Abstract
During rice (Oryza sativa L.) seed development, the primary endosperm nucleus undergoes a series of divisions without cytokinesis, producing a multinucleate cell, known as a syncytium. After several rounds of rapid nuclear proliferation, the syncytium ceases to undergo mitosis; thereafter, the syncytium is partitioned into individual cells by a specific type of cytokinesis called cellularization. The transition between syncytium and cellularization is important in determining the final seed size and is a model for studying the cell cycle and cytokinesis. The involvement of cyclin-dependent kinase (CDK) inhibitors (CKIs) in cell cycle control was investigated here during the transition between syncytium and cellularization. It was found that one of the rice CKIs, Orysa;KRP3, is strongly expressed in the caryopsis at 2 d after flowering (DAF), and its expression is significantly reduced at 3 DAF. The other CKI transcripts did not show such a shift at 2 DAF. In situ hybridization analysis revealed that Orysa;KRP3 is expressed in multinucleate syncytial endosperm at 2 DAF, but not in cellularized endosperm at 3 DAF. Two-hybrid assays showed that Orysa;KRP3 binds Orysa;CDKA;1, Orysa;CDKA;2, Orysa;CycA1;1, and Orysa;CycD2;2. By contrast, Orysa;CDKB2;1 and Orysa;CycB2;2 do not show binding to Orysa;KRP3. Orysa;KRP3 was able to rescue yeast premature cell division due to the dominant positive expression of mutant rice CDKA;1 indicating that Orysa;KRP3 inhibited rice CDK. These data suggest that Orysa;KRP3 is involved in cell cycle control of syncytial endosperm.
Keywords: CDK inhibitor, cellularization, endosperm, KRP, Oryza sativa, syncytium
Introduction
The rice (Oryza sativa L.) endosperm comprises a substantial proportion of the mature seed and contains a large amount of carbohydrates. It is an important source of calories for humans and animals and also provides raw materials for goods and biofuels. Extensive research has been directed at improving the grain size, quality, and yield. Some of the limitations of conventional rice breeding may be overcome by biotechnological engineering. However, significant improvements require an understanding of the molecular processes controlling endosperm development.
Rice seed development begins with double fertilization in which the haploid egg cell and the two polar nuclei in the central cell are fertilized by haploid sperm cells. After double fertilization, the triploid primary endosperm nucleus begins to divide rapidly. Endosperm development proceeds in several distinct phases: syncytium formation, during which the endosperm nuclei undergo many rounds of mitosis without cytokinesis; cellularization during which cell walls form around the endosperm nuclei; differentiation, which includes the formation of transfer cells, aleurone, and starchy endosperm; and maturation, which includes endoreduplication for the accumulation of storage compounds, dormancy, and desiccation (Hoshikawa, 1967a, b, c, 1968; Olsen, 2001, 2004; Brown and Lemmon, 2007; Sabelli and Larkins, 2009).
A major event during endosperm development is the transition between syncytium and cellularization (Brown et al., 1996a). The duration of the syncytial phase and the timing of cellularization are important because they correlate with the extent of nuclear proliferation and may influence seed size and grain weight. Thus, the biotechnological approach to prolong the duration of syncytial nuclear proliferation is a promising strategy for altering seed size. However, little is known about the molecular processes controlling syncytial nuclear proliferation and cellularization.
Cyclin-dependent kinase (CDK) inhibitors (CKIs) bind CDKs and inhibit cell cycle progression. There are two types of CKIs (the INK4 and the Kip/Cip families) in mammals. Plants do not appear to have INK4 CKIs, but proteins related to the Kip/Cip CKIs have been identified and designated as Kip-related proteins (KRPs). Recently, a second group of CKIs, called SIAMESE in Arabidopsis or EL2 in rice, has been identified (Churchman et al., 2006; Peres et al., 2007). The SIAMESE-related genes are not present in animals and fungi; therefore, these genes are most likely unique to plants. In Arabidopsis, seven KRP (Arath;KRP1 to Arath;KRP7) genes were identified and their inhibitory activity against CDK was confirmed both in vitro and in vivo (Wang et al., 1998; De Veylder et al., 2001; Zhou et al., 2002; Verkest et al., 2005). They also arrest the cell cycle in response to specific developmental or environmental cues. Abscisic acid and cold conditions induce Arath;KRP1 expression (Wang et al., 1998). Arath;KRP2 expression is negatively regulated by auxin during early lateral root initiation (Himanen et al., 2002). Arath;KRP6 and Arath:KRP7 were involved in the control of germline proliferation (Kim et al., 2008; Liu et al., 2008; Gusti et al., 2009). In addition to their role in blocking cell cycle progression, KRPs have also been suggested to regulate nuclear DNA endoreduplication. In Arabidopsis, there is functional evidence that KRPs promote endoreduplication (Verkest et al., 2005; Weinl et al., 2005). In monocotyledonous plants, two maize (Zea mays) genes—Zeama;KRP1 and Zeama;KRP2—were characterized and shown to be expressed in developing endosperm. Both the proteins inhibit endosperm Cdc2-related CDK activity in vitro. Zeama;KRP1 was suggested to be involved in endoreduplication during the middle stage of endosperm development (Coelho et al., 2005). In rice, seven KRP (Orysa;KRP1 to Orysa;KRP7) genes were identified in the rice genome database (Barroco et al., 2006; Guo et al., 2007). Overexpression of Orysa;KRP1 reduced cell production and seed filling, which was accompanied by a drop in endoreduplication during the late stage of endosperm development, suggesting a role of Orysa;KRP1 in cell cycle control at the maturation stage of seed development (Barroco et al., 2006).
Three different types of cell cycles occur during endosperm development: (i) mitosis without cytokinesis, resulting in syncytium formation; (ii) mitosis coupled with cell division; and (iii) endoreduplication. Although the latter two cell cycle types have been characterized during the later stages of endosperm development, the molecular factors that control the cell cycle in the syncytium are scarcely known.
The involvement of CDK inhibitors in the altered types of cell cycle control in the early stage of rice endosperm development is investigated here. One of the rice KRPs—Orysa;KRP3—is strongly expressed in the multinucleate syncytial endosperm, and its expression is down-regulated after cellularization. Orysa;KRP3 is able to bind and inhibit rice CDK/cyclin. Our data provide a possible role of Orysa;KRP3 in cell cycle control during the early phase of rice endosperm development.
Materials and methods
Plant materials
Rice plants (Oryza sativa L. cv. Hitomebore) were grown under field conditions in plastic pots filled with soil at Iwate University (Morioka, Japan). Spikelets were marked on the flowering day and subsequently sampled daily following maturity. Different tissues (leaf, stem, root, and panicle) were collected around 10 DAF.
RT-PCR
Total RNA was isolated from plant tissues by the acid guanidinium thiocyanate–phenol–chloroform extraction method (Chomczynski and Sacchi, 1987). First-strand cDNA synthesis was carried out via ReverTra Ace reverse transcriptase (Toyobo, Osaka, Japan) with oligo (dT)15 and random primers. Semi-quantitative PCR was performed with various forward and reverse primers (Table 1). Quantitative real-time RT-PCR was carried out with SYBR Premix Ex Taq II (Takara, Ohtsu, Japan). Samples were analysed in triplicate in a Thermal Cycler Dice Real Time System (Takara). In each case, dissociation curves confirmed the purity of the amplified products. Relative expression levels were calculated according to the 2–ΔΔCT method (Livak and Schmittgen, 2001) using 18S rRNA as the internal control. The primers used for these analyses are listed in Table 1.
Table 1.
List of primers used in this study
| Gene | Locus/Accession no. | Forward primer (5′ to 3′) | Reverse primer (5′ to 3′) | Purpose |
| Orysa;KRP3 | Os11g0614800 | AGCTCTGGCTGGTTCTTGTTGGCAAGCTAG | AGAGGGGAGCTCATCAGAAGCATCCTC | cDNA cloning |
| AGCGCCGGCGATTCGCAGAGAAGTAC | TGCAGACATGGGTGTGAGTTGAGTTGACAC | RT-PCR | ||
| ACATTGCCCTCGACCGCCCGTTGCAAG | TGCAGACATGGGTGTGAGTTGAGTTGACAC | Real-time RT-PCR | ||
| AGCGCCGGCGATTCGCAGAGAAGTAC | GAGCTCATCAGAAGCATCCTC | In situ probe | ||
| Orysa;KRP1 | Os02g0762400 | CGAGAGGAGAGAAACAACTCCATC | CCCCTGTACCTTCCTTTCTAGCTT | RT-PCR |
| CGAGAGGAGAGAAACAACTCCATC | TCAGCTTCGGCTGCTGACCACCGGAGTC | Real-time RT-PCR | ||
| Orysa;KRP4 | Os10g0471700 | ATACGATTAGCACCCCTGGATCTAC | CCATTGAGTCTACAGGCTAACCCT | RT-PCR |
| CAGCAGAGCTGGAAGCGTTCTTCG | CCATTGAGTCTACAGGCTAACCCT | Real-time RT-PCR | ||
| Orysa;KRP5 | Os03g0137800 | CAGCAACATCAGGCTTTCAGAG | AGCTAGGCCTAAAAGAGGTGGTTC | RT-PCR |
| CAGCAACATCAGGCTTTCAGAG | CTAGCAGTCTAGCCTTGTCCATTCGTAC | Real-time RT-PCR | ||
| Orysa;KRP6 | Os09g0459900 | GTCGACATGCCCAGGAAGGCGAAGAAG | ACAGGCGAAGCATCTACTGCTAAC | RT-PCR |
| GCCAAGTACAACTATGACATC | CACATGACATGGAGTAACC | Real-time RT-PCR | ||
| OsACT1 | Os03g0718100 | CCTCGCACCAAGCATGAAGA | CGACTCATCATACTCTCTCCCTTTG | RT-PCR |
| 18S rRNA | AK059783 | ATGATAACTCGACGGATCGC | CCTGGATGTGGTAGCCGTTT | Real-time RT-PCR |
| Orysa;CDKA;1 | Os03g0118400 | CATATGGAGCAGTACGAGAAGGAGGAGAAG | TCATTGTACCATCTCAAGGTCCTTGAAG | cDNA cloning |
| GATTGGGGAGGGCGCCTTCGGGGTGGTGTA | ATGTGGTGGGGCTTCCGCGGGAGGGGTTAG | Mutagenesis | ||
| Orysa;CDKA;2 | Os02g0123100 | CATATGGAGCAGTACGAGAAGGTGGAGAAG | CTACGCCACTTCCAGGTCCTTGAAGTACTC | cDNA cloning |
| Orysa;CDKB2;1 | Os08g0512600 | CATATGGACCTGTACGAGAAGCTGGAGAAG | TCAGTAGAGCTCCTTGTTCACGTCGTTG | cDNA cloning |
| Orysa;CyclinA1:1 | Os01g0233500 | CATATGTCGAGCAACCTAGCAGCCTC | TCAGCATGTTGCGTCGCGAAAGAATTCG | cDNA cloning |
| Orysa;CyclinB2:2 | Os06g0726800 | CATATGGAGAACATGAGATCTGAGAAC | TTACAGTGCCACGCTCTTGAGCAAGAAGAC | cDNA cloning |
| Orysa;CyclinD2;2 | Os07g0620800 | CATATGGGTGTTCTTTGCTTCGGCGCTTC | TCAGATTGGTGTTGTGTTTAATCTCCTC | cDNA cloning |
In situ hybridization
In situ hybridization of sections through developing rice spikelets was performed according to Hirose (2002) with some modifications. Plant materials were fixed in 2% (w/v) paraformaldehyde and 15% (v/v) saturated picric acid in 50 mM sodium phosphate buffer, pH 7.4 overnight at 4 °C, dehydrated through an ethanol series and t-butanol series, and finally embedded in Paraplast Plus (Sigma-Aldrich). Sections (7 μm thick) were used for in situ hybridization. The sections were deparaffinized with xylene and rehydrated through an ethanol series, treated with proteinase K (2 μg ml−1) in 100 mM TRIS-HCl, pH 7.5, 50 mM EDTA at 37 °C for 10 min, followed by post-fixation with 4% paraformaldehyde in 10 mM phosphate buffer, pH 7.2. Subsequently, the sections were washed in distilled water and then dehydrated through an ethanol series. The Orysa;KRP3 template for riboprobe synthesis was amplified by PCR and subcloned into pCR-Blunt vector (Invitrogen). The primers used for PCR are listed in Table 1. Sense and antisense RNA probes were labelled from cDNA inserts in pCR-Blunt with digoxigenin (DIG)-UTP (Roche) by T7 RNA polymerase (Takara). The sections were hybridized with a DIG-labelled RNA probe at 42 °C overnight in a hybridization buffer containing 50% (v/v) formamide, 2× SSC, 1% (w/v) blocking reagent (Roche), 50 mM sodium phosphate, pH 7.4, and 1 mM EDTA. After hybridization, the sections were washed with 2× SSC at 42 °C for 30 min and washed again in 0.5× SSC. The hybridization signals were detected using a DIG nucleic acid detection kit (Roche).
Yeast two-hybrid experiments
Yeast two-hybrid assays were performed using the BD Matchmaker two-hybrid system 3 (Clontech). The open-reading frames of Orysa;KRP3, Orysa;CDKA;1, Orysa;CDKB2;2, Orysa;CycA1;1, Orysa;CycB2;2, and Orysa;CycD2;2 were amplified by PCR with gene-specific primers (Table 1) and cloned into pGBKT7-BD (Clontech) and/or pGADT7-AD (Clontech), respectively. All the constructs were confirmed by sequencing. Different combinations between bait (pGBKT7 DNA-BD-insert) and prey (pGADT7-AD-insert) constructs were transformed into AH109 (Clontech) and their ability to grow on histidine- and adenine-deficient minimal media at 30 °C was assayed.
Expression of Orysa:KRP3 in fission yeast
A dominant positive mutant of the rice CDKA;1 gene (Orysa;CDKA;1.A14F15) was constructed by mutagenizing Orysa;CDKA;1 cDNA. A plasmid containing an Orysa;CDKA;1.A14F15 open-reading frame was amplified by inverse PCR with the mutagenized primer sets (Table 1) and an Orysa;CDKA;1 plasmid as a template. After degradation of the template DNA by DpnI digestion, the mutagenized plasmid was transformed in Escherichia coli. The Orysa;CDKA;1.A14F15 insert was fused to the nmt41 promoter in the fission yeast expression vector pREP42 (Basi et al., 1993). Orysa;KRP3 cDNA was subcloned into the fission yeast expression vector pESP-1 (Stratagene). Plasmids were transformed into the fission yeast strain YN29 h90 leu1 ura4-D18 (Ikemoto et al., 2000) using the electroporation method. All the fission yeast manipulations were performed according to the ESP yeast protein expression system manufacturer's instructions (Stratagene).
Results
Orysa;KRP3 is expressed in the panicle
Seven rice KRP candidates designated Orysa;KRP1 to Orysa;KRP7 were identified in the rice genome database (Barroco et al., 2006; Guo et al., 2007). Among them, Orysa;KRP7 lacks a CDK-binding box, suggesting that this gene may be a pseudogene (Guo et al., 2007).
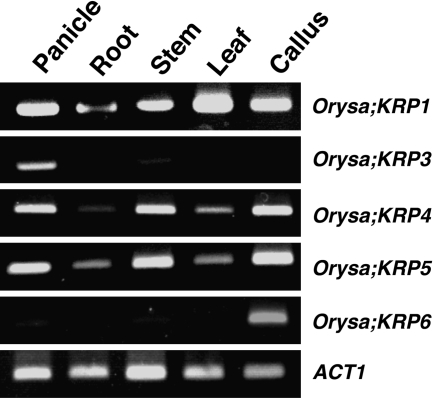
The expression of rice KRP genes was examined in various plant tissues and callus by semi-quantitative RT-PCR (Fig. 1). Orysa;KRP1, Orysa;KRP4, and Orysa;KRP5 were expressed ubiquitously. Orysa;KRP2 transcripts were not detected in any of the tissues or callus examined. Orysa;KRP6 transcripts were detected in the callus but were barely detectable in intact plant tissues.
Fig. 1.
Expression of rice KRP genes in different rice tissues. cDNA prepared from the indicated tissues was subjected to semi-quantitative RT-PCR analysis with gene-specific primers. The rice actin 1 gene (ACT1) was used as a loading control.
Remarkably, Orysa;KRP3 transcripts were detected in the panicle but were barely or not detected in other plant tissues and callus examined. Although different tissue-specific expression patterns of Arabidopsis KRP genes were reported, no Arath;KRP gene was specifically expressed in the panicle (De Veylder et al., 2001). Taken together, the transcript profiles suggest that Orysa;KRP3 might play unique roles in the monocotyledonous panicle.
Orysa;KRP3 expression is dramatically high in the panicle at 2 DAF
For a more detailed characterization of Orysa;KRP3, its cDNA sequence was verified by sequencing a full-length cDNA clone obtained by RT-PCR from a rice panicle RNA. The sequence was confirmed as 100% identical to the predicted cDNA sequence (Os11g0614800) derived from an annotated rice genomic sequence (Tanaka et al., 2008).
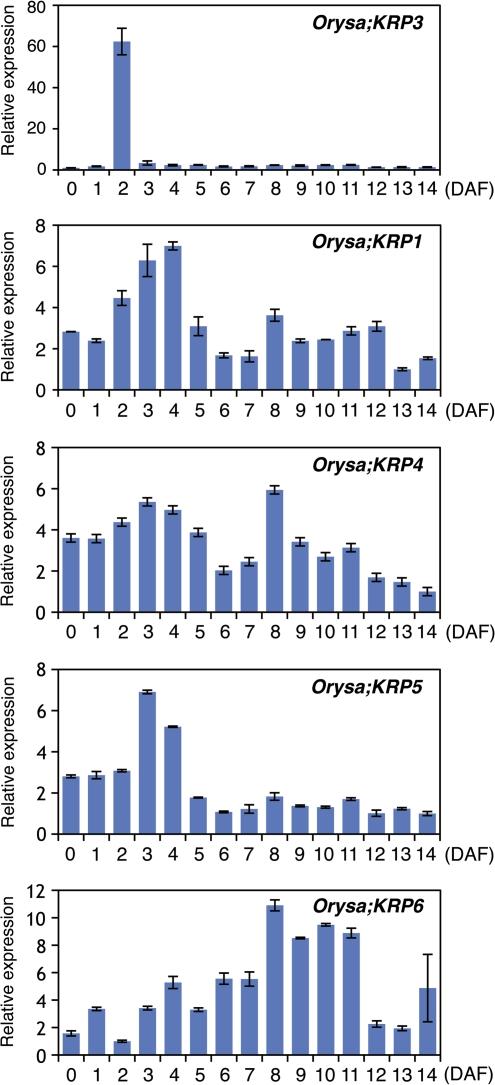
To deduce the possible role of Orysa;KRP3 in the panicle, the transcript levels in developing seeds were measured at different time points from 0 d to 14 d after flowering (DAF) using real-time quantitative RT-PCR (Fig. 2). Orysa;KRP3 transcripts were detected throughout the period of seed development, with a dramatic increase at 2 DAF and a rapid decrease 1–2 d later to a basal level. By contrast, the transcript accumulation profiles of other Orysa;KRP genes did not show any particular change at 2 DAF. The distinct transcript accumulation profile of Orysa;KRP3 suggests a functional diversity between Orysa;KRP3 and other Orysa;KRP members during rice seed development.
Fig. 2.
Expression of rice KRP genes during seed development. Total RNA isolated from 10 seeds at each DAF stage was subjected to real-time quantitative RT-PCR analysis. The values represent the relative amount of transcripts compared to the time point with the lowest transcript level. Error bars indicate mean ±SD of three independent experiments. Data are representative of two independent biological replicates. (This figure is available in colour at JXB online.)
Orysa;KRP3 is expressed in multinucleate syncytial endosperm
A marked increase in the expression of Orysa;KRP3 at 2 DAF implies a specific cell cycle regulation at 2 DAF. It is known that a specific cell cycle event occurs during the early phase of rice seed development: the primary endosperm nucleus undergoes a series of divisions without cytokinesis, producing a multinucleate cell known as a syncytium. The syncytial nuclei are evenly arranged in the peripheral cytoplasm surrounding a large central vacuole. At a specific time point in seed development, mitosis in the peripheral syncytium ceases. Thereafter, the syncytium is partitioned into individual cells by a specific type of cytokinesis called cellularization (Hoshikawa, 1967b; Brown et al., 1996a; Olsen, 2004).
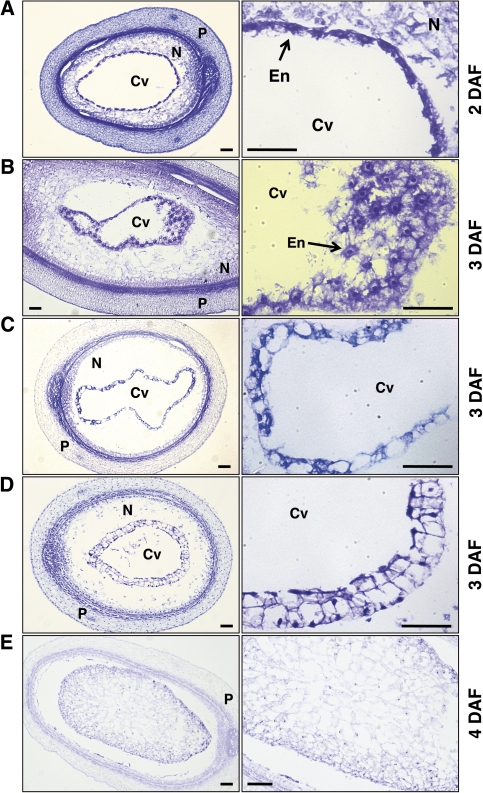
The timing of transition between syncytium formation and cellularization was determined. Figure 3 shows sections of rice seeds at 2–4 DAF stained with toluidine blue. At 2 DAF, numerous syncytial nuclei were observed in the periphery of the central cell (Fig. 3A). At 3 DAF, cellularization was observed. At the beginning of cellularization, a nuclear-based radial microtubule system (RMS) (Brown et al., 1996a; Olsen, 2004; Sabelli and Larkins, 2009) was observed (Fig. 3B). The RMS defines nuclear-cytoplasmic domains, resulting in approximately equally distanced nuclei in a single layer lining the central cell. When neighbouring RMS meet, the first anticlinal wall (perpendicular to the central cell wall) is deposited between each endosperm nucleus, compartmentalizing the cytoplasm lining the central cell (Fig. 3C). In some cases, two layers of walled cells were observed at the periphery of the endosperm, indicating the first mitotic periclinal cell division (Fig. 3D). At 4 DAF, the endosperm was completely cellularized and the central cell cavity was completely filled with cells (Fig. 3E).
Fig. 3.
Morphology of developing rice endosperms collected at 2–4 DAF. Sections of each endosperms were stained with toluidine blue. (A) Two DAF rice caryopses showing syncytial endosperm. (B) Three DAF rice caryopsis showing the radical microtubule system (RMS). (C) Three DAF rice caryopses showing a single layer of cellularized endosperm cells. The initial anticlinal walls compartmentalize the each endosperm nuclei. (D) Three DAF rice caryopses showing two layers of cellularized endosperm cells. (E) Four DAF rice caryopses showing completely cellularized endosperm. Right panels, enlarged views of the left. Cv, central vacuole; N, nucellus; P, pericarp; En, endosperm nucleus. Bars=50 μm.
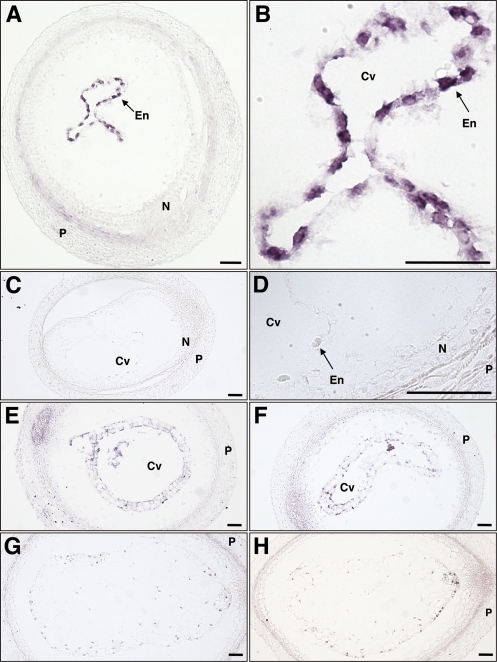
The spatial expression pattern of Orysa;KRP3 in developing rice seeds was examined next by in situ hybridization (Fig. 4). At 2 DAF, the strong expression signal of Orysa;KRP3 was observed in the multinucleate syncytium (Fig. 4A). At higher magnification, Orysa;KRP3 mRNA was detected around the endosperm nuclei arranged at the periphery of the central cell (Fig. 4B). No or a very weak background signal was detected in an equivalent section hybridized with a sense probe (Fig. 4C, D). At 3 DAF, Orysa;KRP3 mRNA was barely detectable in the primary cellularized (the first anticlinal wall formed) cells (Fig. 4E). In completely cellularized endosperm at 4 DAF, the expression of Orysa;KRP3 (Fig. 4G) was not detected. These expression profiles are in agreement with those obtained by the real-time RT-PCR analysis shown in Fig. 2. Collectively, the expression of Orysa;KRP3 is specifically high in the multinucleate syncytium at 2 DAF.
Fig. 4.
In situ localization of Orysa;KRP3 transcripts in sections of developing rice caryopses. (A–D) Caryopses at 2 DAF showing syncytial endosperm. (E, F) Caryopses at 3 DAF showing the beginning of cellularization. (G, H) Caryopses at 4 DAF showing completely cellularized endosperm. (A, B, E, G) Caryopses were hybridized with the Orysa;KRP3 antisense probe. (C, D, F, H) Caryopses were hybridized with the Orysa;KRP3 sense probe. (B) and (D) are enlarged views of (A) and (C), respectively. Cv, central vacuole; N, nucellus; P, pericarp; En, endosperm nucleus. Bars=50 μm.
Orysa;KRP3 binds rice CDKs/cyclins
The ability of Orysa;KRP3 to interact with rice CDKs and cyclins was tested by a GAL4-based yeast two-hybrid system. The plasmid encoding Orysa;KRP3 fused to the GAL4 DNA binding domain was transformed into yeast reporter strains expressing GAL4 activation domain fused CDKs or cyclins. In the absence of adenine and histidine, transformants grow only when those proteins interact. As shown in Table 2, Orysa;KRP3 interacted with Orysa;CDKA;1, Orysa;CDKA;2, Orysa;CycA1;1, and Orysa;CycD2;2. By contrast, Orysa;CDKB2;2 and Orysa;CycB2;2 did not show binding to Orysa;KRP3. In reciprocal two-hybrid assays in which the prey and bait vectors were exchanged, positive interactions were confirmed. These results indicated that Orysa;KRP3 is able to bind several rice CDKs and cyclins.
Table 2.
Interaction of Orysa;KRP3 with different CDKs and Cyclins
| Prey/bait | Orysa;KRP3 |
|
| GAL-BD | GAL-AD | |
| Orysa;CDKA;1 | + | + |
| Orysa;CDKA;2 | + | + |
| Orysa;CDKB2;1 | – | |
| Orysa;CycA1;1 | + | + |
| Orysa;CycB2;2 | – | |
| Orysa;CycD2;2 | + | + |
Orysa;KRP3 inhibits rice CDK activity
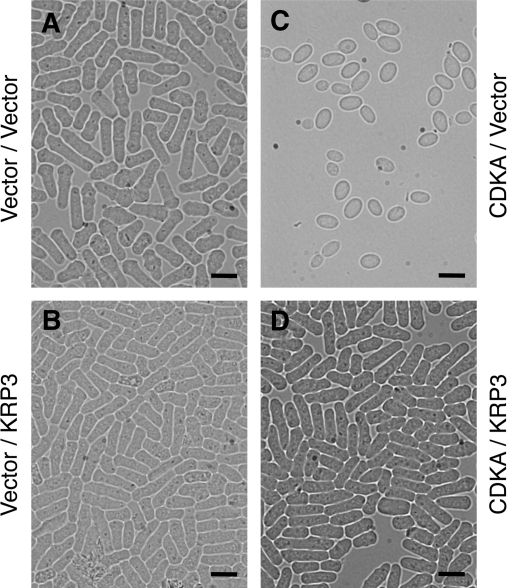
The biological activity of Orysa;KRP3 was assayed in fission yeast in vivo. Orysa;KRP3 cDNA was subcloned into an expression vector under the control of the thiamine-repressible promoter nmt1 and introduced into yeast cells. The cells expressing Orysa;KRP3 exhibited no changes in cell morphology as compared to the control cells transformed with the vector alone (Fig. 5A, B), suggesting that Orysa;KRP3 did not inhibit the function of yeast CDK. The inhibitory effect of Orysa;KRP3 on rice CDK was then tested by coexpressing Orysa;KRP3 and a dominant positive mutant of a rice CDK. The expression of a dominant positive mutant of Arabidopsis CDKA;1 (Arath;CDKA;1.A14F15) caused accelerated cell division, resulting in cells being smaller than the control cells (Hemerly et al., 1995). It has been demonstrated that this reduced size results from an increase in CDK activity in yeast cells (Porceddu et al., 1999). According to the dominant mutations described in Arath;CDKA;1.A14F15, T14 and Y15 of rice CDKA;1 was replaced with A14 and F15 (Orysa;CDKA;1.A14F15), respectively. The expression of the Orysa;CDKA;1.A14F15 gene led to the production of cells that divided at a reduced size (Fig. 5C), compared to the cells transformed with the control vector (Fig. 5A), suggesting increased CDK activity in these cells. However, when Orysa;CDKA;1.A14F15 was co-expressed with Orysa;KRP3, yeast cells exhibited normal cell morphology and cell size compared to the control cells (Fig. 5D). These results indicate that Orysa;KRP3 has the ability to inhibit rice CDK and to change the cell size.
Fig. 5.
Cytological analysis of the yeast cells expressing Orysa;KRP3 and/or the dominant positive Orysa;CDKA;1.A14F15. (A) Wild-type fission yeast cells transformed with empty control vectors. (B) Yeast cells expressing Orysa;KRP3. (C) Yeast cells expressing the dominant positive Orysa;CDKA;1.A14F15. (D) Yeast cells coexpressing Orysa;CDKA;1.A14F15 and Orysa;KRP3. Bars=5 μm.
Discussion
Rice is one of the most important food crops in the world, and improved seed yield has been a major current focus of crop breeding programmes. Alteration of the rate and duration of syncytial nuclear divisions in the endosperm has been proposed as a biotechnological strategy for altering seed size and grain yield (Tiwari et al., 2006). Therefore, it would be desirable to identify the genes specifically expressed in the syncytial endosperm. Thus far, differential screening, mutant screening, microdissection, and microarray experiments have identified the genes that are preferentially expressed in the syncytial endosperm of Arabidopsis and barley (Doan et al., 1996; Luo et al., 2005; Portereiko et al., 2006; Tiwari et al., 2006; Day et al., 2007, 2008; Kang et al., 2008). Despite the identification of syncytial endosperm-preferred genes in Arabidopsis and barley, the rice genes preferentially expressed in the syncytial endosperm have not been identified. In this study, it was found that Orysa;KRP3 is strongly expressed in the multinucleate syncytium at 2 DAF and that its expression is significantly reduced after cellularization. The transcript accumulation profiles of other KRP genes did not show any particularly increased expression at 2 DAF, suggesting that Orysa;KRP3 has distinct roles in cell cycle control during the syncytial stage of endosperm development. It is interesting to note that Arath;KRP4 showed endosperm-preferred expression in Arabidopsis (Day et al., 2008). Although the expression of Arath;KRP4 is not limited in the endosperm; further spatial and temporal expression analyses may provide information about cell cycle control in the endosperm.
The expression of a dominant positive mutant of Arth;CDKA;1.A14F15 or Orysa;CDKA;1.A14F15 in fission yeast results in cells being smaller than control cells (Fig. 5; Hemerly et al., 1995; De Veylder et al., 2001). Because conserved threonine 14 and tyrosine 15 were replaced with alanine and phenylalanine, these mutants were not down-regulated by the phosphorylation of tyrosine 15, resulting in an increase of CDK activity (Porceddu et al., 1999). Similarly, overexpression of the fission yeast cdc25 gene in tobacco results in a significant reduction in cell size (Bell et al., 1993), mostly because cdc25 activates CDK by removing the phosphate group on conserved tyrosine 15 in the catalytic cleft of CDK. By contrast, the overexpression of CKIs in transgenic plants has been shown to inhibit CDK activity and to reduce cell production. Cells in these transgenic plants became larger than those in wild-type plants (De Veylder et al., 2001; Barroco et al., 2006). Similarly, the present study showed that overexpression of Orysa;KRP3 rescued yeast premature cell division due to the expression of Orysa;CDKA;1.A14F15 (Fig. 5). Considering this, along with the results mentioned above, it can be said that Orysa;KRP3 also has the ability to inhibit CDK activity in rice.
In barley, after the initial period of intense proliferation of endosperm nuclei, there is a 2 d cell cycle arrest. During this time, nuclear-based RMS is formed (Brown et al., 1994). Cellularization begins with the formation of a functional phragmoplast at the boundary of the adjacent RMS. Next, the anticlinal wall (perpendicular to the central cell wall) is deposited between each RMS and the syncytium is partitioned into individual cells. Then, the endosperm cells resume cell cycle progression and divide periclinally. The process of cellularization proceeds centripetally until the central cell cavity is completely filled with cells (Olsen, 2001, 2004; Brown and Lemmon, 2007; Sabelli and Larkins, 2009). It is not clear if this cell cycle arrest occurs in rice as well, most likely because the cellularization process is much more rapid in rice: 3–4 d in rice; 6 d in barley (Hoshikawa, 1967b; Brown et al., 1996b; Olsen, 2001). Therefore, it is probable that a similar cell cycle arrest occurs before cellularization in rice. It is also speculated that the variation in the final number of endosperm nuclei in different species is under the control of the cell cycle arrest before cellularization. However, the molecular control mechanism that causes cell cycle arrest just before cellularization is still unknown. It was found that Orysa;KRP3 has the ability to inhibit rice CDK and is strongly expressed in the syncytial endosperm. Furthermore, the expression of Orysa;KRP3 in the late syncytial phase (2 DAF) was much higher than that in the early syncytial phase (0–1 DAF). These results raise the possibility that Orysa;KRP3 determines the final number of endosperm nuclei and the final seed size by regulating cell cycle arrest at the end of the syncytial phase. Therefore, controlling the cell cycle in the syncytial endosperm by manipulation of Orysa;KRP3 could offer the potential to improve the seed size.
In conclusion, our results suggest an important role of Orysa;KRP3 in cell cycle control during the early stage of rice endosperm development. The fact that Orysa;KRP3 accumulates at the syncytial endosperm and that Orysa;KRP3 inhibits CDK activity supports this hypothesis. Although large collections of rice mutants are now available, it is disappointing that no mutant of Orysa;KRP3 was identified. Therefore, rice plants with enhanced or reduced levels of Orysa;KRP3 expression in the syncytial endosperm, engineered for instance by overexpression or knockdown of Orysa;KRP3, can be tested in the future as new transgenic cultivars with large or small grain size or yield.
Acknowledgments
The work was supported in part by a grant from the Ministry of Agriculture, Forestry, and Fisheries of Japan (Green Technology Project EF-1002) and Grants-in-Aid for the 21st Century Center of Excellence Program from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. We are grateful to Dr Tatsuro Hirose, Dr Tetsuya Hori, and Dr Shin-ichiro Kidou for advice on in situ hybridization. We thank Dr Wataru Mitsuhashi, Dr Masaharu Kuroda, and Dr Takehiro Masumura for valuable suggestions and discussions. We also thank Dr Eiki Kuroda, Dr Hiroyuki Shimono, and Dr Setsuo Koike for gifts of various plant materials and the Yeast Genetic Resource Center Japan, supported by the National BioResource Project (YGRC/NBRP), for providing a strain and a plasmid.
Glossary
Abbreviations
- Ct
threshold cycle
- RT-PCR
reverse transcription-polymerase chain reaction
- CDK
cyclin-dependent kinase
- CKI
CDK inhibitor
- KRP
Kip-related protein
- DAF
day after flowering
- RMS
radial microtuble system
References
- Barroco RM, Peres A, Droual AM, et al. The cyclin-dependent kinase inhibitor Orysa;KRP1 plays an important role in seed development of rice. Plant Physiology. 2006;142:1053–1064. doi: 10.1104/pp.106.087056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basi G, Schmid E, Maundrell K. TATA box mutations in the Schizosaccharomyces pombe nmt1 promoter affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene. 1993;123:131–136. doi: 10.1016/0378-1119(93)90552-e. [DOI] [PubMed] [Google Scholar]
- Bell MH, Halford NG, Ormrod JC, Francis D. Tobacco plants transformed with cdc25, a mitotic inducer gene from fission yeast. Plant Molecular Biology. 1993;23:445–451. doi: 10.1007/BF00019293. [DOI] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE. The developmental biology of creal endosperm. Plant Cell Monographs. 2007;8:1–20. [Google Scholar]
- Brown RC, Lemmon BE, Olsen OA. Endosperm development in barley: microtubule involvement in the morphogenetic pathway. The Plant Cell. 1994;6:1241–1252. doi: 10.1105/tpc.6.9.1241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown RC, Lemmon BE, Olsen OA. Development of the endosperm in rice (Oryza sativa L.): cellularization. Journal of Plant Research. 1996a;109:301–313. [Google Scholar]
- Brown RC, Lemmon BE, Olsen OA. Polarization predicts the pattern of cellularization in cereal endosperm. Protoplasma. 1996b;192:168–177. [Google Scholar]
- Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate–phenol–chloroform extraction. Analytical Biochemistry. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Churchman ML, Brown ML, Kato N, et al. SIAMESE, a plant-specific cell cycle regulator, controls endoreplication onset in Arabidopsis thaliana. The Plant Cell. 2006;18:3145–3157. doi: 10.1105/tpc.106.044834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coelho CM, Dante RA, Sabelli PA, Sun Y, Dilkes BP, Gordon-Kamm WJ, Larkins BA. Cyclin-dependent kinase inhibitors in maize endosperm and their potential role in endoreduplication. Plant Physiology. 2005;138:2323–2336. doi: 10.1104/pp.105.063917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, Herridge RP, Ambrose BA, Macknight RC. Transcriptome analysis of proliferating Arabidopsis endosperm reveals biological implications for the control of syncytial division, cytokinin signaling, and gene expression regulation. Plant Physiology. 2008;148:1964–1984. doi: 10.1104/pp.108.128108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day RC, McNoe L, Macknight RC. Evaluation of global RNA amplification and its use for high-throughput transcript analysis of laser-microdissected endosperm. International Journal of Plant Genomics. 2007;2007:61028. doi: 10.1155/2007/61028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Veylder L, Beeckman T, Beemster GT, Krols L, Terras F, Landrieu I, van der Schueren E, Maes S, Naudts M, Inzé D. Functional analysis of cyclin-dependent kinase inhibitors of Arabidopsis. The Plant Cell. 2001;13:1653–1668. doi: 10.1105/TPC.010087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doan DN, Linnestad C, Olsen OA. Isolation of molecular markers from the barley endosperm coenocyte and the surrounding nucellus cell layers. Plant Molecular Biology. 1996;31:877–886. doi: 10.1007/BF00019474. [DOI] [PubMed] [Google Scholar]
- Guo J, Song J, Wang F, Zhang XS. Genome-wide identification and expression analysis of rice cell cycle genes. Plant Molecular Biology. 2007;64:349–360. doi: 10.1007/s11103-007-9154-y. [DOI] [PubMed] [Google Scholar]
- Gusti A, Baumberger N, Nowack M, Pusch S, Eisler H, Potuschak T, De Veylder L, Schnittger A, Genschik P. The Arabidopsis thaliana F-box protein FBL17 is essential for progression through the second mitosis during pollen development. PLoS One. 2009;4:e4780. doi: 10.1371/journal.pone.0004780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemerly A, de Almeida Engler J, Bergounioux C, Van Montagu M, Engler G, Inzé D, Ferreira P. Dominant negative mutants of the Cdc2 kinase uncouple cell division from iterative plant development. EMBO Journal. 1995;14:3925–3936. doi: 10.1002/j.1460-2075.1995.tb00064.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Himanen K, Boucheron E, Vanneste S, de Almeida Engler J, Inzé D, Beeckman T. Auxin-mediated cell cycle activation during early lateral root initiation. The Plant Cell. 2002;14:2339–2351. doi: 10.1105/tpc.004960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirose T, Takano M, Terao T. Cell wall invertase in developing rice caryopsis: molecular cloning of OsCIN1 and analysis of its expression in relation to its role in grain filling. Plant and Cell Physiology. 2002;43:452–459. doi: 10.1093/pcp/pcf055. [DOI] [PubMed] [Google Scholar]
- Hoshikawa K. Studies on the development of endosperm in rice. 3. Observations on the cell division. Japanese Journal of Crop Science. 1967a;36:210–215. [Google Scholar]
- Hoshikawa K. Studies on the development of endosperm in rice. 1. Process of endosperm tissue formation. Japanese Journal of Crop Science. 1967b;36:151–1613. [Google Scholar]
- Hoshikawa K. Studies on the development of endosperm in rice. 2. Process of endosperm tissue formation with special reference to the enlargement of cells. Japanese Journal of Crop Science. 1967c;36:203–209. [Google Scholar]
- Hoshikawa K. Studies on the development of endosperm in rice. 11. Development of starch granules in endosperm tissue. Japanese Journal of Crop Science. 1968;37:207–216. [Google Scholar]
- Ikemoto S, Nakamura T, Kubo M, Shimoda C. S. pombe sporulation-specific coiled-coil protein Spo15p is localized to the spindle pole body and essential for its modification. Journal of Cell Science. 2000;113:545–554. doi: 10.1242/jcs.113.3.545. [DOI] [PubMed] [Google Scholar]
- Kang IH, Steffen JG, Portereiko MF, Lloyd A, Drews GN. The AGL62 MADS domain protein regulates cellularization during endosperm development in Arabidopsis. The Plant Cell. 2008;20:635–647. doi: 10.1105/tpc.107.055137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim HJ, Oh SA, Brownfield L, Hong SH, Ryu H, Hwang I, Twell D, Nam HG. Control of plant germline proliferation by SCF(FBL17) degradation of cell cycle inhibitors. Nature. 2008;455:1134–1137. doi: 10.1038/nature07289. [DOI] [PubMed] [Google Scholar]
- Liu J, Zhang Y, Qin G, et al. Targeted degradation of the cyclin-dependent kinase inhibitor ICK4/KRP6 by RING-type E3 ligases is essential for mitotic cell cycle progression during Arabidopsis gametogenesis. The Plant Cell. 2008;20:1538–1554. doi: 10.1105/tpc.108.059741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Luo M, Dennis ES, Berger F, Peacock WJ, Chaudhury A. MINISEED3 (MINI3), a WRKY family gene, and HAIKU2 (IKU2), a leucine-rich repeat (LRR) KINASE gene, are regulators of seed size in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2005;102:17531–17536. doi: 10.1073/pnas.0508418102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen OA. ENDOSPERM DEVELOPMENT: cellularization and cell fate specification. Annual Review of Plant Physiology and Plant Molecular Biology. 2001;52:233–267. doi: 10.1146/annurev.arplant.52.1.233. [DOI] [PubMed] [Google Scholar]
- Olsen OA. Nuclear endosperm development in cereals and Arabidopsis thaliana. The Plant Cell. 2004;16:S214–227. doi: 10.1105/tpc.017111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peres A, Churchman ML, Hariharan S, et al. Novel plant-specific cyclin-dependent kinase inhibitors induced by biotic and abiotic stresses. Journal of Biological Chemistry. 2007;282:25588–25596. doi: 10.1074/jbc.M703326200. [DOI] [PubMed] [Google Scholar]
- Porceddu A, De Veylder L, Hayles J, Van Montagu M, Inzé D, Mironov V. Mutational analysis of two Arabidopsis thaliana cyclin-dependent kinases in fission yeast. FEBS Letters. 1999;446:182–188. doi: 10.1016/s0014-5793(99)00211-2. [DOI] [PubMed] [Google Scholar]
- Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN. AGL80 is required for central cell and endosperm development in Arabidopsis. The Plant Cell. 2006;18:1862–1872. doi: 10.1105/tpc.106.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabelli PA, Larkins BA. The development of endosperm in grasses. Plant Physiology. 2009;149:14–26. doi: 10.1104/pp.108.129437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka T, Antonio BA, Kikuchi S, et al. The Rice Annotation Project Database (RAP-DB): 2008 update. Nucleic Acids Research. 2008;36:D1028–D1033. doi: 10.1093/nar/gkm978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiwari S, Spielman M, Day RC, Scott RJ. Proliferative phase endosperm promoters from Arabidopsis thaliana. Plant Biotechnology Journal. 2006;4:393–407. doi: 10.1111/j.1467-7652.2006.00189.x. [DOI] [PubMed] [Google Scholar]
- Verkest A, Manes CL, Vercruysse S, Maes S, Van Der Schueren E, Beeckman T, Genschik P, Kuiper M, Inzé D, De Veylder L. The cyclin-dependent kinase inhibitor KRP2 controls the onset of the endoreduplication cycle during Arabidopsis leaf development through inhibition of mitotic CDKA;1 kinase complexes. The Plant Cell. 2005;17:1723–1736. doi: 10.1105/tpc.105.032383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Qi Q, Schorr P, Cutler AJ, Crosby WL, Fowke LC. ICK1, a cyclin-dependent protein kinase inhibitor from Arabidopsis thaliana interacts with both Cdc2a and CycD3, and its expression is induced by abscisic acid. The Plant Journal. 1998;15:501–510. doi: 10.1046/j.1365-313x.1998.00231.x. [DOI] [PubMed] [Google Scholar]
- Weinl C, Marquardt S, Kuijt SJ, Nowack MK, Jakoby MJ, Hulskamp M, Schnittger A. Novel functions of plant cyclin-dependent kinase inhibitors, ICK1/KRP1, can act non-cell-autonomously and inhibit entry into mitosis. The Plant Cell. 2005;17:1704–1722. doi: 10.1105/tpc.104.030486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Y, Wang H, Gilmer S, Whitwill S, Keller W, Fowke LC. Control of petal and pollen development by the plant cyclin-dependent kinase inhibitor ICK1 in transgenic Brassica plants. Planta. 2002;215:248–257. doi: 10.1007/s00425-002-0752-2. [DOI] [PubMed] [Google Scholar]