final concentrations of 20% and 0.1%, respectively, and then resolubilized for electrophoresis on an 8–18% density-gradient SDS-PAGE gel (Laemmli, 1970), together with a marker track of native protein extract prepared as described above. The gel was stained for RNase activity (Blank et al., 1982) and bands of activity in the native sample were marked with a scalpel. The gel was destained, fixed, and restained with Colloidal Coomassie Blue. Bands corresponding to marked bands were excised and submitted for MS–MS sequencing (BSAU Nottingham University).
Results
Stylar RNases in SC and SI diploid Fragaria species
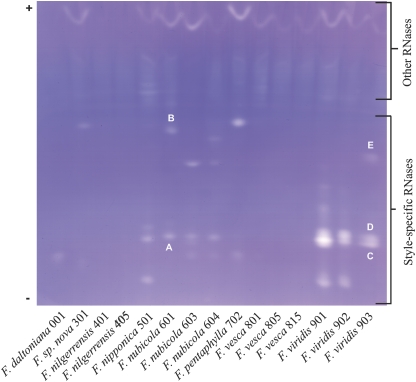
The analysis of the ribonucleases in a range of accessions of diploid Fragaria species of known status allowed the relationship between stylar RNases and (in)compatibility status to be investigated. Figure 1 shows the resulting zymograms. The several uniform bands observed in all accessions in the anodal (acidic) region of the gel are not style-specific (data not shown). In the cathodal region, to which S-RNases, which are basic, should migrate, no activity was observed in accessions of two of the SC species, F. nilgerrensis 401 and 405 and F. vesca 801, 805, and 815, whereas a single band was seen in F. daltoniana 001. In three of the SI accessions F. sp. nova 301, F. nubicola 601, and F. pentaphylla 702, two bands were seen while a complex pattern of three or four bands was revealed in the other SI accessions, F. nipponica 501, F. nubicola 603 and 604, and F. viridis 901, 902, and 903. These RNases showed high polymorphism within and between the species. Thus, the SC species lacked stylar RNases, or in one case showed a single band, but the SI species showed two to four bands. This pattern indicates possible involvement of stylar RNases in mediation of self-incompatibility in diploid Fragaria and the existence of more than one RNase locus. The rest of this paper investigates the nature of (in)compatibility in accessions and progenies of F. nubicola and F. viridis, both SI, and of F. vesca, SC.
Fig. 1.
Ribonuclease zymograms of various diploid Fragaria accessions: F. daltoniana (SC), F. nilgerrensis (SC), F. nipponica (SI), F. nubicola (SI), F. pentaphylla (SI), F. sp. nova 301 (SI), F. vesca (SC), and F. viridis (SI). Gels incorporated Pharmalyte 3-10 and separation was by IEF. Apart from the anodal non-stylar-specific bands, the SC accessions showed no or one band while the SI accessions showed two, three or four. The bands of F. nubicola 601 are annotated A and B and those of F. viridis 903 D, E, and F.
S and T RNase loci and non-S locus in F. nubicola
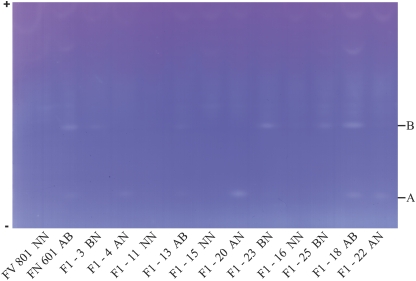
The analysis of the F1 progeny FV×FN (Table 1) for RNases and for self-(in)compatibility helped elucidate the genetic control of the RNase bands in F. nubicola and their possible involvement in the self-incompatibility response. The female parent F. vesca 801 is SC and has no RNases and the male parent F. nubicola 601 is SI and has RNases A and B (Fig. 1). The seedling zymograms showed four RNase classes (Fig. 2), 11 seedlings with the A band only, 6 with B only, 4 with neither, and 9 with both. This segregation cannot be explained by a single locus, as A and B are not allelic, but is consistent with control by two independent loci each segregating approximately 1:1, i.e. 20 A:10 non-A (Chi2=3.72) and 15 B:15 non-B, (Chi2=0) (Table 1). As expected, the four seedlings with no bands – i.e. with the RNase phenotype of F. vesca – were all SC but in each of the three remaining classes both SC and SI seedlings occurred: A only, 6 SC and 5 SI; B only, 1 SC and 5 SI; and AB, 5 SC and 4 SI. In all, 14 of the 26 seedlings with one or other or both RNases were SI. This self-incompatibility indicates that, with the right genetic background, RNases A and B from F. nubicola 601 are each associated with self-incompatibility function, even when a single allele is expressed. Indeed, this correlation, along with the differential pattern of RNases in SI and SC Fragaria species already described and the role of RNases established in the SI Prunoideae and Maloideae, indicates that A and B can be proposed as incompatibility RNases. In the light of this, locus A was labelled as S and locus B as T, using the next letter of the alphabet. This avoids the S and Z notation used for the non-RNase-based complementary two locus system in the Graminae (Lundqvist, 1954; Yang et al., 2008). So the genotype of F. vesca 801 is SnSn TnTn and that of F. nubicola F601 is SaSn TbTn where Sn and Tn are null alleles. As some of the SI seedlings have two active RNases, there appears to be no ‘competitive interaction’ (Lewis, 1943) in the pollen between the corresponding pollen-S and pollen-T factors that would lead to the loss of their SI function. Thus the loci act independently. As amphidiploid Fragaria produced by chromosome doubling of SI hybrids are SC (Evans and Jones, 1967), it appears that competitive interaction does operate between alleles of the same loci in diploid pollen.
Fig. 2.
Ribonuclease zymograms of seedlings of the F1 progeny from F. vesca 801 by F. nubicola 601, showing independent segregation of RNases A and B. Gels incorporated Pharmalyte 3-10 and separation was by IEF. F. vesca 801 lacks RNases whereas F. nubicola 601 is AB; the seedling classes are AB, A, B or neither. Thus A and B are not allelic and are attributable to two loci (later labelled S and T).
That nearly half of the seedlings having one or both RNases are SC may indicate the involvement of an additional, non-S, locus in the expression of the SI response, which was arbitrarily labelled M. This would be an example of complementary action or duplicate recessive epistasis. F. nubicola 601 would be heterozygous Mm and F. vesca 801 would be homozygous recessive mm with the observed segregation of 14 SI:12 SC seedlings in the FV×FN F1 progeny being in accord with the 1:1 expected of Mm versus mm (Chi2=0.14). As the proposed non-RNase factor appears to support self-incompatibility when heterozygous Mm, it should be expressed in the style rather than the pollen since heterozygosity for a pollen factor essential for self-incompatibility would result in self-compatibility, as it would be transmitted to only half the pollen grains. The F1 seedlings from F. vesca×F. nubicola reported by Evans and Jones (1967) were all SI, presumably because that accession of F. nubicola was homozygous MM.
The various sib-crosses attempted among SI F1 hybrids from the different RNase classes allowed the SI function of RNases A and B to be tested. If two hybrids had the same RNase pattern, the cross failed; thus hybrid F1-14×hybrid F1-3 (both BN) and F1-20×F1-4 (both AN) each set no fruit from five flowers pollinated. However, if the hybrids had different single RNases, the cross succeeded; F1-20 (AN)×F1-3 (BN) and F1-3 (BN)×F1-4 (AN) set four and five fruit, respectively, from five flowers. Crosses of one-banded hybrids by two-banded succeeded; thus F1-23 (AN)×F1-10 (AB) and F1-27 (AN)×F1-10 (AB) set three and five fruit, respectively. In contrast, the reciprocals failed, F1-10 (AB)×F1-23 (AN) setting no fruit. The failure of crosses between seedlings with RNases in common supports the conclusion that the RNases are products of incompatibility loci that act independently.
Analysis of the mapping progeny (FV×FN)2 (Table 1) confirmed the genetic control of the RNase bands in F. nubicola by two independent loci. As mentioned, analysis of the stylar RNases of the two grandparents had revealed no stylar-specific RNase bands in SC F. vesca 815 and two bands, A and B, in SI F. nubicola 601 (Fig. 1); and the F1 plants from which the F2 progeny had been raised inherited both RNases from F. nubicola 601 and being SC were presumably homozygous recessive mm for the non-S factor. Analysis of the F2 progeny revealed four classes of seedlings: 17 with only the A band, 14 with only B, 12 with neither, and 19 with both. The range of phenotypes is consistent with the control by the two independent loci already proposed, although the segregations do not fit the expected ratio of 3:1 well, namely 36 A:26 non-A (Chi2=11.63***) and 33 B:29 non-B (Chi2=16.51***) (Table 1). Sargent et al. (2006) reported distorted ratios for many loci in this interspecific F2 progeny and so there is no reason to attribute the aberrant ratios of A and B to incompatibility. A random subset of 14 seedlings tested for self-(in)compatibility all set fruit after selfing and were thus self-compatible, irrespective of their RNase pattern and even when having the AB pattern of the SI grandparent F. nubicola 601; this indicates that they are homozygous recessive mm for the non-S factor, as are the F1 plants from which they derive.
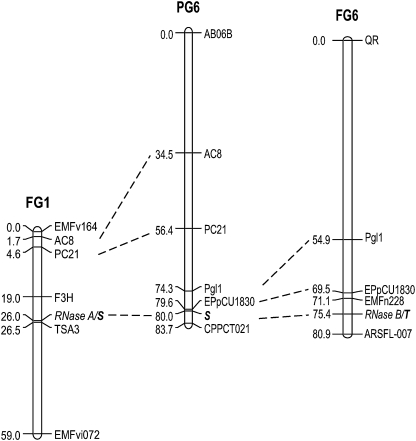
Comparing the segregations of the RNases with those of the markers on the map published for (FV×FN)2 (Sargent et al., 2006) located RNase A (locus S) and RNase B (locus T) loci on two different linkage groups (Fig. 3). Locus A was located on linkage group FG1 flanked by markers F3H and TSA3 and B on FG6 flanked by markers ARSFL007 and EMFn228. As might be expected from the recently reported syntenies between Fragaria and Prunus (Vilanova et al., 2008), in Fragaria locus A was linked with markers PC21 and AC8 and B was linked with EPpCU1830 and Pgl1, but in Prunus these four anchor loci all have homologues on PG6. Regarding Prunus S and Fragaria A and B and the anchor loci, PG6 can be arrived at by inserting the group Pgl1–EPpCU1830–RNase B of FG6 between RNase A and PC21–AC8 of FG1 and coalescing A and B; or, alternatively, FG1 and FG6 can be arrived at by the reverse rearrangement.
Fig. 3.
Mapping of RNase loci A and B (later labelled S and T) to Fragaria linkage groups FG1 and FG6 in F2 progeny of F. vesca 815 by F. nubicola 601 and comparison with Prunus PG6 (Sargent et al., 2006; Vilanova et al., 2008). A is flanked by markers F3H and TSA3 on FG1, B by markers ARSFL-007 and EMFn228 on FG6. In Fragaria, S is linked with PC21 and AC8 and T with EPpCU1830 and Pgl1; in Prunus, homologues of all four map to PG6 on which S is located.
S and T RNase loci and non-S locus in F. viridis
Analysis of two more progenies indicated the existence of a duplicate RNase system and a non-S locus in a different SI Fragaria species.
Progeny FVe×FVi (Table 1) comes from the cross of F. vesca 815, SC and lacking RNase and, though not pointed out earlier, homozygous pgpg for yellow leaves, by F. viridis 903, SI and with three stylar ribonucleases C, D, and E (Fig. 1). The zymograms of the seedlings revealed three RNase phenotypes, C (five seedlings), CE (3), and D (3). This is consistent with there being two independent loci segregating 1:1 in F. viridis, with C being allelic to D at one locus and E to a null allele at the other (both Chi2=0.98) (Table 1). The absence of phenotype DE is not unexpected in view of the small progeny size. All the seedlings, whether having one or two RNases, failed to set fruit on selfing (data not shown) and, since a subset proved fertile when interpollinated, were SI rather than infertile. This contrasts with the F1 progeny of F. vesca 801 by F. nubicola 601 which segregated SI:SC 1:1 and it suggests that F. viridis 903 is homozygous for the non-S factor, MM, and the F1 seedlings are heterozygous.
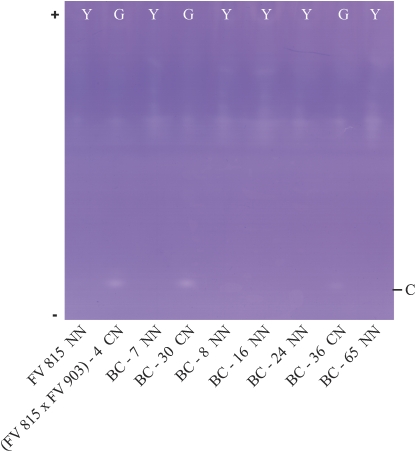
Progeny FVe×(FVe×FVi) (Table 1) comes from back-crossing SC F. vesca with a SI seedling from the F1 that inherited the single RNase C and that is heterozygous Pgpg for yellow leaves. Analysis of this backcross progeny showed that the seedlings segregated for the presence or absence of RNase C (Fig. 4), 23 with C, all of which had green leaves, and 32 without C, all of which were yellow. This approximates to a 1:1 segregation at locus T (Chi2=1.19), locus S being monomorphic null (Table 1). Strikingly, all green plants inherited RNase C whereas all yellow plants were null. This co-segregation of RNase phenotype and leaf colour is consistent with the tight coupling of RNase C with the allele Pg which is known to lie on FG6 (Sargent et al., 2006). Thus we can conclude that RNase C, and its allele D, belong to locus T and that E is likely to belong to locus S. The yellow seedlings, which had no RNase, i.e. were SnSn TnTn, were all SC whereas the green plants, with RNase C, i.e. SnSn TcTn, segregated 16 SI, which are presumably heterozygous Mm, versus 7 SC, presumably homozygous mm. This approximates to the expected 1:1 segregation (Chi2=3.52). Thus it appears that the single RNase allele Tc, which is coupled to Pg, confers self-incompatibility in an appropriate background and that a non-RNase factor, which segregates independently, is necessary for the self-incompatibility reaction.
Fig. 4.
Ribonuclease zymograms of seedlings of the BC progeny from F. vesca 815 by (F. vesca 815 by F. viridis 903)-4, showing segregation of RNase band C. Gels incorporated Pharmalyte 3-10 and separation was by IEF. F. vesca 815 lacks RNases and is homozygous for yellow leaves pgpg and (F. vesca×F. viridis 903)-4 has band C and is heterozygous Pgpg. Y indicates yellow seedlings and G indicates green. All yellow seedlings lack RNase C and are SC. All green seedlings show RNase C and segregate SI:SC 1:1, indicating segregation for an additional factor essential for the expression of self-incompatibility.
S-RNase protein sequences
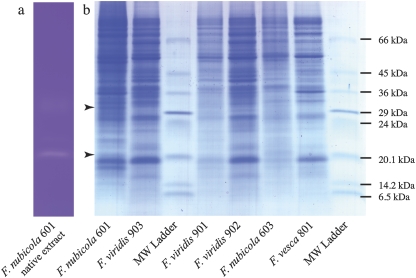
MS–MS sequencing of the two stylar protein bands separated by electrophoresis that corresponded to RNase activity in SI species, at around 21 kDa and 30 kDa (Fig. 5), provided partial sequences of the Fragaria incompatibility RNases. For F. viridis 901, the 21 kDa band yielded the peptide sequences AFDL/IVSVL/IGVEAPK, DPFGL/IVWTL/IVGNK, and VFFDL/IL/IL/IGR and the 30 kDa band FL/IL/IYDTTSK, L/ISDL/IDSL/IL/I, and FDL/IVSV. The 21 kDa band of F. nubicola 601 yielded FDL/IVSVL/IGV and that of F. nubicola 603 AFDL/IVSVL/IGVEAPK, FGL/IVWTL/IVGNK, and AL/ITAL/ITVTL/IGL/IL/IK. None of the peptide sequences matched sequences in the databases. Only one showed some similarity with Prunus S-RNases, namely Fragaria ISDIDSII, if I is assumed instead of L in ambiguous positions, versus Prunus YSDIV/ESPI. Intriguingly, another sequence contained the motif FDLL that is highly conserved in the C4 region of solanaceous S-RNases (Ioerger et al., 1991). The occurrence of peptides common to bands of both sizes indicates differences in glycosylation. As both peptide sizes are not always present on SDS gels, these differences are most likely attributable to the laboratory procedures. The bands at 21 kDa and 30 kDa were absent from F. vesca 801 (Fig. 5), and also from F. vesca 804, 805, and 815 (data not shown), all of which are SC.
Fig. 5.
Identification on 8–18% density gradient SDS gel of stylar proteins associated with RNase activity in accessions of F. nubicola and F. viridis. (a) Part of a gel showing the track of the F. nubicola 601 native protein sample stained for RNase activity. (b) Remaining section of the gel showing concentrated samples stained with Colloidal Coomassie Blue, and Sigma low molecular weight ladder. In (a), the gel has been photo-reduced by 20% to compensate for its expansion after destaining relative to the gel in (b). The bands that correspond to RNase activity at 21 kDa and/or 30 kDa, present in the accessions of SI F. nubicola 601 and 603 and in F. viridis 901, 902, and 903 but absent from SC F. vesca 801, are marked by arrows. The bands for submission for MS–MS sequencing were excised from a similar gel initially stained for RNase activity, and then destained and restained with Colloidal Coomassie Blue.
Discussion
The correlation of RNase phenotypes and self-(in)compatibility status found in species and progenies shows that, in Fragaria, as in other Rosaceae, RNases mediate self-incompatibility. However, in contrast to the single locus control of S-RNases of Maloideae and Prunoideae, the Fragaria RNases are encoded by two genes, S and T, mapping respectively to FG1 and FG6, and both showing elements of synteny with the PG6 on which the S gene lies in Prunus. Surprisingly, a single active allele at either of the two loci is sufficient to confer self-incompatibility. The null alleles of the S and T loci present in SC and SI species are associated with a lack of RNase activity and confer self-compatibility only when homozygous at both loci. Partial protein sequences of S and T RNases showed very little similarity with Prunus S-RNases. The observed self-incompatibility of the F1 plants, raised between SC F. vesca and SI F. viridis accords with the report of Evans and Jones (1967). That half of the F1 plants from crosses of SC F. vesca and SI F. nubicola were SC, in contrast to the findings of Evans and Jones (1967), suggests our accession of F. nubicola, but not theirs, is heterozygous for a non-functional allele at the non-S locus that is essential for the self-incompatibility response. This discovery of S and T loci in a diploid plant reveals additional diversity of RNase-based incompatibility systems and has important implications for the understanding of the self-incompatibility mechanism and its evolution in Rosaceae. The information provided is a useful basis for population studies and should facilitate cloning of S and T RNases.
The self-incompatibility of some seedlings having one or two RNase bands and, presumably, the non-S factor indicates that Sn Tn pollen is rejected by the self styles. By contrast, S-RNase and SFB null alleles in Prunus succeed on self styles (Bošković et al., 1999; Sonneveld et al., 2005). It is remarkable that this SI response, i.e. the failure of the Sn Tn pollen, does not require the active RNases to be cognate; the rejection of the non-cognate pollen indicates the RNases can have unspecific non-cognitive action. As the null allele at one locus is transmissible in a pollen grain if present with an active allele at the other, it seems that the presence of the pollen component suppresses unspecific non-cognitive action by activating cognitive function. The failure of the Sn Tn pollen is sufficient to explain the reported unilateral failure of pollen from SC species on SI species (Evans and Jones, 1967; Sargent et al., 2004) – implicating S-RNases in interspecific unilateral incompatibility in Rosaceae. In the Solanaceae, the S locus has been implicated in unilateral incompatibility (Murfett et al., 1996) and the lethality/failure of SC pollen explained by lack of the pollen component (Golz et al., 2001).
The finding of a duplicate incompatibility RNase system in two diploid SI Fragaria species of the Rosoideae was unexpected. Establishing that the loci were individually sufficient for the SI response depended on the existence of the null alleles at each locus that occurred in our SC×SI interspecific progenies. The nature of these null alleles should become easier to study once S and T are fully sequenced. The apparent rejection of Sn Tn pollen on styles expressing a single active allele at one locus and the lack of intergenic competitive interaction are fundamental to maintaining the self-incompatibility system in Fragaria. Presumably, a fully compatible cross between individuals lacking null alleles would result in 16 genotypic classes, but, assuming all are MM or Mm, a particular class would be compatible, fully or partially, with only nine other classes, whereas the single locus system gives four, intercompatible, classes. In other diploid Rosaceae, incompatibility is attributed to a single locus, but, as already noted, some other diploid Fragaria have complex patterns of stylar ribonucleases, as do some members of other important genera of the Rosoideae, e.g. Rosa and Rubus (R Bošković et al., unpublished data).
The lack of intergenic competitive interaction of S and T alleles in the pollen might be due to their divergence preventing the formation of the heterotetramers proposed by the modified inhibitor model (Luu et al., 2001) so that each pollen component binds to its cognate RNase and protects it from a general inhibitor. However, neither this model nor the sequestration model (Goldraij et al., 2006) can explain the failure of Sn Tn pollen on any SI style. In the modified inhibitor model, lack of the cognate pollen component would leave RNases unprotected from the general inhibitor. In the sequestration model, lack of the cognate pollen component would leave RNases sequestered in the vacuole. Thus in both models, contrary to the experimental evidence just presented, Sn Tn pollen tubes should be protected from their cytotoxic action. The ‘universal’ failure of Sn Tn pollen on SI styles could be explained only by the recently proposed RNase degradation model (Hua et al., 2008). In this, lack of expression of the pollen component would leave non-self S-RNases untargeted for ubiquitination and degradation. Further comparative mapping together with sequence information may indicate how the two locus RNase system arose. The S and T loci may be paralogues resulting from duplication in an ancestor not just of Fragaria but of Prunus too. Pseudogenization is the usual fate of duplicated genes, but in the presence of gene conversion and the balancing selection that would operate at both loci, the probability of fixation and the longevity of duplicated genes increases (Takuno et al., 2008), so that the original function may be partitioned across both copies. In Fragaria and other members of the Rosoideae, comparison of full-length amino acid sequences of S and T RNases should reveal the regions most exposed to diversifying selection acting at both of these loci. In the light of the apparent coalescence in Prunus PG6 of the single copy markers from Fragaria FG1 and FG6, S and T could have become combined within the same chromosome in the rosaceous lineage leading to Prunus; in this line, it may be that an increased rate of gene conversion, relative to the rate of point mutations, led to reduced diversity between the copies and to the loss of one gene. This is consistent with the distinctive shape, flat and unstructured, of the phylogenetic tree of Prunus S-RNases, attributed to a population bottleneck (Ushijima et al., 1998), and with the sharing by phylogenetically unrelated alleles of motifs up to 19 residues long from polymorphic regions of Prunus S-RNases (Ortega et al., 2006). As introns can spread between paralogues via gene conversion (Hankeln et al., 1997), it may be the additional intron in Prunus S-RNases, an autapomorphy (Igic and Kohn, 2001), resulted from gene conversion between S and T in an ancestor of Prunus. Thus, as in the mammalian major histocompatibility complex (MHC) system (Ohta, 1991), gene conversion may have played a role in determining allelic diversity of RNase-based incompatibility systems.
Acknowledgments
Strawberry genetics research at East Malling Research was funded by the UK's Department for Food and Rural Affairs as part of project HH3724SSF, ‘Comparative genomics of rosaceous fruit crops and HNS for sustainable production’. RIB acknowledges the financial support of the Mount Trust. We thank Tineke Sonneveld, Tim Robbins, and Colin Turnbull for helpful comments on the manuscript.
References
- Ballester J, Bošković R, Batlle I, Arús P, Vargas F, de Vicente MC. Location of the self-incompatibility gene on the almond linkage map. Plant Breeding. 1998;117:69–72. [Google Scholar]
- Blank A, Sugiyama RH, Dekker CA. Activity staining of nucleolytic enzymes after sodium dodecyl sulfate–polyacrylamide gel electrophoresis: use of aqueous isopropanol to remove detergent from gels. Analytical Biochemistry. 1982;120:267–275. doi: 10.1016/0003-2697(82)90347-5. [DOI] [PubMed] [Google Scholar]
- Bošković R, Tobutt KR. Correlation of stylar ribonuclease zymograms with incompatibility alleles in sweet cherry. Euphytica. 1996;90:245–250. [Google Scholar]
- Bošković R, Tobutt KR, Duval H, Batlle I, Dicenta F, Vargas F. A stylar ribonuclease assay to detect self-compatible seedlings in almond progenies. Theoretical and Applied Genetics. 1999;99:800–810. [Google Scholar]
- Cheng J, Han Z, Xu X, Li T. Isolation and identification of the pollen-expressed polymorphic F-box genes linked to the S-locus in apple (Malus×domestica) Sexual Plant Reproduction. 2006;19:175–183. [Google Scholar]
- Crane MB, Lawrence WJC. Genetical and cytological aspects of incompatibility and sterility in cultivated fruits. Journal of Pomology. 1929;7:276–301. [Google Scholar]
- De Nettancourt D . Incompatibility in Angiosperms. Berlin: Springer; 1977. [Google Scholar]
- Dirlewanger E, Graziano E, Joobeur T, Garriga-Calderé F, Cosson P, Howad W, Arús P. Comparative mapping and marker-assisted selection in Rosaceae fruit crops. Proceedings of the National Academy of Sciences, USA. 2004;101:9891–9896. doi: 10.1073/pnas.0307937101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- East EM, Mangelsdorf AJ. A new interpretation of the hereditary behavior of self-sterile plants. Proceedings of the National Academy of Sciences, USA. 1925;11:166–171. doi: 10.1073/pnas.11.2.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans WD, Jones JK. Incompatibility in Fragaria. Canadian Journal of Genetics and Cytology. 1967;9:831–836. [Google Scholar]
- Goldraij A, Kondo K, Lee CB, Hancock CN, Sivaguru M, Vazquez-Santana S, Kim S, Philips TE, Cruz-Garcia F, McClure B. Compartmentalization of S-RNase and HT-B degradation in self-incompatible Nicotiana. Nature. 2006;439:805–810. doi: 10.1038/nature04491. [DOI] [PubMed] [Google Scholar]
- Golz JF, Oh H-Y, Su V, Kusaba M, Newbigin E. Genetic analysis of Nicotiana pollen-part mutants is consistent with the presence of an S-ribonuclease inhibitor at the S locus. Proceedings of the National Academy of Sciences, USA. 2001;98:15372–15376. doi: 10.1073/pnas.261571598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hankeln T, Friedl H, Ebersberger I, Martin J, Schmidt ER. A variable intron distribution in globin genes of Chironomus: evidence for recent intron gain. Gene. 1997;205:151–160. doi: 10.1016/s0378-1119(97)00518-0. [DOI] [PubMed] [Google Scholar]
- Hua Z-H, Fields A, T-h Kao. Biochemical models for S-RNase-based self-incompatibility. Molecular Plant. 2008;1:575–585. doi: 10.1093/mp/ssn032. [DOI] [PubMed] [Google Scholar]
- Igic B, Kohn JR. Evolutionary relationships among self-incompatibility RNases. Proceedings of the National Academy of Sciences, USA. 2001;98:13167–13171. doi: 10.1073/pnas.231386798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ioerger TR, Gohlke JR, Xu B, Kao T-h. Primary structural features of the self-incompatibility protein in Solanaceae. Sexual Plant Reproduction. 1991;4:81–87. [Google Scholar]
- Kobel K, Steinegger P, Anliker J. Weitere Untersuchungen über die Befruchtungsverhältnisse der Apfel- und Birnsorten. Landwirtschaftliches Jahrbuch der Schweiz. 1939;53:160–191. [Google Scholar]
- Kohn J. What genealogies of S-alleles tell us. In: Franklin-Tong VE, editor. Self-incompatibility in flowering plants: evolution, diversity and mechanisms. Berlin: Springer; 2008. pp. 103–121. [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Lee H-S, Huang S, T-h Kao. S proteins control rejection of incompatible pollen in Petunia inflata. Nature. 1994;367:560–563. doi: 10.1038/367560a0. [DOI] [PubMed] [Google Scholar]
- Lewis D. Physiology of incompatibility in plants. III. Autopolyploids. Journal of Genetics. 1943;45:171–185. [Google Scholar]
- Lewis D, Crowe LK. Unilateral interspecific incompatibility in flowering plants. Heredity. 1958;12:232–256. [Google Scholar]
- Lundqvist A. Studies on self-sterility in rye, Secale cereale L. Hereditas. 1954;40:278–294. [Google Scholar]
- Luu D-T, Qin X, Laublin G, Yang Q, Morse D, Cappadocia M. Rejection of S-heteroallelic pollen by a dual-specific S-RNase in Solanum chacoense predicts a multimeric SI pollen component. Genetics. 2001;159:329–335. doi: 10.1093/genetics/159.1.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maliepaard C, Alston FH, van Arkel G, et al. Aligning male and female linkage maps of apple (Malus pumila Mill.) using multi-allelic markers. Theoretical and Applied Genetics. 1998;97:60–73. [Google Scholar]
- McClure BA, Franklin-Tong V. Gametophytic self-incompatibility: understanding the cellular mechanisms involved in ‘self’ pollen tube inhibition. Planta. 2006;224:233–245. doi: 10.1007/s00425-006-0284-2. [DOI] [PubMed] [Google Scholar]
- McClure BA, Haring V, Ebert PR, Anderson AE, Simpson RJ, Sakiyama F, Clarke AE. Style self-incompatibility gene products of Nicotiana alata are ribonucleases. Nature. 1989;342:955–957. doi: 10.1038/342955a0. [DOI] [PubMed] [Google Scholar]
- Murfett J, Strabala TJ, Zurek DM, Mou B, Beecher B, McClure BA. S RNase and interspecific pollen rejection in the genus Nicotiana: multiple pollen-rejection pathways contribute to unilateral incompatibility between self-incompatible and self-compatible species. The Plant Cell. 1996;8:943–958. doi: 10.1105/tpc.8.6.943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nier S, Simpson DW, Tobutt KR, Sargent DJ. A genetic linkage map of an inter-specific diploid Fragaria BC1 mapping population and its comparison with the Fragaria reference map (FV×FN) Journal of Horticultural Science and Biotechnology. 2006;81:645–650. [Google Scholar]
- Ohta T. Role of diversifying selection and gene conversion in evolution of major histocompatibility complex loci. Proceedings of the National Academy of Sciences, USA. 1991;88:6716–6720. doi: 10.1073/pnas.88.15.6716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ortega E, Bošković RI, Sargent DJ, Tobutt KR. Analysis of S-RNase alleles in almond (Prunus dulcis): characterization of new sequences, resolution of synonyms and evidence of intragenic recombination. Molecular Genetics and Genomics. 2006;276:413–426. doi: 10.1007/s00438-006-0146-4. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Clarke J, Simpson DW, et al. An enhanced microsatellite map of diploid Fragaria. Theoretical and Applied Genetics. 2006;112:1349–1359. doi: 10.1007/s00122-006-0237-y. [DOI] [PubMed] [Google Scholar]
- Sargent DJ, Geibel M, Hawkins JA, Wilkinson MJ, Battey NH, Simpson DW. Quantitative and qualitative differences in morphological traits revealed between diploid Fragaria species. Annals of Botany. 2004;94:787–796. doi: 10.1093/aob/mch217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sargent DJ, Rys A, Nier S, Simpson DW, Tobutt KR. The development and mapping of functional markers in Fragaria and their transferability and potential for mapping in other genera. Theoretical and Applied Genetics. 2007;114:373–384. doi: 10.1007/s00122-006-0441-9. [DOI] [PubMed] [Google Scholar]
- Sassa H, Hirano H, Nishio T, Koba T. Style-specific self-compatible mutation caused by deletion of the S-RNase gene in Japanese pear (Pyrus serotina) The Plant Journal. 1997;12:223–227. [Google Scholar]
- Sassa H, Mase N, Hirano H, Ikehashi H. Identification of self-incompatibility-related glycoproteins in styles of apple (Malus×domestica) Theoretical and Applied Genetics. 1994;89:201–205. doi: 10.1007/BF00225142. [DOI] [PubMed] [Google Scholar]
- Sonneveld T, Tobutt KR, Vaughan SP, Robbins TP. Loss of pollen-S function in two self-compatible selections of Prunus avium is associated with deletion/mutation of an S haplotype-specific F-box gene. The Plant Cell. 2005;17:37–51. doi: 10.1105/tpc.104.026963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staudt G. The species of Fragaria, their taxonomy and geographical distribution. Acta Horticulturae. 1989;265:23–33. [Google Scholar]
- Takuno S, Nishio T, Satta Y, Innan H. Preservation of a pseudogene by gene conversion and diversifying selection. Genetics. 2008;180:517–531. doi: 10.1534/genetics.108.091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Dandekar AM, Gradziel TM, Tao R, Hirano H. Structural and transcriptional analysis of the self-incompatibility locus of almond: identification of a pollen-expressed F-box gene with haplotype-specific polymorphism. The Plant Cell. 2003;15:771–781. doi: 10.1105/tpc.009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ushijima K, Sassa H, Tao R, Yamane H, Dandekar AM, Gradziel TM, Hirano H. Cloning and characterization of cDNAs encoding S-RNases from almond (Prunus dulcis): primary structural features and sequence diversity of the S-RNases in Rosaceae. Molecular Genetics and Genomics. 1998;260:261–268. doi: 10.1007/s004380050894. [DOI] [PubMed] [Google Scholar]
- Vilanova S, Sargent DJ, Arús P, Monfort A. Synteny conservation between two distantly-related Rosaceae genomes: Prunus (the stone fruits) and Fragaria (the strawberry) BMC Plant Biology. 2008;8:67. doi: 10.1186/1471-2229-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright S. The distribution of self-sterility alleles in populations. Genetics. 1939;24:538–552. doi: 10.1093/genetics/24.4.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B, Thorogood D, Armstead I, Barth S. How far are we from unravelling self-incompatibility in grasses? New Phytologist. 2008;178:740–753. doi: 10.1111/j.1469-8137.2008.02421.x. [DOI] [PubMed] [Google Scholar]