Abstract
Objective
Among the pitfalls of using cell-free fetal DNA in plasma for prenatal diagnosis is quality of the recovered DNA fragments and concomitant presence of maternal DNA (>95%). Our objective is to provide alternative methods for achieving enrichment and high-quality fetal DNA from plasma.
Methods
Cell-free DNA from 31 pregnant women and 18 controls (10 males and 8 females) were size separated using agarose gel electrophoresis. DNA fragments of 100–300, 500–700 and 1,500–2,000 bp were excised and extracted, followed by whole genome amplification (WGA) of recovered fragments. Levels of β-globin and DYS1 were measured.
Results
Distribution of β-globin size fragments was similar among pregnant women and controls. Among control male cases, distribution of size fragments was the same for both β-globin and DYS1. Among maternal cases confirmed to be male, the smallest size fragment (100–300 bp) accounted for nearly 50% (39.76 ± 17.55%) of the recovered DYS1-DNA (fetal) and only 10% (10.40 ± 6.49%) of β-globin (total) DNA. After WGA of plasma fragments from pregnant women, DYS1 sequence amplification was best observed when using the 100–300 bp fragments as template.
Conclusions
Combination of electrophoresis for size separation and WGA led to enriched fetal DNA from plasma. This novel combination of strategies is more likely to permit universal clinical applications of cell-free fetal DNA.
Key Words: Cell-free fetal DNA, Real-time PCR, DYS1, β-Globin, Gel extraction, Whole genome amplification
Introduction
Analysis and quantification of circulating fetal nucleic acids in maternal plasma as a risk-free alternative for prenatal genetic diagnosis has attractive clinical potential. Cell-free fetal (cff) DNA has been shown to exist in plasma and serum of pregnant women as early as 6 weeks of gestation with concentrations rising during pregnancy and peaking prior to parturition [1]. Many laboratories, including ours, have shown the utility of cff DNA as a unique source of genetic material for non-invasive prenatal evaluation of fetal gender, genetic diseases, and aneuploidy [2,3,4,5,6]. In addition, quantitative measurements of plasma DNA have been used to correlate risk among cases with various pregnancy-related complications [7,8,9,10]. Though abundant in plasma, a major limitation to using cff DNA clinically is the concomitant presence of maternal DNA [1].
Fetal DNA likely exists in the form of apoptotic bodies as shown by transmission electron and fluorescent microscopy [11,12]. However, little is known about the biochemical or structural characteristics of cff DNA. Analysis using differentially sized PCR amplicons provides evidence of cff DNA molecules existing as smaller size fragments compared to circulating maternal DNA fragments [13]. Using a combination of gel electrophoresis and real-time PCR, fetal DNA has been shown to display apoptotic characteristics, and to exist shorter in size compared to maternal DNA in plasma [14].
Despite ability to select cff DNA based on fragment size, technical problems relating to quality of DNA collected from plasma hinder broader genetic applications [15]. Whole genome amplification (WGA) of DNA is a promising method that generates large amounts of DNA with improved and equivalent quality from samples of limited quantity, this includes DNA recovered from plasma. Theoretically, WGA is capable of decreasing the stochastic effects resulting from low copy templates [16]. However, reliable WGA presents a significant challenge given circulating DNA in plasma consists of a mixture of differentially sized fragmented apoptotic and necrotic DNA.
We sought to determine efficiency improving purity of recovered fetal DNA based on size fragment enrichment and WGA. Enrichment through agarose gel electrophoresis combined with WGA for analysis of plasma DNA has also enabled better understanding of some of the inherent differences between maternal and fetal circulating DNA, enabling development of improved strategies for molecular analysis cff DNA from maternal plasma.
Materials and Methods
Sample Collection and Processing
Under IRB approval from Baylor College of Medicine, 31 pregnant women (gestational age (GA) = 22.41 ± 6.72), 8 non-pregnant women, and 10 adult males gave informed consent to participate in the study. From each patient, 20 ml of peripheral blood was collected in vacutainers containing 1.5 ml of Solution A (trisodium citrate 22.0 g/l, citric acid 8.0 g/l, and dextrose 24.5 g/l) and processed within 24–48 h. Plasma was first separated from whole blood by centrifugation at 800 g for 10 min. Recovered plasma was then recentrifuged for an additional 10 min at 13,500 g to remove residual intact cells. Supernatant (containing free DNA) was removed and stored at −80°C for up to 1 month in 800-μl aliquots prior to DNA extraction.
DNA Extraction
To extract DNA, 800 μl of plasma from each sample was processed using the QIAmp DNA micro kit (Qiagen Sciences, Germantown, Md., USA) according to the manufacturer's instructions with minor modifications to accommodate the 800-μl sample. Elution of DNA was performed using 100 μl of elution buffer. All DNA sampleswere stored at 4°C prior to analysis.
Size Fractionation Using Agarose Gel Electrophoresis
The extracted DNA (50 μl) was separated on a 1.0% agarose gel. Electrophoresis was carried out at 80 V for 2 h and AmpliSize molecular ruler (Bio-Rad Laboratories, Hercules, Calif., USA) was used to estimate molecular size. Each lane of the gel containing circulatory DNA was then cut with a sterile scalpel blade into three discrete sections, with the molecular weight marker used as a guide. From each of the samples and controls (non-pregnant women, males and water blanks), three sections were cut in the following size range: 100–300, 500–700, and 1,500–2,000 bp. DNA was extracted from the agarose fragments using QIAquick Gel Extraction Kit (Qiagen, Valencia, Calif., USA) and eluted in 50 μl of elution buffer.
Whole Genome Amplification
WGA was performed using GenomePlex™ WGA (Sigma, St. Louis, Mo., USA) according to the manufacturer's instructions with minor modifications. The fragmentation step was eliminated since the circulating DNA is highly fragmented. OmniPlex libraries were created by adding 2 μl of Library Preparation Buffer and 1 μl of Library Stabilization Solution to 10 μl of plasma DNA and 1 μl of Fragmentation Buffer. The reaction was incubated for 2 min at 95°C. The sample was cooled on ice prior to adding 1 μl Library Preparation Enzyme. Thermal cycling was performed in a PE9700 (Applied Biosystems) and incubated at 16°C for 20 min, 24°C for 20 min, 37°C for 20 min, and 75°C for 5 min. Amplification reactions were performed with 7.5 μl of GenomePlex amplification master mix and 12.5 U Jumpstart™ Taq polymerase (Sigma). Cycling conditions in a PE9700 were 95°C for 3 min, and 14 cycles of 95°C for 15 s and 65°C for 5 min. Positive and negative controls included purified human genomic DNA and dH2O respectively.
PCR Analysis
To determine the amount of total and fetal circulating DNA in the original samples and the relative proportions of DNA in each of the gel fractions, β-globin and DYS1 (Y chromosome-specific sequence Y49a) loci were used respectively. Quantitative real-time PCR was performed using the Applied Biosystems 7700 sequence detection system (Foster City, Calif., USA). Quantification of total and fetal DNA as genome equivalents per milliliter of plasma was based on copies of β-globin and DYS1 sequences detected per microliter of plasma. Each 50-μl reaction contained 25 μl of PE-ABI Universal Mastermix and 5 μl of extracted DNA. Primers and probes were used at final concentrations of 300 and 200 nm, respectively. Each reaction plate was run simultaneously with a duplicate calibration curve of titrated DNA (standard curve). Each sample was run in triplicate for both loci with the mean of the values determined using the 7700 software and a standard curve of known DNA concentrations. Cycling conditions for all reactions consisted sequentially of 2 min incubation at 50°C to activate UNGErase, initial denaturation step of 95°C for 10 min, and finally 40 cycles of 95°C for 15 s and 60°C for 60 s. All samples were analyzed blindly with respect to fetal gender. The standard factor of 6.6 pg was used to convert the data to genome equivalents (GEq).
Results
To determine the effect of GA, 31 maternal plasma specimens ranging from 7 to 36 weeks (22.4 ± 6.7) were subjected to quantification of both total (β-globin) and fetal (DYS1) DNA sequences and compared to 18 adult controls (10 males and 8 females). Among the 31 maternal samples, total cell-free DNA levels were nearly double by measurement of β-globin levels (1,380.2 ± 728.2 GEq/ml) compared to both control male (866.1 ± 313.4 GEq/ml) and non-pregnant female (810.0 ± 524.2 GEq/ml) cases (table 1). No significant difference was observed among the two control groups (p = 0.781). As expected, increase in β-globin levels correlated with increase in GA (table 1). Differences between second and third trimester were significant when compared to first trimester and female control (p = 0.026, p = 0.005). However, no significant difference in levels of DYS1 by trimester (fetal origin) was observed among the confirmed male pregnancies by GA (table 1).
Table 1.
Measured levels of β-globin and DYS1 in maternal and control samples
| n | Male fetus | GA, weeks | β-Globin, GEq/ml | DYS1, GEq/ml | |
|---|---|---|---|---|---|
| First trimester | 12 | 6 | 10.7 ± 1.8 | 1,001.2 ± 278.2 | 108.3 ± 92.1 |
| Second trimester | 12 | 6 | 19.6 ± 2.1 | 1,393.2 ± 663.8a | 156.8 ± 147.0 |
| Third trimester | 7 | 4 | 30.0 ± 5.2 | 2,007.7 ± 971.9b | 204.1 ± 200.0 |
| Male control | 10 | NA | NA | 866.1 ± 313.4 | 1,341.1 ± 512.1 |
| Female control | 8 | NA | NA | 810.0 ± 524.2 | NA |
NA = Not applicable.
p = 0.010
p = 0.004.
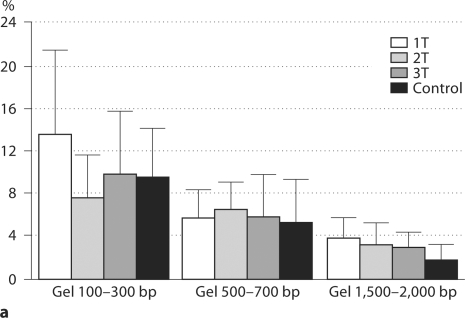
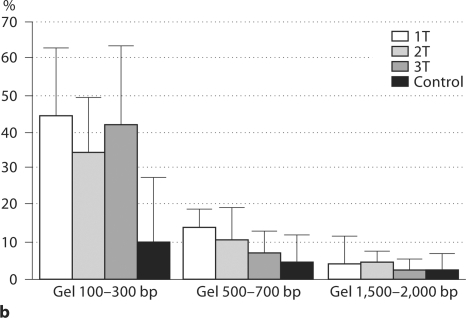
Following quantification, all plasma DNA samples were subjected to agarose gel electrophoresis for fragment size separation. Three discrete agarose sections containing fragments with approximate sizes of 100–300, 500–700, and 1,500–2,000 bp were excised. The DNA was extracted and real-time PCR performed. Proportion of circulating β-globin (adult and fetal) and DYS1 (male or fetal) DNA present in each individual gel fragment was calculated (fig. 1a, b).
Fig. 1.
a Distribution of % β-globin levels (total DNA) based on fragment size in maternal plasma. Graph represents 31 pregnant women divided by first (n = 12; 1T), second (n = 12; 2T), and third (n = 7; 3T) trimesters. Plasma controls included non-pregnant women (n = 8) and adult men (n = 10). No significant difference was found among groups (p > 0.1). b Distribution of % DYS1 (fetal DNA) based on fragment size in maternal plasma. Graph represents 16 pregnant women divided by first (n = 6; 1T), second (n = 6; 2T), and third (n = 4; 3T) trimesters. Plasma controls included adult men (n = 10). Significant difference in DYS1 levels observed at 100–300 bp fragment size between all trimesters (p = 0.0009).
Among both pregnant and control samples (10 non-pregnant women and 10 adult men), the overall distribution was similar for β-globin with the smaller size fragments (100–300 bp) containing the highest percentage of total DNA. Progressive decrease in percentage of total DNA correlated with increase in DNA size (fig. 1a). The quantitative distribution of β-globin and DYS1 DNA based on size fragment was equivalent among male controls (fig. 1a, b). As expected, in female controls no DYS1 sequences were detected (data not showed) indicating that no external source of DNA was present on our experiments. In contrast to adult male samples, DYS1 and β-globin distribution was different among maternal samples (fig. 1b). The proportion of DYS1 was nearly 50% compared to β-globin sequences among all pregnancies in the fragment size range of 100–300 bp (39.8 ± 17.6%). This represents more than 4 times the level of similar size fragments in the control males (9.7 ± 6.8%) (p = 0.0002). Frequency of DYS1 fragments in maternal samples decreased from 10.8 ± 7.3% (500–700 bp) to 3.5 ± 5.0% (1,500–2,000 bp). Thus, in pregnant women, the three measured size fragments account for 25–84% of fetal DNA based on DYS1 sequence. In contrast, the percentage of total DNA by β-globin is only 7–40% among the same size fragments.
WGA was performed in plasma DNA before and after gel extraction. After WGA, DNA levels for β-globin and DYS1 were quantified (table 2). Controls for DYS1 include non-pregnant female in which no DYS1 sequences were detected (data not showed). In plasma DNA samples processed without size fractionation, 100% amplification for both β-globin and DYS1 loci was achieved with similar gain in DNA levels. However, there was variability in the amplification of samples after gel fractionation (table 2). Gel-fractionated samples for β-globin displayed differences in sequence-specific amplification with 86% of the samples being amplifiable and a tendency for higher amplification among bigger fragment size. The average gain after amplification for β-globin in the range of 500–700 bp fragments was 4,687 ± 7,526 in contrast of 1.5 ± 1.1 in the range of 100–300 bp fragments. For DYS1 (fetal DNA) 62% of the total samples were amplifiable after gel fractionation with 100% of amplifiable fragments in the range of 100–300 (43% of samples displayed over 1,300-fold amplification). From the 1,500–2,000 bp fragments, a decrease in the number of amplifiable samples for fetal DYS1 was observed (29% of total samples were amplifiable) with none having more than 400-fold amplification (table 2). For all samples, the 260/280 ratio improved from 0.96 ± 0.22 before WGA to 1.60 ± 0.14 after WGA.
Table 2.
DNA levels following WGA and size fractionation using maternal samples1
| Total DNA | 100–300 bp | 500–700 bp | 1,500–2,000 bp | |
|---|---|---|---|---|
| β-Globin | ||||
| Average of DNA gain | 2,623 ± 1,422 | 1.5 ± 1.1 | 4,867 ± 7,526 | 1,645 ± 2,588 |
| Range of DNA gain | 810–5,158 | 0–3.2 | 1–16,592 | 0–6,634 |
| Positive amplified cases | 7 (100%) | 6 (86%) | 7 (100%) | 5 (71%) |
| Cases with >400-fold amplification | 7 (100%) | 0 (0%) | 4 (57%) | 3 (60%) |
| DYS1 | ||||
| Average of DNA gain | 2,552 ± 3,635 | 1,556 ± 3,009 | 821 ± 2,167 | 4.1 ± 7.5 |
| Range of DNA gain | 1–8,512 | 1–8,229 | 0–5,736 | 0–19 |
| Positive amplified cases | 7 (100%) | 7 (100%) | 4 (57%) | 2 (29%) |
| Cases with >400-fold amplification | 4 (57%) | 3 (43%) | 1 (25%) | 0 (0%) |
Total of seven maternal samples (GA = 23.3±10.9) analyzed by WGA following size fractionation. Each case was confirmed to have a male fetus.
Discussion
Our study supports that the majority of circulatory fetal DNA exists as small molecular size fragments in the presence of maternal derived DNA of larger molecular size as reported by others [13,14]. Chan et al. [13] utilized real-time PCR to detect and quantify DNA using PCR amplicons ranging in size from 107 to 524 bp for fetal DNA (SRY) and 105 to 798 bp for maternal DNA (leptin). The median concentration of SRY with the use of primers producing amplicons longer than 313 bp was below 1%. The median concentration of leptin gene with primers producing an amplicon of 392 was 32%. Li et al. [14] used a combination of gel extraction and real-time PCR to examine DNA in a range of 90–23,000 bp based on detection of GAPDH and SRY. In our study, we selected to do gel fractionation in a range of sizes between 10 and 2,000 bp based on our hypothesis that cff DNA is apoptotic, thus expecting smaller size fragments. Moreover, we demonstrated for the first time that size-fractionated fetal DNA is suitable for WGA, providing a source of enriched fetal DNA.
This study illustrates the molecular size pattern of both fetal and total circulating DNA in maternal circulation as a function of GA before and after gel fractionation. Variability among subjects in the amount of fetal and total DNA was observed. There is an increasing trend among trimesters for both β-globin and DYS1 levels. β-Globin levels display a significant increase as pregnancy progresses, in agreement with studies reported by Lo et al. [1]. Total levels of β-globin in pregnant cases are higher than non-pregnant controls reaching significance in the second and third trimester (p = 0.026, p = 0.005). However, there was no difference in the size fraction distribution of β-globin between pregnancy and controls samples. This result may be expected given the distribution of β-globin mainly represents maternal DNA [1]. Levels of DYS1 also increase according to GA. There is a significant difference and improve of the detection of fetal alleles from maternal samples after gel fractionation with ∼50% of cff DNA localized in the small size range 100–300 bp. These results corroborate our hypothesis that cff DNA, because of its apoptotic origin, is predominantly of small size.
Since gel size fractionation allowed the enrichment of cff DNA, other groups like ours have used that technique as an initial step to decrease the amount of maternal DNA. For example, gel fractionation with MALDI-TOF mass spectrometry allowed detection of single mutation in two achondroplasia-affected pregnancies from maternal plasma [17]. Our group decided to combine gel size fractionation with WGA. Different WGA techniques have been developed to specifically increase the DNA quantities originating from samples with limited DNA contents. There are PCR-based and non-PCR base (isothermal) methodologies. The WGA method used by us involves linker ligation and PCR with universal primers. According to the manufacturer, results are highly reproducible, with 99.8% concordance in SNP detection between amplified and genomic DNA. The method is optimized for use on at least 10 ng of high-quality template DNA, however, single cells and highly degraded DNA can be amplified with high accuracy in contrast to the isothermal methodology that require high-quality DNA. Considering that cff DNA is of low quality and quantity the PCR method was chosen; however, we are aware that this methodology has a risk for bias and uneven distribution of alleles. There are reports indicating that WGA does not replicate telomeric or highly repetitive centromeric regions effectively [18]. In addition, stochastic sampling effects have led to varying results between replicates, with not all alleles being reported in every sample. To obtain the most complete and accurate profile, pooled DNA from separate WGA reactions may be required [19]. In our study, we only performed one round of WGA per sample, perhaps explaining why amplification failed in some cases. Further studies warrant more than one round of WGA per sample to ensure amplification detection of both loci in all samples. Since amplification bias (i.e. allele dropout) appears to be random with respect to an allele, performing multiple amplifications would prevent smaller alleles from being undetected or misrepresented. The ability to increase and improve the amount and quality of starting fetal DNA by using WGA would be invaluable for prenatal diagnostic applications. In fact the 260/280 ratio was improved from 0.96 ± 0.22 to 1.60 ± 0.14 in samples after WGA. Our results suggested that WGA for non-invasive prenatal diagnosis would be most effective after gel extractions since plasma-circulating DNA consists of a mixture of fragmented apoptotic and necrotic DNA in a broad range of sizes that are difficultto amplify uniformly. This was demonstrated by 1,000-fold increase on average of DNA gain when compared DYS1 (average of DNA gain 1,556) to β-globin (average of DNA gain 1.5) in 100–300 bp size population. Using WGA on discrete size populations improves the likelihood of amplifying desired target sequences.
Circulating fetal DNA is stable and highly present as small size fragments (<300 bp). By using agarose gel electrophoresis combined with WGA, we successfully enrich for fetal DNA yielding a population of fetal DNA that is better suited for molecular analysis. The combination of these techniques is a novel approach to generating higher amounts of good quality fetal DNA for high-throughput analysis of genetic abnormalities using plasma-circulating DNA.
Acknowledgements
This research was supported in part by grant NIH/NICHD HDO46623. We thank Linda Martinez, Flor Torres, Deborah Marquez-Do, and Mathew Chapman for technical assistance.
References
- 1.Lo YM, Tein MS, Lau TK, Haines CJ, Leung TN, Poon PM, Wainscoat JS, Johnson PJ, Chang AM, Hjelm NM. Quantitative analysis of fetal DNA in maternal plasma and serum: implications for noninvasive prenatal diagnosis. Am J Hum Genet. 1998;62:768–775. doi: 10.1086/301800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bischoff FZ, Nguyen DD, Marquez-Do D, Moise KJ, Jr, Simpson JL, Elias S. Noninvasive determination of fetal RhD status using fetal DNA in maternal serum and PCR. J Soc Gynecol Investig. 1999;6:64–69. doi: 10.1016/s1071-5576(98)00051-3. [DOI] [PubMed] [Google Scholar]
- 3.Bischoff FZ, Sinacori MK, Dang DD, Marquez-Do D, Horne C, Lewis DE, Simpson JL. Cell-free fetal DNA and intact fetal cells in maternal blood circulation: implications for first and second trimester non-invasive prenatal diagnosis. Hum Reprod Update. 2002;8:493–500. doi: 10.1093/humupd/8.6.493. [DOI] [PubMed] [Google Scholar]
- 4.Bischoff FZ, Dang DX, Marquez-Do D, Martinez D, Horne C, Lewis DE, Simpson JL. Detecting fetal DNA from dried maternal blood spots: another step towards broad scale non-invasive prenatal genetic screening and feasible testing. Reprod Biomed Online. 2003;6:349–351. doi: 10.1016/s1472-6483(10)61856-1. [DOI] [PubMed] [Google Scholar]
- 5.Johnson KL, Dukes KA, Vidaver J, LeShane ES, Ramirez I, Weber WD, Bischoff FZ, Hahn S, Sharma A, Dang DX, Hire LM, Bianchi DW, Simpson JL, Holzgreve W, Elias S, Klinger KW. Interlaboratory comparison of fetal male DNA detection from common maternal plasma samples by real-time PCR. Clin Chem. 2004;50:516–521. doi: 10.1373/clinchem.2003.024380. [DOI] [PubMed] [Google Scholar]
- 6.Minon JM, Gerard C, Senterre JM, Schaaps JP, Foidart JM. Routine fetal RHD genotyping with maternal plasma: a four-year experience in Belgium. Transfusion. 2008;48:373–381. doi: 10.1111/j.1537-2995.2007.01533.x. [DOI] [PubMed] [Google Scholar]
- 7.Hahn S, Huppertz B, Holzgreve W. Fetal cells and cell-free fetal nucleic acids in maternal blood: new tools to study abnormal placentation? Placenta. 2005;26:515–526. doi: 10.1016/j.placenta.2004.10.017. [DOI] [PubMed] [Google Scholar]
- 8.Lo YM, Leung TN, Tein MS, Sargent IL, Zhang J, Lau TK, Haines CJ, Redman CW. Quantitative abnormalities of fetal DNA in maternal serum in preeclampsia. Clin Chem. 1999;45:184–188. [PubMed] [Google Scholar]
- 9.Sugito Y, Sekizawa A, Farina A, Yukimoto Y, Saito H, Iwasaki M, Rizzo N, Okai T. Relationship between severity of hyperemesis gravidarum and fetal DNA concentration in maternal plasma. Clin Chem. 2003;49:1667–1669. doi: 10.1373/49.10.1667. [DOI] [PubMed] [Google Scholar]
- 10.Leung TN, Zhang J, Lau TK, Hjelm NM, Lo YM. Maternal plasma fetal DNA as a marker for preterm labour. Lancet. 1998;352:1904–1905. doi: 10.1016/S0140-6736(05)60395-9. [DOI] [PubMed] [Google Scholar]
- 11.Bischoff FZ, Lewis DE, Simpson JL. Cell-free fetal DNA in maternal blood: kinetics, source and structure. Hum Reprod Update. 2005;11:59–67. doi: 10.1093/humupd/dmh053. [DOI] [PubMed] [Google Scholar]
- 12.Orozco AF, Bischoff FZ, Horne C, Popek E, Simpson JL, Lewis DE. Hypoxia-induced membrane-bound apoptotic DNA particles: potential mechanism of fetal DNA in maternal plasma. Ann NY Acad Sci. 2006;1075:57–62. doi: 10.1196/annals.1368.007. [DOI] [PubMed] [Google Scholar]
- 13.Chan KC, Zhang J, Hui AB, Wong N, Lau TK, Leung TN, Lo KW, Huang DW, Lo YM. Size distributions of maternal and fetal DNA in maternal plasma. Clin Chem. 2004;50:88–92. doi: 10.1373/clinchem.2003.024893. [DOI] [PubMed] [Google Scholar]
- 14.Li Y, Zimmermann B, Rusterholz C, Kang A, Holzgreve W, Hahn S. Size separation of circulatory DNA in maternal plasma permits ready detection of fetal DNA polymorphisms. Clin Chem. 2004;50:1002–1011. doi: 10.1373/clinchem.2003.029835. [DOI] [PubMed] [Google Scholar]
- 15.Jorgez CJ, Dang DD, Simpson JL, Lewis DE, Bischoff FZ. Quantity versus quality: optimal methods for cell-free DNA isolation from plasma of pregnant women. Genet Med. 2006;8:615–619. doi: 10.1097/01.gim.0000241904.32039.6f. [DOI] [PubMed] [Google Scholar]
- 16.Hanson EK, Ballantyne J. Whole genome amplification strategy for forensic genetic analysis using single or few cell equivalents of genomic DNA. Anal Biochem. 2005;346:246–257. doi: 10.1016/j.ab.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 17.Li Y, Page-Christiaens GC, Gille JJ, Holzgreve W, Hahn S. Non-invasive prenatal detection of achondroplasia in size-fractionated cell-free DNA by MALDI-TOF MS assay. Prenat Diagn. 2007;27:11–71. doi: 10.1002/pd.1608. [DOI] [PubMed] [Google Scholar]
- 18.Ballantyne KN, van Oorschot RA, Mitchell RJ. Comparison of two whole genome amplification methods for STR genotyping of LCN and degraded DNA samples. Forensic Sci Int. 2007;166:35–41. doi: 10.1016/j.forsciint.2006.03.022. [DOI] [PubMed] [Google Scholar]
- 19.Nagy M, Rascon J, Massenkeil G, Ebell W, Roewer L. Evaluation of whole genome amplification of low-copy-number DNA in chimerism analysis after allogeneic stem cell transplantation using STR marker typing. Electrophoresis. 2006;27:3028–3037. doi: 10.1002/elps.200500813. [DOI] [PubMed] [Google Scholar]