Abstract
Proliferating cell nuclear antigen (PCNA) is an essential factor in DNA replication and in many other processes in eukaryotic cells. Genetic analysis of Phaseolus coccineus showed the presence of at least two PCNA-like genes in the runner bean genome. Two PCNA genes have previously been found in a few plant species including Arabidopsis, tobacco, and maize. In these species, genes were nearly identical. Two cDNAs of P. coccineus PCNA (PcPCNA1 and PcPCNA-like1) have been identified that differ distinctly from each other. Interestingly, both the genetic organization of PcPCNA1 and PcPCNA-like1 genes and their expression patterns were similar, but these were the only similarities between these genes and their products. The identity between PcPCNA1 and PcPCNA-like1 at the amino acid level was only 54%, with PcPCNA-like1 lacking motifs that are crucial for the activity typical of PCNA. Consequently, these two proteins showed different properties. PcPCNA1 behaved like a typical PCNA protein: it formed a homotrimer and stimulated the activity of human DNA polymerase delta. In addition, PcPCNA1 interacted with a p21 peptide and was recognized by an anti-human PCNA monoclonal antibody PC10. By contrast, PcPCNA-like1 was detected as a monomer and was unable to stimulate the DNA polymerase delta activity. PcPCNA-like1 also could not interact with p21 and was not recognized by the PC10 antibody. Our results suggest that PcPCNA-like1 either is unable to function alone and therefore might be a component of the heterotrimeric PCNA ring or may have other, yet unknown functions. Alternatively, the PcPCNA-like1 gene may represent a pseudogene.
Keywords: DNA polymerase delta, PCNA, Phaseolus coccineus, PRINS, RACE
Introduction
Proliferating cell nuclear antigen (PCNA) was first identified as a factor recognized by an autoantibody present in the sera of patients with autoimmune disorder called systemic lupus erythematosus (Miyachi et al., 1978). It is a homologue of a beta subunit of Escherichia coli DNA polymerase III and a product of bacteriophage T4 gene-45 (Kelman, 1997). The function attributed for PCNA was a processivity factor of DNA polymerase delta required for the synthesis of a new DNA strand (Tan et al., 1986; Bravo et al., 1987; Prelich et al., 1987). It was shown that PCNA with the help of a replication factor C (RF-C) is loaded on DNA, where it forms a trimeric ring structure encircling DNA (Mossi and Hubscher, 1998, Moldvan et al., 2007). These findings were supported by the results of structural studies of yeast and human PCNA (Krishna et al., 1994; Schurtenberger et al., 1998). Afterwards, PCNA was shown to be involved not only in DNA replication but also in DNA repair (Kelman, 1997). In addition, interaction of PCNA with proteins that are involved in many other cellular processes indicates its potential role in chromatin assembly, sister-chromatid cohesion, transcription, and cell cycle regulation (Maga and Hubscher, 2003; Naryzhny, 2008; Stoimenov and Helleday, 2009). Analysis of all known PCNAs suggests that during evolution of eukaryotic organisms, PCNA remained conserved in function, structure, and sequence. Yeast and Drosophila PCNAs were shown to be able to substitute for mammalian PCNA in DNA replication assays (Bauer and Burgers, 1988; Ng et al., 1990). Hashimoto's group demonstrated that recombinant rice PCNA stimulated the enzymatic activity of DNA polymerase delta from human cells (Matsumoto et al., 1994). In other studies, mammalian PCNA stimulated the activity and processivity of two wheat delta-like polymerases (Laquel et al., 1993). Moreover, the formation of a stable complex of purified pea PCNA and human p21/WAF-1 (a p53-dependent protein involved in cell cycle regulation and stress response) was observed (Ball and Lane, 1996). PCNA homologues have been cloned from several groups of eukaryotic organisms such as yeast: budding yeast (Bauer and Burgers, 1990) and fission yeast (Waseem et al., 1992); animals: human (Almendral et al., 1987), rat (Matsumoto et al., 1987), mouse (Yamaguchi et al., 1991), Drosophila (Yamaguchi et al., 1990), and Xenopus (Leibovici et al., 1990) as well as plants: carrot (Hata et al., 1992), maize (Lopez et al., 1995, 1997), periwinkle (Kodama et al., 1991), rice (Suzuka et al., 1991), oilseed rape (Markley et al., 1993), pea (Shimizu and Mori, 1998a), and common bean (Strzalka and Ziemienowicz, 2007).
More detailed studies concerning plant PCNA have been conducted only with a few organisms such as rice and tobacco and concentrated mainly on regulatory elements of PCNA gene expression. Upstream sequences of the rice PCNA gene were shown to mediate expression of the PCNA-GUS chimeric gene in meristems of transgenic tobacco plants (Kosugi et al., 1991). Moreover, two PCNA gene promoter elements essential for meristematic tissue-specific expression were identified (Kosugi et al., 1995). Continuation of this work resulted in the identification of two proteins, PCF1 and PCF2, which specifically bind to cis elements in the rice PCNA gene (Kosugi and Ohashi, 1997). E2F-like sites of the rice and tobacco PCNA promoter were shown to be required for meristematic tissue-specific expression of this gene in actively dividing cells (Kosugi and Ohashi, 2002). Engagement of the E2F site of the tobacco PCNA gene promoter was presented by Hanley–Bowdoin's group who found that the E2F1 + 2 sites contribute to repression of the PCNA promoter in mature tissues, whereas the E2F1 site with transcription activators positively regulates PCNA gene expression in young leaves (Egelkrout et al., 2002).
Most recently, the first analyses of plant PCNA proteins have been reported. Arabidopsis PCNA1 and PCNA2 proteins show very high levels of amino acid sequence similarity and share some common features. Both proteins were shown to be able to form a homotrimeric ring structure while interacting with the C-terminal segment of human p21 (Strzalka et al., 2009). Moreover, protein–protein interaction analysis using yeast two hybrid system revealed that AtPCNA1 and AtPCNA2 could interact with the TLS DNA polymerase eta (Anderson et al., 2008).
In previous studies, an open reading frame (ORF) of the Phaseolus vulgaris PCNA gene was identified (Strzalka and Ziemienowicz, 2007). Here for the first time, the isolation and analysis of two different PCNA cDNAs of Phaseolus coccineus, PcPCNA1 and PcPCNA-like1 is reported.
Materials and methods
Plant material and growth condition
Seeds of runner bean (Phaseolus coccineus L. cultivar KONTRA) were purchased from Plantico Golebiew HiNO Sp. z o.o Poland. The seeds were germinated in darkness at 20 °C in a Petri dish containing water. Samples of embryonic axes were collected from germinating seeds every 24 h, frozen in liquid nitrogen, and stored at –80 °C. In addition, the seeds were germinated and grown in a greenhouse under natural summer light conditions. Ten days after germination, the samples of root, stem, and leaf tissues were collected, frozen in liquid nitrogen, and stored at –80 °C. Moreover, the segments containing the micropylar region of 3–5 mm long seeds (containing micropylum and a part of the embryonic sac including the developing embryo at an early stage of maturation) were collected after pollination and stored as described above.
Cloning of PcPCNA1 and PcPCNA-like1 cDNA using 5′ and 3′ RACE
5′ RACE (rapid amplification of cDNA ends) was carried out using FirstChoice RLM-RACE (Ambion) following the protocol provided by the supplier. Ten μg of total RNA isolated using the Trizol reagent (Invitrogen) from the micropylar region of the seed were treated with calf intestinal alkaline phosphatase. Next, the sample was treated with tobacco acid phyrophosphatase and, subsequently, with RNA ligase to ligate the RNA adapter to the 5′ end of full-length mRNAs. The RNA was reverse transcribed, and two-step PCR amplification was performed. Amplification of PcPCNA1 and PcPCNA-like1 was done using specific reverse primers 5′-PcPCNA1R (5′-TAATATCCTAACCCAACATTCAATAGTG-3′) and 5′-PcPCNAL1R (5′-AC(G/T)GAAAGAA(A/C)AAT(C/T)CTA(A/G)TATCCTAACCC-3′), respectively. Subsequently, nested specific reverse primers for reamplification of the obtained PCR products, 5′-PcPCNA1NR (5′-TTACACTTGAGGTTTCTCTTCTTC-3′) and 5′-PcPCNAL1NR (5′-CTATGATGTAGGGGATGTAATGGG-3′), were used.
3′ RACE was performed similarly using 1 μg of total RNA and specific forward primers: 3′-PcPCNA1F (5′-AACCCTAACCATTTCTAAACGAAACCC-3′) and 3′-PcPCNAL1F (5′-TATTGCTTCCAGACCTCAAAACCCCAAC-3′) for the first PCR amplification, followed by PCR with specific nested primers: 3′-PcPCNA1NF (5′-GGA(C/T)ATTGATAG(C/T)GA(A/G)CA(C/T)CTTGG-3′) and 3′-PcPCNAL1NF (5′-CCCCAACCATTCCTAAACCGCTATC-3′).
The PCR reactions were done in 50 μl volume containing: 1× PCR buffer (10 mM TRIS-HCl, 50 mM KCl, 1.5 mM MgCl2, pH 8.0), 200 μM dNTPs, 1.25 units of SuperTaq DNA polymerase (Ambion) and 2 μM of each primer. The amplification reactions consisted of a preliminary denaturation step at 94 °C for 5 min, followed by 35 cycles of 94 °C for 30 s, 60 °C for 30 s, 72 °C for 2 min, and an incubation at 72 °C for 7 min were performed in a (Biometra) termocycler. The resulting PCR products were purified and cloned into the pTZ57R\T vector (Fermentas) followed by sequencing. The nucleotide sequence data have been deposited in the NCBI GenBank under accession numbers: EF602032 (PcPCNA1) and EF602034 (PcPCNA-like1).
Cloning of PcPCNA1 and PcPCNA-like1 genomic sequences
Amplification of genomic fragments encoding PcPCNA1 and PcPCNA-like1 was performed using genomic DNA extracted using the Genomic Maxi AX Kit (A&A Biotechnology). Gene-specific primers: 3′-PcPCNA1F (5′-AACCCTAACCATTTCTAAACGAAACCC-3′) and gPcPCNA1R (5′-AACTGAATTCCAAATTCGTTGCTCACAG-3′) were used for PcPCNA1 amplification. Amplification of genomic PcPCNA-like1 was done using 3′-PcPCNAL1F (5′-TATTGCTTCCAGACCTCAAAACCCCAAC-3′) and 5′-PcPCNAL1R (5′-AC(G/T)GAAAGAA(A/C)AAT(C/T)CTA(A/G)TATCCTAACC C-3′) primers. The reaction was done in 50 μl volume containing: 1× PCR buffer, 200 μM dNTPs, 1.25 units of SuperTaq DNA polymerase (Ambion), and 2 μM of each primer.
The samples were heated at 94 °C for 5 min and then subjected to 30 cycles of 94 °C for 30 s, 60 °C for 30 s, and 72 °C for 2 min. Then they were incubated at 72 °C for 7 min in a (Biometra) termocycler. The resulting amplified PCR products were purified and cloned into pTZ57R\T vector (Fermentas) followed by sequencing. The nucleotide sequence data have been deposited in the NCBI GenBank under accession numbers: EF602033 (gPcPCNA1) and EF602035 (gPcPCNA-like1).
Real-time RT-PCR
For real-time RT-PCR, total RNA was isolated using the Trizol reagent (Invitrogen). cDNA synthesis was carried out on 1 μg of total RNA using the QuantiTect Reverse Transcription Kit with genomic DNA wipe-out buffer (Qiagen). Real-time PCR reactions were performed in mixtures containing: 1× of SYBR Green PCR Master Mix (SYBR Green qPCR Kit, Finnzymes), 0.5 μM of each primer and 200 ng of cDNA in a final volume of 15 μl. The reactions were performed using control 18S RNA gene-specific primers: 3′-18SRNA_RTPCRF (5′-CCAGGTCCAGACATAGTAAG-3′) and 5′-18SRNA_RTPCRR (5′-GTACAAAGGGCAGGGACGTA-3′) (Duval et al., 2002), PcPCNA1 gene-specific primers 3′-PcPCNA1_RTPCRF (5′-GATATTGGATCTGCAAATATAG-3′) and 5′-PcPCNA1R (5′-TAATATCCTAACCCAACATTCAATAGTG-3′) or PcPCNA-like1 gene-specific primers 3′-PcPCNAL1_RTPCRF (5′-AGAATAAGAAATGGAGGGAC-3′) and 5′-PcPCNAL1_RTPCRR (5′-ATGATGTAGGGGATGTAATG-3′) with annealing temperatures 56 °C, 47 °C, and 55 °C, respectively. All reactions were performed in a PCR machine (Corbett-Research, Rotor-Gene, RG-3000) using the following cycling conditions: 95 °C for 10 min and 40 three-step cycles of 30 s at 95 °C, 45 s of annealing, and 45 s at 72 °C. All PCR reactions were carried out in triplicate. Relative quantification of gene expression was calculated based on the comparative Ct (threshold cycle value) method (ΔCt=Ct gene of interest–Ct 18S RNA). Comparison of gene expression between tested samples was derived by subtracting the leaf ΔCt values from the tested sample ΔCt values to give a ΔΔCt value. Relative gene expression was calculated as 2–ΔΔCt.
DNA isolation and Southern blot analysis
Genomic DNA was extracted from 96 h old seedlings using the Genomic Maxi AX Kit (A&A Biotechnology). The purified DNA (30 μg) was digested with BamHI, BglII, EcoRI, HindIII or XbaI, separated in 0.8% agarose gel, blotted on a positively charged nylon membrane (Roche), following the standard hybridization protocol (Sambrook and Russell, 2001). Hybridization was performed for 16 h at 65 °C with the PcPCNA1 798 bp long DIG-labelled probe generated by PCR. After autoradiography, the probe was stripped off and the blot was hybridized with the 816 bp long PcPCNA-like1 DIG-labelled probe (under the same conditions as used for the PcPCNA1 probe).
PCR
PCR reactions were performed in mixtures containing: 1× PCR buffer (Takara) with 2 mM MgCl2, 200 μM dNTPs, 2 μM of each primer, 1 unit of Takara Taq polymerase, and 50 ng of genomic DNA isolated from P. coccineus seedlings or plasmid pTZ57R\T DNA containing genomic sequence of the PcPCNA1 or PcPCNA-like1 genes in a final volume of 25 μl. The reactions were performed using degenerated primers: PcPCNAF (5′-GTGCAAGGTTC(T/C/A)(C/G)T(T/C)CTGAAGAAGG-3′) and PcPCNAR (5′-C(C/A)(G/A)TCTC(A/T)GCAAT(T/C)TTGTA(T/C)TC-3′). All reactions were performed in a PCR machine (Biometra) using the following cycling conditions: 95 °C for 5 min and 30 three-step cycles of 30 s at 95 °C, 30 s at 55 °C, and 1 min at 72 °C, followed by 5 min at 72 °C.
PRINS (Primed in situ DNA labelling)
Seeds of runner bean were imbibed for 5 h in distilled water at 25 °C with aeration, and then germinated on moistened filter paper in Petri dishes (25 °C) for 16 h. Then they were treated with Hoagland's solution (1.6 g l−1, Sigma-Aldrich) for 5 h (Dolezel et al., 1999). Next, 1–2 cm long roots were collected in iced water and incubated at 0 °C for 24 h, fixed in Carnoy's solution (ethanol and glacial acetic acid, 3:1 v/v) and stored at 4 °C. Permanent squash preparations were made from root meristems as described previously (Schwarzacher and Heslop-Harrison, 2000), with some modifications developed for Lupinus (Naganowska et al., 2003). The slides were stored at –20 °C until used for PRINS. Before performing a PRINS reaction, the slides were dried overnight at 37 °C. Frame-Seal Chambers (MJ Research, Inc.) were stuck to the slides. Gene specific primers: 3′-PcPCNA1F (5′-AACCCTAACCATTTCTAAACGAAACCC-3′) and gPcPCNA1R (5′-AACTGAATTCCAAATTCGTTGCTCACAG-3′) were used for PcPCNA1 amplification. Amplification of PcPCNA-like1 was performed using 3′-PcPCNAL1F (5′-TATTGCTTCCAGACCTCAAAACCCCAAC-3′) and 5′-PcPCNAL1R (5′-C(G/T)GAAAGAA(A/C)AAT(C/T)CTA(A/G)TATCCTAACCC-3′) primers. Reaction mixtures contained a DIG DNA labelling mixture (0.1 mM dATP, dCTP, dGTP, and 0.035 mM DIG-12-dUTP with 0.065 mM dTTP) (Roche), 3 mM MgCl2, 3 units of Taq polymerase (Invitrogen), and 2 μM of each primer. 25 μl of the mixture were put into each frame, and the frames were covered with polyester coverslips. The PRINS reaction mixtures were heated at 91 °C for 5 min and then incubated at 55 °C for 15 min. In the third stage, primer extension reactions were performed at 72 °C for 30 min (MJ Thermal Cycler PTC-200 with a Slide Griddle plate). The reactions were stopped by adding stop buffer (500 mM NaCl, 50 mM EDTA, pH 8.0) followed by incubation at 70 °C for 2 min. Next, the slides were incubated in blocking buffer [0.5% blocking reagent (Roche), 100 mM maleic acid, 150 mM NaCl, pH 7.5] at 37 °C for 30 min, and then in anti-DIG-fluorescein antibody solution (20 μg ml−1) (Roche) for 60 min. Then the slides were placed in washing buffer (100 mM maleic acid, 150 mM NaCl, and 0.05% Tween 20, pH 7.5) at room temperature for 5 min and counterstained with DAPI in Vectashield antifade solution (Vector). The preparations were examined with the OLYMPUS BX60 Research System Microscope. The images were acquired with a black and white CCD camera, interfaced to a PC running the analySIS 3.0 software (Soft Imaging System).
Purification of recombinant PcPCNA1 and PcPCNA-like1 proteins
The open reading frames of PcPCNA1 and PcPCNA-like1 were amplified with specific sets of primers: PcPCNA1ORFf (5′-GGAATTCCATATGTTGGAATTACGTCTCGTGCAAG-3′) and PcPCNA1ORFr (5′-CGGGATCCTTACACTTGAGGTTTCTCTTCTTC-3′), or PcPCNAL1ORFf (5′-GGAATTCCATATGTTGGAAGTCCGTTTCGTGCAAG-3′) and PcPCNAL1ORFr (5′-CGGGATCCCTATGATGTAGGGGATGTAATGGG-3′), respectively. Next, the amplified products were cloned into NdeI/BamHI sites of pET15b expression vector and sequenced. Constructs were introduced into E. coli BL21(DE3) strain. Bacteria were grown at 37 °C in 2.0 l LB medium containing ampicillin (100 μg ml−1) until OD595 0.6 was reached, and production of PcPCNA proteins was induced with 1 mM IPTG at 37 °C. After 4 h of induction, cells were harvested by centrifugation (5 000 g for 15 min at 4 °C) and resuspended in 50 ml of lysis buffer A [50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, 10 mM β-mercaptoethanol, 1 mM PMSF (phenylmethylsulphonyl fluoride), 0.05% Tween 20, pH 8.0] containing lysozyme (1 mg ml−1), RNase A (10 μg ml−1), DNase I (5 μg ml−1), and sonicated (5 pulses for 30 s). All the following procedures were performed at 4 °C. The cells were centrifuged at 40 000 g for 30 min, and the cell lysate was loaded onto a 2 ml Ni-NTA Superflow (Qiagen) column. The unbound proteins were washed with 10 vols of buffer A containing 20 mM imidazole. The bound proteins were eluted with buffer A containing 250 mM imidazole and then dialysed against buffer B (50 mM TRIS-HCl, 0.1 mM EDTA, 10 mM β-mercaptoethanol, 1 mM PMSF, 0.05% Tween 20, pH 7.6). The dialysed fraction was loaded onto a 2 ml Heparin Sepharose 6 Fast Flow (Amersham) column, and the flow-through was collected and loaded onto 1 ml HiTrap Q HP Sepharose (Amersham). The unbound proteins were removed with 10 ml of buffer B. The bound proteins were eluted with a 30 ml linear gradient of 0 to 1 M NaCl in buffer B. The fractions containing the recombinant protein were dialysed against buffer C (50 mM HEPES, 150 mM NaCl, 0.1 mM EDTA, 10 mM β-mercaptoethanol, 1 mM PMSF, 10% glycerol, pH 7.6), then frozen in liquid nitrogen and stored at –80 °C until use. Protein concentration was determined using the Bio-Rad Protein Assay.
Recombinant PCNA2 protein of Arabidopsis thaliana (AtPCNA2) was purified as described previously (Strzalka et al., 2009).
Gel filtration
All the following procedures were performed at 4 °C. The purified proteins (PcPCNA1, PcPCNA-like1, and AtPCNA2) were dialysed against 50 mM sodium phosphate buffer (pH 7.0) containing 150 mM NaCl. The protein sample (0.5 ml; 250 μg) was loaded onto 24 ml Superdex 200 column with the flow rate 0.5 ml min−1. The recorded chromatogram was used for molecular mass calculation of the tested protein, based on previously separated standard proteins: tyroglobulin (670 kDa), γ calf globulin (158 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa).
Complex formation with p21 peptide
Biotinylated synthetic peptide (KRRQTSMTDFYHSKRRLIFS, 2 μg; synthesized by the Protein Analysis service unit at FMI, Basel) was dissolved in DMSO (a final concentration of 0.5 mg ml−1), diluted in 100 μl PBS and incubated with 20 μl of streptavidin-agarose beads (Pierce) for 1 h at room temperature. Unbound peptide was removed by three washing steps with 1 ml of PBS each. The beads with bound peptide were incubated with 1 μg of the recombinant protein (PcPCNA1, PcPCNA-like1, or HsPCNA) at 4 °C for 1 h. Unbound protein was removed by three washings with 1 ml of PBS, and the beads were heated at 94 °C for 5 min in 1× protein sample loading buffer containing SDS and DTT. The samples were separated in 12% polyacrylamide gel during SDS-PAGE (Laemmli, 1970), followed by Commassie staining.
Western blotting and immunodetection
One μg of recombinant PcPCNA1, PcPCNA-like1, and HsPCNA was separated in 12% polyacrylamide gel (SDS-PAGE; Laemmli, 1970) and electrotransferred onto a PVDF (0.2 μm) membrane (Millipore) as described previously (Towbin et al., 1979). After the transfer was completed, the membrane was washed three times for 5 min in 1× PBS supplemented with 0.5% Tween 20 (PBS-T) and blocked with PBS-T containing 5% fat-free milk (PBS-TB) for 30 min. The membrane was incubated overnight at 4 °C with an anti-human PCNA monoclonal antibody (PC10, Sigma, dilution 1:2 000). After washing in PBS-TB, the membrane was incubated for 1 h at room temperature with goat anti-mouse IgG alkaline phosphatase-conjugate (Sigma, dilution 1:10 000). After several washes in PBS-T, immunodetection was performed using the BCIP/NBT substrate (ImmunO, MP Biomedicals) at room temperature.
The DNA polymerase assay
The reaction was carried out according to the previously published protocol (Weiser et al., 1991). A 25 μl volume mixture contained the following components: 50 mM BIS-TRIS, pH 6.5, 1 mM DTT, 0.25 mg ml−1 BSA, 6 mM MgCl2, 20 μM [3H]dTTP 500 (cpm pmol−1), 0.5 μg poly(dA)/oligo(dT) template (10:1), and 40 ng of human polymerase delta (0.54 units), in the absence or presence of 5 μg of the tested protein (BSA, human PCNA, PcPCNA1 or PcPCNA-like1). Reaction mixtures were incubated at 37 °C for 30 min, precipitated with TCA, and the radioactivity of insoluble material was determined in a scintillation counter using CytoScint (ICN) scintillation solution. One unit was defined as 1 pmol of dTMP incorporated into acid-precipitable material during 30 min at 37 °C.
Results
Cloning of cDNA and genomic DNA coding for PcPCNA1 and PcPCNA-like1
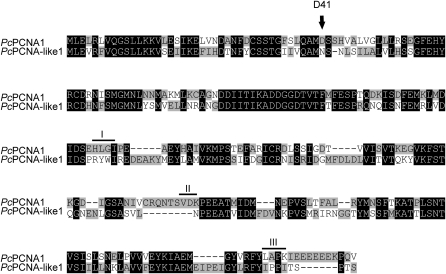
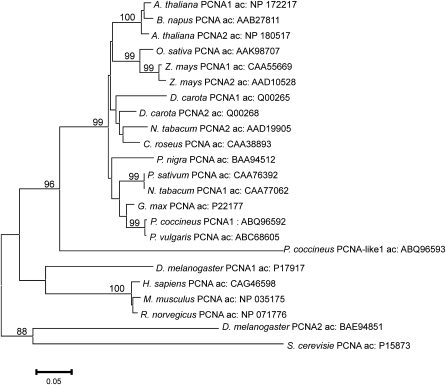
Rapid amplification of 5′ and 3′ cDNA ends (RACE) techniques were employed for the amplification of full-length cDNAs encoding PcPCNA1 and PcPCNA-like1. The primers used for the identification of PcPCNA1 and PcPCNA-like1 cDNAs were designed based on the analysis of Phaseolus coccineus EST fragments deposited in the National Centre for Biotechnology Information (NCBI). The micropylar region of seeds containing an active suspensor was used as a source of total RNA. The identified cDNA sequence of PcPCNA1 contained a 798 bp open reading frame encoding a polypeptide of 265 amino acids. A calculated molecular mass of the polypeptide was 29.45 kDa and pI=4.69. PcPCNA-like1 cDNA consisted of a 816 bp open reading frame encoding a polypeptide of 271 amino acids with a molecular mass of 30.73 kDa and pI=5.0. An alignment of P. coccineus PcPCNA1 and PcPCNA-like1 showed that the identity between these two amino acid sequences was 54.5% (Fig. 1). Alignment analysis of PcPCNA1 against human PCNA (accession number: CAC27344) and pea PCNA (accession number: CAA76392) at the amino acid level demonstrated an identity of 64.5% and 92.9%, respectively, whereas alignment analysis of PcPCNA-like1 against human and pea PCNAs showed an identity of 38.3% and 52.3%, respectively. Analogous evolutionary divergences and similarities of PCNA proteins from various species can also be confirmed, based on the analysis of a PCNA phylogenic tree (see Discussion and Fig. 10).
Fig. 1.
Alignment of amino acid sequences of PcPCNA1 (accession number: ABQ96591) and PcPCNA-like1 (accession number: ABQ96593) proteins. The analysis was done using StretcherP and the graphical representation was made using GeneDoc software. Identical amino acids are denoted as white letters on a black background. The black letters on grey and white backgrounds represent different amino acids. The residue D41 and motifs I (H125L126G127I128), II (V188D189K190), and III (L251A252P253K254) are marked. Dashes represent gaps introduced to maximize identities.
Fig. 10.
Phylogenetic tree of PCNA constructed by the Neighbor–Joining method based on amino acid (aa) sequences from P. coccineus and other selected eukaryotic organisms was created using MEGA 3.1 software (Kumar et al., 2004). The scale bar represents 0.05 substitutions per site, and the numbers next to the nodes are bootstrap values from 100 000 replicates. Values equal to or higher than 80% are shown.
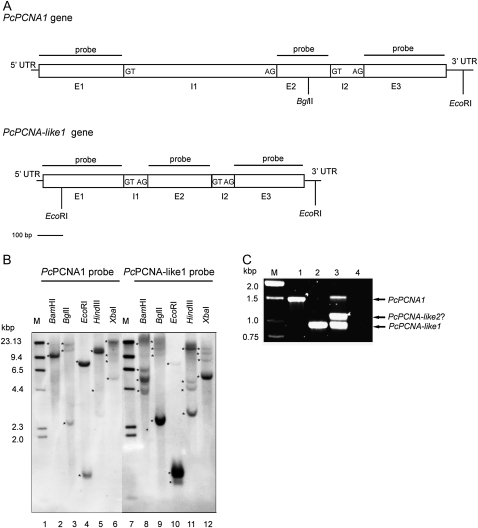
Genomic sequences encoding PcPCNA1 and PcPCNA-like1 have been amplified, cloned, and sequenced. Sequence analysis revealed that both genes contained two introns (Fig. 2A). The PcPCNA1 intron 1 was 633 bp in length, and intron 2 was 137 bp, whereas introns of the PcPCNA-like1 gene were 97 bp and 83 bp. Highly conserved (5′-GT/AG-3′) intron termini were identified in all introns of both PcPCNA1 and PcPCNA-like1 genes (Fig. 2A).
Fig. 2.
Southern blot and PCR analysis of P. coccineus genomic DNA. (A) Structure of PcPCNA1 and PcPCNA-like1 genes. The nucleotide sequences were analysed using WebGene software. Exons (E), introns (I), and 5′-UTR and 3′-UTR regions are marked. The border sequences of introns termini are placed in the boxes. The positions of internal sites recognized by restriction enzymes used in the Southern blotting analysis are marked. (B) Southern blotting results. 30 μg of the genomic DNA isolated from P. coccineus seedlings were digested with BamHI (lanes 2 and 8), BglII (lanes 3 and 9), EcoRI (lanes 4 and 10), HindIII (lanes 5 and 11) or XbaI (lanes 6 and 12), separated in 0.8% agarose gel and subjected to Southern blot procedure with the PcPCNA1 (lanes 2–6) or PcPCNA-like1 (lanes 8–12) probe. Lanes 1 and 7: DNA molecular weight marker II Digoxigenin-labelled (Roche). Stars indicate position of DNA fragments detected with PcPCNA probes. (C) PCR results. PCR was performed using degenerate primers and gDNA isolated from P. coccineus seedlings (lane 3), or plasmid pTZ57R\T DNA containing genomic sequence of the PcPCNA1 gene (lane 1) or of the PcPCNA-like1 gene (lane 2). In negative control, DNA template was omitted (lane 4). Lane M: DNA size standards (1 kb DNA ladder).
Genomic organization and localization of PcPCNA genes
The copy number of PCNA-like sequences and their chromosomal localization in the P. coccineus genome was investigated by Southern blot and PRINS analyses. For Southern blot analysis, two different probes were used; one probe was complementary to the PcPCNA1 open reading frame (ORF) and the second one was complementary to PcPCNA-like1 ORF. At least two bands for both probes were detected during analysis performed with P. coccineus genomic DNA digested (separately) with five restriction enzymes (BamHI, BglII, EcoRI, HindIII, XbaI) (Fig. 2B). The pattern obtained with the PcPCNA-like1 probe was different from and more complex than the pattern obtained with the PcPCNA1 probe. According to the number of bands detected with these probes, at least two different sequences similar to the sequence of the probes used were present in the P. coccineus genome.
Next, PCR analysis of P. coccineus genomic DNA using degenerated primers (designed based on the sequence of PcPCNA1 and PcPCNA-like1 genes) was performed. PCR reaction resulted in the amplification of three DNA fragments with molecular sizes of around 1.5 kb, 1.1 kb, and 0.95 kb (Fig. 2C). Two of these PCR products matched those identified in this work PcPCNA1 and PcPCNA-like1 genes, whereas the third product most likely corresponds to another PcPCNA-like gene (PcPCNA-like2?). Similarity between coding sequences of PcPCNA1 and PcPCNA-like2? genes is close to that between PcPCNA-like1 and PcPCNA-like2? (60% and 57% identity, respectively), whereas PcPCNA1 and PcPCNA-like1 genes share 70.5% of their coding sequences. Thus, PCR analysis revealed the presence of at least one additional PCNA-like gene in the genome of P. coccineus.
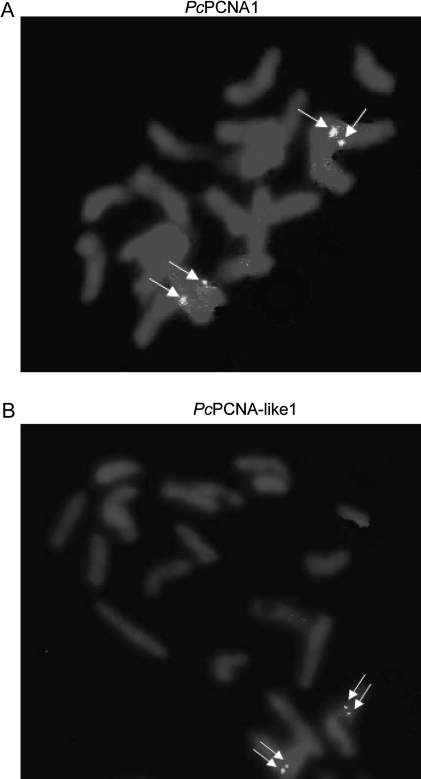
The genomic localization of the PcPCNA1 and PcPCNA-like1 genes was analysed using chromosomes of P. coccineus by primed in situ DNA labelling (PRINS) reactions. PRINS is a method of molecular cytogenetics for detecting DNA sequences in chromosomes of a species studied. It was first described by Bolund's group and, in subsequent years, important applications in human and plants cytogenetics were found (Koch et al., 1989; Abbo et al., 1993; Kubalakova et al., 2001; Kaczmarek et al., 2007). In our experiments, most of the signals after the PRINS reaction were visible as single dots but some of them were visible as double dots (on both chromatids). The signals of PcPCNA1 were observed on two chromatides in one locus, in the centromeric region of a submetacentric chromosome (Fig. 3A). The signals of PcPCNA-like1 were also shown to be localized at one locus, on a short arm of the medium submetacentric chromosome near the centromere (Fig. 3B).
Fig. 3.
PRINS analysis performed on metaphase chromosomes of P. coccineus. The reactions were performed with PcPCNA1 specific primers (A) and with PcPCNA-like1 specific primers (B) in the presence of DIG-12-dUTP and 3 units of Taq polymerase. After the reaction, slides were incubated with anti-DIG-fluorescein antibody and preparations were examined with the OLYMPUS BX60 Research System Microscope. The observed signals are denoted by white arrows. Each pictures shows a single diploid cell (2n=22).
Purification and biochemical characterization of PcPCNA proteins
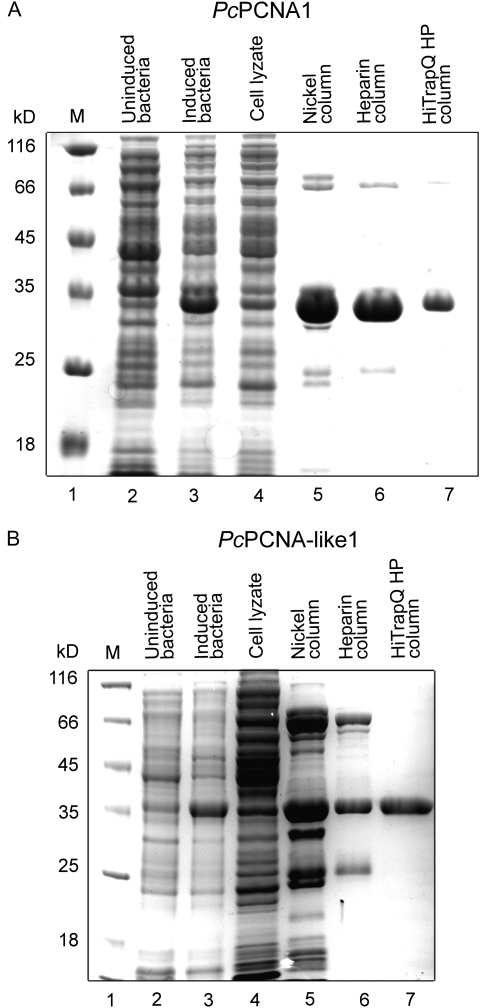
To compare biochemical characteristics of PcPCNA1 and PcPCNA-like1, recombinant proteins were produced in E. coli with short N-terminal His-tags which were shown earlier not to disrupt the PCNA activity (Kimura et al., 2001). These recombinant proteins were purified using a three-step chromatography procedure. First, affinity chromatography on a nickel column was performed, followed by chromatography on heparin and Q-Sepharose columns (see Materials and methods for details). Both proteins were purified to 90% homogeneity (Fig. 4A, B).
Fig. 4.
Purification of the recombinant PcPCNA1 (A) and PcPCNA-like1 (B) proteins. The protein samples were separated on 12% SDS-PAGE gels stained with Coommassie dye. The level of production of PcPCNA1 and PcPCNA-like1 and the subsequent purification steps are shown. Lane 1: molecular mass marker; lanes 2 and 3: non-induced and induced BL21(DE3)[pET15bPcPCNA1] and BL21(DE3)[pET15bPcPCNA-like1]; lane 4: cell lyzates; lane 5: elution from nickel column; lane 6: heparin column flow-through; lane 7: elution from HiTrap Q HP sepharose (1 μg of purified protein).
Biochemical characterization of the purified recombinant PcPCNA1 and PcPCNA-like1 proteins included analysis of their native structure, stimulation of the DNA polymerase delta activity, ability to interact with the p21 peptide, and reactivity with anti-PCNA antibody.
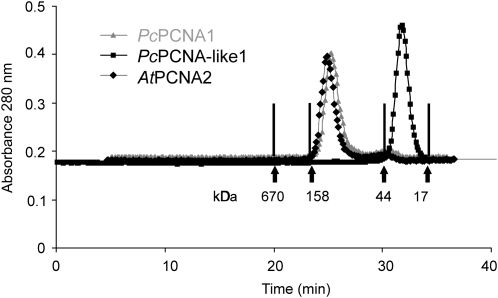
For analysing the native structure of the PcPCNA proteins, gel filtration chromatography on Superdex 200 was employed. This analysis demonstrated that the PcPCNA1 protein was present in solution mainly as a homo-oligomer (most likely a homotrimer), as its native molecular mass was estimated to be around 118 kDa (Fig. 5). The estimated mass is close to a theoretically calculated molecular mass of 93 kDa for three PcPCNA1 molecules and similar to the estimated native molecular mass of Arabidopsis PCNA2 (∼124 kDa; Fig. 5) which has been shown to form a homotrimer (Strzalka et al., 2009). However, for PcPCNA-like1, the formation of the trimeric structure was not observed, as it migrated with a molecular mass of around 25 kDa, suggesting that this protein was in a monomeric form (Fig. 5).
Fig. 5.
Analysis of native structures of the PcPCNA1 and PcPCNA-like1 proteins. 250 μg of PcPCNA1, PcPCNA-like1, and AtPCNA2 protein were separately filtered through Superdex 200. The arrows indicate the retention time of proteins used as molecular mass standards: thyroglobulin (670 kDa), γ calf globulin (158 kDa), ovalbumin (44 kDa), and myoglobin (17 kDa).
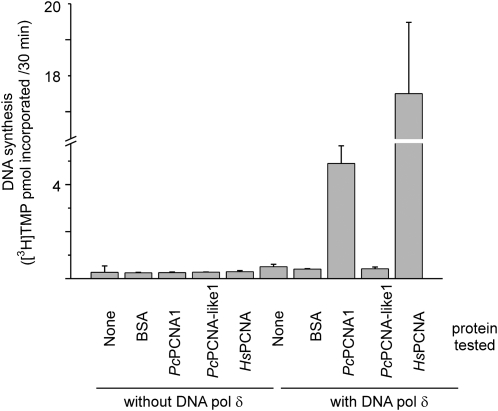
To assess biological function of recombinant PcPCNAs in the DNA replication process, an in vitro polymerase activity assay was used. Analysis of the stimulatory effect on the activity of polymerase delta was performed using human DNA polymerase delta with human PCNA as a positive control, BSA as a negative control, and PcPCNA1 and PcPCNA-like1 as tested proteins. PcPCNA1 exhibited a distinct ability to stimulate the processivity of human polymerase delta, similarly to human PCNA (HsPCNA; Fig. 6). By contrast, the PcPCNA-like1 protein was not able to stimulate the activity of polymerase delta. A similar effect was also observed for the negative control (BSA), as expected. No endogenous polymerase activity was detected in the proteins tested (Fig. 6). The observed stimulatory effect was dose-dependent and could also be observed if low amounts of the tested proteins were used (data not shown).
Fig. 6.
Test for the stimulatory effect of PCNA proteins on DNA polymerase delta activity. The polymerase activity assay was performed using poly(dA)/oligo(dT) as a template, [3H]TTP and 0.54 units of human DNA polymerase delta either alone or in the presence of 5 μg of the tested proteins: human PCNA (HsPCNA), PcPCNA1, PcPCNA-like1 or BSA (negative control). In addition, these proteins were assayed in the absence of human DNA polymerase delta. The radioactivity of the acid-precipitable material, measured in cmp, was then converted to pmol of [3H]TMP incorporated during 30 min of the reaction.
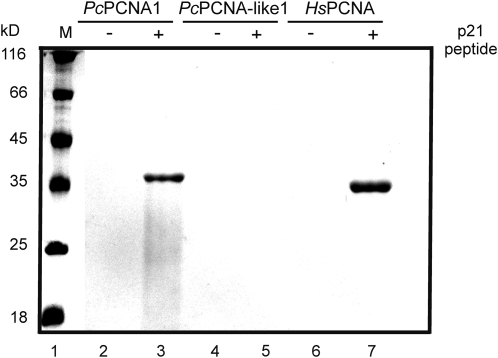
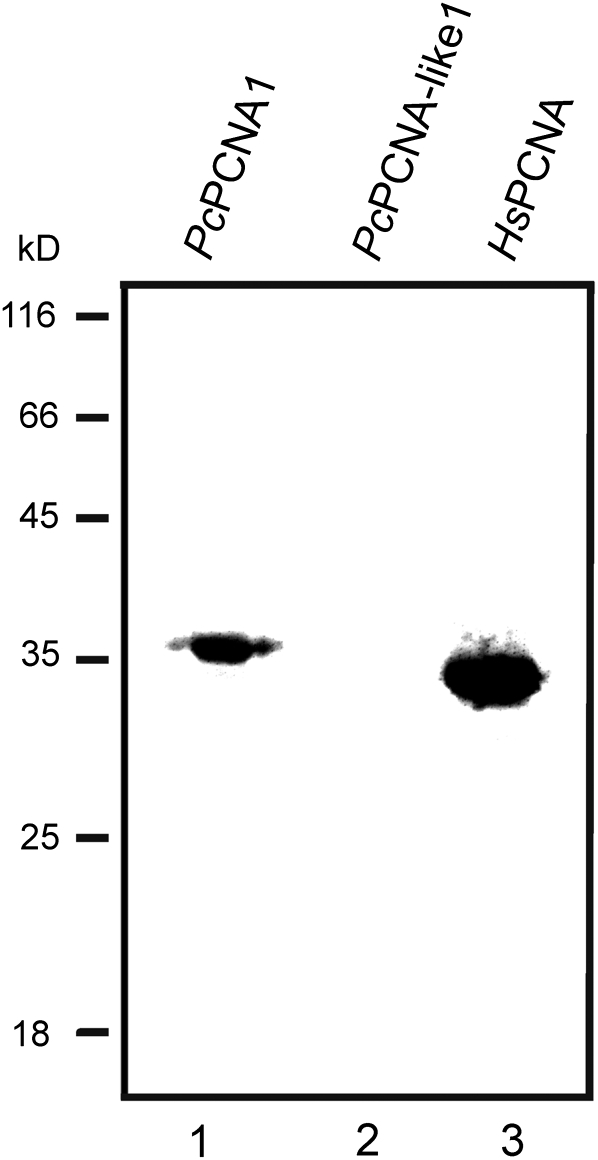
Moreover, PcPCNA proteins were tested for their ability to interact with the p21 peptide, as well as to be recognized by the anti-PCNA antibody. To study the interaction of PcPCNA proteins with a fragment of the human p21 protein, an affinity-precipitation assay was applied, using p21-streptavidin–agarose beads. Analysis of PcPCNA1 and PcPCNA-like1 interactions with the p21 peptide showed that only PcPCNA1 was able to bind to the p21 peptide specifically, similarly to human PCNA protein (Fig. 7). By contrast, no binding of PcPCNA-like1 to the p21 peptide was observed (Fig. 7). In addition, analysis of the PcPCNAs recognition by an anti-human PCNA monoclonal antibody (PC10) was performed. This antibody had been shown previously to be able to recognize plant (pea) PCNA (Ball and Lane, 1996). Our experiment performed with runner bean and human PCNA proteins showed that HsPCNA and PcPCNA1, but not PcPCNA-like1, was recognized by the PC10 antibody (Fig. 8).
Fig. 7.
Analysis of PcPCNA1 and PcPCNA-like1 binding to p21 peptide. Biotinylated p21 peptide was attached to streptavidin–agarose beads and incubated with 1 μg of the recombinant protein followed by extensive washing. The samples were then separated in 12% SDS-PAGE and stained with Coommassie dye. Lane 1: molecular mass marker; lane 2: streptavidin–agarose beads without p21, reacted with PcPCNA1; lane 3: beads with p21, reacted with PcPCNA1; lane 4: beads without p21, reacted with PcPCNA-like1; lane 5: beads with p21 reacted with PcPCNA-like1; lane 6: beads without p21, reacted with HsPCNA; lane 7: beads with p21, reacted with HsPCNA.
Fig. 8.
Western blot analysis of PcPCNA proteins with anti-human PCNA monoclonal antibody (PC10). One microgram of PcPCNA1, PcPCNA-like1, and HsPCNA was separated in 12% polyacrylamide gel and subjected to Western blot analysis using PC10 antibodies. Lane 1: PcPCNA1; lane 2: PcPCNA-like1; lane 3: HsPCNA. The positions of molecular mass markers are indicated.
Expression of PcPCNA genes at early stages of plant development
A real-time RT-PCR technique was used to evaluate the relative levels of PcPCNA1 and PcPCNA-like1 transcripts in germinating embryos (embryonic axes) and plant organs: roots, stems, leaves, and the micropylar region of seeds. These tissues were chosen for the analysis of PcPCNA gene expression since intensive cell proliferation accompanied by DNA replication is expected to occur in germinating embryos as well as in the micropylar region of developing seeds which contains the developing embryo at the early stages of maturation, whereas mature plant organs predominantly contain differentiated, non-dividing cells.
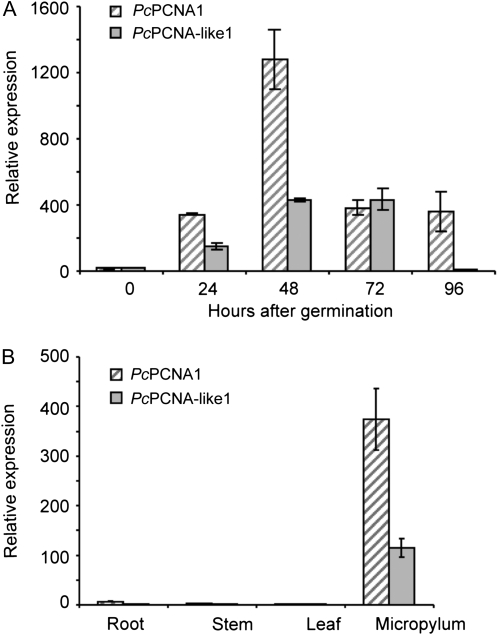
To test changes in the levels of these transcripts in germinating embryos, a time-course experiment was employed (Fig. 9A). The level of the PcPCNA1 transcript was low at time 0 (dry embryo). During the 24 h of germination, it rapidly increased by a factor of several hundred to reach the maximum level after 48 h. Then, between 72 h and 96 h after the start of germination, the level of the transcript decreased to the level that had been observed after 24 h. Next, the PcPCNA1 transcript level was evaluated for P. coccineus plant organs. The expression of this gene in root, stem, and leaf tissues was at a very low level (Fig. 9B). Analysis of the PcPCNA1 gene expression in the micropylar region of the seed showed that its transcript was present at a level comparable to the levels observed in germinating embryos at 24, 72, and 96 h of germination (Fig. 9A, B).
Fig. 9.
Real-time RT-PCR analysis of PcPCNA genes expression. (A) Relative changes in the PcPCNA1 and PcPCNA-like1 transcript levels during the early stages of germination from 0 h to 96 h. (B) Relative expression of PcPCNA1 and PcPCNA-like1 genes in P. coccineus root, stem, leaf, and micropylum. The figure presents data obtained in one of three independent experiments, and is representative for the observed changes.
Analysis of the PcPCNA-like1 gene expression revealed that the transcript was present at a very low level in the dry embryo (time 0; Fig. 9A). After 24 h of seed germination, the transcript level increased by several hundred fold and reached a stable level at 48 h and 72 h of germination, followed by a decrease to a lower level at 96 h of germination. Analysis of root, stem, and leaf tissues showed that expression of the PcPCNA-like1 gene was at a very low level, whereas the transcript level in the micropylar region was comparable to the one observed in 24 h germinating embryos (Fig. 9A, B).
Discussion
The purpose of this work was to characterize PCNA coding genes of Phaseolus coccineus. PCNA is an important factor involved in many cellular processes: DNA replication, DNA repair, and cell cycle regulation. However, most data published on PCNA originate from studies on animal organisms (including human) and yeasts. Although the quantity of new experimental data that broaden our knowledge about the role of PCNA in plant cells has increased during recent years, many aspects of its function in plants still remains obscure.
During the course of this study, two putative PCNA coding cDNAs were identified. The degree of identity at the amino acid level was over 50%. The theoretically calculated molecular mass and the isoelectric point of both proteins were similar (the molecular mass was around 30 kDa and the pI value ∼5), corresponding to PCNA from other organisms (human, accession number: CAC27344; mouse, accession number: P17918; rat, accession number: NP_071776; pea, accession number: CAA76392; Arabidopsis, accession number: Q9M7Q7). It is interesting that the level of identity between amino acid sequences of identified PcPCNA1 and PcPCNA-like1 was lower than that between PcPCNA1 and human PCNA. In this context, it is especially striking that if identified PcPCNA1 and PcPCNA-like1 act as eukaryotic PCNA proteins, one could expect higher identity between these two proteins than the identity between PCNA proteins originating from evolutionary distant organisms.
PCNA, as an important replication factor and cell cycle regulator, was shown to have several conserved motifs and residues, such as the residue D41 responsible for the stimulation of DNA polymerase delta and the efficient stimulation of the RF-C ATPase activity (Ayyagari et al., 1995; Fukuda et al., 1995), a motif I (mammals: Q125L126G127I128, plants: H125L126G127I128) that is essential for binding of p21 and polymerase delta (Gulbis et al., 1996; Jonsson et al., 1998; Zhang et al., 1998), a motif II (V188D189K190) conserved within plants and vertebrates (Jonsson et al., 1998), and a motif III (L251A252P253K254) responsible for proper folding of PCNA (Jonsson et al., 1998). By analysing the linear amino acid sequence of both PcPCNA1 and PcPCNA-like1 proteins, all the above listed motifs characteristic for PCNA were identified in the PcPCNA1 protein (Fig. 1). In contrast to PcPCNA1, none of the motifs was found in PcPCNA-like1. The lack of these motifs in PcPCNA-like1 may be responsible for different biochemical properties of this protein compared with PcPCNA1.
Interestingly, it could be shown that human PCNA and PcPCNA1 effectively stimulated the activity of human DNA polymerase delta, whereas PcPCNA-like1 did not exhibit any stimulatory effect on this enzyme. Although human PCNA used in the same dose as PcPCNA1 had a greater stimulating impact on the activity of human DNA polymerase delta as compared to PcPCNA1, such a phenomenon is not surprising, a similar observation was also reported when the biological activity of the yeast proliferating cell nuclear antigen was tested using human DNA polymerase delta (Bauer and Burgers, 1988). PCNA function is much conserved among eukaryotes; however, there might be some subtle differences in human/plant/yeast PCNA and human polymerase delta interactions resulting from slight differences in the protein surface charge and structure.
Gel filtration analysis of the purified recombinant PcPCNA1 and PcPCNA-like1 proteins performed under native conditions clearly showed that PcPCNA1 formed a homotrimer, and this feature is known to be necessary for its biological activity. Contrary to PcPCNA1, PcPCNA-like1 could only be detected as a monomer. This finding indicates that this protein is unlikely to function as a sliding clamp itself. However, it cannot be excluded that, although PcPCNA-like1 was not able to form a homotrimeric ring, it might be involved in the formation of a heterotrimeric ring around DNA. Such a phenomenon was described previously for archaeons Sulfolobus solfataricus and Aeropyrum pernix, in which three different PCNA proteins were found (Dionne et al., 2003; Imamura et al., 2007). Despite the low sequence similarities (less than 25% identity), PCNAs from S. solfataricus and A. pernix showed some analogous features. In both species, PCNAs formed a heterotrimeric ring structure. However, in the case of S. solfataricus, none of these proteins could itself form a homotrimer (trimer formation occurred only in the presence of three different PCNA proteins; Dionne et al., 2003), whereas A. pernix PCNA2 could form a trimeric structure both by itself (a homotrimer) and with PCNA1 and PCNA3 proteins (a heterotrimer), while neither PCNA1 nor PCNA3 of A. pernix could form a homotrimer (Imamura et al., 2007). Moreover, it was shown that archaeal PCNA monomers exhibited different substrate interaction specificities, indicating that each PCNA is responsible for attracting different replication-related proteins to the replication fork (Dionne et al., 2003; Imamura et al., 2007). Similar features may characterize Phaseolus coccineus PCNAs as well. On the other hand, Sakaguchi's group identified Drosophila melanogaster DmPCNA2 showing 51.7% identity to DmPCNA1 (Ruike et al., 2006); and such a low similarity was also observed for PcPCNA1 and PcPCNA-like1. DmPCNA1 showed all features typical of PCNA, similarly to PcPCNA1 (Henderson et al., 1994). DmPCNA2 contains D41 and motif III, but its motifs I and II are incomplete. However, DmPCNA2, in contrast to PcPCNA-like1, was capable of forming a homotrimer and stimulating the DNA pol delta activity. Differences in the expression pattern of DmPCNA1 and DmPCNA2 genes in response to UV treatment suggested that DmPCNA2 may function as an independent sliding clamp of DmPCNA1 during DNA repair (Ruike et al., 2006). In another organism containing two PCNA genes, Toxoplasma gondii, both gene products also contain D41 and motif III and are able to form homotrimers (Guerini et al., 2000). However, only TgPCNA1 probably serves as the major replisomal PCNA, whereas TgPCNA2 probably exhibits a different function (Guerini et al., 2005). In fact, no actual (specific) function could be shown for TgPCNA2, since disruption of its gene did not influence the DNA polymerase activity, the response to chemical mutagens or the recombination frequency (Guerini et al., 2000). Recent studies on Arabidopsis PCNA1 and PCNA2 revealed that these proteins showed very high similarity in their amino acid sequence as well as their ability to interact with Arabidopsis DNA polymerase eta and human p21 (Anderson et al., 2008; Strzalka et al., 2009). However, only AtPCNA2, but not the AtPCNA1 gene, was able to trigger restoration of normal UV resistance and mutation kinetics in the yeast rad30 mutant expressing the Arabidopsis POLH gene (yeast Rad30 and Arabidopsis POLH genes encode DNA polymerase eta; Anderson et al., 2008). In addition, AtPCNA1 and AtPCNA2 genes showed slightly different expression patterns in response to the exposure of Arabidopsis plants to heavy metal cadmium ions which cause genotoxic effects (Liu et al., 2009). These findings indicate that in eukaryotic cells the second PCNA protein may indeed exert functions different from those of PCNA1.
In addition to a stimulatory function that PCNA exerted on DNA polymerase delta during DNA replication and repair, this protein is also involved in the regulation of the cell cycle through its interaction with the p21/WAF1 protein. p21 is known to function as a p53-dependent cyclin-dependent kinase inhibitor, thus enabling cells to survive exposure to DNA damaging factors such as UV radiation (Maeda et al., 2002). Following the previously published results of Ball and Lane (1990) who demonstrated that the p21 peptide was able to precipitate pea PCNA from a crude extract, p21 interactions were analysed with PcPCNA proteins and showed that PcPCNA1 was co-precipitated with the p21 peptide, thus confirming their interaction, whereas the PcPCNA-like1 protein did not interact with the peptide most likely due to the lack of motif I in PcPCNA-like1. Another piece of evidence confirming differences between PcPCNA1 and PcPCNA-like1 in the structure and function was provided by Western blot analysis with the anti-human PCNA monoclonal antibody (PC10). This antibody can not only be used for the detection of human PCNA, it was also shown to recognize PCNA proteins originating from other animal organisms such as mouse and rat and from plants (e.g. pea; Ball and Lane, 1996). An epitope recognized by the PC10 antibody can be found in PCNA isolated from mammalian organisms and in PCNA from plant species, including soybean, Arabidopsis thaliana, and P. coccineus (PcPCNA1; Table 1). Although sequences of this epitope are not identical, they differ only in 1 or 2 amino acids. On the contrary, the sequence of the PcPCNA-like1 epitope is only similar but not identical to sequences of other PCNA epitopes, and a degree of differentiation is obviously too high for PcPCNA-like1 to be recognized by the PC10 antibody. Since PcPCNA-like1 does not exhibit any features required for its function as the typical PCNA, it is likely that the PcPCNA-like1 protein possesses another as yet unknown function in P. coccineus cells. Further experiments need to be done to shed more light on the functions of PcPCNA1 and PcPCNA-like1 by analysing their localization in the plant cell. These proteins (or at least PcPCNA1) are expected to function in the plant cell nucleus, although no obvious conserved nuclear localization signals (NLS) could be found in the amino acid sequence of these proteins.
Table 1.
PC10 epitope sequences of PCNA proteins from selected eukaryotic organisms
| Sequence of PC10 epitope | PCNA proteinsa |
| SDYEMKLMDL | Homo sapiens PCNA |
| SDYEMKLMDL | Mus musculus PCNA |
| SDYEMKLMDL | Rattus norvegicus PCNA |
| SDFEMKLMDI | Pisum sativum PCNA |
| SDFEMKLMDI | Glycine max PCNA |
| ADFEMKLMDI | Arabidopsis thaliana PCNA1 and PCNA2 |
| SDFEMKLMDI | Phaseolus. coccineus PCNA1 |
| SNFEMRLVDI | Phaseolus coccineus PCNA-like1 |
PC10 epitopes of human (accession number: CAC27344), mouse (accession number: P17918), rat (accession number: P04961), pea (accession number: CAA76392), soybean (accession number: P22177), Arabidopsis (accession numbers: NP172217 and NP180517), and runner bean PCNA1 (accession number: ABQ96591) proteins as well as PC10-like epitope of runner bean PCNA-like1 (accession number: ABQ96593) protein were compared.
Analysis of the relative expression of PcPCNA1 and PcPCNA-like1 genes using real-time RT-PCR gave us the opportunity to study expression patterns of both genes in P. coccineus plants at the early stages of plant development and in mature plant organs. It was observed that these patterns were generally similar; analogous observations have been reported for other plant species. The data from the AtGenExpress atlas show that in non-stressed A. thaliana plants both AtPCNA1 and AtPCNA2 genes have a similar expression pattern (Schmid et al., 2005). No significant differences in the expression pattern of maize ZmPCNA1 and ZmPCNA2 genes could be observed by Hussey's group, although the level of each transcript between the samples tested was slightly varied (Lopez et al., 1997), similar to our observations for PcPCNA1 and PcPCNA-like1. It was found that at the beginning of germination, PcPCNA1 and PcPCNA-like1 transcripts were present at low levels, whereas the expression of both genes was up-regulated during the first stage of germination and then down-regulated during the late phase of germination. Studying the expression pattern of PcPCNA1 and PcPCNA-like1 in plant organs, it was noticed that in root, stem, and leaf tissues, the level of both transcripts was very low, contrary to their level in the micropylar region where these genes were actively expressed. The observed increase in PcPCNA1 and PcPCNA-like1 expression in the embryonic axis during seed germination and in the developing embryo from the micropylar region of developing seeds was related to intensive cell proliferation. As cell proliferation is accompanied by DNA replication, an increase in PcPCNAs expression is most likely due to the resumption of DNA replication. The observed decrease in the level of PcPCNA transcripts at the later stages of germination when young seedlings are formed is most likely due to the shift in the ratio between dividing (meristematic) and non-dividing cells towards the latter ones. Low expression levels of PcPCNA genes in mature plant organs confirm a correlation between PCNA expression and cell proliferation/DNA replication as these organs predominantly contain non-dividing cells. PCNA expression at the early stages of seed germination could also be related to DNA repair that occurs throughout the nearly entire period of germination and decreases significantly before cell proliferation begins. However, PcPCNAs expression due to DNA repair may be low and comparable to the level in dry embryos because of a small number of cells in the embryo at this stage.
A correlation between PCNA gene expression and cell proliferation was also observed in other plant species. ZmPCNA1 and ZmPCNA2 genes were expressed in root and shoot tips as well as in young spikelets and cobs but not in leaves, old spikelets, and pollen. These results were confirmed by the analysis of PCNA expression in rice. It has been shown that the transcript was intensively produced in roots and in root tips but not in mature leaves where it was undetectable (Kimura et al., 2001). Also, the data presented by Shimizu and Mori who studied levels of PCNA transcripts in dormant auxiliary buds confirmed the correlation between PCNA gene expression and cell proliferation. They demonstrated that, before decapitation, the level of the transcript in dormant auxiliary buds was very low, whereas after decapitation, PCNA gene expression in pea was remarkably up-regulated, which correlated with bud growth and thus, with cell proliferation (Shimizu and Mori, 1998a, b).
Many attempts were undertaken in order to estimate the number of PCNA genes in plant genomes. However, due to plant genome complexity, the clearest results were obtained for Arabidopsis thaliana and Oryza sativa, and they originated as a result of the completion of the genome sequencing projects. In our studies by employing a PCR technique using degenerated primers, the presence of at least three PCNA-like genes in the genome of P. coccineus was demonstrated. In addition, Southern blot analysis revealed that, in the genome of P. coccineus, there are at least two sequences that are highly similar to PcPCNA1 as well as to PcPCNA-like1 cDNAs. However, high similarity between nucleotide fragments coding for the ORF of PcPCNA1 and PcPCNA-like1 could cause recognition of the PcPCNA-like1 sequence by the PcPCNA1 probe and vice versa. Based on these results, it is suggested that more than two sequences or genes similar to the PcPCNA1 or PcPCNA-like1 genes are present in the genome of P. coccineus. As gene duplication is a common mechanism and source of the evolutionary variability of eukaryotic genomes, it cannot be excluded that if PcPCNA gene duplication occurred, the PcPCNA-like1 gene evolved separately from the PcPCNA1 gene. As result, the PcPCNA-like1 protein might have lost some functions of the ancestral PcPCNA but retained and even gained other functions that are still unknown. It is theoretically possible that, after duplication, some functions of the ancestral PcPCNA were split into presently existing PcPCNA1 and PcPCNA-like1. The mechanisms responsible for the preservation of duplicate genes have been debated for more than 70 years. Recently, Lynch and Force (2000) have proposed a new explanation: subfunctionalization–suggesting that, after duplication, two gene copies specialize to perform complementary functions. Two PCNA genes are present in the genomes of some but not all plant species, for example, in the genome of Arabidopsis thaliana (Fig. 10). The AtPCNA1 gene located on chromosome 1 and the AtPCNA2 gene localized on chromosome 2 encode almost identical proteins. Moreover, both proteins have motifs characteristic for PCNA. Studies conducted on maize and carrot showed the presence of two PCNA genes and did not exclude the presence of more than two PCNA genes (Hata et al., 1992; Lopez et al., 1997). In all the cases known so far of plants containing two PCNA genes, high levels of the PCNA proteins amino acid sequence identity were observed: A. thaliana, 96.6%; N. tabacum, 97.0%; and Z. mays, 98.5%. Lower identity was observed only for D. carota, 63.0%, but this is mainly due to the presence of a >100 amino-acid-long C-terminal tail in DcPCNA2; the identity level between the first 264 aa of DcPCNA1 and DcPCNA2 being 87.6%. In animals, one copy of the PCNA gene was found in the rat genome (Matsumoto et al., 1987), whereas one PCNA gene and several pseudogenes are present in mouse and human genomes (Almendral et al., 1987; Ku et al., 1989; Travali et al., 1989; Yamaguchi et al., 1991). Most pseudogenes are not functional and expressed, although a few exceptions from this rule are known, for example the Makorni1-p1 pseudogene that is expressed in mouse cells and regulates the expression of the functional Makorin1 gene by increasing the stability of a gene transcript (Hirotsune et al., 2003). It cannot be excluded that in the genome of P. coccineus there are PCNA pseudogenes and PcPCNA-like1 might be such a pseudogene. It is presumed that PcPCNA-like1 is a functional gene because its cDNA contains a full ORF, by contrast to Makorni1-p1 that has premature stop codons (Hirotsune et al., 2003). Based on its genetic structure, PcPCNA-like1 may definitely be excluded as a processed pseudogene. Analysis of PcPCNA1 and PcPCNA-like1 genomic sequences showed that both genes contain two introns and three exons. It would be interesting to study in the future whether the PcPCNA1 or PcPCNA-like1 introns play a role in the regulation of the expression of PcPCNAs genes. Such a phenomenon occurred in the human PCNA gene and, in this case, introns 1 and 4 were shown to regulate HsPCNA gene expression (Ottavio et al., 1990; Alder et al., 1992). The possibility cannot be exclude that PcPCNA-like1 gene is expressed only at the transcript level, as some, although few, untranslated transcript containing ORF have been found in eukaryotic cells, i.e. Sry in the testes of adult mice (Capel et al., 1993) and 22k48 cDNA of the HIRA gene in human cells (Pizzuti et al., 1999). However, in contrast to the linear expressed form, unexpressed Sry transcript exists in a circular form, whereas 22k48 is composed of several tandemly arranged repeat elements. Such features have not been found for PcPCNA-like1. On the other hand, a few functionally transcribed and translated pseudogenes are known, for example, PsiCx43 and CRIPTO3 (Kandouz et al., 2004; Sun et al., 2008). If PcPCNA-like1 is an expressed pseudogene, it encodes protein exerting functions different from those typical for PCNA. Alternatively, PcPCNA-like1 may represent a pseudogene that encodes non-functional PCNA protein. It would also be the first PCNA pseudogene ever discovered in plants.
Finally, the PRINS technique was used to study the chromosomal localization of PcPCNA1 and PcPCNA-like1 genes. Using gene-specific starters (to eliminate the cross-reactivity between these two investigated sequences), it was demonstrated that both PcPCNA1 and PcPCNA-like1 localize in the submetacentric chromosomal region(s). However, due to the PRINS resolution, the copy number of PcPCNA1 and PcPCNA-like1 genes cannot be estimated. Even considering the data from both Southern blot and PRINS analyses, it is impossible to discriminate between three possibilities: (i) only one copy of the PcPCNA1 and PcPCNA-like1 gene is present in the genome of P. coccineus; or (ii) additional members of PCNA-like sequences are also present besides the identified PcPCNA1 and PcPCNA-like1 genes; or (iii) several copies of both PCNA genes are present in the genome of P. coccineus. If the latter possibility is true, these additional copies of the particular PcPCNA gene localize within the region of detected spots on chromosomes.
To conclude: two PCNA-like genes have been identified in the genome of Phaseolus coccineus. Although these genes show a number of common structural features and their expression patterns analysed at the transcript level are relatively similar, they encode two distinct proteins. The recombinant PcPCNA1 protein showed biochemical features typical for all known PCNA proteins, allowing it to function in the DNA replication/repair and cell cycle regulation processes. None of these features was observed in PcPCNA-like1. Since the PcPCNA-like1 gene most probably encodes a functional protein, the PcPCNA-like1 protein must exert as yet unknown functions, different from those of PcPCNA1.
Acknowledgments
We thank Professor Ulrich Hübscher for the generous gift of human PCNA and DNA polymerase delta, Professor Barbara Hohn for critical reading of the manuscript, and Dr Valentina Titova for language revision. We also thank Magdalena Manczak and Magdalena Plaminiak for their technical assistance. This project was supported by the Polish Ministry of Science and Higher Education (project no. PB 2 P04A 063 30 to AZ) and by the University of Lethbridge, Canada (internal grant to AZ).
Glossary
Abbreviations
- cpm
counts per minute
- PCNA
proliferating cell nuclear antigen
- PRINS
primed in situ DNA labelling
- RACE
rapid amplification of cDNA ends
- SDS-PAGE
sodium dodecyl sulphate-polyacrylamide gel electrophoresis
- TLS
translesion synthesis
References
- Abbo S, Miller T, King I. Primed-induced in situ hybridization to plant chromosomes. Genome. 1993;36:815–817. doi: 10.1139/g93-107. [DOI] [PubMed] [Google Scholar]
- Alder H, Yoshinouchi M, Prystowsky MB, Appasamy P, Baserga R. A conserved region in intron 1 negatively regulates the expression of the PCNA gene. Nucleic Acids Research. 1992;20:1769–1775. doi: 10.1093/nar/20.7.1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almendral JM, Huebsch D, Blundell PA, Macdonald-Bravo H, Bravo R. Cloning and sequence of the human nuclear protein cyclin: homology with DNA-binding proteins. Proceedings of the National Academy of Sciences, USA. 1987;84:1575–1579. doi: 10.1073/pnas.84.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson H, Vonarx EJ, Pastushok L, et al. Arabidopsis thaliana Y-family DNA polymerase η catalyses translesion synthesis and interacts functionally with PCNA2. The Plant Journal. 2008;55:895–908. doi: 10.1111/j.1365-313X.2008.03562.x. [DOI] [PubMed] [Google Scholar]
- Ayyagari R, Impellizzeri KJ, Yoder BL, Gary SL, Burgers PM. A mutational analysis of the yeast proliferating cell nuclear antigen indicates distinct roles in DNA replication and DNA repair. Molecular and Cellular Biology. 1995;15:4420–4429. doi: 10.1128/mcb.15.8.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ball KL, Lane DP. Human and plant proliferating-cell nuclear antigen have a highly conserved binding site for the p53-inducible gene product p21WAF1. European Journal of Biochemistry. 1996;237:854–861. doi: 10.1111/j.1432-1033.1996.0854p.x. [DOI] [PubMed] [Google Scholar]
- Bauer GA, Burgers PM. The yeast analog of mammalian cyclin/proliferating-cell nuclear antigen interacts with mammalian DNA polymerase delta. Proceedings of the National Academy of Sciences, USA. 1988;85:7506–7510. doi: 10.1073/pnas.85.20.7506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer GA, Burgers PM. Molecular cloning, structure and expression of the yeast proliferating cell nuclear antigen gene. Nucleic Acids Research. 1990;18:261–265. doi: 10.1093/nar/18.2.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bravo R, Frank R, Blundell PA, Macdonald-Bravo H. Cyclin/PCNA is the auxiliary protein of DNA polymerase-delta. Nature. 1987;326:515–517. doi: 10.1038/326515a0. [DOI] [PubMed] [Google Scholar]
- Capel B, Swain A, Nicolis S, Hacker A, Walter M, Koopman P, Goodfellow P, Lovell-Badge R. Circular transcripts of the testis-determining gene Sry in adult mouse testis. Cell. 1993;73:1019–1030. doi: 10.1016/0092-8674(93)90279-y. [DOI] [PubMed] [Google Scholar]
- Dionne I, Nookala RK, Jackson SP, Doherty AJ, Bell SD. A heterotrimeric PCNA in the hyperthermophilic archaeon Sulfolobus solfataricus. Molecular Cell. 2003;11:275–282. doi: 10.1016/s1097-2765(02)00824-9. [DOI] [PubMed] [Google Scholar]
- Dolezel J, Cihalikova J, Weiserova J, Lucretti S. Cell cycle synchronization in plant root meristems. Methods in Cell Science. 1999;21:95–107. doi: 10.1023/a:1009876621187. [DOI] [PubMed] [Google Scholar]
- Duval FD, Renard M, Jaquinod M, Biou V, Montrichard F, Macherel D. Differential expression and functional analysis of three calmodulin isoforms in germinating pea (Pisum sativum L.) seeds. The Plant Journal. 2002;32:481–493. doi: 10.1046/j.1365-313x.2002.01409.x. [DOI] [PubMed] [Google Scholar]
- Egelkrout EM, Mariconti L, Settlage SB, Cella R, Robertson D, Hanley-Bowdoin L. Two E2F elements regulate the proliferating cell nuclear antigen promoter differently during leaf development. The Plant Cell. 2002;14:3225–3236. doi: 10.1105/tpc.006403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fukuda K, Morioka H, Imajou S, Ikeda S, Ohtsuka E, Tsurimoto T. Structure–function relationship of the eukaryotic DNA replication factor, proliferating cell nuclear antigen. Journal of Biological Chemistry. 1995;270:22527–22534. doi: 10.1074/jbc.270.38.22527. [DOI] [PubMed] [Google Scholar]
- Guerini MN, Que X, Reed SL, White MW. Two genes encoding unique proliferating-cell-nuclear-antigens are expressed in Toxoplasma gondii. Molecular and Biochemical Parasitology. 2000;109:121–131. doi: 10.1016/s0166-6851(00)00240-1. [DOI] [PubMed] [Google Scholar]
- Guerini MN, Behnke MS, White MW. Biochemical and genetic analysis of the distinct proliferating cell nuclear antigens of Toxoplasma gondii. Molecular and Biochemical Parasitology. 2005;142:56–65. doi: 10.1016/j.molbiopara.2005.03.020. [DOI] [PubMed] [Google Scholar]
- Gulbis JM, Kelman Z, Hurwitz J, O'Donnell M, Kuriyan J. Structure of the C-terminal region of p21(WAF1/CIP1) complexed with human PCNA. Cell. 1996;87:297–306. doi: 10.1016/s0092-8674(00)81347-1. [DOI] [PubMed] [Google Scholar]
- Hata S, Kouchi H, Tanaka Y, Minami E, Matsumoto T, Suzuka I, Hashimoto J. Identification of carrot cDNA clones encoding a second putative proliferating cell-nuclear antigen, DNA polymerase delta auxiliary protein. European Journal of Biochemistry. 1992;203:367–371. doi: 10.1111/j.1432-1033.1992.tb16559.x. [DOI] [PubMed] [Google Scholar]
- Henderson DS, Banga SS, Grigliatti TA, Boyd JB. Mutagen sensitivity and suppression of position effect variegation result from mutation in mus209, the Drosophila gene encoding PCNA. The EMBO Journal. 1994;13:1450–1459. doi: 10.1002/j.1460-2075.1994.tb06399.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirotsune S, Yoshida N, Chen A, Garrett L, Sugiyama F, Takahashi S, Yagami K, Wynshaw-Boris A, Yoshiki A. An expressed pseudogene regulates the messenger-RNA stability of its homologous coding gene. Nature. 2003;423:91–96. doi: 10.1038/nature01535. [DOI] [PubMed] [Google Scholar]
- Imamura K, Fukunaga K, Kawarabayasi Y, Ishino Y. Specific interactions of three proliferating cell nuclear antigenes with replication-related proteins in Aeropyrum pernix. Molecular Microbiology. 2007;64:308–318. doi: 10.1111/j.1365-2958.2007.05645.x. [DOI] [PubMed] [Google Scholar]
- Jonsson ZO, Hindges R, Hubscher U. Regulation of DNA replication and repair proteins through interaction with the front side of proliferating cell nuclear antigen. The EMBO Journal. 1998;17:2412–2425. doi: 10.1093/emboj/17.8.2412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kandouz M, Bier A, Carystinos GD, Alaoui MA, Batist G. Connexin43 pseudogene is expressed in tumor cells and inhibits growth. Oncogene. 2004;23:4763–4770. doi: 10.1038/sj.onc.1207506. [DOI] [PubMed] [Google Scholar]
- Kaczmarek A, Naganowska B, Wolko B. PRINS and C-PRINS: promising tools for the physical mapping of the lupin genome. Cellular and Molecular Biology Letters. 2007;12:16–24. doi: 10.2478/s11658-006-0056-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelman Z. PCNA: structure, functions and interactions. Oncogene. 1997;14:629–640. doi: 10.1038/sj.onc.1200886. [DOI] [PubMed] [Google Scholar]
- Kimura S, Suzuki T, Yanagawa Y, Yamamoto T, Nakagawa H, Tanaka I, Hashimoto J, Sakaguchi K. Characterization of plant proliferating cell nuclear antigen (PCNA) and flap endonuclease-1 (FEN-1), and their distribution in mitotic and meiotic cell cycles. The Plant Journal. 2001;28:643–653. doi: 10.1046/j.1365-313x.2001.01184.x. [DOI] [PubMed] [Google Scholar]
- Koch JE, Kolvraa S, Petersen KB, Gregersen N, Bolund L. Oligonucleotide-priming methods for the chromosome-specific labelling of alpha satellite DNA in situ. Chromosoma. 1989;98:259–265. doi: 10.1007/BF00327311. [DOI] [PubMed] [Google Scholar]
- Kodama H, Ito M, Ohnishi N, Suzuka I, Komamine A. Molecular cloning of the gene for plant proliferating-cell nuclear antigen and expression of this gene during the cell cycle in synchronized cultures of Catharanthus roseus cells. European Journal of Biochemistry. 1991;197:495–503. doi: 10.1111/j.1432-1033.1991.tb15937.x. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y, Murakami T, Arai Y. Upstream sequences of rice proliferating cell nuclear antigen (PCNA) gene mediate expression of PCNA-GUS chimeric gene in meristems of transgenic tobacco plants. Nucleic Acids Research. 1991;19:1571–1576. doi: 10.1093/nar/19.7.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Suzuka I, Ohashi Y. Two of three promoter elements identified in a rice gene for proliferating cell nuclear antigen are essential for meristematic tissue-specific expression. The Plant Journal. 1995;7:877–886. doi: 10.1046/j.1365-313x.1995.07060877.x. [DOI] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. PCF1 and PCF2 specifically bind to cis elements in the rice proliferating cell nuclear antigen gene. The Plant Cell. 1997;9:1607–1619. doi: 10.1105/tpc.9.9.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosugi S, Ohashi Y. E2F sites that can interact with E2F proteins cloned from rice are required for meristematic tissue-specific expression of rice and tobacco proliferating cell nuclear antigen promoters. The Plant Journal. 2002;29:45–59. doi: 10.1046/j.1365-313x.2002.01196.x. [DOI] [PubMed] [Google Scholar]
- Krishna TS, Kong XP, Gary S, Burgers PM, Kuriyan J. Crystal structure of the eukaryotic DNA polymerase processivity factor PCNA. Cell. 1994;79:1233–1243. doi: 10.1016/0092-8674(94)90014-0. [DOI] [PubMed] [Google Scholar]
- Ku DH, Travali S, Calabretta B, Huebner K, Baserga R. Human gene for proliferating cell nuclear antigen has pseudogenes and localizes to chromosome 20. Somatic Cell and Molecular Genetics. 1989;15:297–307. doi: 10.1007/BF01534969. [DOI] [PubMed] [Google Scholar]
- Kubalakova M, Vrana J, Cihalikova J, Lysak MA, Dolezel J. Localisation of DNA sequences on plant chromosomes using PRINS and C-PRINS. Methods in Cell Science. 2001;23:71–82. [PubMed] [Google Scholar]
- Kumar S, Tamura K, Nei M. MEGA3: integrated software for molecular evolutionary genetics analysis and sequence alignment. Briefings in Bioinformatics. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- Laemmli UK. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- Laquel P, Litvak S, Castroviejo M. Mammalian proliferating cell nuclear antigen stimulates the processivity of two wheat embryo DNA polymerases. Plant Physiology. 1993;102:107–114. doi: 10.1104/pp.102.1.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leibovici M, Gusse M, Bravo R, Mechali M. Characterization and developmental expression of Xenopus proliferating cell nuclear antigen (PCNA) Developmental Biology. 1990;141:183–192. doi: 10.1016/0012-1606(90)90113-w. [DOI] [PubMed] [Google Scholar]
- Liu W, Zhou Q, Li P, Gao H, Han YP, Li XJ, Yang YS, Li Y. DNA mismatch repair related gene expression as potential biomarkers to assess cadmium exposure in Arabidopsis seedlings. Journal of Hazardous Materials. 2009;167:1007–1013. doi: 10.1016/j.jhazmat.2009.01.093. [DOI] [PubMed] [Google Scholar]
- Lopez I, Khan S, Vazquez-Ramos J, Hussey PJ. Molecular cloning of a maize cDNA clone encoding a putative proliferating cell nuclear antigen. Biochimica et Biophysica Acta. 1995;1260:119–121. doi: 10.1016/0167-4781(94)00192-6. [DOI] [PubMed] [Google Scholar]
- Lopez I, Khan S, Vazquez J, Hussey PJ. The proliferating cell nuclear antigen (PCNA) gene family in Zea mays is composed of two members that have similar expression programmes. Biochimica et Biophysica Acta. 1997;1353:1–6. doi: 10.1016/s0167-4781(97)00072-9. [DOI] [PubMed] [Google Scholar]
- Lynch M, Force A. The probability of duplicate gene preservation by subfunctionalization. Genetics. 2000;154:459–473. doi: 10.1093/genetics/154.1.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maeda T, Chong MT, Espino RA, Chua PP, Cao JQ, Chomey EG, Luong L, Tron VA. Role of p21Waf-1 in regulating the G1 and G2/M checkpoints in ultraviolet-irradiated keratinocytes. Journal of Investigative Dermatology. 2002;119:513–521. doi: 10.1046/j.1523-1747.2002.01828.x. [DOI] [PubMed] [Google Scholar]
- Maga G, Hubscher U. Proliferation cell nuclear antigen (PCNA): a dancer with many partners. Journal of Cell Science. 2003;116:3051–3060. doi: 10.1242/jcs.00653. [DOI] [PubMed] [Google Scholar]
- Markley NA, Bonham-Smith PC, Moloney MM. Molecular cloning and expression of a cDNA encoding the proliferating cell nuclear antigen from Brassica napus (oilseed rape) Genome. 1993;36:459–466. doi: 10.1139/g93-063. [DOI] [PubMed] [Google Scholar]
- Matsumoto K, Moriuchi T, Koji T, Nakane PK. Molecular cloning of cDNA coding for rat proliferating cell nuclear antigen (PCNA)/cyclin. The EMBO Journal. 1987;6:637–642. doi: 10.1002/j.1460-2075.1987.tb04802.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto T, Hata S, Suzuka I, Hashimoto J. Expression of functional proliferating-cell nuclear antigen from rice (Oryza sativa) in Escherichia coli. Activity in association with human DNA polymerase delta. European Journal of Biochemistry. 1994;223:179–187. doi: 10.1111/j.1432-1033.1994.tb18981.x. [DOI] [PubMed] [Google Scholar]
- Miyachi K, Fritzler MJ, Tan EM. Autoantibody to a nuclear antigen in proliferating cells. Journal of Immunology. 1978;121:2228–2234. [PubMed] [Google Scholar]
- Moldvan GL, Pfander B, Jentosch S. PCNA, the maestro of the replication fork. Cell. 2007;129:665–679. doi: 10.1016/j.cell.2007.05.003. [DOI] [PubMed] [Google Scholar]
- Mossi R, Hubscher U. Clamping down on clamps and clamp loaders, the eukaryotic replication factor C. European Journal of Biochemistry. 1998;254:209–216. [PubMed] [Google Scholar]
- Naganowska B, Dolezel J, Swiecicki W. Development of molecular cytogenetics and physical mapping of ribosomal RNA genes in Lupinus. Biologia Plantarum. 2003;46:211–215. [Google Scholar]
- Naryzhny SN. Proliferating cell nuclear antigen: a proteomics view. Cellular and Molecular Life Sciences. 2008;65:3789–3808. doi: 10.1007/s00018-008-8305-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ng L, Prelich G, Anderson CW, Stillman B, Fisher PA. Drosophila proliferating cell nuclear antigen. Structural and functional homology with its mammalian counterpart. Journal of Biological Chemistry. 1990;265:11948–11954. [PubMed] [Google Scholar]
- Ottavio L, Chang CD, Rizzo MG, Travali S, Casadevall C, Baserga R. Importance of introns in the growth regulation of mRNA levels of the proliferating cell nuclear antigen gene. Molecular and Cellular Biology. 1990;10:303–309. doi: 10.1128/mcb.10.1.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pizzuti A, Novelli G, Ratti A, et al. Isolation and characterization of a novel transcript embedded within HIRA, a gene deleted in DiGeorge syndrome. Molecular Genetics and Metabolism. 1999;67:227–235. doi: 10.1006/mgme.1999.2868. [DOI] [PubMed] [Google Scholar]
- Prelich G, Tan CK, Kostura M, Mathews MB, So AG, Downey KM, Stillman B. Functional identity of proliferating cell nuclear antigen and a DNA polymerase-delta auxiliary protein. Nature. 1987;326:517–520. doi: 10.1038/326517a0. [DOI] [PubMed] [Google Scholar]
- Ruike T, Takeuchi R, Takata K, Oshige M, Kasai N, Shimanouchi K, Kanai Y, Nakamura R, Sugawara F, Sakaguchi K. Characterization of a second proliferating cell nuclear antigen (PCNA2) from Drosophila melanogaster. The FEBS Journal. 2006;273:5062–5073. doi: 10.1111/j.1742-4658.2006.05504.x. [DOI] [PubMed] [Google Scholar]
- Sambrook J, Russell D. Molecular cloning: a laboratory manual. 3rd edn. Cold Spring Harbor NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU. A gene expression map of Arabidopsis thaliana development. Nature Genetics. 2005;37:501–506. doi: 10.1038/ng1543. [DOI] [PubMed] [Google Scholar]
- Schurtenberger P, Egelhaaf SU, Hindges R, Maga G, Jonsson ZO, May RP, Glatter O, Hubscher U. The solution structure of functionally active human proliferating cell nuclear antigen determined by small-angle neutron scattering. Journal of Molecular Biology. 1998;275:123–132. doi: 10.1006/jmbi.1997.1435. [DOI] [PubMed] [Google Scholar]
- Schwarzacher T, Heslop-Harrison J. Practical in situ hybridization. BIOS Scientific Publishers; 2000. Preparation of chromosome; pp. 51–71. [Google Scholar]
- Shimizu S, Mori H. Analysis of cycles of dormancy and growth in pea axillary buds based on mRNA accumulation patterns of cell cycle-related genes. Plant and Cell Physiology. 1998a;39:255–262. doi: 10.1093/oxfordjournals.pcp.a029365. [DOI] [PubMed] [Google Scholar]
- Shimizu S, Mori H. Changes in protein interactions of cell cycle-related genes during the dormancy-to-growth transition in pea axillary buds. Plant and Cell Physiology. 1998b;39:1073–1079. doi: 10.1093/oxfordjournals.pcp.a029304. [DOI] [PubMed] [Google Scholar]
- Stoimenov I, Helleday T. PCNA on the crossroad of cancer. Biochemical Society Transactions. 2009;37:605–613. doi: 10.1042/BST0370605. [DOI] [PubMed] [Google Scholar]
- Strzalka W, Ziemienowicz A. Molecular cloning of Phaseolus vulgaris cDNA encoding proliferating cell nuclear antigen. Journal of Plant Physiology. 2007;164:209–213. doi: 10.1016/j.jplph.2006.04.009. [DOI] [PubMed] [Google Scholar]
- Strzalka W, Oyama T, Tori K, Morikawa K. Crystal structure of the Arabidopsis thaliana proliferation cell nucear antigen 1 and 2 proteins complexed with the human p21 C-terminal segment. Protein Science. 2009;18:1072–1080. doi: 10.1002/pro.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun C, Orozco O, Olson DL, Choi E, Garber E, Tizard R, Szak S, Sanicola M, Carulli JP. CRIPTO3, a presumed pseudogene is expressed in cancer. Biochemical and Biophysical Research Communications. 2008;377:215–220. doi: 10.1016/j.bbrc.2008.09.113. [DOI] [PubMed] [Google Scholar]
- Suzuka I, Hata S, Matsuoka M, Kosugi S, Hashimoto J. Highly conserved structure of proliferating cell nuclear antigen (DNA polymerase delta auxiliary protein) gene in plants. European Journal of Biochemistry. 1991;195:571–575. doi: 10.1111/j.1432-1033.1991.tb15739.x. [DOI] [PubMed] [Google Scholar]
- Tan CK, Castillo C, So AG, Downey KM. An auxiliary protein for DNA polymerase-delta from fetal calf thymus. Journal of Biological Chemistry. 1986;261:12310–12316. [PubMed] [Google Scholar]
- Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proceedings of the National Academy of Sciences, USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Travali S, Ku DH, Rizzo MG, Ottavio L, Baserga R, Calabretta B. Structure of the human gene for the proliferating cell nuclear antigen. Journal of Biological Chemistry. 1989;264:7466–7472. [PubMed] [Google Scholar]
- Waseem NH, Labib K, Nurse P, Lane DP. Isolation and analysis of the fission yeast gene encoding polymerase delta accessory protein PCNA. The EMBO Journal. 1992;11:5111–5120. doi: 10.1002/j.1460-2075.1992.tb05618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiser T, Gassmann M, Thommes P, Ferrari E, Hafkemeyer P, Hubscher U. Biochemical and functional comparison of DNA polymerases alpha, delta, and epsilon from calf thymus. Journal of Biological Chemistry. 1991;266:10420–10428. [PubMed] [Google Scholar]
- Yamaguchi M, Hayashi Y, Hirose F, Matsuoka S, Moriuchi T, Shiroishi T, Moriwaki K, Matsukage A. Molecular cloning and structural analysis of mouse gene and pseudogenes for proliferating cell nuclear antigen. Nucleic Acids Research. 1991;19:2403–2410. doi: 10.1093/nar/19.9.2403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi M, Nishida Y, Moriuchi T, Hirose F, Hui CC, Suzuki Y, Matsukage A. Drosophila proliferating cell nuclear antigen (cyclin) gene: structure, expression during development, and specific binding of homeodomain proteins to its 5′-flanking region. Molecular and Cellular Biology. 1990;10:872–879. doi: 10.1128/mcb.10.3.872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Sun Y, Hsu H, Zhang L, Zhang Y, Lee MY. The interdomain connector loop of human PCNA is involved in a direct interaction with human polymerase delta. Journal of Biological Chemistry. 1998;273:713–719. doi: 10.1074/jbc.273.2.713. [DOI] [PubMed] [Google Scholar]