Abstract
The crystal structure of a putative dipeptidase (Phdpd) from Pyrococcus horikoshii OT3 was solved using X-ray data at 2.4 Å resolution. The protein is folded into two distinct entities. The N-terminal domain consists of the general topology of the α/β fold, and the C-terminal domain consists of five long mixed strands, four helices, and two 310 helices. The structure of Phdpd is quite similar to reported structures of prolidases from P. furiosus (Zn-Pfprol) and P. horikoshii (Zn-Phdpd), where Zn ions are observed in the active site resulting in an inactive form. However, Phdpd did not contain metals in the crystal structure and showed prolidase activity in the absence of additional Co ions, whereas the specific activities increased by 5 times in the presence of a sufficient concentration (1.2 mM) of Co ions. The substrate specificities (X-Pro) of Phdpd were broad compared with those of Zn-Phdpd in the presence of Co ions, whose relative activities are 10% or less for substrates other than Met-Pro, which is the most favorable substrate. The binding constants of Zn-Phdpd with three metals (Zn, Co, and Mn) were higher than those of Phdpd and that with Zn was higher by greater than 2 orders, which were determined by DSC experiments. From the structural comparison of both forms and the above experimental results, it could be elucidated why the protein with Zn2+ ions is inactive.
1. Introduction
Prolidases (proline-specific dipeptidase) are peptidases with specificity for X-Pro dipeptides. X-Pro substrates contain N-terminal residues that are hydrophobic/uncharged (Ala-, Ile-, Leu-, Val-), basic (His-), aromatic (Phe-, Tyr-), or sulphur-containing (Met-) [1]. Prolidases only cleave dipeptides with proline at the C terminus (NH2-X-/-Pro-COOH). This modification or truncation process can develop either cotranslationally or posttranslationally after the action of an endoproteinase. Prolidase is widespread in nature and has been isolated from different mammalian tissues [2–4] as well as from bacteria, such as the species of Lactobacillus [1, 5] and Xanthomonas [6]. While the physiological role of prolidase in bacteria is unclear, a deficiency of this enzyme in humans results in abnormalities of the skin and other proline-rich collagenous tissues [7]. In contrast with other endopeptidases and exopeptidases, prolidase is thought to be involved in the terminal degradation of intracellular proteins, and may also function in the recycling of proline. Prolidase also has biotechnological applications; it has a potential use in the dairy industry as a cheese-ripening agent [8] because the degradation of proline-containing peptides in cheese reduces bitterness. Prolidases are also capable of detoxifying organophosphorus nerve agents such as sarin and soman [9].
The crystal structure of prolidase has been solved only from Pyrococcus furiosus [10], where the main subunit is a “pita-bread” fold containing a metal active center like aminopeptidase P from E. coli [11] and methionine aminopeptidase from P. furiosus [12]. Two Zn atoms in the active site of the solved crystalline structure have been found [10], which are included as an impurity in the crystallization medium. However, the native prolidase from P. furiosus requires two Co ions per molecule in the active center for full catalytic activity. When Co ions are replaced by Zn ions, the protein does not show any enzymatic activity [13]. The structure of the prolidase containing Co ions with full activities remains to be solved.
Recently, the structure of prolidase from Pyrococcus horikoshii OT3 (Project ID, PH1149), which has 80% sequence identities with that from P. furiosus, has been deposited in the Protein Data Bank (1WY2). This protein also has Zn ions in the active center as observed in the P. furiosus enzyme. Furthermore, when the structure of a protein annotated as a putative dipeptidase from P. horikoshii (Project ID, PH0974), having 36% sequence identities with PH1149, was solved, no metal ions were found in the active center. The protein showed substrate specific activities for a dipeptide of Met-Pro, which is a feature of X-Pro dipeptidase (prolidase).
In this paper, the structure of PH0974 (Phdpd) is described in detail, and the difference in the structure of PH1149 with Zn ions (Zn-Phdpd) will be discussed. In addition, the differences in both proteins in the binding feature of Co or Zn ions and in substrate-specific activities were examined in order to clarify the enzymatic function of this enzyme.
2. Materials and Methods
2.1. Expression and Purification
The gene was amplified by a polymerase chain reaction (PCR) using P. horikoshii OT3 genomic DNA as a template (Project ID: PH0974). Recombinant plasmid was constructed by the super-rare-cutter system (Hayashizaki et al., manuscript in preparation). E. coli BL21-CodonPlus (DE3)-RIL cells were transformed with the recombinant plasmid and grown at 37°C in LB medium containing 50 μg mL−1 ampicillin for 20 hours. The cells were harvested by centrifugation at 6500 rpm for 5 minutes, suspended in 20 mM Tris-HCl, pH 8.0 (Buffer A) containing 0.5 M NaCl and 5 mM 2-mercaptethanol and disrupted by sonication. The cell lysate was heated at 90°C for 13 minutes. After heat treatment, denaturated proteins were removed by centrifugation (15,000 rpm, 30 minutes), and the supernatant solution was used as the crude extract for purification. The crude extract was desalted using a HiPrep 26/10 desalting column (Amersham-Biosciences) and applied onto a Super Q TOYOPEARL 650 M column (Tosoh) equilibrated with Buffer A. The protein was eluted with a linear gradient of 0–0.3 M NaCl in Buffer A. The protein was desalted with HiPrep 26/10 desalting column with Buffer A and subjected to a RESOURCE Q column (Amersham Biosciences) equilibrated with Buffer A. The protein was eluted with a linear gradient of 0–0.3 M NaCl in Buffer A. The buffer of the fractions containing the protein was exchanged using the HiPrep 26/10 desalting column to 10 mM sodium phosphate, pH 7.0 and applied onto a Bio-Scale CHT-20-I column (BIO-RAD) equilibrated with the same buffer. The protein was eluted with a linear gradient of 10–200 mM sodium phosphate, pH 7.0. The fractions containing protein were pooled, concentrated by ultrafiltration (VIVASPIN, 5 k cut) and loaded onto a HiLoad 16/60 Superdex 75 pg column (Amersham Biosciences) equilibrated with Buffer A containing 0.2 M NaCl. The purified protein showed a single band on SDS-PAGE. The concentration of the protein was estimated from the absorbance at 280 nm assuming E 1% 1 cm = 10.14. This protein is abbreviated as Phdpd.
Prolidase from P. horikoshii OT3 (Project ID: PH1149) was expressed and purified using similar methods. This protein also showed a single band on SDS-PAGE. The concentration of the protein was estimated from the absorbance at 280 nm assuming E 1% 1 cm = 7.81. This protein is abbreviated as Zn-Phdpd.
2.2. Crystallization
The protein concentration of Phdpd subjected to crystallization was 20 mg mL−1 in 100 mM Tris buffer at pH 8.0 containing 0.2 M sodium chloride. Single crystals were grown using polyethylene glycol by the hanging drop vapor diffusion method. The reservoir contained 0.1 M buffer solution (Cacodylate—NaOH pH 6.5), 40% (w/v) polyethylene glycol 400, and 0.02 M magnesium acetate. Each well was filled with 500 μL of reservoir solution. Protein solution consisted of 1 μL of a 20 mg mL−1 protein solution and 1 μL of reservoir solution. The protein crystal used for data collection grew to the size of 0.1 × 0.1 × 0.1 mm after 8–10 days.
2.3. Data Collection and Processing
Diffraction data for Phdpd were collected using a Rigaku R-AXIS V imaging-plate detector at the BL26B1 beamline, SPring-8, Japan. The crystals were flash-frozen in nitrogen-gas stream at 100 K during data collection. The oscillation angle used was 1.0° and the crystal-to-detector distance was set to 350 mm. Three data sets for the MAD (Multiwavelength Anomalous Dispersion) phasing were collected from a single selenomethionone-labelled crystal. Three wavelengths, corresponding to the maximum f′′ (peak), the minimum f′ (edge) and a reference wavelength (remote), were selected for the selenomethionine-labelled crystal, based on the fluorescence spectrum of the Se atom in the crystals. The wavelength was set to 1.0 Å for native crystals. The diffraction data were processed and scaled with the HKL 2000 package [14].
2.4. Phase Determination and Refinement
The structure was solved by MAD method [15]. Se-atom positions were obtained with the program SOLVE and the initial electron density map was calculated by SOLVE and RESOLVE [16]. Phase calculation resulted in an overall figure of merit of 0.45 for data in the range of 20–2.6 Å resolutions. The program ARP/wARP [17] was used to automatically build a partial model of the dimeric enzyme based on the amino acid sequence to the MAD-phased electron density map at 2.6 Å and placed approximately 50% of the entire residues. Combined solvent flattening and histogram matching, as implemented in DM were used to improve the phases. Most of the secondary structure elements were interpretable in the improved map. Unambiguous parts and side chains could be added during the refinement, without noncrystallographic symmetry (NCS) restraints. The rest of the residues was built manually with QUANTA (Accelrys San Diego, Calif, USA). All crystallographic refinements were carried out using CNS 1.1 [18, 19]. Solvent molecules were gradually included into the structure at stereochemically preferred positions and with difference densities higher than 2.8σ (F 0-F c) and 0.8σ (2F 0-F c). A summary of the statistics for structure determination of Phdpd is given in Table 1 and a ribbon diagram of the structure in Figure 1(a).
Table 1.
Data collection and refinement statistics of Phdpd.
| Native | MAD (Se) | |
|---|---|---|
| Data collection and phasing | ||
| Wavelength (Å) | 1.0000 | 0.97929, 0.97950, 1.0000 |
| Temperature (K) | 100 | 100 |
| Space group | P212121 | P212121 |
| Unit cell dimensions | ||
| a (Å) | 57.9 | 57.8 |
| b (Å) | 88.8 | 88.8 |
| c (Å) | 147.6 | 147.1 |
| V m (Å3 Da−1) | 2.4 | 2.3 |
| No. of molecules in asu | 2 | 2 |
| Resolution (Å) | 2.4 (2.5-2.4) | 2.3 (2.38–2.3) |
| No. of observations | 182 560 | 192 019, 190 610, 177 795 |
| No. of unique reflections | 29 349 (2954) | 34 548, 34 494, 33 486 |
| Completeness (%) | 95.9 (98.6) | 99.3 (99.9), 99.2 (99.9), 96.5 (98.4) |
| R merge (%)(a) | 11.2 (37) | 7.0 (34.4), 5.7 (30.9), 5.2 (28.7) |
| FOM(Solve/Resolve)(b) | — | 0.45/0.57 |
|
| ||
| Refinement | ||
| Resolution range (Å) | 20.0–2.4 | |
| R work (c) | 21.0 | |
| R free (d) | 26.5 | |
| No. of protein atoms | 5664 | |
| No. of solvent atoms | 310 | |
| Ramachandran plot(e) (%) in most favored | 93.8 | |
| Allowed | 5.7 | |
| Generously | 0.5 | |
| PDB code | 2HOW | |
(a) R merge = ∑|I i − 〈I〉|/∑I i, where I is the observed intensity and 〈I〉 is the average intensity from observations of symmetry related reflections, respectively.
(b)FOM, figure of merit.
(c) R work = ∑|F obs − F calc|/∑|F obs|, where F obs and F calc are the observed and calculated structure factors, respectively.
(d) R free is the R factor for a subset of 5% of the reflections that were omitted from refinement.
(e)As calculated by PROCHECK [23].
The values within the parentheses refer to the last shell.
Figure 1.
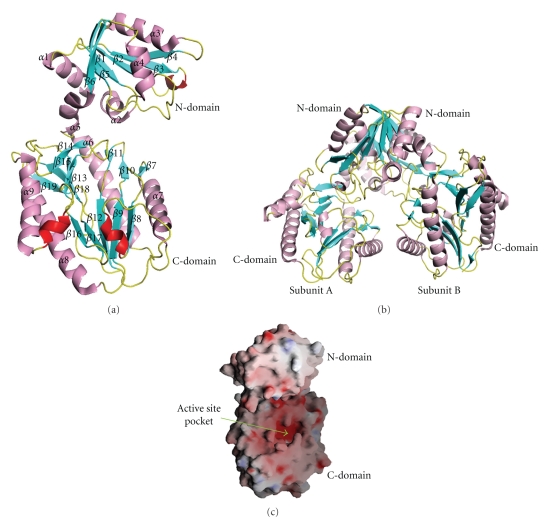
(a) Ribbon diagram of Phdpd (monomer form) showing N and C-terminal domains. α-helix, 310-helices, β-strands, and loops colored magenta, red, cyan, and yellow, respectively. The figure was made by the programs MOLSCRIPT and Raster3D [20, 21]. (b) Dimer form of Phdpd. (c) Electrostatic potential surface of the Phdpd and its active site pocket. The positively and the negatively charged surface regions are noted in blue and red, respectively.
2.5. Analytical Ultracentrifugation
Sedimentation equilibrium experiments were carried out using a Beckmann Optima mode XL-A at 20°C with an An-60 Ti rotor at a speed of 13 K rpm. Prior to the measurements, the protein solutions were dialyzed overnight against the respective buffer at 4°C. The experiments at three different protein concentrations between 0.93 and 0.31 mg mL−1 were performed in Beckman 4-sector cells. The buffer used was 20 mM Tris, pH 8.0, including 100 mM NaCl. The partial specific volume of 0.751 cm3 g−1 used for Phdpd was based on the amino acid compositions of the protein [22]. Analysis of the sedimentation equilibrium was performed using the program “XLAVEL” (Beckman, version 2.0).
2.6. Assay of Enzyme Activity
Proline dipeptidase activity was measured by a modification of the colorimetric ninhydrin method [24] using Met-Pro.HCl, Val-Pro.HCl, Gly-Pro.HCl, Ala-Pro.HCl, Phe-Pro.HCl, Glu-Pro.HCl, and Lys-Pro.HCl as substrates. Aminopeptidase and endopeptidase activities were measured by Met-MCA.TosOH (Tosylate form of L-methionine 4-methyl-coumaryl-7-amide) [25] and FRETS-25Xaa [26], respectively, as substrates. The FRETS-25Xaa is a fluorescence resonance energy transfer substrate (FRETS) library for determining endopeptidase specificity (Peptide Institute, Inc.). All assays were carried out at 100°C in 50 mM MOPS (3-[N-morpholino]propanesulfonic acid) buffer of pH 7.0, containing 1.2 mM CoCl2.
2.7. DSC Experiments
DSC (differential scanning calorimetry) was carried out using a VP-capillary DSC platform (MicroCal, USA) at a scan rate of 100 deg h−1. The protein concentration in the measurements was fixed at 0.01 mM in 50 mM Tris, pH7.8. Metals (ZnCl2, CoCl2, and MnCl2) were dissolved in the buffer. The sample was filtered through a 0.22-μm pore size membrane.
3. Results and Discussion
3.1. Quality of the Model
The structure of Phdpd was determined by the MAD method at 2.3 Å resolution. The asymmetric unit contains two molecules, which are related by a two-fold noncrystallographic symmetry (NCS). The native structure was refined to an R-factor of 21% (R free = 26.5%) at 2.4 Å resolution. The root mean square deviations (rmsds) from ideal geometry for the bond lengths and bond angles were 0.008 Å and 1.4°, respectively. All residues are within allowed regions in the Ramachandran plot (93.8% in the most favored region). The stereochemistry of the refined structure was analyzed with the program PROCHECK [23]. The program LSQKAB [27] from CCP4 was used to calculate rms deviations for the superposition of molecules. A summary of the data collections, refined model and the relevant geometrical parameters is given in Table 1.
3.2. Description of the Structure
The final refined model consists of two complete polypeptide chains from Met1 to Leu356 and 310 ordered water molecules. Each of the monomer subunits has an N terminal domain (residues 1–120), an α-helical linker (residues 121–130) and a C terminal domain (residues 131–356). The overall topology of Phdpd and a view of the Cα backbone are shown in Figures 1(a) and 4(a). The N-terminal domain is composed of a central β-sheet with six β-strands (strand order: β4, β3, β2, β1, β5, and β6) and of five α-helices around the central β-sheet. The strands of β1 to β3 are in antiparallel relationship and the other strands are in parallel directions. The C-terminal domain is comprised of long mixed stranded β-sheets (β7–β19) with four α-helices (α6–α9), lying on the outside of the surface. The α-helices α6 and α8 run parallel to the nearby β-sheet, while helices α7 and α9 are in antiparallel relationship on the outside surface. This domain should be a catalytic domain, which is similar to the reported structures of the “pita-bread” fold [10, 12, 28–33].
Figure 4.
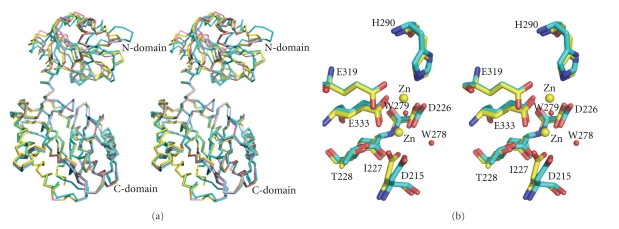
(a) Stereoview of the superposition of Phdpd (Cyan) with Zn-Phdpd (Yellow) and Zn-Pfprol (Pink) structures. (b) Stereoview of the conserved active site residues superimposed between the Phdpd (cyan) and Zn-Phdpd (yellow). Two active site metal atoms of Zn-Phdpd and two water molecules (W278 and W279) of Phdpd are shown in yellow and red color spheres, respectively. The side chains of the conserved active site residues are labeled according to Phdpd sequence numbering.
The active center of Phdpd can be assumed from the analogy of the structure of a “pita-bread” folded enzyme [34]. The putative active site pocket is located between two 310 helixes (residues 191–195 and 281–284) (two red color helices in Figure 1(a)) and in a deep groove of the inner surface as shown in Figure 1(c). The active site is strongly curved by the central β-sheet of the C-terminal domain and stabilized by four helices (α6–α9) that cover the outside surface of the deep pocket. The N and C terminal domains are linked by an α5 helix (residues 121–130) spanning between β6 and α6.
3.3. Substrate Specificity
Sequence identities of Phdpd and Zn-Phdpd from P. horikoshii are 36% (131/357), but those of Zn-Phdpd and the prolidase (Zn-Pfprol) from P. furiosus are quite high, 80% (279/348). Therefore, Zn-Phdpd has been assigned as a prolidase, although Phdpd is a putative dipeptidase. We then examined the enzyme functions of Phdpd and Zn-Phdpd from P. horikoshii. Proline dipeptidase activities (X-Pro) of both proteins were the highest for the dipeptide Met-Pro among the substrates examined (Table 2). The specific activity of Phdpd for the substrate Met-Pro was about 3 times that of Zn-Phdpd. In the case of Phdpd, the catalytic efficiencies for the peptide containing nonpolar amino acids were higher than those for the peptide containing polar amino acids such as Lys and Glu. The activity for Gly-Pro was the lowest. The substrate specificity of Phdpd was broad compared with that of Zn-Phdpd whose relative activities are 10% or less for substrates other than Met-Pro. The substrate specificities of Zn-Phdpd are quite similar to the reported results for Zn-Pfprol as shown in Table 2. The effect of metal ions on the dipeptidase activities of Phdpd and Zn-Phdpd was examined using Met-Pro as a substrate. As shown in Table 3, the relative activity of Phdpd in the presence of 1.2 mM MnCl2 was higher than that in 1.2 mM CoCl2 , but that of Zn-Phdpd was about half. When the metal ions were not added, Phdpd had 20% relative activity, but Zn-Phdpd had none.
Table 2.
Substrate specificity of three prolidases from P. horikoshii and P. furiosus.
| Substrates | Relative activity (%) | ||
|---|---|---|---|
| Phdpd | Zn-Phdpd | Zn-Pfprol(f) | |
| Met-Pro.HCl(a) | 100 | 100 | 100 |
| Val-Pro.HCl(a) | 53 | 4 | 10 |
| Ala-Pro.HCl(a) | 35 | 7 | 17 |
| Glu-Pro.HCl(a) | 28 | 5 | (e) |
| Phe-Pro.HCl(a) | 24 | 10 | 24 |
| Lys-Pro.HCl(a) | 17 | 0 | 10 |
| Gly-Pro.HCl(a) | 2 | (e) | 1 |
| Met-MCA.TosOH(b) | <0.1 | (e) | (e) |
| FRETS-25Xaa(c) | 0(d) | (e) | (e) |
(a)The specific activity in the presence of 1.2 mM CoCl2 is expressed as a percentage of the activity compared to that obtained with Met-Pro. The average values of three experiments are listed.
(b) V max was compared to that obtained with Val-Pro.
(c)FRETS is a fluorescence resonance energy transfer substrate library for determining endopeptidase specificity (Peptide Institute, Inc.).
(d)The endopeptidase activity was not detectable.
(d) Not examined.
(e) Reported results [13].
Table 3.
Effect of metal ions on dipeptidase (Met-Pro) activity of Phdpd and Zn-Phdpd.
| Metal (1.2 mM) | Relative activity (%) | |
|---|---|---|
| Phdpd | Zn-Phdpd | |
| CoCl2 | 100 | 100 |
| MnCl2 | 155 | 53 |
| ZnCl2 | 1 | 1 |
| no metal | 20 | 1 |
Zn-Phdpd was not inhibited in the presence of cacodylate ion from 0.4 μM to 40 mM concentrations.
Specific activity of Phdpd in the presence of 1.2 mM Co ions was 3 times that of Zn-Phdpd.
Kinetic parameters for Val-Pro of Phdpd in the presence of 1.2 mM CoCl2 were determined to be 5.0 mM, 807 μmol min−1 mg−1, 541 s−1, and 108 mM−1 s−1 for K m, V max, k cat, and k cat/K m, respectively. The value of K m was similar to that reported for the prolidase from P. furiosus (Zn-Pfprol), but the other kinetic parameters of Phdpd were several times greater [13].
The methionine aminopeptidase activity of Phdpd was examined for Met-AMC. It was detectable, but the V max value was less than 0.1% of that for the dipeptide Val-Pro. Furthermore, the endopeptidase activity was also examined with the substrate FRETS-25Xaa (Peptide Insititute, Inc.) which contains 475 combinations (25 × 19 = 475) of tripeptides except for cysteine. No endopeptidase activity was detected, even when 30 times the enzyme concentration was used compared with the assay for the dipeptide Val-Pro. These results indicate that Phdpd can be called prolidase.
Because a cacodylate ion has been found the near active sites of Zn-Phdpd, the dipeptidase activity using Met-Pro as a substrate was measured in the presence of cacodylate ion from 0.4 μM to 40 mM. The results indicate that Zn-Phdpd is not inhibited by cacodylate ion which was included in the crystalline buffer.
3.4. Changes in Denaturation Temperatures by the Addition of Metal Ions
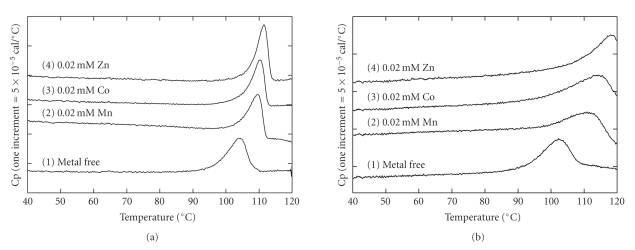
The heat stability of a protein is enhanced by ligand binding. Using DSC, the binding constant between a protein and a ligand can be estimated from the shift in the denaturation temperature for thermal unfolding of a protein in the presence of a ligand relative to the denaturation temperature in the absence of the ligand [35]. Figure 2(a) shows the DSC curves of Phdpd in the presence of metals whose concentrations are twice that of the protein where two metal-binding sites in the protein are saturated. The peak temperature of the DSC curve in the absence of the metal was 104.4°C and was lower than those in the presence of metals (Table 4), indicating that these metals can tightly bind to Phdpd. The difference in peak temperatures between the proteins in the absence and presence of metals indicates that Zn ion most strongly binds to the protein, followed by Co and Mn ions (Figure 2(a), Table 4). In the case of Zn-Phdpd, as shown in Figure 2(b), the order of binding strength for three kinds of metals to the protein was similar to that of Phdpd, but the strength seemed to be considerably higher than that of Phdpd: the differences in peak temperatures between the proteins in the absence and presence of 0.02 mM Co were 5.9 and 13.2°C for Phdpd and Zn-Phdpd, respectively (Table 4). The DSC curves of Phdpd and Zn-Phdpd, which were dialyzed in 50 mM Tris buffer at pH 7.8 including 1 mM EDTA overnight, were similar to those of samples without the metal ion. This suggests that original proteins hardly bound metal ions such as Zn, Co, and Mn.
Figure 2.
DSC curves of Phdpd and Zn-Phdpd in the presence of metal ions. (a) and (b) represent the DSC curves of Phdpd and Zn-Phdpd, respectively. All protein concentrations were 0.01 mM. DSC curves 1, 2, 3, and 4 were measured in the presence of 0 M metal, 0.02 mM Zn, 0.02 mM Co, and 0.02 mM Mn, respectively, in 50 mM Tris buffer at pH 7.8. The scan rate was 100 K h−1.
Table 4.
Changes in denaturation temperatures for Pfdpd and Zn-Pfdpd in the presence of metal ions at pH 7.8.
| (mM) | Phdpd | Zn-Phdpd | ||
|---|---|---|---|---|
| T m (°C) | ΔT m | T m (°C) | ΔT m | |
| Zn | ||||
| 0.05 | 118.4 | 16.9 | ||
| 0.02 | 111.6 | 7.2 | 118.4 | 16.9 |
| 0.01 | 110.9 | 6.5 | 118.4 | 16.9 |
|
| ||||
| Co | ||||
| 0.05 | 110.7 | 6.3 | 116.5 | 15.0 |
| 0.02 | 110.3 | 5.9 | 114.7 | 13.2 |
| 0.01 | 109.9 | 5.5 | 112.9 | 11.4 |
|
| ||||
| Mn | ||||
| 0.05 | 110.1 | 5.7 | 114.5 | 13.0 |
| 0.02 | 109.7 | 5.3 | 112.2 | 10.7 |
| 0.01 | 109 | 4.6 | 109.8 | 8.3 |
The denaturation temperatures (T m) were obtained from the peak temperatures of DSC curves. The T m values of Pfdpd and Zn-Pfdpd in the absence of metals were 104.4 and 101.5°C, respectively. All protein concentrations were 0.01 mM.
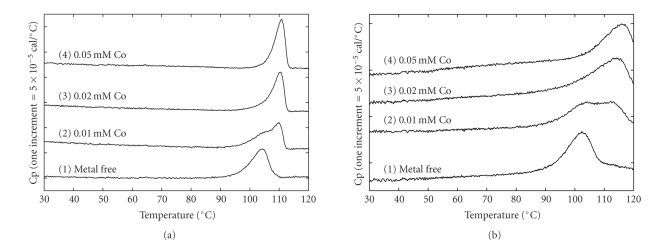
Reheating the DSC curves of Phdpd and Zn-Phdpd did not show any excess heat capacities, indicating that heat denaturation of both proteins is irreversible. Therefore, it might be difficult to strictly analyze the binding constants from the shifts in peak temperature due to ligand binding. After an error margin had been agreed upon, we calculated the binding constants using estimated DSC parameters and changes in denaturation temperatures because these are considerably more reliable. In the presence of 0.02 mM Zn ions, the binding constants of Phdpd and Zn-Phdpd were 1.2 × 107 and 1.6 × 109 M−1, respectively. The results suggest that the binding constants of Zn-Phdpd with Zn ions were roughly higher than 2 orders compared to those of Phdpd (Figures 3(a) and 3(b)).
Figure 3.
DSC curves of Phdpd and Zn-Phdpd in various concentrations of Co ion. (a) and (b) represent the DSC curves of Phdpd and Zn-Phdpd, respectively. All protein concentrations were 0.01 mM. DSC curves 1, 2, 3, and 4 were measured in the presence of 0, 0.01, 0.02, and 0.05 mM Co, respectively, in 50 mM Tris buffer at pH 7.8. The scan rate was 100 Kh−1.
On the other hand, methionine aminopeptidase from E. coli, which has a “pita-bread” fold with two active metal sites, has been reported to be maximally stimulated with the addition of one equivalent of Co2+ or Fe2+, and the first metal ion binds with a binding constant of 3–5 × 106 M−1, while the second one binds at 0.4 × 103 M−1 based on the changes in the absorption spectra during titration [36].
3.5. Comparison of Structures Near Active Sites of Phdpd with Those of Zn-Phdpd
The crystal structure and amino acid sequence of the prolidase from P. horikoshii (Zn-Phdpd) (PDB ID: 1WY2) are quite similar to those of that from P. furiosus (Zn-Pfprol) (PDB ID: 1PV9). Both proteins have two Zn ions in the active sites resulting in the absence of function. On the other hand, Phdpd with the sequence identity of 38% to Zn-Pfprol and Zn-Phdpd showed prolidase activity in the absence of additional Co ions (Table 3). Therefore, to elucidate why the proteins containing Zn ions do not have prolidase activity without the addition of Co ions, the structures of Phdpd, Zn-Phdpd, and Zn-Pfprol were compared.
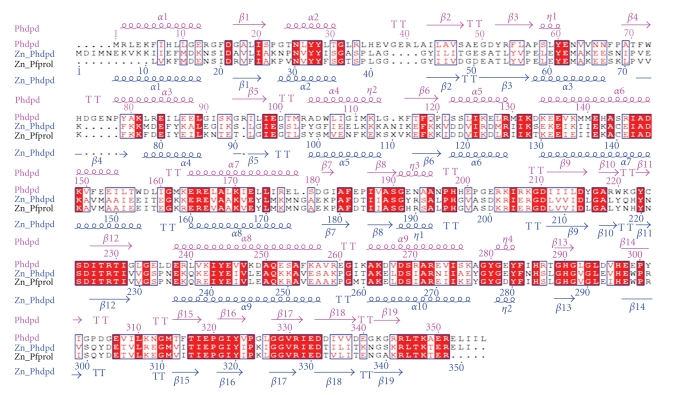
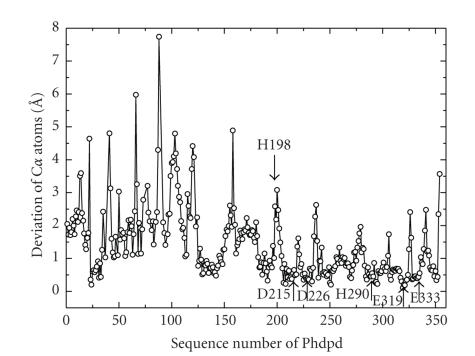
Figure 4(a) shows a stereoview of the superposition of Phdpd with Zn-Phdpd and Zn-Pfprol structures. Furthermore, structure-based sequence alignment of the three proteins and rms deviation of Cα atoms between Phdpd and Zn-Phdpd are shown in Figures 5 and 6, respectively. Comparison of Phdpd with Zn-Phdpd and Zn-Pfprol reveals major differences in folding, size, insertions and positioning of secondary structure elements in the N-terminal domain (Figure 5). In particular, the 310 helix η1 is replaced by an α3 helix (residues: 57 to 67) in both Zn-Phdpd and Zn-Pfprol (Figure 5). The rms deviation from Cα superposition of the whole, N- and C-terminal domains was calculated separately as follows: 1.4, 2.3, and 1.0 Å for the superposition between Zn-Pfprol and Phdpd, respectively; and 1.5, 2.0, and 1.1 Å for that between Zn-Phdpd and Phdpd, respectively (Figure 4(a)). Rms deviation values of five conserved residues (Asp215, Asp226, His290, Glu319, and Glu333) and the neighboring two residues (Ile227 and Thr228) belong to the lowest group of rms deviation values as shown in Figure 6, suggesting that these seven residues are considerably important in the active center. Figure 4(b) shows a stereoview of the conserved active site residues superimposed between the Phdpd and Zn-Phdpd. A cacodylate ion was found close to the active site of Zn-Phdpd, but not in that of Phdpd although both proteins were crystallized in the buffer containing cacodylate ions. A structural comparison of Phdpd with Zn-Phdpd reveals that the metal-coordination sphere and stereochemical organization of the active site are slightly altered due to Zn2+ binding as shown in Figure 4(b). Five water molecules (wat133, wat268, wat278, wat279 and wat290) are located around the active site pocket in Phdpd, all of which form a hydrogen-bonding network. The wat279 molecule nearly occupies the place of one of zinc ions in Zn-Phdpd. The water molecule w279 also creates similar coordination distances with conserved active site residues of Phdpd and Zn-Phdpd. The coordination distances of Asp226, Glu333, Ile227, and Thr228 of Phdpd are slightly different from those of Zn-Phdpd (Table 5). These observations might be correlated with the differences in binding with Zn and Co ions in the active site pocket.
Figure 5.
Structure-based sequence alignment of dipeptidase (PDB ID: 2HOW from P. horikoshii OT3 (Phdpd) with prolidase from P. furiosus (Zn-Pfprol) and dipeptidase (PDB ID: 1WY2) from P. horikoshii OT3 (Zn-Phdpd). Above and below the alignment are shown the secondary structure elements. Alignment was performed using CLUSTAL W [37], and the figure was produced using ESPript [38].
Figure 6.
Deviations of Cα atoms after the Cα superposition between Phdpd and Zn-Phdpd. Downward and upward arrows represent the positions of H198, D215, D226, H290, E319, and E333 which are important residues in the active site.
Table 5.
Comparison of the active site coordination distances between Phdpd and Zn-Phdpd. The distances of Zn1 and Zn2 in Phdpd were calculated after the superposition between atoms of the active site residues of Phdpd and Zn-Phdpd.
| Distances in Phdpd (Å) | Distances in Zn-Phdpd (Å) | ||||||
|---|---|---|---|---|---|---|---|
| Residues | Zn1 | Zn2 | W278 | W279 | Residues | Zn1 | Zn2 |
|
| |||||||
| Asp215 OD1 | 2.3 | 3.9 | Asp212 OD1 | 2.0 | |||
| Asp215 OD2 | 3.4 | 2.8 | Asp212 OD2 | 2.6 | |||
| Asp226 OD1 | 2.3 | 2.9 | Asp223 OD1 | 2.0 | |||
| Asp226 OD2 | 2.0 | 3.4 | Asp223 OD2 | 2.1 | |||
| His290 NE2 | 2.5 | 3.5 | His287 NE2 | 2.1 | |||
| Glu319 OE1 | 3.1 | 2.8 | Glu316 OE1 | 3.2 | |||
| Glu319 OE2 | 2.3 | 3.1 | Glu316 OE2 | 2.2 | |||
| Glu333 OE1 | 2.2 | 3.6 | Glu330 OE1 | 2.0 | |||
| Glu333 OE2 | 1.9 | 3.2 | Glu330 OE2 | 2.1 | |||
| Ile227 O | 3.5 | Ile224 O | 4.2 | ||||
| Thr228 OG1 | 3.0 | Thr225 OG1 | 3.5 | ||||
| W278 | 2.6 | 1.8 | 3.6 | Zn1 | 3.2 | ||
| W279 | 4.2 | 5.1 | |||||
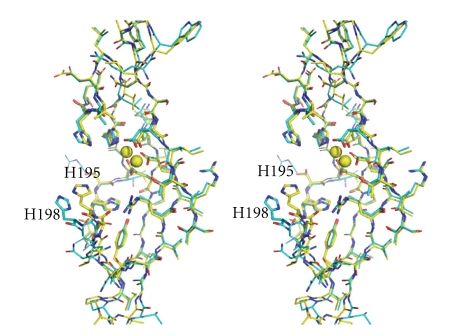
It has been proposed that methionine aminopeptidase and aminopeptidase P, which involve the “pita-bread” fold that contains Co or Mn ions in the active site, have a common reaction mechanism [34]. The important active site residues interacting with substrates are conserved in the three proteins described above. One of them, His198 of Phdpd corresponds to His79 of methioneine aminopeptidase from E. coli, which interacts with the nitrogen atoms of the scissile peptide bonds. Mutation of this residue of methionine aminopeptidase and aminopeptidase P has been reported to lead to variants with negligible activities [39, 40]. As shown in Figures 6 and 7, the position of His198 of Phdpd is remarkably different from that of the corresponding His195 of Zn-Phdpd: the rms deviation value of Cα atoms of both proteins was 2.59 Å as represented by an arrow to His198 in Figure 7. Relocation of this His residue tends to decrease the volume of the active site pocket. These results indicate that the absence of activity of Zn-Phdpd containing Zn ions might be caused by changes in coordination geometry of the metal ions and/or relocation of an important active side residue.
Figure 7.
Stereoview of selected active site pocket residues of Phdpd (cyan) superimposed on Zn-Phdpd (yellow). Active site pocket residues are shown by the ball and stick model and the metal ions are shown as spheres. His195 of Zn-Phdpd corresponds to His198 of Phdpd.
3.6. Structure of the Dimer and Dimer Interface
Analytical centrifugation results showed that Phdpd exists as a dimer in solution, and the association constant of monomer/dimer is 1.6 × 106 M. As shown in a ribbon diagram of Figure 1(b), the dimer form of Phdpd is an assembly of two monomers, related by a noncrystallographic 2-fold axis. The asymmetric unit also contains two molecules in the crystal as well as a dimer in solution. The dimeric enzyme has an overall globular shape of approximately 55 Å × 80 Å × 61 Å with a depression at its center. The accessible surface area of the monomer subunit is 16037 and 15955 Å2 for the respective subunits A and B. The area buried due to a dimer formation was 2310 Å2 and 7.2% of the total surface area. The buried surface area of Phdpd was remarkably smaller than those of Zn-Phdpd and Zn-Pfprol; especially, the difference in buried area of nonpolar atoms was remarkably great between them as shown in Table 6. When the buried area is divided into nonpolar (C/S) and polar (N/O) atoms, the hydrophobic interaction of dimer formation (ΔG HP) can be estimated using the following equation:
Table 6.
Comparison of hydrophobic interaction due to dimer formation of A and B subunits for Phdpd, Zn-Phdpd, and Zn-Pfprol.
| Phdpd | Zn-Phdpd | Zn-Pfprol | Δ(=Phdpd-Zn-Phdpd) | Δ(=Phdpd-Zn-Pfprol) | |
|---|---|---|---|---|---|
| ΔASAnp = (monomer* − dimer) | 1396 | 2341 | 2263 | −945 | −867 |
| ΔASAp = (monomer* − dimer) | 914 | 1067 | 1070 | −153 | −156 |
| Total buried area (Å2) | 2310 | 3408 | 3333 | ||
| ΔG HP (kJ/mol) | 191 | 333 | 320 | −142 | −129 |
| (1) |
where ΔASAnonpolar and ΔASApolar represent the difference in ASA (accessible surface area) due to dimer formation of the nonpolar and polar atoms of all residues, respectively. Parameters α and β have been determined to be 0.154 and −0.026 kJ mol−1 Å−2, respectively, using the stability/structure database of mutant human lysozymes [41]. The great differences in ΔASA values of nonpolar atoms between Phdpd and Zn-Phdpd (or Zn-Pfprol) resultantly indicate that hydrophobic interaction (ΔG HP) due to dimer formation of Zn-Phdpd and Zn-Pfprol is remarkably higher than that of Phdpd (Table 6).
3.7. Prolidases from P. horikoshii
One of two prolidases from P. horikoshii, Zn-Phdpd, has considerably high sequence identities with the prolidase (Zn-Pfprol) from P. furiosus, but the corresponding gene to Phdpd is not found in the genome of P. furiosus. In the process of BLAST searches [42], we found that a hypothetical protein (PH1902) from P. horikoshii can be annotated as X-pro dipeptidase from its 91% sequence identity with the X-pro dipeptidase from Pyrococcus absysi, which has identities of 26% and 28% with Phdpd and Zn-Phdpd, respectively. The sequence of PH1902 has 29% identity with Zn-Pfprol but has higher identity with the other two prolidases from P. furiosus (76 and 58%). Furthermore, PepQ-3 X-pro aminopeptidase and PepQ-2 cobalt-dependent proline dipeptidase from Pyrococcus abyssi have high sequence identities with Phdpd (76%) and Zn-Phdpd (84%), respectively, and two different X-pro aminopeptidases from Thermococcus kodakarensis have 71 and 63% identities with Phdpd and Zn-Phdpd, respectively. There are several prolidases in each genome.
Although the physiological role of prolidases in a cell remains to be solved, the substrate specificity of Phdpd is broader and its function is more effective than that of Zn-Phdpd. The active site structures of both proteins are changed to active or inactive forms depending on the binding metals. When both active and inactive structures of each prolidase are solved in the future, the role of metal ions on the function of metalloaminopeptidases could be more clearly elucidated.
4. Conclusions
The enzyme assay of Project ID PH0974 (Phdpd) of P. horikoshii indicated that Phdpd has the function of X-pro dipeptidase (prolidase). The crystal structure of Phdpd was solved at 2.4 Å resolution, and there are no metal ions in the active site. Furthermore, DSC experiments suggest that there are big differences in binding constants with Zn between Phdpd and Zn-Phdpd. In order to elucidate why the proteins containing Zn ions do not have the prolidase activity without the addition of Co ions, the three structures of Phdpd, Zn-Phdpd, and Zn-Pfprol were compared. The conclusions were (1) the coordination geometry in the active site of Phdpd was slightly different from that of Zn-Phdpd and (2) the important His residue of Zn-Phdpd, which seems to interact with the nitrogen atoms of the scissile peptide bonds, considerably moved resulting in decreasing the volume of the active site pocket due to Zn binding.
Acknowledgments
The first author thanks the National Synchrotron Radiation Research Center, Taiwan for providing financial support and thanks the beamlines BL12B2, BL26B1 and BL26B2 of SPring-8 for excellent facilities and assistances, and also thanks the Bioinformatics Center, Pondicherry University, Pondicherry, India for provided the computer systems for analysis. This work was supported by RIKEN Structural Genomics/proteomics Initiative (RSGI), the “National Project on Protein Structural and Functional Analyses ” funded by the Ministry of Education, Culture, Sports, Science and Technology of Japan.
Abbreviations
- Phdpd:
X-Pro dipeptidase (prolidase) of Project ID PH0974 from P. horikoshii
- Zn-Phdpd:
X-Pro dipeptidase (prolidase) of Project ID PH1149 from P. horikoshii
- Zn-Pfprol:
Prolidase from P. furiosus
- DSC:
Differential scanning calorimetry.
References
- 1.Booth M, Jennings PV, Fhaolain IN, O'Cuinn G. Prolidase activity of Lactobacillus lactis subsp. cremoris AM2: partial purification and characterization. Journal of Dairy Research. 1990;57:245–254. [Google Scholar]
- 2.Browne P, O'Cuinn G. The purification and characterization of a proline dipeptidase from guinea pig brain. The Journal of Biological Chemistry. 1983;258(10):6147–6154. [PubMed] [Google Scholar]
- 3.Endo F, Tanoue A, Nakai H, et al. Primary structure and gene localization of human prolidase. The Journal of Biological Chemistry. 1989;264(8):4476–4481. [PubMed] [Google Scholar]
- 4.Sjöström H, Norén O, Josefsson L. Purification and specificity of pig intestinal prolidase. Biochimica et Biophysica Acta. 1973;327(2):457–470. doi: 10.1016/0005-2744(73)90429-4. [DOI] [PubMed] [Google Scholar]
- 5.Fernández-Esplá MD, Martín-Hernández MC, Fox PF. Purification and characterization of a prolidase from Lactobacillus casei subsp. casei IFPL 731. Applied and Environmental Microbiology. 1997;63(1):314–316. doi: 10.1128/aem.63.1.314-316.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Suga KT, Kabashima T, Ito K, et al. Prolidase from Xanthomonas maltophilia: purification and characterization of the enzyme. Bioscience, Biotechnology and Biochemistry. 1995;59(11):2087–2090. doi: 10.1271/bbb.59.2087. [DOI] [PubMed] [Google Scholar]
- 7.Scriver RC, Smith RJ, Phang JM. Disorders of proline and hydroxyproline metabolism. In: Stanbury JB, Wyngaarden DS, Fredrickson JL, Goldstein MS, Brown, editors. The Metabolic Basis of Inherited Diseases. New York, NY, USA: McGraw Hill; 1983. [Google Scholar]
- 8.Bockelmann W. The proteolytic system of starter and non-starter bacteria: components and their importance for cheese ripening. International Dairy Journal. 1995;5(8):977–994. [Google Scholar]
- 9.Cheng T-C, Liu L, Wang B, et al. Nucleotide sequence of a gene encoding an organophosphorus nerve agent degrading enzyme from Alteromonas haloplanktis . Journal of Industrial Microbiology and Biotechnology. 1997;18(1):49–55. doi: 10.1038/sj.jim.2900358. [DOI] [PubMed] [Google Scholar]
- 10.Maher MJ, Ghosh M, Grunden AM, et al. Structure of the prolidase from Pyrococcus furiosus . Biochemistry. 2004;43(10):2771–2783. doi: 10.1021/bi0356451. [DOI] [PubMed] [Google Scholar]
- 11.Graham SC, Lee M, Freeman HC, Guss JM. An orthorhombic form of Escherichia coli aminopeptidase P at 2.4 Å resolution. Acta Crystallographica D. 2003;59(5):897–902. doi: 10.1107/s0907444903005870. [DOI] [PubMed] [Google Scholar]
- 12.Tahirov TH, Oki H, Tsukihara T, et al. Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus . Journal of Molecular Biology. 1998;284(1):101–124. doi: 10.1006/jmbi.1998.2146. [DOI] [PubMed] [Google Scholar]
- 13.Ghosh M, Grunden AM, Dunn DM, Weiss R, Adams MW. Characterization of native and recombinant forms of an unusual cobalt-dependenct proline dipeptidase (prolidase) from the hyperthermophilic archaeon Pyrococcus furiosus . Journal of Bacteriology. 1998;180:4781–4789. doi: 10.1128/jb.180.18.4781-4789.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods in Enzymology. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 15.Hendrickson WA, Horton JR, LeMaster DM. Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD): a vehicle for direct determination of three dimensional structure. The EMBO Journal. 1990;9(5):1665–1672. doi: 10.1002/j.1460-2075.1990.tb08287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Terwilliger TC, Berendzen J. Automated MAD and MIR structure solution. Acta Crystallographica D. 1999;55(4):849–861. doi: 10.1107/S0907444999000839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Perrakis A, Morris R, Lamzin VS. Automated protein model building combined with iterative structure refinement. Nature Structural Biology. 1999;6(5):458–463. doi: 10.1038/8263. [DOI] [PubMed] [Google Scholar]
- 18.Brünger AT, Kuriyan J, Karplus M. Crystallographic R factor refinement by molecular dynamics. Science. 1987;235(4787):458–460. doi: 10.1126/science.235.4787.458. [DOI] [PubMed] [Google Scholar]
- 19.Brünger AT, Adams PD, Clore GM, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallographica D. 1998;54(5):905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 20.Kraulis PJ. A program to produce both detailed and schematic plots of protein structures. Journal of Applied Crystallography. 1991;24, part 5:947–950. [Google Scholar]
- 21.Merritt EA, Bacon DJ. Raster3D: photorealistic molecular graphic. Methods in Enzymology. 1997;277:505–524. doi: 10.1016/s0076-6879(97)77028-9. [DOI] [PubMed] [Google Scholar]
- 22.Durchschlag H. In: Thermodynamic Data for Biochemistry and Biotechnology. chapter 3. Hinz H-J, editor. Berlin, Germany: Springer; 1986. p. 45. [Google Scholar]
- 23.Laskowski RA, McArthur MW, Moss DS, Thornton JM. PROCHECK: a program to check the stereochemical quality of protein structures. Journal of Applied Crystallography. 1993;26:283–291. [Google Scholar]
- 24.Yaron A, Mlynar D. Aminopeptidase-P. Biophys Res Commun. 1968;32:658–663. doi: 10.1016/0006-291x(68)90289-1. [DOI] [PubMed] [Google Scholar]
- 25.Yang G, Kirkpatrick RB, Ho T, et al. Steady-state kinetic characterization of substrates and metal-ion specificities of the full-length and N-terminally truncated recombinant human methionine aminopeptidases (type 2) Biochemistry. 2001;40(35):10645–10654. doi: 10.1021/bi010806r. [DOI] [PubMed] [Google Scholar]
- 26.Tanskul S, Oda K, Oyama H, Noparatnaraporn N, Tsunemi M, Takada K. Substrate specificity of alkaline serine proteinase isolated from photosynthetic bacterium, Rubrivivax gelatinosus KDDS1. Biochemical and Biophysical Research Communications. 2003;309(3):547–551. doi: 10.1016/j.bbrc.2003.08.035. [DOI] [PubMed] [Google Scholar]
- 27.Kabsch WA. Solution for the best rotation to relate two sets of vectors. Acta Crystallographica A. 1976;32:922–923. [Google Scholar]
- 28.Padmanabhan B, Paehler A, Horikoshi M. Structure of creatine amidinohydrolase from Actinobacillus. Acta Crystallographica D. 2002;58(8):1322–1328. doi: 10.1107/s0907444902010156. [DOI] [PubMed] [Google Scholar]
- 29.Bazan JF, Weaver LH, Roderick SL, Huber R, Matthews BW. Sequence and structure comparison suggest that methionine aminopeptidase, prolidase, aminopeptidase P, and creatinase share a common fold. Proceedings of the National Academy of Sciences of the United States of America. 1994;91(7):2473–2477. doi: 10.1073/pnas.91.7.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Liu S, Widom J, Kemp CW, Crews CM, Clardy J. Structure of human methionine aminopeptidase-2 complexed with fumagillin. Science. 1998;282(5392):1324–1327. doi: 10.1126/science.282.5392.1324. [DOI] [PubMed] [Google Scholar]
- 31.Wilce MCJ, Bond CS, Dixon NE, et al. Structure and mechanism of a proline-specific aminopeptidase from Escherichia coli . Proceedings of the National Academy of Sciences of the United States of America. 1998;95(7):3472–3477. doi: 10.1073/pnas.95.7.3472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Roderick SL, Matthews BW. Structure of the cobalt-dependent methionine aminopeptidase from Escherichia coli: a new type of proteolytic enzyme. Biochemistry. 1993;32(15):3907–3912. doi: 10.1021/bi00066a009. [DOI] [PubMed] [Google Scholar]
- 33.Hoeffken HW, Knof SH, Bartlett PA, Huber R, Moellering H, Schumacher G. Crystal structure determination, refinement and molecular model of creatine amidinohydrolase from Pseudomonas putida. Journal of Molecular Biology. 1988;204(2):417–433. doi: 10.1016/0022-2836(88)90586-4. [DOI] [PubMed] [Google Scholar]
- 34.Lowther WT, Mathews BW. Metalloamonopeptidases: common functional themes in disparate structural surrounding. Chemical Reviews. 2002;102:4581–4607. doi: 10.1021/cr0101757. [DOI] [PubMed] [Google Scholar]
- 35.Plotnikov V, Rochalski A, Brandts M, et al. An autosampling differential scanning calorimeter instrument for studying molecular interactions. Assay Drug Dev Technol. 2002;1(1, part 1):83–90. doi: 10.1089/154065802761001338. [DOI] [PubMed] [Google Scholar]
- 36.D'souza VM, Bennett B, Copik AJ, Holz RC. Divalent metal binding properties of the methionyl aminopeptidase from Escherichia coli . Biochemistry. 2000;39(13):3817–3826. doi: 10.1021/bi9925827. [DOI] [PubMed] [Google Scholar]
- 37.Thompson JD, Higgins DG, Gibson TJ. ClustalW: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gouet P, Robert X, Courcelle E. ESPript/ENDscript: extracting and rendering sequence and 3D information from atomic structures of proteins. Nucleic Acids Research. 2003;31(13):3320–3323. doi: 10.1093/nar/gkg556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Griffith EC, Su Z, Niwayama S, Ramsay CA, Chang Y-H, Liu JO. Molecular recognition of angiogenesis inhibitors fumagillin and ovalicin by methionine aminopeptidase 2. Proceedings of the National Academy of Sciences of the United States of America. 1998;95(26):15183–15188. doi: 10.1073/pnas.95.26.15183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lowther WT, Orville AM, Madden DT, Lim S, Rich DH, Matthews BW. Escherichia coli methionine aminopeptidase: implications of crystallographic analyses of the native, mutant, and inhibited enzymes for the mechanism of catalysis. Biochemistry. 1999;38(24):7678–7688. doi: 10.1021/bi990684r. [DOI] [PubMed] [Google Scholar]
- 41.Funahashi J, Takano K, Yutani K. Are the parameters of various stabilization factors estimated from mutant human lysozymes compatible with other proteins? Protein Engineering. 2001;14(2):127–134. doi: 10.1093/protein/14.2.127. [DOI] [PubMed] [Google Scholar]
- 42.Altschul SF, Madden TL, Schäffer AA, et al. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25(17):3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]